Abstract
In this study, transcriptomics and metabolomics data were integrated in order to examine the regulation of glucosinolate (GS) biosynthesis in Arabidopsis (Arabidopsis thaliana) and its interface with pathways of primary metabolism. Our genetic material for analyses were transgenic plants overexpressing members of two clades of genes (ALTERED TRYPTOPHAN REGULATION1 [ATR1]-like and MYB28-like) that regulate the aliphatic and indole GS biosynthetic pathways (AGs and IGs, respectively). We show that activity of these regulators is not restricted to the metabolic space surrounding GS biosynthesis but is tightly linked to more distal metabolic networks of primary metabolism. This suggests that with similarity to the regulators we have investigated here, other factors controlling pathways of secondary metabolism might also control core pathways of central metabolism. The relatively broad view of transcripts and metabolites altered in transgenic plants overexpressing the different factors underlined novel links of GS metabolism to additional metabolic pathways, including those of jasmonic acid, folate, benzoic acid, and various phenylpropanoids. It also revealed transcriptional and metabolic hubs in the “distal” network of metabolic pathways supplying precursors to GS biosynthesis and that overexpression of the ATR1-like clade genes has a much broader effect on the metabolism of indolic compounds than described previously. While the reciprocal, negative cross talk between the methionine and tryptophan pathways that generate GSs in Arabidopsis has been suggested previously, we now show that it is not restricted to AGs and IGs but includes additional metabolites, such as the phytoalexin camalexin. Combining the profiling data of transgenic lines with gene expression correlation analysis allowed us to propose a model of how the balance in the metabolic network is maintained by the GS biosynthesis regulators. It appears that ATR1/MYB34 is an important mediator between the gene activities of the two clades. While it is very similar to the ATR1-like clade members in terms of downstream gene targets, its expression is highly correlated with that of the MYB28-like clade members. Finally, we used the unique transgenic plants obtained here to show that AGs are likely more potent deterrents of the whitefly Bemisia tabaci compared with IGs. The influence on insect behavior raises an important question for future investigation of the functional aspect of our initial finding, which pointed to enriched expression of the MYB28-like clade genes in the abaxial domain of the Arabidopsis leaf.
Glucosinolates (β-thioglucoside-N-hydroxysulfates; GSs) are nitrogen- and sulfur-containing plant-specialized metabolites. The GS-myrosinase system serves as a major chemical defense mechanism against insects, bacteria, and fungi (Raybould and Moyes, 2001). In Arabidopsis (Arabidopsis thaliana), there are at least 37 different GSs (Reichelt et al., 2002), with side chains derived mainly from Met (aliphatic glucosinolates [AGs]) and Trp (indole glucosinolates [IGs]; see Fig. 5 below for pathway schemes). The biosynthesis of GSs starts with amino acid side chain elongation that forms a chain-elongated α-keto acid that could either be subjected to further elongation cycles or used in the generation of the GS-defining core structure (or glucone). Glucone formation is a five-step pathway that starts with the formation of an aldoxime through oxidation of the precursor amino acids by cytochrome P450 monooxygenases, members of the CYP79 family. In the last step of the pathway, sulfation of desulfoglucosinolates occurs and, thereafter, secondary modification of the side chain takes place (e.g. oxidation, elimination, and alkylation).
Figure 5.
