Abstract
Calcineurin B-like (CBL) proteins represent a unique family of calcium sensors in plant cells. Sensing the calcium signals elicited by a variety of abiotic stresses, CBLs transmit the information to a group of serine/threonine protein kinases (CBL-interacting protein kinases [CIPKs]), which are currently known as the sole targets of the CBL family. Here, we report that the CBL3 member of this family has a novel interaction partner in addition to the CIPK proteins. Extensive yeast two-hybrid screenings with CBL3 as bait identified an interesting Arabidopsis (Arabidopsis thaliana) cDNA clone (named AtMTAN, for 5′-methylthioadenosine nucleosidase), which encodes a polypeptide similar to EcMTAN from Escherichia coli. Deletion analyses showed that CBL3 utilizes the different structural modules to interact with its distinct target proteins, CIPKs and AtMTAN. In vitro and in vivo analyses verified that CBL3 and AtMTAN physically associate only in the presence of Ca2+. In addition, we empirically demonstrated that the AtMTAN protein indeed possesses the MTAN activity, which can be inhibited specifically by Ca2+-bound CBL3. Overall, these findings suggest that the CBL family members can relay the calcium signals in more diverse ways than previously thought. We also discuss a possible mechanism by which the CBL3-mediated calcium signaling regulates the biosynthesis of ethylene and polyamines, which are involved in plant growth and development as well as various stress responses.
Plant cells use calcium ion (Ca2+) as a second messenger in mediating a number of various signal transduction pathways. Changes in the cytosolic concentrations of free Ca2+ ([Ca2+]cyt) precede a wide range of cellular and developmental processes as well as responses to biotic and abiotic stimuli (White and Broadley, 2003). This raises an intriguing question: how can a simple element like Ca2+ be involved in such a large number of diverse signal transduction pathways and yet manage to produce a stimulus-specific response? Recent progress in this research area has begun to provide some useful explanations. It seems that the specificity of Ca2+ signaling pathways can be achieved at multiple levels.
First, the Ca2+ signal itself is so complex that it can actually convey diverse information. According to a recently formulated concept, “Ca2+ signatures” are represented not only by the concentrations of Ca2+ but also by temporal and spatial parameters, which consist of frequency, duration, and subcellular localization of the transient increases in [Ca2+]cyt (Evans et al., 2001; Rudd and Franklin-Tong, 2001; Sanders et al., 2002). In addition to the Ca2+ flux and reflux across the plasma membrane, the rates at which the cytosolic Ca2+ enters and exits intracellular compartments, including the endoplasmic reticulum, Golgi apparatus, vacuole, and nucleus, can also contribute to generation of the Ca2+ signatures with distinct temporal and spatial information (Bootman et al., 2001; Sanders et al., 2002). Such complexity of the Ca2+ parameters, therefore, allows plant cells to produce distinct Ca2+ signatures in response to disparate stimuli.
Next, additional levels of the specificity in the Ca2+ signaling cascades can be attributed to the existence of many Ca2+-binding proteins in plant cells, which possess different characteristics such as Ca2+-binding affinity, expression pattern, and subcellular localization. These Ca2+-binding proteins sense and transduce the changes in the Ca2+ parameters to their distinct target proteins, thereby channeling them into disparate signaling pathways. Therefore, it is conceivable that specificity in the Ca2+ signal transduction pathways can be largely determined by a specific Ca2+ signature generated by a particular stimulus and the availability of a distinct set of Ca2+ sensors. To date, three major families of Ca2+ sensors in plants have been most extensively studied: calcium-dependent protein kinase (CDPK), calmodulin (CaM), and calcineurin B-like protein (CBL).
The CDPK family, which consists of 34 genes in the Arabidopsis (Arabidopsis thaliana) genome, can be classified as a sensor responder, because it consists of the C-terminal CaM-like Ca2+ sensor and the N-terminal kinase responder. There are approximately 34 CDPK genes in the Arabidopsis genome (Hrabak et al., 2003). Meanwhile, CaMs are classified as sensor relays, because they have no enzymatic activities themselves. Upon Ca2+ binding, CaMs undergo conformational changes and thereby associate mainly by hydrophobic interaction with a number of diverse target proteins, such as NAD kinase, Glu decarboxylase, Ca2+-ATPase, protein kinases, and transcription factors (Yang and Poovaiah, 2003).
The most recently identified is the CBL family, which is most similar to the regulatory B subunit of the protein phosphatase calcineurin in animals and does not have enzymatic activities like CaMs (Liu and Zhu, 1998; Kudla et al., 1999). The CBL family members, consisting of 10 genes in both Arabidopsis and rice (Oryza sativa; Luan et al., 2002; Kolukisaoglu et al., 2004), were predicted to contain three to four EF-hand motifs (Kudla et al., 1999; Nagae et al., 2003). As the sensor relays, the CBL family members are currently known to interact exclusively with a group of Ser/Thr protein kinases called CIPKs (for CBL-interacting protein kinases), thereby mediating the calcium signals elicited by various stimuli, including cold, salinity, low K+ concentration, high pH, abscisic acid, and osmotic stress (Shi et al., 1999; Halfter et al., 2000; Kim et al., 2000, 2003; Albrecht et al., 2001, 2003; Guo et al., 2002; Kolukisaoglu et al., 2004; Jeong et al., 2005; D'Angelo et al., 2006; Li et al., 2006; Xu et al., 2006; Fuglsang et al., 2007; Quan et al., 2007). Furthermore, analyses of Arabidopsis and rice mutant plants demonstrated that CBLs are also involved in plant responses to drought, Glc, and gibberellic acid (Cheong et al., 2003; Pandey et al., 2004; Hwang et al., 2005). At present, however, it is not known whether or not the responses are mediated through the CIPK members as well.
Judging from the number of the CBL-mediated stimuli identified so far and the target diversity exhibited by another sensor relay CaM, as mentioned above, it is reasonable to think that CBLs associate with a variety of proteins in addition to CIPKs to mediate more diverse signals. Therefore, in this study, we tried to investigate the possibility of novel CBL interactors that do not belong to the CIPK family. To this end, we carried out extensive yeast two-hybrid screening of Arabidopsis cDNA libraries using a member of the CBL family, CBL3, as bait to seek out new interaction partners. Through the screening, a novel CBL3-interacting cDNA clone was identified and designated AtMTAN because it encodes a polypeptide similar to the 5′-methylthioadenosine nucleosidase (MTAN; EC 3.2.2.16) in Escherichia coli. In fact, we demonstrated that AtMTAN possesses the MTAN activity, which can be inhibited by CBL3 in a Ca2+-dependent manner. Interestingly, AtMTAN did not interact with other CBL family members such as CBL1 and CBL4. Taken together, our findings strongly suggest that each of the CBL family members may have distinct target proteins along with CIPKs, which allows CBLs to control more diverse cellular processes in plant cells.
RESULTS
Isolation of a Novel CBL3 Interactor, AtMTAN
To investigate whether or not the CBL Ca2+ sensors have interaction partners other than CIPKs, we extensively screened the Arabidopsis λACT cDNA expression libraries CD4-10 and CD4-22 obtained from the Arabidopsis Biological Resource Center via a yeast two-hybrid system using CBL3 as bait. The CBL3 bait (pGBT.CBL3 or BD.CBL3) was created by cloning the complete coding region of CBL3 cDNA into the GAL4 DNA-binding domain vector pGBT9.BS (BD). A total of 74 positive clones were obtained from the screening, and their sequence analysis revealed that most of them represent, as anticipated, the previously known CIPK members CIPK1, CIPK2, CIPK3, CIPK6, CIPK9, CIPK11, CIPK12, CIPK15, and CIPK21 (Kolukisaoglu et al., 2004; Jeong et al., 2005). Isolation of these CIPK genes indicated that the yeast two-hybrid screening had been carried out efficiently and successfully.
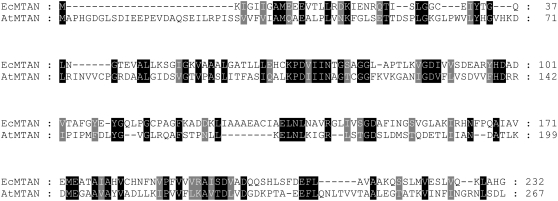
Fortunately, however, we were also able to discover five positive clones that do not belong to the CIPK family. Although the novel clones contained different sizes of inserts varying from 903 to 932 bp in length, we found that they all derived from a single gene (At4g38800) and contained an open reading frame (804 bp) encoding a polypeptide of 267 amino acids with an estimated molecular mass of 28.5 kD. Through GenBank searches, we realized that the polypeptide shows sequence similarity (25% identity) to MTAN in E. coli (Fig. 1). Therefore, the novel CBL3-interacting protein isolated from Arabidopsis was designated AtMTAN and subjected to further analysis. Comparison of the full-length AtMTAN cDNA sequence with its genomic counterpart revealed that the AtMTAN gene consists of eight exons and seven introns in the Arabidopsis genome (data not shown).
Figure 1.
Sequence alignment of E. coli MTAN and Arabidopsis MTAN. Amino acid sequences of EcMTAN (accession no. NP_414701) and AtMTAN (accession no. NP_195591) were aligned using the Lasergene MegAlign program (DNASTAR) and modified with GeneDoc software. Identical amino acids are shaded black, and amino acids with similar characteristics are shaded gray. Dashes represent gaps to maximize the alignment.
