Abstract
OsbZIP23 is a member of the basic leucine zipper (bZIP) transcription factor family in rice (Oryza sativa). Expression of OsbZIP23 is strongly induced by a wide spectrum of stresses, including drought, salt, abscisic acid (ABA), and polyethylene glycol treatments, while other stress-responsive genes of this family are slightly induced only by one or two of the stresses. Transactivation assay in yeast demonstrated that OsbZIP23 functions as a transcriptional activator, and the sequences at the N terminus (amino acids 1–59) and a region close to the C terminus (amino acids 210–240) are required for the transactivation activity. Transient expression of OsbZIP23-green fluorescent protein in onion (Allium cepa) cells revealed a nuclear localization of the protein. Transgenic rice overexpressing OsbZIP23 showed significantly improved tolerance to drought and high-salinity stresses and sensitivity to ABA. On the other hand, a null mutant of this gene showed significantly decreased sensitivity to a high concentration of ABA and decreased tolerance to high-salinity and drought stress, and this phenotype can be complemented by transforming the OsbZIP23 back into the mutant. GeneChip and real-time polymerase chain reaction analyses revealed that hundreds of genes were up- or down-regulated in the rice plants overexpressing OsbZIP23. More than half of these genes have been annotated or evidenced for their diverse functions in stress response or tolerance. In addition, more than 30 genes that are possible OsbZIP23-specific target genes were identified based on the comparison of the expression profiles in the overexpressor and the mutant of OsbZIP23. Collectively, these results indicate that OsbZIP23 functions as a transcriptional regulator that can regulate the expression of a wide spectrum of stress-related genes in response to abiotic stresses through an ABA-dependent regulation pathway. We propose that OsbZIP23 is a major player of the bZIP family in rice for conferring ABA-dependent drought and salinity tolerance and has high potential usefulness in genetic improvement of stress tolerance.
Plants can initiate a number of molecular, cellular, and physiological changes in response and adaptation to the ever-changing environment during their growth and development. The phytohormone abscisic acid (ABA) controls various aspects of plant growth throughout development (Finkelstein et al., 2002; Himmelbach et al., 2003). It prevents embryos from precocious germination, modulates seedling growth under normal conditions, and regulates seed maturation (Busk and Pages, 1998; Ladyzhenskaya, 2001; Cheng et al., 2002; Chen et al., 2006). ABA also plays important roles in the accumulation of storage proteins and other reserve materials and in the acquisition of desiccation tolerance. During vegetative growth, one of the major roles of ABA is to mediate adaptive responses to various environmental stresses. It has a critical role in the plant adaptation or acclimation to abiotic stresses, such as high salinity, drought, low temperature, and mechanical wounding. The pathways leading to stress adaptation can be divided into two major categories: ABA-dependent and ABA-independent pathways. Underlying the ABA-mediated stress responses is the transcriptional regulation of stress-responsive gene expression dependent on ABA (Shinozaki and Yamaguchi-Shinozaki, 2000; Ladyzhenskaya, 2001; Fedoroff, 2002; Finkelstein et al., 2002; Jakoby et al., 2002; Xiong et al., 2002; Zhu, 2002; Himmelbach et al., 2003).
During the response and adaptation to diverse abiotic stresses, many stress-related genes are induced and the levels of a variety of stress resistance-related functional proteins are accumulated. Numerous genes have been reported to be up-regulated under stress conditions in vegetative tissues (Seki et al., 2002; Zhu, 2002). As a trigger of gene expression, transcription factors play important regulatory roles in almost every aspect of life in plants, such as plant growth, development, and responses to abiotic and biotic stresses. Basic leucine zipper (bZIP) is a big transcription factor family in plants, and the members of this family have diverse roles, especially in plant stress-responsive and hormone signal transduction (Uno et al., 2000; Jakoby et al., 2002; Rodriguez-Uribe and O'Connell, 2006).
In Arabidopsis (Arabidopsis thaliana), there are many bZIP genes identified (Jakoby et al., 2002; Iida et al., 2005). All of the putative bZIP transcription factors in Arabidopsis have been classified into 10 subfamilies (Jakoby et al., 2002). To date, quite a few bZIP transcription factors have been functionally characterized. Among those are HY5, which mediates plant light response (Chattopadhyay et al., 1998; Osterlund et al., 2000), ATB2, which is involved in Suc-specific sugar sensing and signaling (Rook et al., 1998), and AtbZIP60, which regulates the endoplasmic reticulum stress response in a manner unique to plants (Iwata and Koizumi, 2005). Special notice has been given to the third subfamily (also known as the ABI5 subfamily), as many members of this subfamily were identified for their roles in conferring ABA sensitivity and/or stress responses. Previously reported members of the third subfamily in Arabidopsis were designated ABA-responsive element (ABRE)-binding factors, abbreviated as ABFs (i.e. ABF1–ABF4) or AREBs (i.e. AREB1–AREB3; Choi et al., 2000; Uno et al., 2000; Kim et al., 2004). These transcription factors are highly homologous to ABI5, a genetically identified ABA signaling component that plays an essential role in seed germination and ABA-triggered postgermination developmental arrest processes in Arabidopsis (Lopez-Molina et al., 2003; Bensmihen et al., 2005). ABF-encoding genes (ABF1–ABF4) are expressed mainly in vegetative tissues, and their expression is induced by various abiotic stresses (Choi et al., 2000; Uno et al., 2000). Additionally, the distinct stress induction patterns of ABF genes suggest that they may function in different stress response pathways (e.g. ABF1 in cold, ABF2 and ABF3 in salt, and ABF4 in cold, salt, and drought signaling pathways; Uno et al., 2000; Jakoby et al., 2002; Kang et al., 2002).
Rice (Oryza sativa) has become a model plant for monocotyledon species. About 100 putative bZIP sequences (88 and 109 gene models in the sequenced indica and japonica genomes, respectively) were predicted in the rice genome (Guo et al., 2005), and they can also be classified into 10 subfamilies (http://greenphyl.cines.fr/cgi-bin/greenphyl.cgi). Distinct expression profiles of this family in rice indicated that bZIP proteins may also play diverse functions in rice, including developmental and physiological functions during floral transition, panicle and seed development, light signaling, and abiotic stress tolerance (Nijhawan et al., 2008). Several members of bZIP family in rice have been identified for their functions potentially related to biotic or abiotic stress response or signaling. For example, LIP19 is induced by low temperature and may function as a molecular switch in cold signaling in rice (Shimizu et al., 2005). OsBZ8, another bZIP gene of the family, is rapidly induced by ABA and shows stronger expression in salt-tolerant cultivars than in salt-sensitive cultivars (Nakagawa et al., 1996; Mukherjee et al., 2006). Another two rice bZIP transcription factors, RF2a and RF2b, which can interact with each other at their C-terminal regions, are involved in regulation of the rice tungro bacilliform virus promoter and in the development of rice tungro disease (Yin et al., 1997; Zhu et al., 2002; Dai et al., 2004; Liu et al., 2007). Compared with the intensive research of the third subfamily in Arabidopsis, only two genes of this subfamily have been studied in rice. The first one is TRAB1, which has been thoroughly characterized for the biochemical aspects of the protein (Hobo et al., 1999; Kagaya et al., 2002; Kobayashi et al., 2005), although its biological function remains unclear. TRAB1 was identified by yeast two-hybrid screening in a search for proteins that interact with VP1/ABI3, a transcription factor that is required for ABA-regulated gene expression during seed development (Hobo et al., 1999). TRAB1 is localized in the nucleus and can be activated via ABA-dependent phosphorylation, and its Ser-102 residue is critical for this function and this residue is phosphorylated in response to ABA or hyperosmotic stress (Kagaya et al., 2002; Kobayashi et al., 2005). Another gene named OsABI5 encodes a protein that can bind to ABRE (G-box) and was suggested to be involved in ABA signal transduction and stress responses (Zou et al., 2007, 2008).