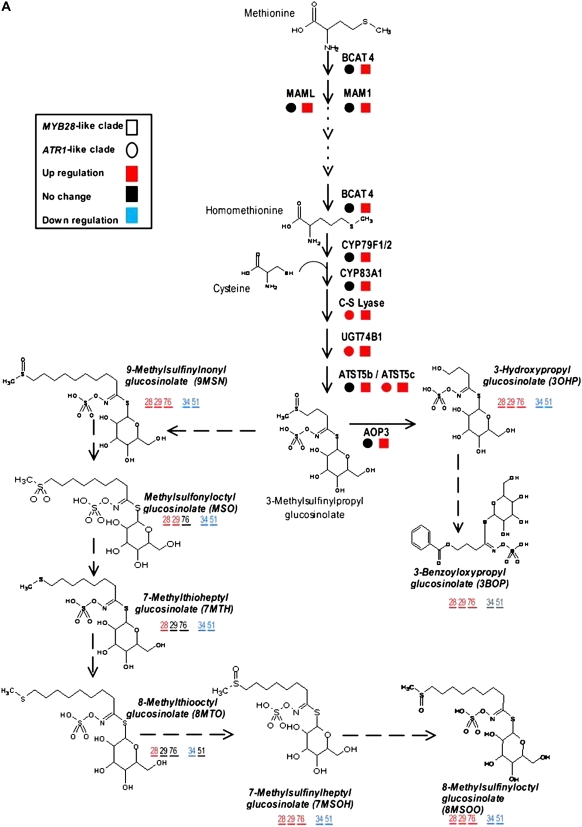
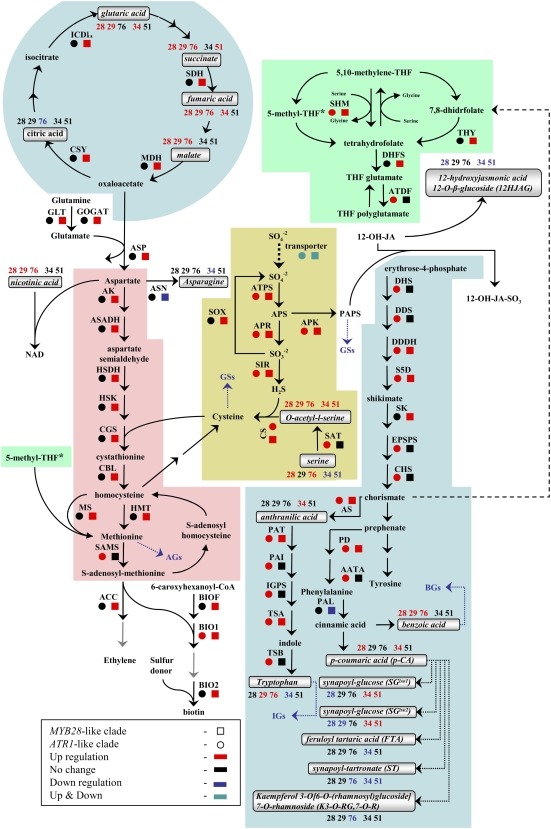
Gene expression and metabolite levels in the proximal network of Met- and Trp-derived GS pathways. The biosynthesis pathway of Met-derived GSs starting from Met (A) and the biosynthesis pathway of Trp-derived GSs starting from Trp and including branches such as IAA and camalexin biosynthesis (B). The mRNA expression analysis was done with joined values obtained in overexpression plants of each clade, while metabolomic analysis was done separately for each line (see “Materials and Methods”). All metabolites were measured under normal growth conditions except for camalexin (in green), which was detected after AgNO3 treatment (see “Materials and Methods”). All known enzymatic reactions are marked with black arrows, while the predicted reactions are marked with dotted arrows. Colored squares and circles represent statistically significant changes in gene expression of the overexpression plants belonging to the MYB28-like and ATR1-like clade genes, respectively. Putatively identified compounds are marked with boldface and italic characters, and the colored numbers represent statistically significant changes in the corresponding overexpression lines. Underlined metabolites were positioned in the pathway based on results obtained by isotope feeding experiments and predictions (see explanation of the DLEMMA approach in Supplemental Data Set S1). Detailed information regarding each gene can be found in Supplemental Table S4. See also Figure 7 for metabolite profiles of the proximal network metabolites.
The formation of GSs is interconnected to the metabolism of key plant metabolites; recent work showed that the Trp-derived indole aldoxime (IAOx) is a key branching point between the biosynthesis of IGs, the plant hormone indole-3-acetic acid (IAA), and the phytoalexin camalexin (Glawischnig et al., 2004; Halkier and Gershenzon, 2006). Camalexin is synthesized from Trp via IAOx by CYP79B2 and CYP79B3, while CYP71B15 catalyzes the final step in its biosynthesis (Schuhegger et al., 2006). Recently, Nafisi et al. (2007) provided evidence that CYP71A13 catalyzes the conversion of IAOx to indole-3-acetonitrile (IAN) in camalexin synthesis. Auxin is a crucial plant hormone that regulates many aspects of plant growth and development (Woodward and Bartel, 2005). Inhibition of flux through any of the three reactions downstream to IAOx results in decreased levels of IGs and increased levels of IAA (Grubb and Abel, 2006). Several lines of evidence suggest that there is also a direct metabolic link between IGs and IAA: IGs can be degraded into IAN, which in turn can be hydrolyzed by nitrilases into IAA. Since GSs are sulfur-containing compounds that have amino acid skeletons, their biosynthesis is also strongly linked to primary metabolism.