AtMTAN Interacts Specifically with CBL3 But Not with CBL1 and CBL4
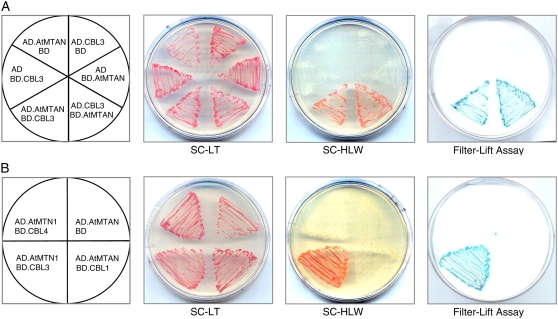
To test whether the complete form of AtMTAN maintains the interaction with CBL3 in yeast cells, we created the pGAD.AtMTAN (or AD.AtMTAN) construct by cloning just the coding region (open reading frame) of AtMTAN cDNA into the yeast expression vector pGAD.GH (AD), which contains the GAL4 activation domain. As shown in Figure 2A (the left half circle), the Y190 yeast cells carrying both AD.AtMTAN and BD.CBL3 grew well on the selection medium (SC-HLW) and exhibited β-galactosidase activity, demonstrating expression of the reporter genes HIS3 and LacZ, respectively. However, the yeast cells cotransformed with either BD.CBL3 and AD or BD and AD.AtMTAN failed to express the reporter genes. These results indicated that CBL3 and AtMTAN indeed interact in the yeast two-hybrid system.
Figure 2.
Yeast two-hybrid assays. A, AtMTAN interacts with CBL3 in a vector-independent manner. B, Interaction specificity between AtMTAN and CBL3. The first panels at left show the arrangement of the Y190 yeast cells carrying the indicated pGBT and pGAD plasmids. The second and third panels display yeast growth on synthetic complete medium lacking Leu and Trp (SC-LT) and synthetic complete medium lacking His, Leu, and Trp (SC-HLW), respectively. The last panels show β-galactosidase activity (filter-lift assay). [See online article for color version of this figure.]
In addition, we carried out vector-swapping analysis by constructing the pGAD.CBL3 (AD.CBL3) and pGBT.AtMTAN (BD.AtMTAN) plasmids. As shown in Figure 2A (the right half circle), the yeast cells possessing AD.CBL3 and BD.AtMTAN expressed the two reporter genes as well, revealing that the interaction between CBL3 and AtMTAN occurs regardless of the vectors expressing the two proteins. Meanwhile, we were also interested in determining whether the AtMTAN protein could interact with other members of the CBL family. Figure 2B shows that AtMTAN interacts only with CBL3 but not with other CBL family members such as CBL1 and CBL4. The specificity observed in the CBL3-AtMTAN interaction suggests the possibility that each of the CBL family members may have distinct interaction partners other than the CIPK family.
Interaction Domains in AtMTAN and CBL3
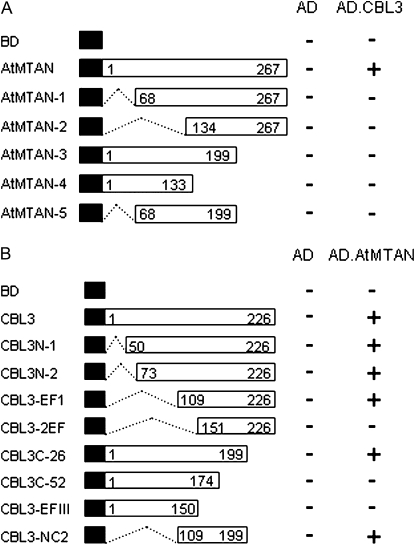
To delimit the AtMTAN region necessary for the interaction with CBL3, we created a series of deletion constructs by cloning AtMTAN fragments into the BD vector. These constructs were then introduced into yeast cells carrying either AD or AD.CBL3. Interactions were determined by monitoring the expression of the HIS3 and LacZ reporter genes by the transformed yeast cells. None of the AtMTAN deletion mutants interacted with CBL3 (Fig. 3A), suggesting that the complete form of AtMTAN is required for the interaction with CBL3.
Figure 3.
Structural requirement of AtMTAN and CBL3 for their interaction. A, Deletion mutants of AtMTAN. Different regions of AtMTAN were cloned into the pGBT9.BS vector and transformed into Y190 yeast cells carrying pGAD.GH or pGAD.CBL3. B, Identification of the CBL3 region involved in the interaction with AtMTAN. A series of CBL3 deletion mutants were created in the pGBT9.BS vector and cotransformed into yeast cells with either pGAD.GH or pGAD.AtMTAN. Yeast growth on the selection medium (SC-HLW) was scored as growth (+) or no growth (−). Black boxes indicate the binding domain of the GAL4 transcription factor. Numbers in the white boxes indicate the beginning and the ending positions of each protein fragment.
We also determined the structural requirement of CBL3 for the interaction with AtMTAN. As shown in Figure 3B, N-terminal deletions down to the 108th amino acid residue of CBL3 did not interfere with its affinity toward MTAN, and removal of the last 27 amino acid residues (CBL3C-1) from the C-terminal end of CBL3 still maintained the interaction with AtMTAN. However, further deletions from either the N- or C-terminal end completely abolished the interaction. Moreover, the CBL3-2EF mutant containing the 91 amino acids between the 109th and 199th amino acid residues of CBL3 retained the ability to interact with AtMTAN. Taken together, these results strongly suggest that the 91-amino acid region of CBL3 harbors all of the sequence information required and is sufficient for the interaction with AtMTAN.
It is interesting that CBL3 is known to require its entire sequence in order to interact with the previously known interactors, the CIPK family members (Kim et al., 2000). Based on these findings, it seems apparent that CBL3 uses different structural modules depending on its interaction partners.
Expression Patterns of AtMTAN
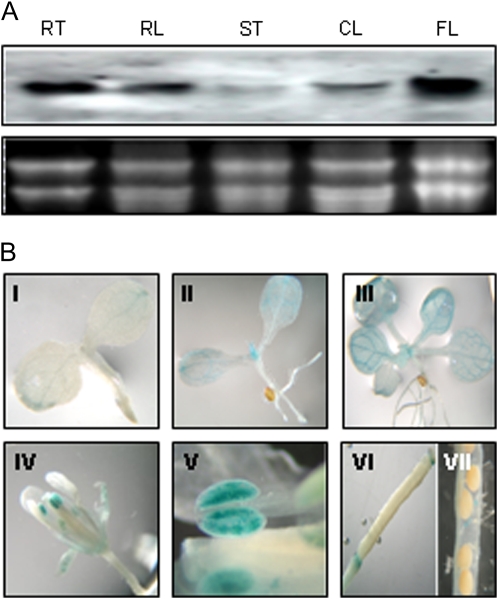
To determine the spatial expression pattern of the AtMTAN gene, we carried out RNA gel-blot analysis using total RNA prepared from the various organs of 5-week-old Arabidopsis wild-type plants (ecotype Columbia [Col-0]). The AtMTAN gene was expressed basically in all organs tested (Fig. 4A), although there were significant differences in the expression levels: the flowers expressed the highest level of the AtMTAN transcripts, and other organs displayed lower levels of AtMTAN expression in the order roots, rosette leaves, cauline leaves, and stems.
Figure 4.
Expression patterns of the AtMTAN gene. A, RNA gel-blot analysis. Each lane was loaded with 10 μg of total RNA isolated from various organs of 5-week-old Arabidopsis plants: root (RT), rosette leaf (RL), stem (ST), cauline leaf (CL), and flower (FL). B, Histochemical GUS assay of AtMTAN promoter-GUS transgenic plants. I, Three-day-old seedling; II, 1-week-old seedling; III, 2-week-old seedling; IV, flower; V, anther; VI, pistil; VII, silique.
We generated transgenic Arabidopsis plants that carry a fusion construct of the GUS reporter gene driven by the AtMTAN promoter (pBI.AtMTAN) in order to further analyze AtMTAN gene expression in terms of plant development. Histochemical staining of the transgenic plants showed that GUS activity began to appear at the tip of the cotyledons of the 3-d-old seedlings and expanded into the rest as the plants grew (Fig. 4B). GUS activity was also detected in the shoot apex and the leaves of young seedlings, particularly in the vascular tissues. In the mature plants, strong GUS activity was mainly found in sepals and anthers, whereas no GUS activity was observed in the remaining flower parts, such as pistils and petals. It is noteworthy that only the mature anthers, not the immature ones, exhibited the GUS activity, suggesting that AtMTAN is involved in pollen development.
CBL3 Physically Interacts with AtMTAN in a Ca2+-Dependent Manner in Vitro
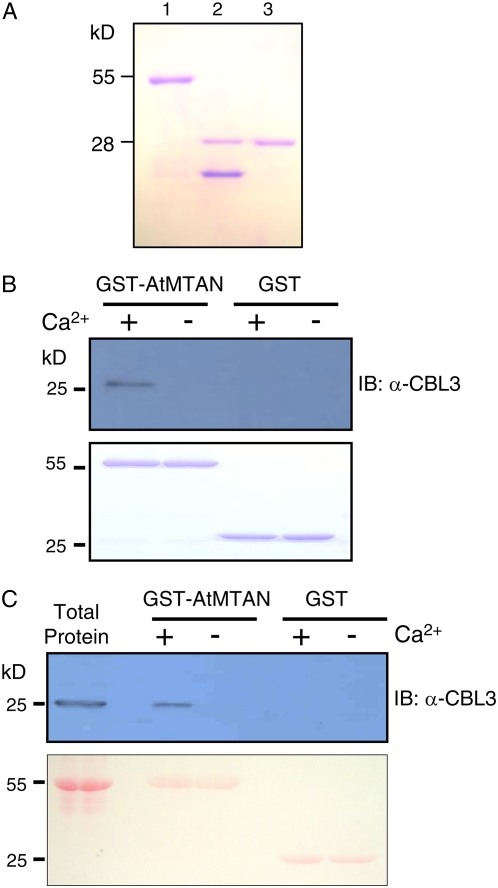
To verify the CBL3-AtMTAN interaction demonstrated in the yeast two-hybrid system in vitro, we first expressed and purified both CBL3 and AtMTAN proteins from E. coli using the glutathione S-transferase (GST) gene fusion system (Amersham Biosciences). Figure 5A shows an approximately 28-kD AtMTAN protein band that was originally purified as a GST fusion form and subsequently digested with thrombin to remove the GST protein. Similarly, the CBL3 protein was purified and used to produce polyclonal antibodies from rabbit (Jeong et al., 2005).