A preliminary analysis of the gene expression profiles of the rice genome under various stress conditions revealed that most of the genes in the third (or ABI5) bZIP subfamily were more or less induced by ABA, drought, or high salinity (Y. Xiang, N. Tang, H. Du, H. Ye, and L. Xiong, unpublished GeneChip data). We noticed that a member named OsbZIP23, with the systematic name adopted from the sequence analysis of the bZIP family in rice (Nijhawan et al., 2008), has relatively low sequence similarity to reported bZIP members such as TRAB1 and OsABI5 but has much more strongly induced expression by a wide spectrum of stresses than other members in this family. Here, we report the identification and functional analysis of OsbZIP23 by investigating the ABA sensitivity and stress tolerance of transgenic plants overexpressing OsbZIP23 and a T-DNA insertion mutant of this gene. Expression profiling analysis of the overexpression and mutant plants was also performed to deduce the putative target genes regulated by OsbZIP23. Our results indicate OsbZIP23 might be a major player in the bZIP family for conferring ABA-dependent stress tolerance in rice.
RESULTS
Isolation and Sequence Analysis of OsbZIP23
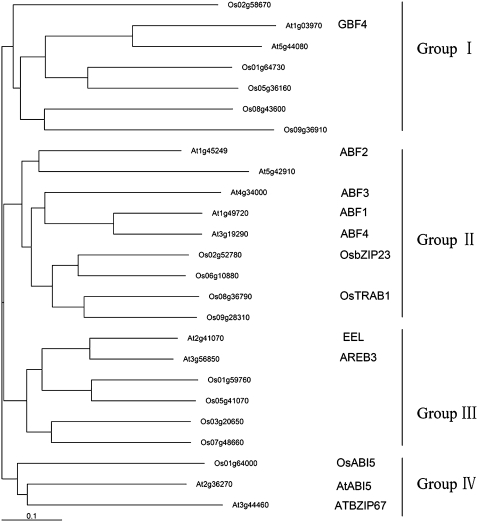
In our GeneChip analysis of drought-resistant upland rice IRAT109 (japonica) under abiotic stresses (Y. Xiang, N. Tang, H. Du, H. Ye, and L. Xiong, unpublished data), we noticed one gene, designated OsbZIP23 according to Nijhawan et al. (2008), that encodes a putative bZIP transcription factor (annotation identifier in The Institute for Genomic Research database: LOC_Os02g52780) and was strongly induced by drought, high-salinity, and ABA treatment (Supplemental Table S1). The induction level of this gene was significantly higher than that of the two reported genes, TRAB1 and OsABI5, and the other members of the bZIP family in rice. Therefore, we isolated the full-length cDNAs of OsbZIP23 from upland rice IRAT109 for further functional analysis. Comparison of the cDNA sequences of OsbZIP23 from IRAT109 and Zhengshan 97 (a drought-sensitive indica rice) revealed a deletion of 9 bp (three AGC repeats corresponding to amino acids 14–16 at the N terminus of the predicted protein) in IRAT109 (Supplemental Fig. S1), but this deletion also exists in the drought-sensitive japonica rice Nipponbare. Protein sequence analysis suggested that OsbZIP23 belongs to the third subfamily of bZIP in rice, and it has all of the features of a typical bZIP transcription factor of this subfamily: a bZIP domain and five conserved domains that are predicted to be phosphorylation sites involved in stress or ABA signaling. Phylogenetic analysis of the members of the third subfamily in Arabidopsis and rice suggested that these bZIP proteins can be further classified into four groups (I–IV; Fig. 1). Although OsbZIP23 and TRAB1 are in the same group (group II), they have only moderate similarity of protein sequence. An unknown bZIP sequence of rice (LOC_Os06g10880) is the closest homolog to OsbZIP23. Interestingly, four reported bZIP proteins (ABF1–ABF4) in Arabidopsis were grouped with OsbZIP23 (Fig. 1). The recently reported bZIP transcription factor OsABI5, with its name following ABI5 in Arabidopsis (Zou et al., 2007), was classified into a separate group (group IV) and has rather low sequence similarity to OsbZIP23.
Figure 1.
Phylogenetic tree of the third bZIP subfamily members from Arabidopsis and rice. Alignment of conserved bZIP protein sequences and phylogenetic analysis were performed by the program ClustalX 1.83 and Phylip 3.63, respectively.
Expression Profile of OsbZIP23 under Different Stresses
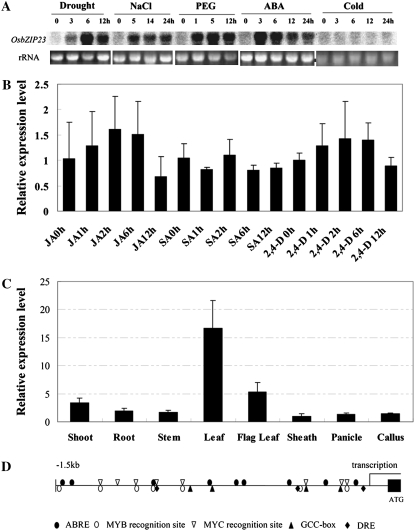
To speculate on the physiological and functional relevance of the OsbZIP23 gene, we further checked its expression profile under different abiotic stresses and various chemical treatments. In RNA gel-blot analysis, the transcript level of OsbZIP23 was rapidly and strongly induced by drought, high-salinity, polyethylene glycol (PEG), and ABA treatment but remained unchanged after low-temperature treatment (Fig. 2A), which agrees with the microarray data very well (Supplemental Table S1). The transcript level of this gene was also checked in a few chemical treatments, including jasmonic acid (JA), salicylic acid (SA), and 2,4-dichlorophenoxyacetic acid (2,4-D), by real-time PCR analysis. The results showed that the expression level of this gene was not obviously affected by any of these treatments (Fig. 2B). We also investigated whether the expression of this gene has any tissue specificity. The results suggested it was relatively higher in seedling and flag leaves than in other tissues checked (Fig. 2C). The strong induction of this gene by abiotic stresses prompted us check its promoter sequence (1,500 bp upstream the transcription start site) by searching the prompter sequence against the PLACE database (http://www.dna.affrc.go.jp/PLACE/). The promoter sequence indeed contains many putative stress response-related cis-elements, such as ABRE (nine hits for the core sequence of ABRE), DRE (three hits), MYC recognition site (eight hits), MYB recognition site (seven hits), and GCC box (four hits; Fig. 2D). The numbers of these response-related cis-elements are apparently higher than those of the other stress-responsive genes in this family, such as TRAB1 (five ABRE, no DRE, seven MYC recognition sites, and five MYB recognition sites) and OsABI5 (no ABRE, no DRE, six MYC recognition sites, and five MYB recognition sites).
Figure 2.
Expression analysis of the OsbZIP23 gene. A, Northern-blot analysis of OsbZIP23 expression level under drought, high-salinity, and low-temperature stress and ABA and PEG treatment. B, Real-time PCR analysis of OsbZIP23 in JA, SA, and 2,4-D treatments. C, Expression level of OsbZIP23 in different tissues by real-time PCR analysis. D, Distribution of major stress-related cis-elements in the promoter region of OsbZIP23.
OsbZIP23 Is Located in the Nucleus and Has Transactivation Activity in Yeast
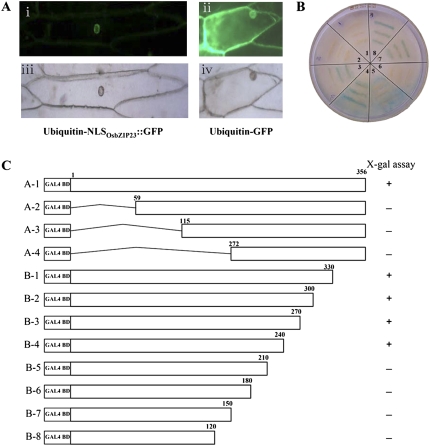
To confirm that OsbZIP23 is a putative transcription factor, the OsbZIP23-GFP fusion construct was used for in vivo protein targeting in onion epidermal cells by bombarding transformation. The results showed that GFP signal was detected only in the nucleus of the OsbZIP23-GFP-transformed cell (Fig. 3A), while the control (transformation of GFP construct) showed ubiquitous distribution of GFP signal in the cell, suggesting that OsbZIP23 is a nucleus-localized protein. A putative nuclear localization signal was predicted in the N terminus of the protein by WoLF PSORT (http://wolfpsort.seq.cbrc.jp/). A segment of 60 amino acids containing the nuclear localization signal was fused to GFP, and the fused protein was also located in the nucleus of transformed onion epidermal cells (data not shown).