The facts that GSs are derived from several different amino acids and that the intersection of their metabolism with other metabolic pathways produces key compounds in plants (e.g. IAA) suggest complex regulation of their production. Such a regulatory network should be able to modulate levels of each metabolite either coordinately or separately, as required by developmental and environmental signals (Celenza et al., 2005). The altered tryptophan regulation1D (atr1D) mutant is a dominant overexpression allele of the MYB transcription factor ATR1 (MYB34). In atr1D, transcript levels of both the Trp biosynthesis genes ANTHRANILATE SYNTHASE1 (ASA1) and TRYPTOPHAN SYNTHASE β-SUBUNIT1 (TSB1) and of the cytochrome P450 genes CYP79B2, CYP79B3, and CYP83B1 are induced in specific seedling tissues (Bender and Fink, 1998; Smolen and Bender, 2002; Smolen et al., 2002; Celenza et al., 2005). The CYP79B2 and CYP79B3 enzymes catalyze the formation of the Trp-derived IAOx, while CYP83B1 converts IAOx to the next intermediate in the IG pathway (1-aci-nitro-2-indolyl-ethane). All five genes mentioned above are induced in plants overexpressing ATR1 under the control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter (Celenza et al., 2005), while expression of CYP79F1, encoding a key enzyme in AG biosynthesis, is not altered in these plants. The overexpression of ATR1, therefore, results in a dramatic increase in the accumulation of IGs (but not AGs) and in the formation of double the amount of IAA compared with wild-type plants (Celenza et al., 2005). Interestingly, neither atr1D nor 35S∷ATR1 plants display obvious high-IAA phenotypes, such as elongated hypocotyls, leaf epinasty, or adventitious rooting, while the atr1D/cyp83B1 double mutant exhibits enhanced adventitious rooting compared with the single cyp83B1 mutant (Smolen and Bender, 2002). Moreover, in atr1-2, a loss of ATR1 function suppresses the cyp83B1 mutant adventitious rooting phenotype. The atr1-2 mutant does not exhibit any morphological abnormalities and shows a decrease in levels of IGs and reduced expression of CYP79B2, CYP79B3, and CYP83B1 but not of ASA1 and TSB1 genes (in adult leaves). Expression of ATR1 is elevated in the IG-deficient cyp83B1 and cyp79B2/cyp79B3 mutants, and this points to a mechanism in which IG levels are restored to the required levels by induced up-regulation of ATR1 activity (Celenza et al., 2005).
The cyp83B1 mutant plants also exhibit elevated expression of Trp synthesis genes and of IG-biosynthesis CYP genes, while the atr1-2 mutation suppresses this induction (predominantly of the Trp synthesis genes). Smolen and Bender (2002) demonstrated that ATR1 is highly responsive to exogenously applied plant signaling molecules such as methyl jasmonate, brassinolide, abscisic acid, and cytokinin, which induce its expression, while 1-aminocyclopropane-1-carboxylic acid, IAA, and salicylic acid repress it. The atr2D mutant confers constitutively activated expression of MYB synthesis genes and corresponds to a mutation in a basic helix-loop-helix transcription factor (Smolen and Bender, 2002). The atr2D/atr1D double mutant exhibits additive effects on Trp regulation; thus, ATR1 and ATR2 may possibly take part in different pathways activating Trp genes.
Like ATR1, overexpression of OBP2, a different type of transcription factor (DOF, for DNA binding with one finger), positively regulates IG and auxin biosynthesis (Skirycz et al., 2006). Expression of ATR1, TSB2, putative myrosinase-binding proteins, and MAM-1, which catalyzes the condensing reactions of the first two Met elongation cycles in short-chain AG biosynthesis, is also induced in these plants, altogether leading to 2- to 3-fold increases in IG levels. The concentration of auxin is increased in OBP2-overexpressing plants, and they display a strong apical dominance, reduced height, short hypocotyls, and a reduced number of lateral roots. Levels of OBP2 transcripts are increased upon external application of methyl jasmonate, auxin, mechanical wounding, and by generalist herbivore feeding. It was suggested that the primary effect of OBP2 is on CYP83B1 and that OBP2 plays a role in biotic and abiotic stress responses, possibly as part of a network regulating GS biosynthesis in Arabidopsis.