Figure 5.
AtMTAN forms a complex with CBL3 only in the presence of calcium in vitro. A, Purification of the recombinant AtMTAN protein expressed in E. coli. Lanes 1 to 3 contain the GST-AtMTAN fusion protein, the thrombin-digested forms of GST and AtMTAN, and purified AtMTAN, respectively. B, Pull-down assay. The GST-AtMTAN fusion protein was used as a bait to pull down the prey CBL3 in the presence (+) or absence (−) of calcium. For a negative control, GST was used as bait. The top panel shows an immunoblot probed with rabbit anti-CBL3 antibody, and the bottom panel shows a Coomassie Brilliant Blue-stained SDS-PAGE gel indicating the amount of bait proteins used in each pull-down assay. C, Affinity purification of CBL3 by GST-AtMTAN from the Arabidopsis flower total protein extract. Proteins retrieved from the plant extract by GST-AtMTAN and GST (control) affinity beads were analyzed with immunoblot assay using the anti-CBL3 antibody as a probe (top). Total protein extract (10 μg) from the Arabidopsis flowers was loaded in the first lane to show the size of the CBL3 protein. The bottom panel shows the immunoblot stained with Ponceau S indicating the amount of bait proteins used as affinity reagents. [See online article for color version of this figure.]
Pull-down assays were performed by incubating GST-AtMTAN (bait) with the cleaved form of CBL3 (prey) in the presence or absence of Ca2+. We then carried out immunoblot analyses to determine whether the prey CBL3 was pulled down by GST beads. The anti-CBL3 antibody was purified by the CBL3 antigen and used as a probe. As shown in Figure 5B, GST-AtMTAN successfully pulled down CBL3 only in the presence of Ca2+, whereas the GST protein alone did not retrieve the prey CBL3 regardless of Ca2+.
To further corroborate the interaction between CBL3 and AtMTAN, we tried to isolate Arabidopsis CBL3 using the GST-AtMTAN protein as an affinity reagent. For this affinity purification experiment, total protein extract was prepared from Arabidopsis flowers; CBL3 was expressed most highly in the pollen grains (data not shown). The immunoblot in Figure 5C demonstrates that the plant CBL3 protein, approximately 26 kD in size, was purified by the beads carrying the GST-AtMTAN protein only in a Ca2+-dependent manner. In contrast, no protein bands were detected in the lanes of the GST control beads. Taken together, these results strongly suggest that the CBL3 Ca2+-binding protein requires Ca2+ in order to interact with AtMTAN.
CBL3 and AtMTAN Associate with Each Other in Plant Cells
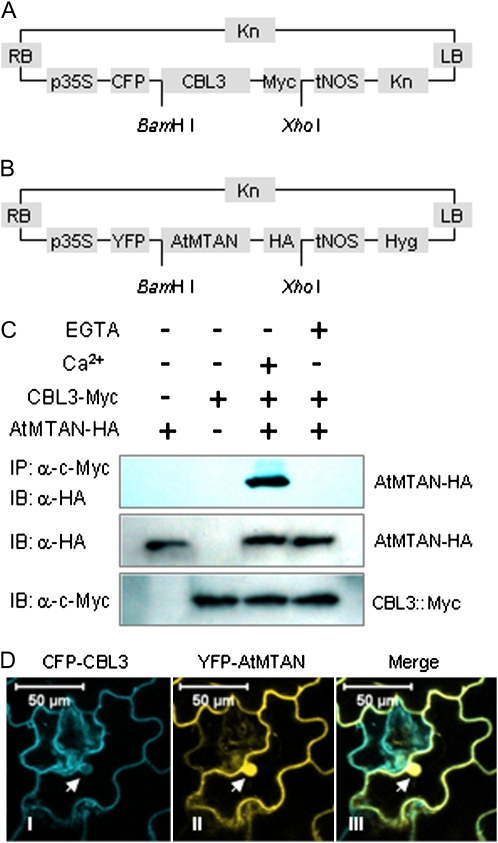
It is obvious that CBL3 and AtMTAN interact in vitro as well as in the yeast two-hybrid system. To confirm their interaction in vivo, we created the following two chimeric constructs. First, cyan fluorescent protein (CFP) and c-Myc tag (Myc) were fused in-frame with the N- and C-terminal ends of CBL3, respectively. The chimeric gene was placed under transcriptional control of the 35S cauliflower mosaic virus promoter and then cloned into the binary vector pBINPLUS (van Engelen et al., 1995), thereby generating 35S∷CFP-CBL3-Myc (Fig. 6A). Similarly, the second construct was created by fusing yellow fluorescent protein (YFP) and hemagglutinin tag (HA) to the N- and C-terminal ends of AtMTAN, eventually producing 35S∷YFP-AtMTAN-HA (Fig. 6B).
Figure 6.
In vivo interaction of CBL3 and AtMTAN. A, Schematic diagram of 35S∷CFP-CBL3-Myc. B, Schematic diagram of 35S∷YFP-AtMTAN-HA. C, Coimmunoprecipitation assay. Total protein extracts prepared from tobacco leaves transiently expressing CFP-CBL3-Myc and/or YFP-AtMTAN-HA were immunoprecipitated with anti-Myc mouse monoclonal antibody in the buffers containing either 1 mM CaCl2 or 2 mM EGTA. The immunoprecipitated (IP) proteins were separated by SDS-PAGE and subjected to immunoblot (IB) analysis with the anti-HA antibody (top). The presence of the transiently expressed proteins in the total protein extracts was determined with immunoblot analysis using either anti-HA (middle) or anti-Myc (bottom) antibodies. D, Colocalization of CBL3 and AtMTAN1 in tobacco cells. Confocal laser scanning images of the tobacco epidermal cells transiently expressing the two fusion proteins display the subcellular localization of the CFP-tagged CBL3 (I) and YFP-tagged AtMTAN (II) proteins. The two fluorescence images are merged (III). Arrows indicate the nucleus. Bars = 50 μm.
The 35S∷CFP-CBL3-Myc and 35S∷YFP-AtMTAN-HA constructs were introduced, either alone or in combination, into tobacco (Nicotiana benthamiana) leaves by the Agrobacterium tumefaciens-mediated infiltration method. Total proteins were extracted from the tobacco leaves at 3 d after infiltration and were verified to contain the HA-tagged AtMTAN (Fig. 6C, middle) and the Myc-tagged CBL3 (Fig. 6C, bottom) proteins via immunoblot analyses. The protein extracts were then subjected to coimmunoprecipitation assay in the presence or absence of Ca2+. Immunoprecipitations were carried out using anti-Myc mouse monoclonal antibodies followed by immunoblot analysis with anti-HA rabbit polyclonal antibodies. The coimmunoprecipitation assay showed that HA-tagged AtMTAN was coimmunoprecipitated with Myc-tagged CBL3 only in the reaction sample containing the two fusion proteins and Ca2+ (Fig. 6C, top). This result clearly suggests that CBL3 and AtMTAN indeed associate in vivo and that the association depends on Ca2+, as demonstrated by the in vitro interaction assays.
Because the fusion proteins transiently expressed in the tobacco leaves were also tagged with either CFP or YFP, we visualized their subcellular localization using confocal laser scanning microscopy and then attempted to determine whether they are colocalized in the plant cells by merging the two confocal images. According to the fluorescence images (Fig. 6D), CBL3 appeared to be localized at the plasma membrane and in the cytoplasm, where AtMTAN was also found. AtMTAN was additionally observed in the nucleus. The merged image clearly displays a color change, indicating that the CBL3 and AtMTAN proteins are colocalized mainly outside of the nucleus. In contrast, the control proteins, CFP and YFP, appeared to be present throughout the cell, including the nucleus and the cytoplasm, as shown in Supplemental Fig. S1.
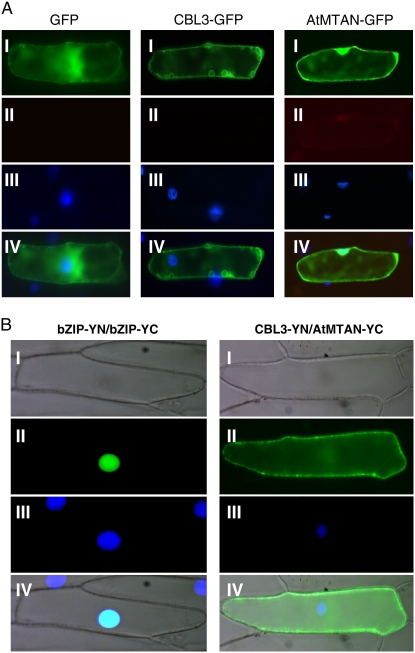
To further investigate the subcellular localization of CBL3 and AtMTAN, we fused GFP to the C-terminal ends of CBL3 and AtMTAN and created CBL3-GFP and AtMTAN-GFP chimeric genes, which were placed under the control of the cauliflower mosaic virus 35S promoter (pMD.CBL3 and pMD.AtMTAN). Each of the chimeric constructs was transiently expressed in onion (Allium cepa) epidermal cells via the particle bombardment procedure and subjected to fluorescence microscopy. As shown in Figure 7A, the GFP fusion proteins individually exhibited fluorescence localization patterns basically similar to those of the confocal images above (Fig. 6D). Apparently, CBL3 seemed to be mainly localized outside of the nucleus. The GFP control, however, displayed similar intensities of fluorescence throughout the cytoplasm and the nucleus.