Figure 3.
Subcellular localization and transactivation analysis of OsbZIP23. A, Nuclear localization of OsbZIP23. The OsbZIP23∷EGFP fusion protein, driven by the maize ubiquitin promoter, was transiently expressed in onion epidermal cells and visualized by fluorescence microscopy (A-i). EGFP alone, driven by the maize ubiquitin promoter, was used as a negative control (A-ii). A-iii and A-iv show photographs of A-i and A-ii, respectively, without UV light. B, Transactivation assay of OsbZIP23 in the yeast strain Mav203. Sections 1 to 5, Five controls provided in the kit; section 6, negative control; sections 7 and 8, OsbZIP23 fusion with GAL4-DB in the vector pDEST32. C, Transactivation assay of truncated OsbZIP23. Fusion proteins of the GAL4 DNA-binding domain and different portions of OsbZIP23 were checked for their transactivation activity in the yeast strain. + and − indicate positive and negative, respectively, for the transactivation activity.
To check if OsbZIP23 has transcriptional activity, the open reading frame of OsbZIP23 was fused to the GAL4 DNA-binding domain (GAL4-DB) in the vector pDEST32 and the construct was transformed into yeast. Colony-lift filter assay showed that the reporter gene LacZ was expressed only in the yeast cell transformed with the GAL4-DB-OsbZIP23 construct (Fig. 3B), indicating that OsbZIP23 had transcriptional activity in yeast. Transactivation activity assay of a series of shortened OsbZIP23 from both the N and C termini revealed that the amino acid sequences at the N terminus (amino acids 1–59) and a region close to the C terminus (amino acids 210–240) are required for the transactivation activity of the fusion protein in yeast (Fig. 3C). However, deletion from the C terminus up to amino acid 240 did not abolish the transactivation activity of the fusion protein in yeast (Fig. 3C). Nevertheless, no difference in the transactivation activity was detected for the OsbZIP23 sequences from indica and japonica rice, even though there is a polymorphism (amino acids 14−16) in the N-terminal region.
Increases in ABA Sensitivity of OsbZIP23-Overexpressing Plants
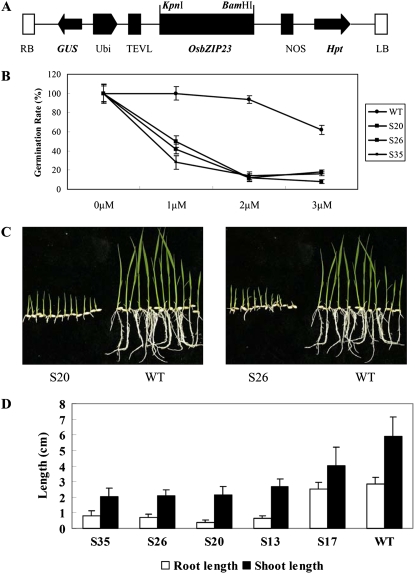
Since OsbZIP23 was strongly induced by ABA, we tested if OsbZIP23 is involved in ABA sensitivity of rice, which is an important aspect of ABA-dependent regulation. To confirm this, an overexpression construct with the OsbZIP23 gene under the control of the maize (Zea mays) ubiquitin promoter (Fig. 4A) was transformed into rice Zhonghua 11. The expression level and copy number of T-DNA was checked by northern- and Southern-blot analyses (Supplemental Figs. S2 and S3). ABA sensitivity was examined for two independent homozygous T2 overexpression lines (S20 and S26, containing a single copy of the T-DNA). The results indicated that OsbZIP23-overexpressing lines were hypersensitive to ABA at the germination stage (Fig. 4B). At 6 d after seed imbibition, transgenic seeds had only about 50% of the relative germination rate (ratio of germination rate in the ABA treatment to that in water control) with 1 μm ABA in the medium, while the wild type showed nearly 100% of the relative germination rate under the same conditions. In the treatment with 2 μm ABA, the relative germination rate of the wild type was about 90%, but the relative germination rate of overexpression lines was reduced to about 15%. Under the same dose of ABA, the germinated wild-type plantlets continued to grow, albeit at a slower rate than on ABA-free medium, but transgenic plantlets grew very slowly. Such a hypersensitivity of OsbZIP23-overexpressing lines was also confirmed at the postgermination stage. In this experiment, transgenic and wild-type seeds were geminated on normal Murashige and Skoog (MS) medium, and the seedlings were then transferred to medium containing 3 μm ABA. The shoot and root length were measured at 10 d after being transferred. The results showed that OsbZIP23 overexpressors had significantly (P < 0.05) shorter roots and shoots than the wild type (Fig. 4, C and D), while on normal MS medium all transgenic plants showed no difference from the wild type (Supplemental Fig. S4).
Figure 4.
ABA sensitivity of transgenic rice overexpressing OsbZIP23. A, Overexpression construct for rice transformation. Hpt, Hygromicin phosphotransferase; LB, left border; NOS, NOS terminator; RB, right border; TEVL, TEV leading signal sequence; Ubi, maize ubiquitin promoter. B, Germination rate of OsbZIP23-overexpressing transgenic seeds on MS medium containing 0, 1, 2, or 3 μmol L−1 ABA measured at 6 d after initiation. C and D, Growth performance (C) and shoot and root length (D) of OsbZIP23 overexpressors grown on MS medium containing 3 μmol L−1 ABA at the postgermination stage. S13, S20, S26, and S35 are OsbZIP23-overexpressing lines; S17 is a negative control of the transgenic plant. WT, Wild type.
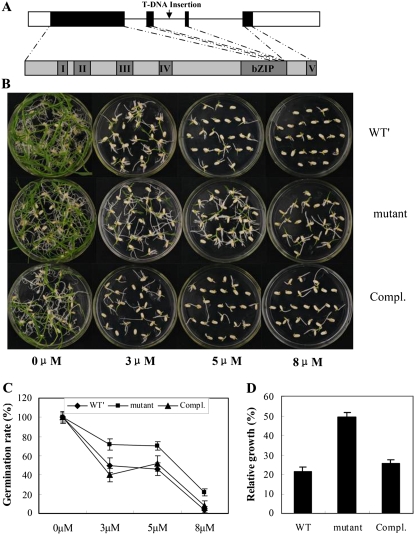
Decreased ABA Sensitivity of the Osbzip23 Mutant
The function of OsbZIP23 was further investigated by loss-of-function mutant analysis. A mutant line of OsbZIP23 in the background of Zhonghua 11, with a T-DNA inserted in the second intron (Fig. 5A), was obtained from the Rice Mutant Database (http://rmd.ncpgr.cn/). After being confirmed for the T-DNA insertion site and the abolishment of OsbZIP23 expression (Supplemental Fig. S5), homozygous mutant plants derived from a single T-DNA insertion were checked for an ABA sensitivity phenotype. In a seed germination experiment with application of ABA in the medium, the mutant had significantly higher germination rates (about 80% and 20% in the medium with 3 μm and 8 μm ABA, respectively) than that in WT′ (wild-type plants segregated from a heterozygous Osbzip23 mutant; about 50% and 0% in the medium with 3 μm and 8 μm ABA, respectively; Fig. 5, B and C). In the normal medium, the germination rates of mutant and WT′ plants had no significant difference. We also checked the ABA sensitivity of the mutant in the medium containing ABA at the postgermination stage. The mutant also showed a significantly higher relative growth (50% of the shoot length on normal medium) than WT′ (30%; Fig. 5D). After a DNA fragment containing genomic sequence of the OsbZIP23 gene and its native promoter was retransformed into the mutant, the germination rate of the retransformed transgenic seeds in the ABA-containing medium was decreased to a level similar to that in WT′ (Fig. 5, B–D), indicating that the decreased ABA sensitivity of the mutant can be complemented by the OsbZIP23 gene. These results indicated that OsbZIP23 plays a critical role in mediating ABA sensitivity in rice.