Another factor, IQ-DOMAIN1 (IQD1), encodes a basic nuclear protein that modulates the expression of several GS pathway genes (Levy et al., 2005). Overexpression of IQD1 results in increased expression of IG-biosynthesis CYP genes, while genes encoding enzymes related to AG biosynthesis (CYP79F1 and CYP79F2) and GS degradation (myrosinase-encoding TGG1) are reduced in expression. Gain- and loss-of-function iqd1 alleles result in significant but mild changes in the accumulation of both AGs and IGs. Expression of IQD1 seems to be independent of the classical plant hormone signaling pathways, but mechanical stimuli, including aphid feeding, cause a moderate increase of its transcripts. IQD1 is a member of a large family of plant proteins containing calmodulin-binding motifs, and it was suggested that it may integrate early wound- and pathogen/elicitor-induced changes in cytoplasmic Ca2+ concentrations to coordinate an array of defense responses, including GS production. A different factor that influences GS levels is TERMINAL FLOWER2 (TFL2), which encodes the Arabidopsis homolog of the animal HETEROCHROMATIN PROTEIN1 controlling heterochromatin structure. Phenotypes of the tfl2 mutant alleles include early flowering, short stature, stunted rosette leaves, increased branching, reduced leaf GSs, increased IGs in roots, altered seed GS levels, altered IAA levels, altered Trp metabolism, temperature sensitivity, increased resistance to a fungal pathogen, and reduced levels of sinapine and sinapoyl esters (phenylpropanoid derivatives) compared with wild-type plants (Kim et al., 2004; Bennett et al., 2005). It is currently not clear what are the direct or indirect consequences of TFL2 activity on developmental programs and metabolic pathways that could explain this array of phenotypes.
Recently, five proteins with sequence similarity to ATR1, members of the Arabidopsis R2R3-MYB superfamily, were reported to act as transcriptional activators of GS biosynthesis. Hirai et al. (2007) showed that MYB28 and MYB29 proteins are involved in the regulation of AG production. While MYB28 induces the basal production of AGs, MYB29 might have an additional function by inducing AG accumulation upon methyl jasmonate treatment. Gigolashvili et al. (2007b) reported that MYB28 expression was induced by mechanical stimuli and by Glc. They suggested that MYB28 regulates AG biosynthesis and controls the response to biotic challenges. Both MYB28 and MYB29 and a third protein belonging to the same clade (MYB76) were studied by Sonderby et al. (2007). As demonstrated by the previous studies for MYB28 and MYB29, they also showed that overexpression of MYB76 increased the production of AGs and their biosynthetic genes in the leaves. This increase in AGs was evident not only in leaves but also in seeds. AG level analysis of knockout line leaves showed that MYB29 and MYB76 might control short-chained AGs, while MYB28 might control both short- and long-chained products. In another study, additional members of the same MYB subclade, MYB51 and MYB122, were shown to act as activators of the IG biosynthetic pathway, together with ATR1/MYB34 (Gigolashvili et al., 2007a). Overexpression of ATR1/MYB34 in the myb51 mutant background could complement the phenotype at the chemical level and showed a strong high-auxin phenotype, while overexpression of MYB122 in the same background resulted in a high-auxin phenotype but did not elevate IG levels. As for MYB28, MYB51 expression was induced by mechanical stimuli (touch or wounding), but ATR1/MYB34 expression was not induced by the same treatments.