Figure 7.
Subcellular localization of the CBL3 and AtMTAN proteins and their association in a plant cell. A, Fluorescence images. The indicated plasmids were introduced into the onion epidermal cells via the biolistic particle-delivery system. Following a 4-h incubation at 23°C, the onion cells were analyzed with a fluorescence microscope. I and II show images of GFP and autofluorescence, respectively. III shows nuclei visualized by 4′,6-diamidino-2-phenylindole (DAPI) staining. IV shows the merged image. B, BiFC assay. The CBL3-YN and AtMTAN-YC constructs were coexpressed in onion epidermal cells. The bZIP63-YN and bZIP63-YC plasmids were used as a positive control. I and II show images of bright field and YFP, respectively. III shows the DAPI-stained nuclei. The merged images are shown in IV.
In the case of CBL1, which has the consensus myristoylation motif (MGXXXS/T) at the N terminus, the reporter GFP gene should be placed at the C terminus in order for the CBL1-GFP protein to be targeted correctly (Batistic et al., 2008). Unlike CBL1, however, CBL3 does not possess the N-myristoylation motif. Therefore, it was predicted that the localization pattern of CBL3 would be the same regardless of whether the CFP tag is placed at the N or C terminus. In fact, as shown in Supplemental Fig. S2, the C-terminally tagged construct (CBL3-CFP) and the N-terminally tagged construct (CFP-CBL3) exhibited the same localization pattern, supporting the prediction. The α-TIP-YFP (At1g73190) fusion protein, used as a control for the colocalization assays, showed that the tonoplast appeared as small circles in tobacco epidermal cells and perinuclear regions, with vacuolar membranous invagination surrounding the nucleus (Hunter et al., 2007; Kim et al., 2007).
We next tested whether CBL3 and AtMTAN physically associate with each other in living plant cells using the bimolecular fluorescence complementation (BiFC) assay, which reveals the subcellular localization of protein interaction in the normal cellular environment (Walter et al., 2004). CBL3 and AtMTAN were fused to the N-terminal YFP fragment (YN) in the pUC-SPYNE vector and the C-terminal YFP fragment (YC) in the pUC-SPYCE vector, respectively, thereby creating the CBL3-YN and AtMTAN-YC fusion constructs. The onion epidermal cells bombarded with the two fusion constructs (CBL3-YN and AtMTAN-YC) showed YFP fluorescence strongly in the membrane structure and weakly in the cytoplasm (Fig. 7B, right column). However, no fluorescence was detected from the onion cells bombarded with the following combinations: CBL3-YN/pUC-SPYCE and AtMTAN-YC/pUC-SPYCE (data not shown). A member of the basic Leu zipper (bZIP) transcription factor family in Arabidopsis, bZIP63 (At5g28770), was used as a positive control (Fig. 7B, left column), which is known to form a homodimer in the nucleus of plant cells (Siberil et al., 2001). Taken together, these results strongly suggest that CBL3 and AtMTAN form a complex outside of the nucleus.
AtMTAN Possesses the MTAN Activity Specifically Modulated by CBL3 in a Ca2+-Dependent Manner
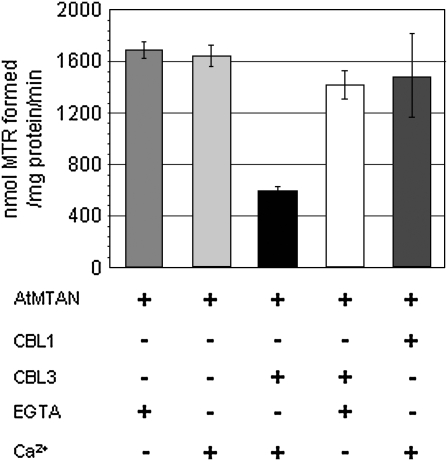
Sequence analysis of the deduced amino acids, as mentioned above, revealed that AtMTAN is similar to EcMTAN, which catalyzes the hydrolysis of the N9-C1′ bond of MTAN to 5′-methylthioribose (MTR) and adenine. Therefore, we performed the MTAN activity assay using the AtMTAN protein prepared with the GST fusion expression and purification system in order to determine empirically whether AtMTAN is able to cleave the ribosidic bond of MTA. As shown in Figure 8, AtMTAN efficiently converted MTA into MTR (approximately 1.6 × 103 nmol MTR formed mg−1 AtMTAN min−1), indicating that AtMTAN indeed has the MTAN activity.
Figure 8.
MTAN activity of AtMTAN is inhibited by Ca2+-bound CBL3. The MTAN activity is represented with the amount of MTR generated from the hydrolysis of MTA by the action of 1 mg of AtMTAN for 1 min at 37°C. + and − indicate the presence and absence, respectively, of the indicated components in the reaction samples. Each value represents the average of three independent experiments. Error bars indicate se values.
Because AtMTAN associates with CBL3 in a Ca2+-dependent manner, we examined whether the enzyme activity of AtMTAN is altered as a result of the association with CBL3. Our enzyme assays (Fig. 8) showed that CBL3 markedly lowered the enzyme activity of AtMTAN in the presence of Ca2+ (approximately 65% decrease). Without Ca2+, however, CBL3 failed to decrease the AtMTAN enzyme activity. It is obvious that such a decrease in enzyme activity was not due to Ca2+, because AtMTAN maintained almost the same levels of enzyme activity regardless of the Ca2+ concentration. Furthermore, it should also be noted that CBL1, another member of the CBL family, which does not interact with AtMTAN, did not exert any substantial effect on the AtMTAN enzyme activity. Taken together, these results clearly suggest that the MTAN activity of AtMTAN is specifically inhibited by the association with Ca2+-bound CBL3.
DISCUSSION
The plant CBL Ca2+ sensors are known to target CIPK family members to mediate the Ca2+ signals induced by a diverse array of external stimuli, such as cold, salinity, low K+ concentration, high pH, abscisic acid, and osmotic stress. Upon interaction with Ca2+-bound CBLs, CIPKs become active so that they can exert the kinase activity (Halfter et al., 2000). Because no other CBL interactors besides the CIPK family members were reported previously, it was believed that CBLs target only the CIPK members. In this study, however, we found a novel CBL target protein that does not belong to the CIPK family. Therefore, it appears that the CBL family can have various interaction partners with distinct biochemical properties, which makes it possible for CBLs to modulate more diverse cellular and physiological processes. This finding provides a new and critical insight into our understanding of the CBL-mediated Ca2+ signaling pathways, which are more complicated than previously known.
CBL3 Targets AtMTAN in Addition to the CIPK Family Members, Which Confers an Additional Level of Complexity on the CBL-Mediated Ca2+ Signaling Pathway
Through the extensive yeast two-hybrid screening performed in this study, we found that one CBL family member, CBL3, can interact not only with multiple CIPK family members (Kolukisaoglu et al., 2004; Jeong et al., 2005) but also with AtMTAN. Using the pull-down assays and coimmunoprecipitation analyses (Figs. 5 and 6C), we confirmed that the CBL3-AtMTAN interaction observed in the yeast two-hybrid system occurs in vitro as well as in vivo. In addition, these analyses also revealed that CBL3 requires Ca2+ to form a complex with AtMTAN. Previously, we showed that Ca2+ is required for the CBL3-CIPK11 association (Jeong et al., 2005). Therefore, it seems that CBL3 undergoes conformational change upon binding with Ca2+, thereby gaining the three-dimensional structure capable of associating with its target proteins, including CIPKs and AtMTAN. In fact, the crystal structure of Ca2+-bound CBL2, the most closely related isoform of CBL3, was markedly different from that of the unbound form (Nagae et al., 2003).
In order for the CBL3-AtMTAN interaction to take place in Arabidopsis cells, both proteins should be present together in the same place. Therefore, we investigated whether the CBL3 and AtMTAN proteins display some overlaps in their expression patterns and subcellular localizations. First, northern-blot and promoter-GUS analyses (Fig. 4) revealed that the expression pattern of the AtMTAN gene roughly coincides with that of the CBL3 gene (Kudla et al., 1999). Second, image analyses of the fluorescent fusion proteins transiently expressed in tobacco (Fig. 6D) and onion (Fig. 7A) epidermal cells showed that CBL3 and AtMTAN can be colocalized mainly at the plasma membrane. In fact, the BiFC assays (Fig. 7B) further verified that the two proteins can physically associate with each other in onion epidermal cells. Taken together, these results strongly suggest that CBL3 can deliver the cytosolic Ca2+ signals to the AtMTAN protein as well as the previously known CIPK family members in Arabidopsis. Meanwhile, we also learned through GenBank search that the Arabidopsis genome contains an additional MTAN-like gene (At4g34840), which encodes a polypeptide very similar to AtMTAN (59% sequence identity). Recently, we cloned a full-length cDNA of the gene and demonstrated that the AtMTAN-like protein also interacts with CBL3 in the yeast two-hybrid system (data not shown).
AtMTAN interacted only with CBL3 but not with other CBL family members tested in this study (Fig. 2). This interaction specificity between the CBL3 and AtMTAN proteins led us to speculate that other CBL family members may also have as yet unidentified distinct interaction partners in addition to the CIPK members. In this case, the CBL family can target a much larger number of various proteins that possess different biochemical properties. Such target diversity provides the CBL members with the molecular mechanisms by which they can be involved in regulating more diverse cellular and physiological processes. In this context, it is important to identify novel interaction partners for each CBL member in order to fully understand the CBL-mediated Ca2+ signaling pathways in plant cells. Our present findings clearly indicate that the Ca2+ signaling pathways mediated by the CBL family are more complicated then previously known; therefore, they represent a new insight into CBL-mediated Ca2+ signaling in higher plants.