Figure 5.
ABA sensitivity of the Osbzip23 mutant and the complementation line of the mutant at germination and postgermination stages. A, Schematic exon-intron structure of the OsbZIP23 gene and the T-DNA insertion position. B, Germination performance of the Osbzip23 mutant and the complementation line on MS medium containing 0, 3, 5, and 8 μmol L−1 ABA at 6 d after initiation. C, Germination rate corresponding to B. D, Relative growth of the mutant and complementation line grown on MS medium containing 5 μmol L−1 ABA at the postgermination stage. WT′, The wild type segregated from the heterozygous Osbzip23 mutant.
Stress Tolerance of OsbZIP23 Transgenic Plants
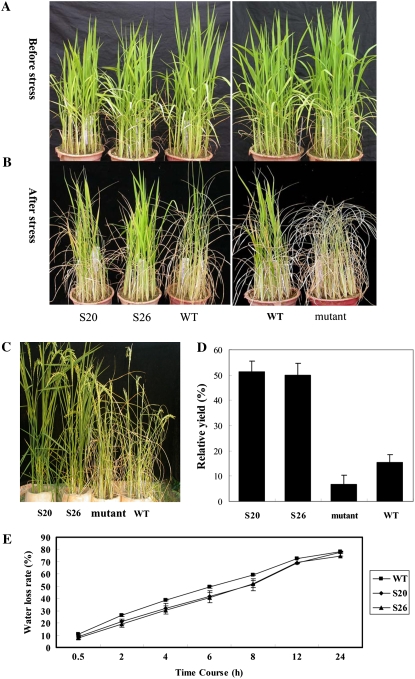
The strong induction of OsbZIP23 expression by drought and high-salinity stresses suggested that this gene might be involved in stress resistance. Homozygous plants of the two independent T2 overexpression lines (S20 and S26) and the mutant of this gene were selected for drought resistance testing. At seedling stage, the overexpression lines showed increased and decreased drought resistance, respectively, compared with the wild type (Fig. 6, A and B). We evaluated the drought stress tolerance at reproductive stage in PVC pipes under the rain-off shelter according to the well-established drought stress method (Yue et al., 2006; Xiao et al., 2007). The results indicated that overexpressing OsbZIP23 can also significantly improve drought resistance of transgenic rice at the reproductive stage (Fig. 6C). Relative grain yield per plant, one of the best criteria for evaluating drought resistance at the reproductive stage (Yue et al., 2006), was very significantly (P < 0.01) higher in the two overexpression lines (approximately 50%) than in the wild type (15.3%) or the mutant (6.6%; Fig. 6D; Supplemental Table S2).
Figure 6.
Drought stress tolerance testing of OsbZIP23 transgenic rice. A, Vigorous seedlings before drought stress treatment. B, Plant phenotype after medium drought stress treatment (with water withheld for 5 d followed by recovery). C, Phenotype of OsbZIP23 transgenic plants and wild-type (WT) plants under severe drought stress at flowering stage. D, Relative yield per plant after being stressed by drought at flowering stage. E, Water loss rate in the leaves cut from the OsbZIP23 overexpressors and the wild type grown under normal conditions.
Since OsbZIP23 is expressed predominantly in leaves (Fig. 2C), we further checked if water loss rate in leaves was affected in the transgenic plants during the dehydration process. The results showed that the leaves from the overexpression plants had significantly lower (P < 0.05) rates of water loss than wild-type plants (Fig. 6E). But the mutant showed no significant difference of water loss rate compared with the wild type (data not shown). This result prompted us to check the density and aperture of stomata in the overexpression and mutant plants, but no significant differences were observed under normal and drought stress conditions (data not shown).
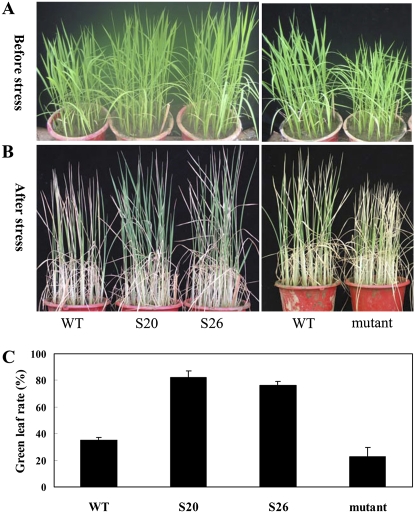
To evaluate salt tolerance of the transgenic rice, four-leaf plants were irrigated with water containing 200 mm NaCl, and leaf death rate was checked at 10 d after treatment. The results showed that the overexpressors had significantly more green leaf area left (approximately 80%) than the wild-type control (approximately 38%) or the mutant (approximately 20%; Fig. 7), suggesting that overexpression of OsbZIP23 in rice can also improve salt tolerance.
Figure 7.
Salt stress tolerance testing of the OsbZIP23 overexpressors and the mutant line. A, Vigorously growing seedlings before salt stress treatment. B, Plant phenotype at 7 d after salt stress (200 mm NaCl) treatment. C, Percentage of green leaves of salt-stressed plants. WT, Wild type.
Overexpression or knockout of this gene had no significant effect on changing cold tolerance (data not shown). In addition, there was no significant difference in plant morphology (such as root depth and volume, plant height, and numbers of tillers and spikelets) between the transgenic lines and the wild type under normal growth conditions (data not shown), although spikelet fertility of transgenic plants was slightly lower than in the wild type (Supplemental Table S2), a phenomenon that has been observed very frequently in the early generations of transgenic rice produced by the tissue culture method. In general, overexpressing OsbZIP23 had no detrimental effect on the growth and development of rice.
Dramatic Changes of the Expression Profiles in the OsbZIP23-Overexpressing and Mutant Plants
Toward identification of the target genes regulated by OsbZIP23 and thus elucidation of the molecular mechanism of stress tolerance mediated by this gene, we further checked the genome-wide expression profile changes in the OsbZIP23-overexpressing and knockout plants using the Affymetrix GeneChip. Two independent OsbZIP23-overexpressing lines, the mutant line, and the wild type (Zhonghua 11) were used to check the expression profiles under the same growth conditions at the four-leaf stage.
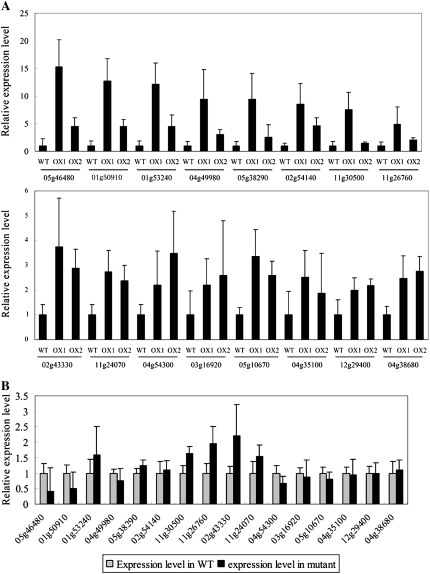
A total of 795 genes showed greater than 2-fold higher (P < 0.01) expression levels in both the OsbZIP23-overexpressing lines and in the wild type (Supplemental Table S3). Even though a limited number of genes with greater than 2-fold changes are statistically significant, we still chose 2-fold as an experiential threshold because those genes with greater than 2-fold changes can seldom be confirmed by real-time PCR. Noticeably, nearly half of the up-regulated genes have been annotated or evidenced for stress response or adaptation (Supplemental Table S3). Examples are genes encoding stress-related functional proteins, such as dehydrin family proteins (four genes), late embryogenesis abundant (LEA) proteins (five genes), seed storage/lipid transfer proteins (14 genes), amino acid metabolism or transportation proteins (33 genes), cell membrane stability-related proteins (eight genes), and stress-related regulatory factors such as transcription factors (25 genes), protein kinase (seven genes), and phosphatase (10 genes). Real-time PCR was performed to confirm the expression of 16 up-regulated genes with their annotations related to stress tolerance, and the results suggested that all of the chosen genes showed elevated expression levels in the overexpression lines (Fig. 8A), although the absolute values of the fold changes showed some variation for some genes between microarray and real-time PCR analysis. On the other hand, all 16 genes showed decreases or no significant changes in expression levels in the mutant based on real-time PCR analysis (Fig. 8B). Checking of additional genes (Supplemental Fig. S6) by real-time PCR further consolidated the GeneChip data. We also checked the stress responsiveness of the up-regulated genes in the overexpression lines in our previous microarray analysis of rice under abiotic stresses (Zhou et al., 2007; Y. Xiang, N. Tang, H. Du, H. Ye, and L. Xiong, unpublished data). Preliminary results suggested that 134, 56, and 90 of these genes were induced by drought, salt, and ABA, respectively (Supplemental Table S3).