Array analysis of mutant plants altered in leaf polarity led to the initial finding of this study, in which expression of the MYB28, MYB29, and MYB76 genes (here termed the MYB28-like clade) was shown to be enriched in the abaxialized leaf tissue. Phylogenetic analysis showed that these MYB factors are closely related to a second clade (here termed the ATR1-like clade) that included the previously described ATR1/MYB34 IG pathway regulator, MYB51, and MYB122 genes. To circumvent the functional redundancy in each of these two clades and to examine their effect on GS metabolism we generated transgenic Arabidopsis plants in which expression of members of either clade (MYB28, MYB29, MYB76 and ATR1/MYB34, MYB51) was simultaneously down-regulated. Metabolic profiling of these plants showed that levels of AGs and IGs were altered and suggested that genes belonging to the ATR1-like and MYB28-like clades are activators of the IG and AG biosynthetic pathways, respectively. While this study was in progress, other groups showed that single mutant lines of these factors are altered in AG and IG metabolism (see above). We subsequently overexpressed the different GS regulators in Arabidopsis and used the transgenic plants for a detailed analysis of the transcriptome and metabolome. Overexpression of these factors resulted in severe morphological alterations and had a profound effect on gene expression and metabolism. The metabolic changes included those associated with either feeding precursors to GS biosynthesis, mainly primary metabolism (i.e. the distal networks), or the proximal networks (i.e. metabolism of GSs and related branching pathways), starting from their amino acid precursors (Trp and Met). To complement the data on transcript and metabolite changes, we used a bioinformatic approach in which the correlation between expression of the various GS regulators and genes of the proximal and distal networks during more than 200 perturbations was evaluated. Surprisingly, ATR1/MYB34, which is phylogenetically related to the ATR1-like clade and showed very similar metabolic and expression effects when overexpressed in transgenic plants (like the ATR1-like clade genes), was strongly correlated in expression to the MYB28-like clade genes. Taken together, the data showed that these transcription factors might have downstream target genes in both primary and secondary metabolism. The data also provided new insights to how several members of these two clades are temporally expressed in a way that allows for the accumulation of particular metabolic pathway products. As a result of this study, we obtained a unique set of Arabidopsis plants that produce either AGs or IGs to high levels and used this genetic material to demonstrate that AGs are more potent deterrents to the whitefly Bemisia tabaci than IGs. Finally, the effect on insect behavior is discussed with relation to the spatial, abaxialized expression of the GS regulators.
RESULTS
Identification of Arabidopsis R2R3-MYB Genes Regulating GS Biosynthesis through Array Analysis of Plants Altered in Leaf Polarity
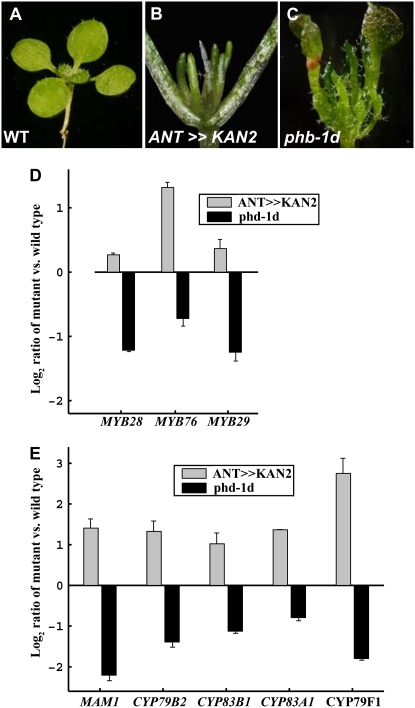
Uniform expression of the GARP domain transcription factor KANADI2 (KAN2; Fig. 1, A and B) in both sides of Arabidopsis plants results in radial abaxialized leaves (Eshed et al., 2001). Conversely, the dominant mutation in the PHABULOSA (PHB) gene, phb-1d, results in radial adaxialized leaves (McConnell et al., 2001; Fig. 1, A and C). To identify transcripts preferentially expressed in either abaxial or adaxial leaf domains, GeneChip expression profiles in seedlings of the two opposing types were compared with similarly sized wild-type seedling tissue and with each other (Fig. 1A; see “Materials and Methods”). Analysis of this data set uncovered a small clade of three R2R3-MYB transcription factors (MYB28, MYB29, and MYB76), with expression of all three enriched in the abaxialized tissues (Fig. 1D). All three proteins (referred to here as the MYB28-like clade) are part of a larger MYB cluster encompassing a total of six members, including also MYB51, MYB122, and ATR1/MYB34 (referred to here as the ATR1-like clade; Stracke et al., 2001). A possible association of these clade members and the regulation of GS biosynthesis were implicated from the similar expression enrichment of genes associated with GS metabolism in abaxial tissues (Fig. 1E). Moreover, plants expressing the GUS reporter gene driven by the upstream regions of all of these MYB genes had expression patterns similar to those described previously for GS-related genes, such as IQD1 (Levy et al., 2005), OBP2 (Skirycz et al., 2006), CYP79F1 and CYP79F2 (Reintanz et al., 2001; Tantikanjana et al., 2001), CYP79B2 (Mikkelsen et al., 2000), and UGT74B1 (Grubb and Abel, 2006; Supplemental Fig. S1).
Figure 1.