CBL3 Uses Different Modules in Interacting with CIPKs and AtMTAN
The Arabidopsis genome was predicted to contain 10 CBLs and 25 CIPK genes (Kolukisaoglu et al., 2004). Each of the CBL family members interacted specifically with multiple CIPKs at different affinities in the yeast two-hybrid system (Kim et al., 2000; Luan et al., 2002; Batistic and Kudla, 2004; Kolukisaoglu et al., 2004; Jeong et al., 2005). Previous deletion analyses have revealed the structural platforms involved in the CBL-CIPK interaction. It appeared that all three EF-hand motifs in the CBL family members are responsible for direct interaction with the NAF (or FISL) motif, a stretch of 21 amino acids conserved in the C termini of the CIPK family members (Halfter et al., 2000; Kim et al., 2000; Albrecht et al., 2001). In particular, almost the entire sequence of CBL3 is required for interaction with CIPK members (Kim et al., 2000).
In this study, we dissected the structural requirement in CBL3 for interaction with AtMTAN and found that, unlike the CBL3-CIPK interaction, only the 91-amino acid region of CBL3 spanning from the 109th to the 199th amino acid residues is required and sufficient for the AtMTAN interaction (Fig. 3B). Therefore, it should be noted that different parts of CBL3 are used to associate with the different interaction partners, CIPKs and AtMTAN. Meanwhile, we also determined the AtMTAN region necessary for the interaction with CBL3. Because small deletions from either the N- or C-terminal end of AtMTAN completely abolished the ability to associate with CBL3 (Fig. 3A), it seems that almost all sequence information in AtMTAN is required.
CBL3 Can Act as Either Activator or Inhibitor Depending on Its Target Proteins
Our previous in vitro interaction assays demonstrated that CBL1 and CBL3 physically interact with CIPK1 and CIPK11, respectively, in a Ca2+-dependent manner (Shi et al., 1999; Jeong et al., 2005). As mentioned above, their interaction is mediated by the NAF motif in the C termini of CIPK family members to which the CBL proteins directly bind (Kim et al., 2000; Albrecht et al., 2001). Meanwhile, Halfter et al. (2000) showed that CIPK24 (also known as SOS2) requires Ca2+-bound CBL4 (also known as SOS3) to phosphorylate the synthetic peptide substrate p3. Without Ca2+-bound CBL4 (SOS3), CIPK24 (SOS2) was inactive, because the intramolecular interaction between the kinase domain and the NAF-containing regulatory C terminus prevented the active site of CIPK24 from binding with the substrate (Guo et al., 2001). These findings together indicated that the Ca2+-bound CBL proteins act as activators that enhance the kinase activity of the CIPK family members by disrupting the intramolecular interaction.
As a matter of fact, several recent lines of genetic evidence demonstrated that activation of the CIPK enzyme by the Ca2+-bound CBL proteins is an important process for Arabidopsis plants to respond properly to the environmental stresses that elicit changes in the cellular Ca2+ signatures. For example, the kinase activity of CIPK23 should be activated by the interaction with either CBL1 or CBL9 in order for the plants to successfully cope with the low concentration of K+ in the soil. The activated CIPK23 is known to phosphorylate the C-terminal region of the K+ transporter AKT1, which results in an increase of the transport activity (Li et al., 2006; Xu et al., 2006; Cheong et al., 2007). Another example is the well-known SOS pathway. The term SOS stands for “salt overly sensitive,” which was used to describe the phenotype of the Arabidopsis mutants with reduced salt tolerance (Zhu et al., 1998). A series of subsequent analyses indicated that CBL4 (SOS3) is required to activate CIPK24 (SOS2) in a Ca2+-dependent manner, which in turn phosphorylates and promotes SOS1, a Na+/H+ antiporter, to get rid of the excess Na+ in the plant cells (Qiu et al., 2002; Quintero et al., 2002). In addition, it should be noted that the kinase activity of CIPK24 (SOS2) can be also activated by CBL10 in the presence of Ca2+ (Kim et al., 2007; Quan et al., 2007). Although both CBL4 (SOS3) and CBL10 are involved in the salt stress response, they carry out the mission in different organs of Arabidopsis plants: the CBL4-CIPK24 (SOS3-SOS2) complex functions primarily in the root, whereas the CBL10-CIPK24 complex works mainly in the shoot.
We found in this study that CBL3 physically interacts with AtMTAN in a Ca2+-dependent manner, thereby lowering the MTAN activity of the AtMTAN protein (Fig. 8). Contrary to the activator role of the CBL family observed in the CBL-CIPK complex, this finding clearly indicates that Ca2+-bound CBL3 can act as inhibitor, which decreases the enzyme activity of the target protein AtMTAN. How can CBL3 exert the inhibitory effect on the AtMTAN enzyme activity? Although the exact mechanism is currently unknown and remains to be investigated, we speculate that it has something to do with the fact that AtMTAN forms a homodimer, which was revealed by our recent yeast two-hybrid assay and in vitro interaction analysis (data not shown). In fact, MTAN purified to homogeneity from Lupinus luteus seeds has shown that the nucleosidase consists of two identical subunits (Guranowski et al., 1981). Therefore, it is conceivable that the association of AtMTAN with CBL3 may result in disrupting or destabilizing the AtMTAN homodimer, which is probably important for the optimal enzyme activity. Anyway, it is obvious that Ca2+-bound CBL3 can act as either activator or inhibitor depending on the interaction partners. Furthermore, it is likely that the rest of the CBL family members may also display dual activities on their distinct target proteins.
Modulation of the AtMTAN Enzyme Activity by Ca2+-Bound CBL3 May Influence Ethylene and Polyamine Biosynthesis as Well as the Met Salvage Pathway in Arabidopsis
The plant hormone ethylene is synthesized from S-adenosyl-l-methionine (SAM) via the intermediate 1-aminocyclopropane-1-carboxylic acid (ACC). ACC synthase catalyzes the conversion of SAM to ACC and MTA, which is the rate-limiting step in the biosynthesis of ethylene (Yang and Hoffmann, 1984). In fact, the in vivo activity of ACC synthase was shown to be markedly increased in plant tissues producing high levels of ethylene, including submerged deepwater rice (Cohen and Kende, 1987) and ripening avocado (Persea americana) fruit (Kushad et al., 1988). Moreover, expression of two rice ACC synthase genes, OsACS1 and OsACS5, was enhanced upon submergence treatments (Zarembinski and Theologis, 1997; Zhou et al., 2001, 2002). It should be noted that MTA is a potent inhibitor of ACC synthase, indicating that ACC synthase is subject to feedback inhibition by the by-product MTA (Hyodo and Tanaka, 1986).
SAM can be alternatively channeled into the polyamine biosynthetic pathway. Polyamines, including putrecine (Put), spermidine (Spd), and spermine (Spm), are small polycationic nitrogenous organic compounds that play important roles in stress response, seed development, senescence, cell proliferation, and differentiation (Walden et al., 1997; Martin-Tanguy, 2001; Kakkar and Sawhney, 2002; Urano et al., 2005). In Arabidopsis, polyamine synthesis is initiated by Arg decarboxylase (EC 4.1.1.19), which catalyzes the synthesis of Put from Arg. Spd is synthesized from Put, to which an aminopropyl group from decarboxylated SAM (dSAM) is added by Spd synthase (EC 2.5.1.16). SAM decarboxylase (EC 4.1.1.50) removes carbon dioxide from SAM and produces dSAM, which serves as the aminopropyl donor in Spd and Spm synthesis. Spm synthase (EC 2.5.1.22) generates Spm using Spd and dSAM as substrates. The reactions catalyzed by Spd and Spm synthases also produce MTA as a by-product, which can act as a feedback inhibitor of the two enzymes (Bagni and Tassoni, 2001). Therefore, it is reasonable to think that high levels of MTA concentration in the plant cells should display a negative effect on the biosynthetic rates of not only ethylene but also polyamines such as Spd and Spm.
In this context, MTA should be actively metabolized and maintained at low concentrations for the efficient biosynthesis of ethylene and polyamines. In fact, the plant tissues producing high levels of ethylene and polyamines exhibited dramatically enhanced enzymatic activity of MTAN (Adams and Yang, 1977; Kushad et al., 1988). MTAN is the first enzyme in the Met salvage pathway, also known in plants as the Yang cycle (Miyazaki and Yang, 1987). In brief, MTR kinase phosphorylates MTR to yield 5′-methylthioribose-1-phosphate, which subsequently undergoes enzymatic isomerization, dehydration, and oxidative decarboxylation to 2-keto-4-methylthiobutyrate, the immediate precursor of Met. SAM synthase converts Met to SAM, which can be used to generate MTA. Therefore, this Met cycle eventually allows high rates of ethylene and polyamine biosynthesis without net consumption of Met. In addition, it also shows how important the MTAN activity is for Met salvaging, let alone for the ethylene and polyamine biosynthesis mentioned above.
We found in this study that the CBL3 calcium sensor can target AtMTAN in a Ca2+-dependent manner, revealing a novel CBL3-mediated calcium signaling pathway in addition to the well-known CBL-CIPK network in plants. The CBL3-AtMTAN interaction resulted in inhibition of the enzymatic activity of AtMTAN, which hydrolyzes MTA and thereby plays a critical role in determining the activities of ACC, Spd, and Spm synthases. Our finding, therefore, shows that CBL3-mediated calcium signaling can be channeled at least in part to AtMTAN in the Met cycle and eventually end up modulating the biosynthesis of ethylene and polyamines, which are involved in plant growth and development as well as various stress responses. Moreover, it also provides a possible mechanism by which the calcium signals are transduced to regulate ethylene and polyamine biosynthesis. Further molecular genetic and biochemical analyses will clarify the biological function of the CBL3-AtMTAN interaction in plants.