Figure 8.
Relative expression levels of 16 stress-related genes in the OsbZIP23 overexpressors (A) and mutant (B) detected by real-time PCR. OX1 and OX2, Two independent OsbZIP23-overexpressing lines. The putative stress-related functions of these 16 representative genes include LEA protein (Os05g46480, Os01g50910, and Os04g49980), dehydrin (Os11g26760), lipid-transfer protein 1 precursor (Os11g24070), transmembrane amino acid transporter protein (Os04g38680), heat shock protein (Os02g54140 and Os03g16920), protein phosphatase (Os05g38290 and Os04g35100), ABA- or wound-inducible protein (Os12g29400, Os04g54300, Os11g30500, and Os01g53240), zinc finger protein (Os05g10670), and homeobox domain-containing protein (Os02g43330).
A total of 1,017 genes showed decreased expression (greater than 2-fold) in the OsbZIP23-overexpressing lines compared with the wild type (Supplemental Table S4), suggesting that overexpression of OsbZIP23 can also suppress the expression of a large number of genes. The predicted functions of these down-regulated genes are extremely diverse, and there was no obvious predominance of certain categories. In contrast to the high proportion of up-regulated genes in the overexpression lines that were induced by stresses (Supplemental Table S3), very limited numbers of the down-regulated genes were responsive (neither induced nor suppressed) to the stresses based on our previous microarray data (Supplemental Table S4).
In the mutant, 86 and 141 genes were up- and down-regulated, respectively, compared with the wild type. The stress responsiveness of these genes was also checked in our previous GeneChip data. Among the 86 up-regulated genes, nine, 28, and five genes were induced by drought, ABA, and salt, respectively, while 23, nine, and 14 genes were down-regulated by drought, ABA, and salt stress, respectively (Supplemental Table S5). Among the 141 down-regulated genes, 50, 23 and seven genes were induced by drought, ABA, and high-salinity stress, respectively, and 37, 12, and 20 genes were down-regulated by drought, ABA, and salt stress, respectively (Supplemental Table S6). Some of these expression level-altered genes have also been annotated or evidenced with direct or indirect functions in stress response or adaptation, although most of them are annotated as having unknown functions in the database.
By comparing the expression profile changes between the overexpressor and the mutant of OsbZIP23, 37 genes showed reverse direction of expression level changes (Supplemental Table S7). Among them, 24 genes were up-regulated in the overexpressor but down-regulated in the mutant, and 13 genes were down-regulated in the overexpressor but up-regulated in the mutant. These genes might be regulated specifically by OsbZIP23.
Taken together, the expression profiling analysis revealed dramatic changes of expression profiles in both the overexpressor and the mutant of OsbZIP23, and a significant portion of the expression level-altered genes are related to stress response and adaptation. The altered ABA sensitivity and stress tolerance of the overexpressor and the mutant may have resulted from cumulative effect of the expression changes of these genes.
DISCUSSION
The bZIP transcription factors play important roles in diverse biologic processes, such as seed mutation, flower development, and stress responses. In Arabidopsis, quite a few members of the third subfamily of the bZIP family have been reported for their roles in regulating ABA-mediated stress responses. These bZIP transcription factors include ABI5 (Carles et al., 2002; Lopez-Molina et al., 2002; Bensmihen et al., 2005), ABF1 to ABF4 (Choi et al., 2000; Kang et al., 2002; Kim et al., 2004), and EEL (Bensmihen et al., 2002). In rice, however, only two members in this subfamily, TRAB1 (Hobo et al., 1999; Kagaya et al., 2002) and OsABI5 (Zou et al., 2007, 2008), have been reported, but their biological functions are largely unclear. Here, we report the isolation and characterization of a rice bZIP transcription factor, OsbZIP23, which belongs to the third subfamily of the bZIP family. Our data suggest that OsbZIP23 may be major player in the bZIP family in mediating ABA-dependent pathways for conferring stress tolerance, on the basis of the following arguments.
First, the OsbZIP23 gene has stronger stress-induced expression than other members of the family. OsbZIP23 is strongly induced by drought and high-salinity stresses and by PEG and ABA treatment (Fig. 2A; Supplemental Table S1). Meanwhile, more stress-related cis-acting elements, including ABRE, DRE, MYBRS, MYCRS, and GCC boxes, are present in the 1.5-kb promoter region of OsbZIP23 than in other members of this family (Fig. 2D). ABRE, MYBRS, and MYCRS can be recognized by the AREB/ABF, MYB, and MYC transcription factors, respectively. These cis-elements and the corresponding transcription factors have important roles in ABA signaling and abiotic stress responses (Yamaguchi-Shinozaki and Shinozaki, 2005, 2006). The GCC boxes are known as recognition sites for WRKY and ERF transcription factors and have been suggested to be involved in responses and adaptation to plant abiotic or biotic stresses (Eulgem et al., 2000; Brown et al., 2003). Such an enriched presence of various stress responsive cis-acting elements in the OsbZIP23 promoter may suggest a critical role of this gene in stress tolerance. In addition, phylogenetic analysis showed that OsbZIP23 is more closely related to the reported Arabidopsis homologs ABF1 to ABF4 than to the reported rice homolog OsABI5. ABF1 to ABF4 have been characterized for their critical roles in ABA-dependent signaling and stress tolerance (Choi et al., 2000; Kang et al., 2002; Kim et al., 2004). The strong induction of the OsbZIP23 gene by ABA and drought and salinity stresses and the high similarity of OsbZIP23 to ABF1 to ABF4 together suggest that OsbZIP23 may play critical roles in ABA-dependent signaling and stress tolerance in rice.
Second, overexpression and knockout of the OsbZIP23 gene resulted in significant changes in ABA sensitivity and stress resistance. The OsbZIP23-overexpressing rice plants showed significantly increased sensitivity to exogenous ABA at both germination and postgermination stages. To the contrary, the null mutant of this gene showed significantly decreased sensitivity to ABA at both germination and postgermination stages. Importantly, the phenotype of decreased ABA sensitivity in the mutant can be complemented by the OsbZIP23 gene under the control of its native promoter. The overexpression and null mutants of OsbZIP23 also showed opposite phenotypes in stress tolerance. The seedlings of the OsbZIP23 overexpression lines showed increased drought and salt tolerance, but the mutant plants performed slightly poorer (but still significantly) than the wild type under the stress conditions. This is also true for the drought resistance tested at the heading stage. Considering the facts that there are quite a few closely related members in the third subfamily of bZIP in rice (Fig. 1) and that a few other members in this subfamily are also induced by ABA and/or stresses, we originally suspected that there is a functional redundancy for OsbZIP23. But the mutant of the OsbZIP23 gene showed quite a strong phenotype (especially for ABA sensitivity), suggesting that it is a major (if not dispensable) player for mediating ABA-dependent stress responses. In rice, OsABI5 is so far the only bZIP transcription factor that was characterized for its role in stress tolerance through a transgenic approach. OsABI5-overexpressing plants were sensitive to ABA but also to high-salinity stress and PEG treatment, while the transgenic plants of antisense OsABI5 exhibited increased tolerance to salt and PEG treatment (Zou et al., 2007, 2008), suggesting a negative regulatory role of OsABI5 in stress tolerance, which is apparently distinct from the positive regulatory role of OsbZIP23. We also tested the TRAB1-overexpressing rice plants and the trab1 mutant as well for stress tolerance, but no significant change was observed (data not shown).
Third, overexpression of OsbZIP23 can change the expression levels of a large number of genes. It is not very surprising to detect expression changes of a certain number of genes in an overexpressor of a transcription factor. However, the numbers of the genes with expression level changes in the OsbZIP23 overexpressor and mutant are indeed unexpected. Nearly 800 genes showed greater than 2-fold up-regulation in the overexpression lines. Actually, more than 2,000 genes showed up- or down-regulation in the overexpressors at a threshold of 1.5-fold, although most of the genes with 1.5- to 2-fold (some of them are significant at P < 0.05) changes cannot be confirmed by real-time PCR (data not shown). On the other hand, knockout of this gene also resulted in expression changes of hundred of genes. Importantly, many of the up- or down-regulated genes in the OsbZIP23 overexpressor and mutant are predicted or shown to be involved in stress tolerance. Since OsbZIP23 has transactivation activity in yeast and many genes in rice can be affected in their expression levels in the OsbZIP23 overexpressor and mutant, we speculate that OsbZIP23 functions as a major transcriptional activator among the bZIP family to regulate genes involved in ABA sensitivity and stress tolerance in rice.
The results of leaf water-loss experiments indicated that OsbZIP23 overexpressors have stronger water-holding capability than wild-type plants, but the results of scanning electron microscopy suggested that stomata may not be involved in the drought resistance in this case, since neither density nor aperture of stomata showed differences between the overexpressor and the wild type. Nevertheless, the GeneChip profiling analysis provided informative clues for dissecting the molecular mechanism of the improved stress tolerance of transgenic rice plants overexpressing OsbZIP23. Among the up-regulated genes in OsbZIP23-overexpressing plant lines, a large number of genes have been reported or predicted to be involved in stress tolerance. We noticed that more than 10 genes encoding dehydrin or LEA proteins were significantly up-regulated in the overexpressors. Dehydrin family proteins have been reported for their roles in the plant-protective reactions against dehydration (Nylander et al., 2001; Lee et al., 2005). LEA proteins have been implicated in many stress responses of plants (Moons et al., 1997; Goyal et al., 2005; RoyChoudhury et al., 2007; Tunnacliffe and Wise, 2007) and can be regulated by bZIP transcription factors through ABRE cis-elements (Carles et al., 2002; Ditzer and Bartels, 2006; Kobayashi et al., 2008). There are five LEA-encoding genes up-regulated in OsbZIP23 overexpressor, including OsLEA3-1 (LOC_05g46480). In a recent publication, we confirmed that overexpression of OsLEA3-1 in rice can significantly improve drought resistance (in terms of relative yield under drought stress) under field conditions (Xiao et al., 2007). Meanwhile, a few genes encoding AP2 domain-containing proteins and several ABA-responsive genes, such as RD22-like and ABA/WDS-like-responsive, and heat shock protein genes were also up-regulated in OsbZIP23-overexpressing plants. Some of these types of genes have been implicated in stress response or tolerance (Kizis et al., 2001; Song et al., 2005). Many genes for seed storage or lipid transfer proteins were also up-regulated in OsbZIP23-overexpressing plants, and these proteins may help in maintaining cell membrane stability of plants under adverse environments (Brick et al., 1987; Hong et al., 2008). A large number of genes up-regulated in the overexpressors have been annotated with putative functions in photosynthesis and amino acid metabolism. Up-regulation of these genes in the transgenic plants might contribute to the energy supply of the plants under adverse conditions. We also noticed that a few genes encoding putative ion transporters were also up-regulated in the overexpressors, but whether or how these up-regulated genes contribute to the improved salt tolerance requires further evidence.
Although more than 1,000 genes showed decreased expression in the overexpression lines, only a small portion of them might be related to abiotic stress according to their annotations. It cannot be excluded that some of these down-regulated genes may also contribute to the improved stress tolerance or other growth or developmental aspects, since a majority of these genes are unknown in their functions. The expression profiling analysis of the mutant has also provided some informative clues on the stress tolerance conferred by the OsbZIP23 gene. For example, most of those genes up-regulated in the OsbZIP23 overexpression plants were down-regulated or not changed (because of partial functional redundancy) from their expression levels in the mutant. Many of the genes down-regulated in the mutant are also stress responsive and/or annotated with putative functions in stress response or tolerance.
Even though a full scenario of the molecular mechanism of the stress resistance conferred by OsbZIP23 remains to be completely revealed, the expression changes of all the well-annotated or evidenced stress-related genes in the overexpression or mutant line suggest that enhanced protection ability at the cellular level (such as osmotic adjustment, stabilization of functional proteins, and cell membrane) may be a major contribution to the stress tolerance conferred by OsbZIP23.
In conclusion, this study showed that OsbZIP23 belongs to the third subfamily of bZIP transcription factors and positively regulates ABA sensitivity and stress resistance in rice. Alteration of OsbZIP23 expression can change the expression levels of more than 1,000 genes, and many of these genes are involved in stress responses or tolerance. Our results also suggest that the OsbZIP23 gene has high potential usefulness in improving the stress resistance of crops.
MATERIALS AND METHODS
Generation of Transgenic Plants and Knockout Mutant
The full-length cDNAs of OsbZIP23 were amplified from upland rice (Oryza sativa IRAT109) by RT-PCR with the following primers: 5′-TAAGGTACCATCCCACCTCTCCTCAGGTT-3′ and 5′-TAAGGATCCGGCAGCTTCACCATCCTACT-3′ (the underlined sequences are for restriction sites KpnI and BamHI, respectively). The sequence-confirmed PCR fragment was digested by KpnI and BamHI and ligated into the binary expression vector pCAMBIA1301U, which was also digested with KpnI and BamHI, thus allowing the genes to be driven by a maize (Zea mays) ubiquitin promoter. The construct was introduced into japonica cv Zhonghua 11 by Agrobacterium-mediated transformation (Hiei et al., 1994).
Mutant seeds of OsbZIP23 were obtained from the Rice Mutant Database (http://rmd.ncpgr.cn/; Zhang et al., 2006). For mutant complementation, the bacterial artificial chromosome clone A0022C06 from the bacterial artificial chromosome library of japonica rice Nipponbare was digested with BamHI, and the target fragment containing the OsbZIP23 gene and its promoter was ligated into pCAMBIA2301. The complementation construct was introduced into the mutant lines by Agrobacterium-mediated transformation.
Stress Tolerance Testing of Transgenic Plants
The planting and drought treatment in the PVC pipes (1 m in length and 0.2 m in diameter) was essentially the same as described previously (Yue et al., 2006; Xiao et al., 2007). For each homozygous T2 line, 24 plants were divided into two groups for drought stress and normal growth treatments, respectively, and planted individually in the PVC pipes placed under plastic tents (length × width × height, 26 × 6 × 3.6 m) with foldable roofs. The wild-type plants were inserted every six transgenic plants for comparison. Drought stress was initiated at the panicle development stage by discharging water through a hole near the bottom of the pipes. After 2 weeks from water deprivation, a period that can normally cause serious drought stress (almost all leaves became completely rolled and some of the leaves died) in this environment, plants were reirrigated to allow recovery at the flowering and seed maturation stages. For drought stress testing at the seedling stage, homozygous T2 transgenic and wild-type plantlets were growing in barrels filled with a mixture of sand and soil (1:1). At the five-leaf stage, watering was stopped and then resumed 5 d later. For high-salinity stress testing, homozygous T2 transgenic and wild-type plantlets were growing in barrels filled with paddy soil. At the five-leaf stage, the plants were irrigated with water containing 200 mm NaCl and kept for growth for 10 d. After that, we investigated the green leaf area. To measure the water loss rate under dehydration conditions, leaves of homozygous transgenic plants at the five-leaf stage were cut and exposed to air at room temperature (approximately 24°C). The leaves were weighed at 0, 0.5, 2, 4, 6, 8, 12, and 24 h after being cut down.