MATERIALS AND METHODS
Yeast Two-Hybrid Screening and Assays
The Arabidopsis (Arabidopsis thaliana) λACT cDNA expression libraries (CD4-10 and CD4-22) were obtained from the Arabidopsis Biological Resource Center and used in the yeast two-hybrid screening, which was performed basically according to Durfee et al. (1993). Briefly, the plasmid libraries were produced from the phage libraries by in vivo excision and used to transform the Y190 strain expressing CBL3 protein (bait). Transformants were plated onto synthetic medium that lacks Leu, Trp, and His (SC-Leu-Trp-His) in order to screen the interacting colonies. Colonies that appeared within a 5-d incubation were selected for further analyses. For yeast two-hybrid interaction assays, genes of interest were first cloned into either the activation domain (pGAD.GH) or the DNA-binding domain (pGBT9.BS) vector. Then, the two plasmids were introduced into yeast strain Y190 carrying two reporter genes by the lithium acetate method (Schiestl and Gietz, 1989). Yeast cells carrying both plasmids were selected on synthetic medium lacking Leu and Trp (SC-Leu-Trp). The yeast cells were then streaked on the SC-Leu-Trp-His plate to determine the expression of the HIS3 nutritional reporter. The β-galactosidase expression of the His+ colonies was analyzed by filter-lift and/or quantitative assays as described previously (Ok et al., 2005).
Plant Materials and RNA Gel-Blot Analysis
Arabidopsis (ecotype Col-0) plants were grown at 23°C in a growth chamber under long-day conditions (16-h-light/8-h-dark cycle). For RNA gel-blot analysis, total RNA (10 μg) prepared from roots, stems, leaves, and flowers was resolved by electrophoresis on 1.2% agarose gels, transferred to Hybond N membranes (Amersham Biosciences), and hybridized with the 32P-labeled specific probe. The membranes were autoradiographed with Kodak XAR film.
Analysis of AtMTAN Promoter-GUS Expression
The AtMTAN promoter-GUS construct (pBI.AtMTAN) was transformed into Agrobacterium tumefaciens strain GV3101 and introduced into Arabidopsis plants by the floral dip method (Clough and Bent, 1998). Transformants were selected as described previously (Kim et al., 2003). Histochemical GUS assays of the transgenic plants were performed according to the protocol described by Jefferson et al. (1987).
Purification of GST Fusion Proteins from Escherichia coli
GST fusion proteins were purified according to the protocols described previously (Ok et al., 2005). Briefly, E. coli BL21 cells carrying a GST fusion construct were grown at 37°C overnight and were subcultured until the optical density at 600 nm reached 0.5 to 0.6. Following a 3-h induction with 0.3 mm isopropyl-β-d-thiogalactopyranoside at 20°C, the cell lysate was prepared in ice-cold lysis buffer (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 1 mm phenylmethylsulfonyl fluoride [PMSF], 5 mm dithiothreitol [DTT], 5 mm EDTA, and 1 mm EGTA). Glutathione-Sepharose 4B beads were used to retrieve the GST fusion protein. Ice-cold washing buffer (50 mm Tris-HCl, pH 7.4, and 100 mm NaCl) was used to wash the beads. Protein concentration was determined according to Bradford (1976).
Pull-Down Assay and Immunoblot Analysis
Pull-down assay and immunoblot analysis were performed as described previously (Shi et al., 1999). Briefly, GST fusion proteins attached to the glutathione-Sepharose 4B beads were incubated at 4°C with prey proteins lacking the GST protein in the binding buffer (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 0.05% Tween 20, and 1 mm PMSF) supplemented with either 0.2 mm CaCl2 or 1 mm EGTA. Pull-down samples were resolved by SDS-PAGE and transferred onto polyvinylidene fluoride membranes (Immobilon-P; Millipore) to detect via immunoblot analysis the prey proteins, which were precipitated by the GST fusion bait proteins.
Affinity Purification of CBL3 from Arabidopsis Total Proteins
Total protein preparation and affinity purification were performed as described previously (Shi et al., 1999). Briefly, total proteins were prepared from 100 mg of the mature flowers of 6-week-old Arabidopsis plants (ecotype Col-0) using 300 μL of extraction buffer (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 5 mm DTT, 0.05% Tween 20, 1 mm PMSF, 5 mg mL−1 leupeptin, and 5 mg mL−1 aprotinin) supplemented with either 0.2 mm CaCl2 or 1 mm EGTA. The GST-AtMTAN protein immobilized on the GST beads was added to the total protein extract. Following 4 h of incubation, the beads were washed six times with the binding buffer described above and then subjected to immunoblot analysis to detect the Arabidopsis CBL3 protein.
Agrobacterium Infiltration, Coimmunoprecipitation, and Colocalization Assays
For transient expression, the leaves of 3- to 4-week-old tobacco (Nicotiana benthamiana) plants were infiltrated with Agrobacterium GV3101 carrying either CFP or YFP fusion constructs as described by Brandizzi et al. (2002). Briefly, the bacterial cells were cultured at 28°C with agitation until the stationary phase (approximately 24 h). For coexpression of two different constructs, 0.5 mL of each bacterial culture was mixed and pelleted. The pellet was washed once and resuspended in 1 mL of infiltration buffer (50 mm MES, pH 5.6, 0.5% [w/v] Glc, 2 mm Na3PO4, and 100 μm acetosyringone). The bacterial suspension was applied to the abaxial epidermis of the tobacco leaves with a 1-mL plastic syringe by gently pressing the nozzle.
The coimmunoprecipitation assay was carried out as described previously (Serino et al., 1999) with minor modifications. Briefly, total proteins were extracted from the tobacco leaves at 3 d after the Agrobacterium infiltration with binding buffer (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 5 mm DTT, 0.1% Nonidet P-40, 1 mm PMSF, 1× Complete Protease Inhibitors, and either 2 mm EGTA or 1 mm CaCl2). After a preclearing step, anti-c-Myc mouse monoclonal IgG (1 μg; Santa Cruz Biotechnology) was added to the total protein extract (1 mg) and incubated for 16 h at 4°C. The immune complexes were retrieved with Protein A/G PLUS-Agarose (Santa Cruz Biotechnology) and then subjected to immunoblot analysis in which anti-HA rabbit polyclonal IgG was used as a probe.
For the colocalization assay, the tobacco leaves transiently expressing the fluorescence proteins were analyzed with a confocal laser scanning microscope (LSM 510 META; Carl Zeiss) and Zeiss LSM 510 software (Zeiss LSM Image Examiner) to capture live fluorescent cell images. CFP excitation was performed with a diode laser at 405 nm, and emission was detected with a 420- to 480-nm band-pass filter. Meanwhile, YFP was excited with an argon laser at 514 nm, and emission wavelength was captured with a 530- to 600-nm band-pass filter.
Subcellular Localization of GFP Fusion Proteins and BiFC Assays
The plasmid constructs of interest were introduced into onion (Allium cepa) epidermal cells by particle bombardment as described previously (Ok et al., 2005). Particle bombardments were performed using the Biolistic PDS-1000/He system (Bio-Rad) with 1,100-pounds per square inch rupture discs under a vacuum of 26 inches of mercury. After bombardment, tissues were maintained on Parafilm-sealed half-strength Murashige and Skoog plates at 23°C for 24 h and imaged with the Olympus BX51 fluorescence microscope. Nuclei were stained with 1 μg mL−1 4′,6-diamidino-2-phenylindole (Sigma) in phosphate-buffered saline for 5 min at room temperature and examined with a fluorescence microscope at an excitation wavelength of 350 nm. For GFP imaging, an excitation wavelength of 450 to 490 nm was used, and emission was detected with a 515- to 565-nm band-pass filter.
MTAN Assay
As described by Schlenk (1983), [3H]MTA was synthesized from S-adenosyl-l-[methyl-3H]Met (0.55 mCi mL−1, 63.6 mCi mmol−1), which was purchased from Perkin-Elmer. The standard nucleosidase activity assay was carried out according to Cornell et al. (1996) with minor modifications. Briefly, the assays were initiated by adding 20 μL of AtMTAN (100 pg) into 180-μL reaction samples containing 50 mm HEPES (pH 7.0), 0.5% bovine serum albumin, [3H]MTA (specific activity, 1.41 × 1011 cpm μmol−1), and 2 mm EGTA or 1 mm CaCl2. Depending on the reaction conditions, either CBL1 or CBL3 was also included. After incubation at 37°C for 30 min, reactions were terminated by the addition of 20 μL of 3 m TCA and centrifuged at 10,000g for 5 min to precipitate proteins. The supernatant (200 μL) was applied to a 2-mL AG50-X8 cation-exchange column (Bio-Rad; 100–200 mesh, hydrogen form) and eluted into a 20-mL vial containing 10 mL of LSC cocktail solution cocktail (Perkin-Elmer). Radioactivity of the eluted [3H]MTR was counted in a scintillation counter (Beckman LS6500).