Plant Materials and Stress Treatment
To detect the transcript levels of genes, IRAT109 rice plants were grown in the greenhouse with a 14-h-light/10-h-dark cycle. Two-week-old seedlings were treated with chemical or abiotic stress. Chemical treatments were conducted by spraying leaves with 0.1 mm ABA followed by sampling at 0, 3, 6, 12, and 24 h, or spraying leaves with 0.1 mm JA, 0.1 mm SA, or 0.1 mm 2,4-D followed by sampling at 0, 1, 2, 6, and 12 h, or irrigating the plants with 20% PEG 6000 followed by sampling at 0, 1, 5, and 12 h. Abiotic stress treatments for gene expression level analysis were essentially the same as in our previous report (Xiang et al., 2007). For cold stress, seedlings were transferred to a growth chamber at 4°C and sampled at 0, 3, 6, 12, and 24 h after treatment. Drought stress was realized by putting intact plants in the air without water supply, and plant leaves were sampled at 0, 3, 6, 12, and 24 h after treatment. The 2-week-old seedlings were irrigated with 200 mm NaCl solution for salt stress and sampled at 0, 5, 14, and 24 h after treatment.
RNA Isolation and Quantitative PCR
Total RNA was isolated from rice leaves using TRIzol reagent (Invitrogen). For real-time PCR analysis, first-strand cDNAs were synthesized from DNaseI-treated total RNA using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Real-time PCR was performed on an optical 96-well plate with an ABI PRISM 7500 real-time PCR system (Applied Biosystems). Each reaction contained 10 μL of 2× SYBR Green Master Mix Reagent (Applied Biosystems), 1.0 μL of cDNA samples, and 200 nm gene-specific primer in a final volume of 20 μL. The thermal cycle used was as follows: 95°C for 3 min, then 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min. The rice Actin1 gene (accession no. X16280) was used as an internal control with primers 5′-TGGCATCTCTCAGCACATTCC-3 and 5′-TGCACAATGGATGGGTCAGA-3′. The relative expression levels were determined as described previously (Livak and Schmittgen, 2001).
Subcellular Localization and Transactivation Assay
The subcellular localization vector was constructed by replacing the GUS fragment of pCAMBIA1391Xb (CAMBIA) with a ubiquitin promoter-GFP cassette. The coding region of OsbZIP23 cDNA was amplified from IRAT109 rice using the following primer pair: 5′-TAAGGTACCATCCCACCTCTCCTCAGGTT-3′ (the underlined part is for the KpnI site) and 5′-TAAAAGCTTGATCAGAGACGGGAACCTGA-3′ (the underlined part is for the BamHI site). The amplified fragment was inserted upstream of and in frame with GFP. The procedure of bombarding onion (Allium cepa) epidermal cells was conducted as described by Dai et al. (2007). The expression of the OsbZIP23-GFP fusion protein in onion epidermal cells was observed by confocal microscopy at 36 h after bombardment. For transactivation assay, the open reading frame of OsbZIP23 generated by PCR was fused in frame with the yeast GAL4 DNA binding domain in the vector pDEST32 by the Gateway Recombination Cloning method (Invitrogen). The fused gene was expressed in yeast strain Mav203 (Invitrogen). The transformed yeast strain was plated on SD/Leu− medium and cultured for 3 d, and colony-lift filter assay (X-gal assay) was performed as described by the manufacturer (Invitrogen).
GeneChip Analysis
RNA samples of transgenic plants of overexpressors, the T-DNA insertion mutant, and the wild type were extracted using TRIzol (Invitrogen) as described by the manufacturer. Three independent samples (replicates) were used for each line. Probe labeling and chip hybridization were carried out through the Affymetrix custom service (CapitalBio) by following the standard protocol (http://www.affymetrix.com/support/technical/manual/expression_manual.affx). Normalization was performed according to the standard Affymetrix protocol to allow the comparison of the samples for each set of experiments. The Excel add-in for significance analysis of microarrays was used to identify differentially expressed genes between the control and transgenic plants. Partial up- or down-regulated genes from GeneChip analysis were confirmed by real-time PCR analysis using the rice Actin1 gene as an internal control for the calibration of relative expression. Stress responsiveness of the genes that were up- or down-regulated in the overexpression or mutant plant were checked in our previous expression profiling analysis of rice under various stress conditions (Zhou et al., 2007; Y. Xiang, N. Tang, H. Du, H. Ye, and L. Xiong, unpublished data).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AK072062.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. A 9-bp deletion in the cDNA sequence of OsbZIP23 from japonica rice IRAT109 compared with indica rice Zhengshan 97 (only the partial cDNA sequence containing the polymorphism is shown).
Supplemental Figure S2. Northern-blot analysis of the expression level of OsbZIP23 in transgenic plants under normal growth conditions.
Supplemental Figure S3. Southern-blot analysis of T-DNA copy number of the OsbZIP23 transgene (DNA probe of a hygromicin phosphotransferase gene was used for hybridization).
Supplemental Figure S4. Phenotype of OsbZIP23 transgenic plants and wild-type plants grown on normal MS medium for 10 d.
Supplemental Figure S5. Identification of the Osbzip23 mutant.
Supplemental Figure S6 Confirmation of additional genes with expression level changes in the OsbZIP23 overexpressors (A) and mutant (B) by real-time PCR (this result is an addition to the data in Figure 8).
Supplemental Table S1. Expression level changes of the third subfamily bZIP genes after drought, high-salinity, low-temperature, and ABA treatments.
Supplemental Table S2. Grain yield and spikelet fertility of the OsbZIP23-overexpressing transgenic plants (T2) and the mutant under normal and drought stress conditions.
Supplemental Table S3. Up-regulated genes in transgenic rice plants overexpressing OsbZIP23 and their expression level changes after drought, high-salinity stress, or ABA treatment by GeneChip analysis
Supplemental Table S4. Down-regulated genes in transgenic rice plants overexpressing OsbZIP23 and their expression level changes after drought, high-salinity stress, or ABA treatment by GeneChip analysis.
Supplemental Table S5. Up-regulated genes in the Osbzip23 mutant and their expression level changes after drought, high-salinity stress, or ABA treatment by GeneChip analysis.
Supplemental Table S6. Down-regulated genes in the Osbzip23 mutant and their expression level changes after drought, high-salinity stress, or ABA treatment by GeneChip analysis.
Supplemental Table S7. Genes with expression levels reversely regulated between the OsbZIP23 overexpressor and the mutant.
Supplemental Table S8. Primers used for real-time PCR analysis.