Construction of Plasmids
The following plasmids were constructed as described previously (Kim et al., 2000): pGBT.CBL1, pGBT.CBL3, pGBT.CBL4, pGBT.CBL3N-1, pGBT.CBL3N-2, pGBT.CBL3-EFI, pGBT.CBL3-2EF, pGBT.CBL3-C26, pGBT.CBL3-C52, and pGBT.CBL3-EFIII. The pGEX.CBL1 was created as reported previously (Kudla et al., 1999). To make pGBT.CBL3-NC, we performed PCR on CBL3 cDNA using CBL3-15 and CBL3-20 primers and cloned the amplified fragment into the EcoRI/SalI site of pGBT9.BS. The pGAD.CBL3 and pGEX.CBL3 plasmids were generated as described previously (Jeong et al., 2005).
To create pGAD.AtMTAN, pGBT.AtMTAN, and pGEX.AtMTAN plasmids, the coding region of the AtMTAN cDNA was PCR amplified with primers M-1 and M-2. The PCR product was digested with EcoRI/SalI and ligated into pGAD.GH, pGBT9.BS, and pGEX-4T-3, respectively. The plasmids pGBT.AtMTAN-1 and pGBT.AtMTAN-2 were constructed by cloning each of the PCR products amplified with the M-11/M-2 and M-8/M-2 primer sets into the EcoRI/SalI sites of the pGBT9.BS plasmid. Similarly, primer sets M-1/M-9 and M-1/M-10 were used to generate pGBT.AtMTAN-3 and pGBT.AtMTAN-4, respectively. To produce the pGBT.AtMTAN-5 plasmid, primers M-11 and M-10 were used to amplify the middle region of the AtMTAN cDNA. The resulting PCR product was then cloned into the EcoRI/SalI sites of the pGBT9.BS vector. To make the pBI.AtMTAN construct, the 5′ flanking DNA region between −661 and −1 relative to the translation start codon (ATG) of the AtMTAN gene was amplified with M-PF and M-PR primers using the Arabidopsis (Col-0) genomic DNA as template. After digestion with XbaI and BamHI, the PCR fragment was cloned into the pBI101.1 binary vector (Clontech). For creation of the AtMTAN-GFP chimeric construct (pMD.AtMTAN), primers M-3 and M-7 were used to PCR amplify the AtMTAN coding region without a stop codon. Following digestion with XbaI/BamHI, the PCR product was cloned into the pMD1 binary vector that contains a GFP reporter gene (Sheen et al., 1995). In a similar way, the pMD.CBL3 plasmid was created using CBL3-28 and CBL3-29 primers. For BiFC assays, the CBL3-YN and AtMTAN-YC fusion constructs were created by cloning the coding regions of CBL3 and AtMTAN, which were amplified with the primer sets CBL3-30/CBL3-31 and M-14/M-15 into the BamHI/SmaI sites of pUC-SPYNE and pUC-SPYCE, respectively.
The 35S∷CFP-CBL3-Myc and 35S∷YFP-AtMTAN-HA constructs were made as follows. First, fluorescent protein genes (eCFP, catalog no. 6076-1; eYFP, catalog no. 6005-1; BD Clontech) were amplified by GFP-F and GFP-R primers and then cloned into the SmaI/BamHI sites of the p35S∷FAST vector (Ge et al., 2005), thereby creating p35S∷CFP-FAST and p35S∷YFP-FAST, respectively. Next, the CBL3-Myc and AtMTAN-HA regions were amplified from the CBL3-YN and AtMTAN-YC constructs by the primer sets CBL3-30/MYC-R and M-14/HA-R and then ligated into the BamHI/SalI sites of p35S∷CFP-FAST and p35S∷YFP-FAST, respectively, thereby producing p35S∷CFP-CBL3-Myc-FAST and p35S∷YFP-AtMTAN-HA-FAST. Using these two constructs as templates, the 35S∷CFP-CBL3-Myc and 35S∷YFP-AtMTAN-HA regions were amplified with ASC-F and ASC-R primers and then cloned into the AscI site of the pBINPLUS (Kn) and pBINPLUS (Hyg) binary vectors, respectively. To create the p35S∷CBL3-CFP and p35S∷α-TIP-YFP constructs, the coding regions of CBL3 and α-TIP (At1g73190) were amplified by the primer sets CBL3-F/CBL3-R and TIP-F /TIP2-R, respectively. Each of the resulting PCR products was digested with KpnI and then ligated into the KpnI/SmaI sites of p35S∷CFP-FAST and p35S∷YFP-FAST vectors, respectively. All PCRs were carried out using Pfu DNA polymerase (Stratagene) to enhance fidelity. All of the constructs above were verified by DNA sequencing.
Oligonucleotide Primers Used in the Plasmid Construction
Primers used in this study are listed below, with restriction enzyme sites underlined. Three additional bases, which were chosen randomly by considering their effect on melting temperature and on dimer and stem-loop formation, were included at the 5′ end of the primers for efficient digestion by restriction enzymes: M-1, 5′-TATGAATTCTATGGCTCCTCATGGAGATG-3′; M-2, 5′-ATTGTCGACTTAAAGGTCCGAAAGGTTTC-3′; M-3, 5′-ATATCTAGAATGGCTCCTCATGGAGATGG-3′; M-7, 5′-AGAGGATCCAAGGTCCGAAAGGTTTCTCC-3′; M-8, 5′-ATAGAATTCATCTGATGTTGTGTTTCATG-3′; M-9, 5′-AACGTCGACTCAGATACAAGGAATACATC-3′; M-10, 5′-TTAGTCGACCTTTAGCGTAGCATCATTGG-3′; M-11, 5′-ATTGAATTCCGTGCATAAAGATCTTCGAA-3′; M-14, 5′-CGCGGATCCATGGCTCCTCATGGAGATGG-3′; M-15, 5′-AAGGTCCGAAAGGTTTCTCCCATTG-3′; M-PF, 5′-TATTCTAGAGCTACCTTTCTGCCGTCAAC-3′; M-PR, 5′-ACAGGATCCAACCTTATCCCTCTCTTCCG-3′; CBL3-15, 5′-TTTGAATTCGCGTGCTCTCTCTGTCTTTC-3′; CBL3-20, 5′-AATGTCGACTAGAAGGGAAGGATGCCTGA-3′; CBL3-28, 5′-AAATCTAGAATGTCGCAGTGCATAGACGG-3′; CBL3-29, 5′-AAAGGATCCGGTATCTTCCACCTGGGAGT-3′; CBL3-30, 5′-CGCGGATCCATGTCGCAGTGCATAGACGG -3′; CBL3-31, 5′-GGTATCTTCCACCTGCGAGTGGAAC-3′; GFP-F, 5′-GTACCCGGGATGGTGAGCAAGGGCGAGG-3′; GFP-R, 5′-TTTGGATCCCCTTGTACAGCTCGTCCATGCCG-3′; MYC-R, 5′-ACCCTCGAGTGAACAGCTCCTCGCCCTTGCTC-3′; HA-R, 5′-GCGCTCGAGGAAGTTCACCTTGATGCCGTTCTTC-3′; ASC-F, 5′-ATTGGCGCGCCACACAGGAAACAGCTATGACCA-3′; ASC-R, 5′-TATGGCGCGCCAGTCACGACGTTGTAAAACGAC-3′; CBL3-F, 5′-CGGGGGTACCATGTCGCAGTGCATAGACGGTTTC-3′; CBL3-R, 5′-GGTATCTTCCACCTGCGAGTGGAACAC-3′; TIP-F, 5′-ACGGGGTACCATGGCAACATCAGCTCGTAGAGCATACGG-3′; and TIP-R, 5′-GTAATCTTCAGGGGCCAAGGGCTGGTGTAC-3′.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AT4G38800.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Subcellular localization patterns of the control fluorescent proteins.
Supplemental Figure S2. Confirmation of the CBL3 localization pattern using either the N- or C-terminal tagged CFP constructs.
Supplementary Material
Acknowledgments
We are grateful to the Arabidopsis Biological Resource Center (Ohio State University) for Arabidopsis cDNA libraries. We also thank Dr. Jörg Kudla (Institut für Botanik und Botanischer Garten, Universität Münster) for providing the vectors pUC-SPYCE and pUC-SPYNE for BiFC assays.
This work was supported by the Korea Science and Engineering Foundation, funded by the Korean government (grant no. R01–2006–000–11164–0), and by grants from the Plant Signaling Network Research Center, the Korea Science and Engineering Foundation, and the Korea Research Foundation, funded by the Korean government (Basic Research Promotion Fund; grant no. KRF–2007–313–C00686).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kyung-Nam Kim (knkim@sejong.ac.kr).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Adams DO, Yang SF (1977) Methionine metabolism in apple tissue: implication of S-adenosylmethionine as an intermediate in the conversion of methionine to ethylene. Plant Physiol 60 892–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht V, Ritz O, Linder S, Harter K, Kudla J (2001) The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J 20 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht V, Weinl S, Blazevic D, D'Angelo C, Batistic O, Kolukisaoglu U, Bock R, Schulz B, Harter K, Kudla J (2003) The calcium sensor CBL1 integrates plant responses to abiotic stresses. Plant J 36 457–470 [DOI] [PubMed] [Google Scholar]
- Bagni N, Tassoni A (2001) Biosynthesis, oxidation and conjugation of aliphatic polyamines in higher plants. Amino Acids 20 301–317 [DOI] [PubMed] [Google Scholar]
- Batistic O, Kudla J (2004) Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta 219 915–924 [DOI] [PubMed] [Google Scholar]
- Batistic O, Sorek N, Schultke S, Yalovsky S, Kudla J (2008) Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell 20 1346–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootman MD, Collins TJ, Peppiatt CM, Prothero LS, MacKenzie L, De Smet P, Travers M, Tovey SC, Seo JT, Berridge MJ, et al (2001) Calcium signalling: an overview. Semin Cell Dev Biol 12 3–10 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Frangne N, Marc-Martin S, Hawes C, Neuhaus JM, Paris N (2002) The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell 14 1077–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Kim KN, Pandey GK, Gupta R, Grant JJ, Luan S (2003) CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell 15 1833–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Pandey GK, Grant JJ, Batistic O, Li L, Kim BG, Lee SC, Kudla J, Luan S (2007) Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J 52 223–239 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Cohen E, Kende H (1987) In vivo 1-aminocyclopropane-1-carboxylate synthase activity in internodes of deepwater rice: enhancement by submergence and low oxygen levels. Plant Physiol 84 282–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell KA, Swarts WE, Barry RD, Riscoe MK (1996) Characterization of recombinant Escherichia coli 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase: analysis of enzymatic activity and substrate specificity. Biochem Biophys Res Commun 228 724–732 [DOI] [PubMed] [Google Scholar]
- D'Angelo C, Weinl S, Batistic O, Pandey GK, Cheong YH, Schultke S, Albrecht V, Ehlert B, Schulz B, Harter K, et al (2006) Alternative complex formation of the Ca-regulated protein kinase CIPK1 controls abscisic acid-dependent and independent stress responses in Arabidopsis. Plant J 48 857–872 [DOI] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ (1993) The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev 7 555–569 [DOI] [PubMed] [Google Scholar]
- Evans NH, McAinsh MR, Hetherington AM (2001) Calcium oscillations in higher plants. Curr Opin Plant Biol 4 415–420 [DOI] [PubMed] [Google Scholar]
- Fuglsang AT, Guo Y, Cuin TA, Qiu Q, Song C, Kristiansen KA, Bych K, Schulz A, Shabala S, Schumaker KS, et al (2007) Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19 1617–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Dietrich C, Matsuno M, Li G, Berg H, Xia Y (2005) An Arabidopsis aspartic protease functions as an anti-cell-death component in reproduction and embryogenesis. EMBO Rep 6 282–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Halfter U, Ishitani M, Zhu JK (2001) Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13 1383–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xiong L, Song CP, Gong D, Halfter U, Zhu JK (2002) A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev Cell 3 233–244 [DOI] [PubMed] [Google Scholar]
- Guranowski AB, Chiang PK, Cantoni GL (1981) 5′-Methylthioadenosine nucleosidase: purification and characterization of the enzyme from Lupinus luteus seeds. Eur J Biochem 114 293–299 [DOI] [PubMed] [Google Scholar]
- Halfter U, Ishitani M, Zhu JK (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA 97 3735–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, et al (2003) The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter PR, Craddock CP, Di Benedetto S, Roberts LM, Frigerio L (2007) Fluorescent reporter proteins for the tonoplast and the vacuolar lumen identify a single vacuolar compartment in Arabidopsis cells. Plant Physiol 145 1371–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang YS, Bethke PC, Cheong YH, Chang HS, Zhu T, Jones RL (2005) A gibberellin-regulated calcineurin B in rice localizes to the tonoplast and is implicated in vacuole function. Plant Physiol 138 1347–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyodo H, Tanaka K (1986) Inhibition of 1-aminocyclopropane-1-carboxylic acid synthase activity by polyamines, their related compounds and metabolites of S-adenosylmethionine. Plant Cell Physiol 27 391–398 [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong HJ, Jwa NS, Kim KN (2005) Identification and characterization of protein kinase that interact with the CBL3 calcium sensor in Arabidopsis. Plant Sci 169 1125–1135 [Google Scholar]
- Kakkar RK, Sawhney VK (2002) Polyamine research in plants: a changing perspective. Physiol Plant 116 281–292 [Google Scholar]
- Kim BG, Waadt R, Cheong YH, Pandey GK, Dominguez-Solis JR, Schultke S, Lee SC, Kudla J, Luan S (2007) The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J 52 473–484 [DOI] [PubMed] [Google Scholar]
- Kim KN, Cheong YH, Grant JJ, Pandey GK, Luan S (2003) CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 15 411–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KN, Cheong YH, Gupta R, Luan S (2000) Interaction specificity of Arabidopsis calcineurin B-like calcium sensors and their target kinases. Plant Physiol 124 1844–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolukisaoglu U, Weinl S, Blazevic D, Batistic O, Kudla J (2004) Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol 134 43–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J, Xu Q, Harter K, Gruissem W, Luan S (1999) Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc Natl Acad Sci USA 96 4718–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushad MM, Yelenosky G, Knight R (1988) Interrelationship of polyamine and ethylene biosynthesis during avocado fruit development and ripening. Plant Physiol 87 463–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Kim BG, Cheong YH, Pandey GK, Luan S (2006) A Ca(2)+ signaling pathway regulates a K(+) channel for low-K response in Arabidopsis. Proc Natl Acad Sci USA 103 12625–12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhu JK (1998) A calcium sensor homolog required for plant salt tolerance. Science 280 1943–1945 [DOI] [PubMed] [Google Scholar]
- Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W (2002) Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell (Suppl) 14 S389–S400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Tanguy J (2001) Metabolism and function of polyamines in plants: recent development (new approaches). Plant Growth Regul 34 135–148 [Google Scholar]
- Miyazaki JH, Yang SF (1987) The methionine salvage pathway in relation to ethylene and polyamine biosynthesis. Physiol Plant 69 366–370 [Google Scholar]
- Nagae M, Nozawa A, Koizumi N, Sano H, Hashimoto H, Sato M, Shimizu T (2003) The crystal structure of the novel calcium-binding protein AtCBL2 from Arabidopsis thaliana. J Biol Chem 278 42240–42246 [DOI] [PubMed] [Google Scholar]
- Ok SH, Jeong HJ, Bae JM, Shin JS, Luan S, Kim KN (2005) Novel CIPK1-associated proteins in Arabidopsis contain an evolutionarily conserved C-terminal region that mediates nuclear localization. Plant Physiol 139 138–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GK, Cheong YH, Kim KN, Grant JJ, Li L, Hung W, D'Angelo C, Weinl S, Kudla J, Luan S (2004) The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 16 1912–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99 8436–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan R, Lin H, Mendoza I, Zhang Y, Cao W, Yang Y, Shang M, Chen S, Pardo JM, Guo Y (2007) SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19 1415–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Ohta M, Shi H, Zhu JK, Pardo JM (2002) Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA 99 9061–9066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd JJ, Franklin-Tong VE (2001) Unravelling response-specificity in Ca2+ signalling pathways in plant cells. New Phytol 151 7–33 [DOI] [PubMed] [Google Scholar]
- Sanders D, Pelloux J, Brownlee C, Harper JF (2002) Calcium at the crossroads of signaling. Plant Cell (Suppl) 14 S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD (1989) High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet 16 339–346 [DOI] [PubMed] [Google Scholar]
- Schlenk F (1983) Methylthioadenosine. Adv Enzymol Relat Areas Mol Biol 54 195–265 [DOI] [PubMed] [Google Scholar]
- Serino G, Tsuge T, Kwok S, Matsui M, Wei N, Deng XW (1999) Arabidopsis cop8 and fus4 mutations define the same gene that encodes subunit 4 of the COP9 signalosome. Plant Cell 11 1967–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J, Hwang S, Niwa Y, Kobayashi H, Galbraith DW (1995) Green-fluorescent protein as a new vital marker in plant cells. Plant J 8 777–784 [DOI] [PubMed] [Google Scholar]
- Shi J, Kim KN, Ritz O, Albrecht V, Gupta R, Harter K, Luan S, Kudla J (1999) Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 11 2393–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siberil Y, Doireau P, Gantet P (2001) Plant bZIP G-box binding factors: modular structure and activation mechanisms. Eur J Biochem 268 5655–5666 [DOI] [PubMed] [Google Scholar]
- Urano K, Hobo T, Shinozaki K (2005) Arabidopsis ADC genes involved in polyamine biosynthesis are essential for seed development. FEBS Lett 579 1557–1564 [DOI] [PubMed] [Google Scholar]
- van Engelen FA, Molthoff JW, Conner AJ, Nap JP, Pereira A, Stiekema WJ (1995) pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res 4 288–290 [DOI] [PubMed] [Google Scholar]
- Walden R, Cordeiro A, Tiburcio AF (1997) Polyamines: small molecules triggering pathways in plant growth and development. Plant Physiol 113 1009–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40 428–438 [DOI] [PubMed] [Google Scholar]
- White PJ, Broadley MR (2003) Calcium in plants. Ann Bot (Lond) 92 487–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH (2006) A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125 1347–1360 [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffmann NE (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol 35 155–189 [Google Scholar]
- Yang T, Poovaiah BW (2003) Calcium/calmodulin-mediated signal network in plants. Trends Plant Sci 8 505–512 [DOI] [PubMed] [Google Scholar]
- Zarembinski TI, Theologis A (1997) Expression characteristics of OS-ACS1 and OS-ACS2, two members of the 1-aminocyclopropane-1-carboxylate synthase gene family in rice (Oryza sativa L. cv. Habiganj Aman II) during partial submergence. Plant Mol Biol 33 71–77 [DOI] [PubMed] [Google Scholar]
- Zhou Z, de Almeida Engler J, Rouan D, Michiels F, Van Montagu M, Van Der Straeten D (2002) Tissue localization of a submergence-induced 1-aminocyclopropane-1-carboxylic acid synthase in rice. Plant Physiol 129 72–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Vriesen W, Van Caeneghem W, Van Montagu M, Van Der Straeten D (2001) Rapid induction of a novel ACC synthase gene in deepwater rice seedlings upon complete submergence. Euphytica 121 137–143 [Google Scholar]
- Zhu JK, Liu J, Xiong L (1998) Genetic analysis of salt tolerance in Arabidopsis: evidence for a critical role of potassium nutrition. Plant Cell 10 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.