Supplementary Material
This work was supported by the National Special Key Project of China on Functional Genomics of Major Plants and Animals, the National Program on the Development of Basic Research, the National Natural Science Foundation of China, the Ministry of Education of China (grant no. NO 707045), the European Union FP6 INCO-MPC2 project (grant no. INCOCT–2005–015468), and the Rockefeller Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Lizhong Xiong (lizhongx@mail.hzau.edu.cn).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bensmihen S, Giraudat J, Parcy F (2005) Characterization of three homologous basic leucine zipper transcription factors (bZIP) of the ABI5 family during Arabidopsis thaliana embryo maturation. J Exp Bot 56 597–603 [DOI] [PubMed] [Google Scholar]
- Bensmihen S, Rippa S, Lambert G, Jublot D, Pautot V, Granier F, Giraudat J, Parcy F (2002) The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14 1391–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick J, Pohorecky LA, DeTurck K (1987) Cardiac lipase: effect of ethanol and stress. Life Sci 40 1897–1901 [DOI] [PubMed] [Google Scholar]
- Brown RL, Kazan K, McGrath KC, Maclean DJ, Manners JM (2003) A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol 132 1020–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busk PK, Pages M (1998) Regulation of abscisic acid-induced transcription. Plant Mol Biol 37 425–435 [DOI] [PubMed] [Google Scholar]
- Carles C, Bies-Etheve N, Aspart L, Leon-Kloosterziel KM, Koornneef M, Echeverria M, Delseny M (2002) Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J 30 373–383 [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Ji F, Xie H, Liang J, Zhang J (2006) The regulator of G-protein signaling proteins involved in sugar and abscisic acid signaling in Arabidopsis seed germination. Plant Physiol 140 302–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, Koshiba T, Sheen J (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275 1723–1730 [DOI] [PubMed] [Google Scholar]
- Dai M, Hu Y, Zhao Y, Liu H, Zhou DX (2007) A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol 144 380–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S, Zhang Z, Chen S, Beachy RN (2004) RF2b, a rice bZIP transcription activator, interacts with RF2a and is involved in symptom development of rice tungro disease. Proc Natl Acad Sci USA 101 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzer A, Bartels D (2006) Identification of a dehydration and ABA-responsive promoter regulon and isolation of corresponding DNA binding proteins for the group 4 LEA gene CpC2 from C. plantagineum. Plant Mol Biol 61 643–663 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5 199–206 [DOI] [PubMed] [Google Scholar]
- Fedoroff NV (2002) Cross-talk in abscisic acid signaling. Sci STKE 2002 RE10. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14 S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal K, Walton LJ, Tunnacliffe A (2005) LEA proteins prevent protein aggregation due to water stress. Biochem J 388 151–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A, He K, Liu D, Bai S, Gu X, Wei L, Luo J (2005) DATF: a database of Arabidopsis transcription factors. Bioinformatics 21 2568–2569 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6 271–282 [DOI] [PubMed] [Google Scholar]
- Himmelbach A, Yang Y, Grill E (2003) Relay and control of abscisic acid signaling. Curr Opin Plant Biol 6 470–479 [DOI] [PubMed] [Google Scholar]
- Hobo T, Kowyama Y, Hattori T (1999) A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc Natl Acad Sci USA 96 15348–15353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JK, Choi HW, Hwang IS, Kim DS, Kim NH, Choi du S, Kim YJ, Hwang BK (2008) Function of a novel GDSL-type pepper lipase gene, CaGLIP1, in disease susceptibility and abiotic stress tolerance. Planta 227 539–558 [DOI] [PubMed] [Google Scholar]
- Iida K, Seki M, Sakurai T, Satou M, Akiyama K, Toyoda T, Konagaya A, Shinozaki K (2005) RARTF: database and tools for complete sets of Arabidopsis transcription factors. DNA Res 12 247–256 [DOI] [PubMed] [Google Scholar]
- Iwata Y, Koizumi N (2005) An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc Natl Acad Sci USA 102 5280–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7 106–111 [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Hobo T, Murata M, Ban A, Hattori T (2002) Abscisic acid-induced transcription is mediated by phosphorylation of an abscisic acid response element binding factor, TRAB1. Plant Cell 14 3177–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kang JY, Cho DI, Park JH, Kim SY (2004) ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J 40 75–87 [DOI] [PubMed] [Google Scholar]
- Kizis D, Lumbreras V, Pages M (2001) Role of AP2/EREBP transcription factors in gene regulation during abiotic stress. FEBS Lett 498 187–189 [DOI] [PubMed] [Google Scholar]
- Kobayashi F, Maeta E, Terashima A, Kawaura K, Ogihara Y, Takumi S (2008) Development of abiotic stress tolerance via bZIP-type transcription factor LIP19 in common wheat. J Exp Bot 59 891–905 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Murata M, Minami H, Yamamoto S, Kagaya Y, Hobo T, Yamamoto A, Hattori T (2005) Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J 44 939–949 [DOI] [PubMed] [Google Scholar]
- Ladyzhenskaya EP (2001) Pathways of the abscisic acid hormonal signal transduction across the plant cell plasma membrane. Membr Cell Biol 14 699–713 [PubMed] [Google Scholar]
- Lee SC, Lee MY, Kim SJ, Jun SH, An G, Kim SR (2005) Characterization of an abiotic stress-inducible dehydrin gene, OsDhn1, in rice (Oryza sativa L.). Mol Cells 19 212–218 [PubMed] [Google Scholar]
- Liu Y, Dai S, Beachy RN (2007) Role of the C-terminal domains of rice (Oryza sativa L.) bZIP proteins RF2a and RF2b in regulating transcription. Biochem J 405 243–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)).Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH (2003) AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev 17 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32 317–328 [DOI] [PubMed] [Google Scholar]
- Moons A, De Keyser A, Van Montagu M (1997) A group 3 LEA cDNA of rice, responsive to abscisic acid, but not to jasmonic acid, shows variety-specific differences in salt stress response. Gene 191 197–204 [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Choudhury AR, Gupta B, Gupta S, Sengupta DN (2006) An ABRE-binding factor, OSBZ8, is highly expressed in salt tolerant cultivars than in salt sensitive cultivars of indica rice. BMC Plant Biol 6 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H, Ohmiya K, Hattori T (1996) A rice bZIP protein, designated OSBZ8, is rapidly induced by abscisic acid. Plant J 9 217–227 [DOI] [PubMed] [Google Scholar]
- Nijhawan A, Jain M, Tyagi AK, Khurana JP (2008) Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol 146 333–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander M, Svensson J, Palva ET, Welin BV (2001) Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol Biol 45 263–279 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405 462–466 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Uribe L, O'Connell MA (2006) A root-specific bZIP transcription factor is responsive to water deficit stress in tepary bean (Phaseolus acutifolius) and common bean (P. vulgaris). J Exp Bot 57 1391–1398 [DOI] [PubMed] [Google Scholar]
- Rook F, Gerrits N, Kortstee A, van Kampen M, Borrias M, Weisbeek P, Smeekens S (1998) Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J 15 253–263 [DOI] [PubMed] [Google Scholar]
- RoyChoudhury A, Roy C, Sengupta DN (2007) Transgenic tobacco plants overexpressing the heterologous lea gene Rab16A from rice during high salt and water deficit display enhanced tolerance to salinity stress. Plant Cell Rep 26 1839–1859 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31 279–292 [DOI] [PubMed] [Google Scholar]
- Shimizu H, Sato K, Berberich T, Miyazaki A, Ozaki R, Imai R, Kusano T (2005) LIP19, a basic region leucine zipper protein, is a Fos-like molecular switch in the cold signaling of rice plants. Plant Cell Physiol 46 1623–1634 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3 217–223 [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnacliffe A, Wise MJ (2007) The continuing conundrum of the LEA proteins. Naturwissenschaften 94 791–812 [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Huang Y, Xiong L (2007) Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol 144 1416–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Huang Y, Tang N, Xiong L (2007) Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor Appl Genet 115 35–46 [DOI] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK (2002) Cell signaling during cold, drought, and salt stress. Plant Cell (Suppl) 14 S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2005) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci 10 88–94 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57 781–803 [DOI] [PubMed] [Google Scholar]
- Yin Y, Zhu Q, Dai S, Lamb C, Beachy RN (1997) RF2a, a bZIP transcriptional activator of the phloem-specific rice tungro bacilliform virus promoter, functions in vascular development. EMBO J 16 5247–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue B, Xue W, Xiong L, Yu X, Luo L, Cui K, Jin D, Xing Y, Zhang Q (2006) Genetic basis of drought resistance at reproductive stage in rice: separation of drought tolerance from drought avoidance. Genetics 172 1213–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li C, Wu C, Xiong L, Chen G, Zhang Q, Wang S (2006) RMD: a rice mutant database for functional analysis of the rice genome. Nucleic Acids Res 34 D745–D748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang X, Jiao Y, Qin Y, Liu X, He K, Chen C, Ma L, Wang J, Xiong L, et al (2007) Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Mol Biol 63 591–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Ordiz MI, Dabi T, Beachy RN, Lamb C (2002) Rice TATA binding protein interacts functionally with transcription factor IIB and the RF2a bZIP transcriptional activator in an enhanced plant in vitro transcription system. Plant Cell 14 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M, Guan Y, Ren H, Zhang F, Chen F (2007) Characterization of alternative splicing products of bZIP transcription factors OsABI5. Biochem Biophys Res Commun 360 307–313 [DOI] [PubMed] [Google Scholar]
- Zou M, Guan Y, Ren H, Zhang F, Chen F (2008) A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol 66 675–683 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.