Detection of the MYB-type GS regulators in the abaxial leaf domain. A to C, Genotypes used for array analysis included the wild type (WT; A), a transgenic plant expressing the KAN gene under the control of the ANT promoter (abaxialized; B), and a plant with a dominant mutation in the PHB gene (adaxialized; C). Microarrays were used to identify abaxial-enriched genes differentially expressed between the adaxial and abaxial leaf domains (see “Materials and Methods”). D and E, Differential mRNA expression levels between the two leaf domains for the three MYB transcription factors (D) and for known markers of GS biosynthesis (E). The mRNA levels are presented as log2 ratios versus wild-type levels, and in all cases there was a statistically significant difference (P < 0.05) between the genotypes representing either leaf domain.
To further characterize the association between abaxial cell types, the MYB factors, and the GS biosynthetic pathways, functional analysis of these MYBs was carried out. Notably, during the course of this study, several reports characterizing the ATR1-like and MYB28-like clade members were published (Hirai et al., 2007; Gigolashvili et al., 2007a, 2007b, 2008; Sonderby et al., 2007; see introduction). These reports highlighted the importance of these MYB factors in the regulation of GS biosynthesis. In this study, we describe novel, additional aspects regarding the effect of this set of transcription factors on the Arabidopsis transcriptome and metabolome and further link their function to insect repellants directed at phloem cells and the lower (abaxial) leaf surface.
Overexpression of Synthetic MicroRNAs Targeting the Two MYB Factor Groups Results in Plant Growth Retardation and a Reduction in GSs and Expression of Their Biosynthetic Genes
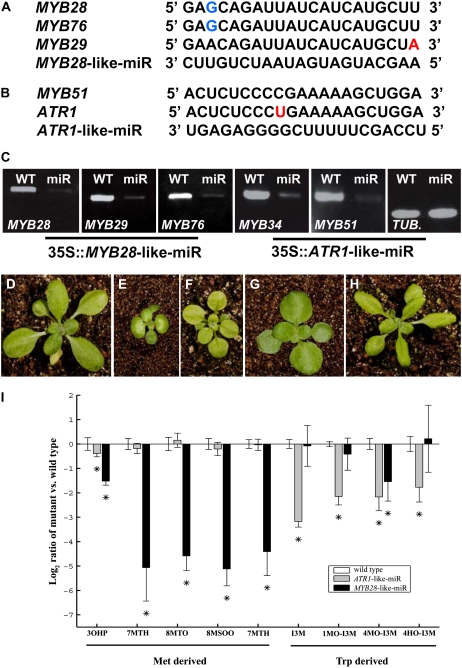
To investigate the consequences of reduced expression of genes of the two clades, we first examined homozygous knockout lines of four of the six genes (apart from ATR1 and MYB122). None of these knockout lines showed a clear developmental phenotype, possibly due to redundancy of the members of either clade (data not shown). Generating a multiple knockout mutant line of the MYB28-like clade was problematic, due to the fact that MYB29 and MYB76 are closely linked in tandem on chromosome 5. Therefore, we generated transgenic lines using a recently reported method in which synthetic microRNAs (miRs) are overexpressed (here under the control of the 35S CaMV promoter; Alvarez et al., 2006). The first synthetic miR (termed MYB28-like-miR) was designed to conegatively regulate the MYB28-like clade members (MYB28, MYB29, and MYB76; Fig. 2A), while the second (termed ATR1-like-miR) was designed to generate a double MYB51/ATR1(MYB34) mutant line (i.e. targeting the two ATR1-like clade genes; Fig. 2B). To verify whether the synthetic miR had successfully down-regulated its target genes, reverse transcription (RT)-PCR was used to monitor steady-state mRNA levels of genes of the two clades relative to the wild type. In all cases, the target genes were down-regulated more then 3-fold by the corresponding miR (Fig. 2C).
Figure 2.
Reduced expression of multiple members of the two clades of MYB genes by use of synthetic miRs and its effects. A, A sequence region unique to the MYB28-like clade (MYB28, MYB29, and MYB76) permitted the design of a synthetic miR (MYB28-like-miR) that targets all three genes. Blue letters represent a G-U wobble, and the red letter represents a mismatch. B, The ATR1-like-miR, designed to target MYB51 and ATR1 simultaneously. The red letter represents a mismatch. C, Down-regulation of the different MYB gene expression as detected by RT-PCR experiments in the wild type (WT; left in each panel) and transgenic lines (right in each panel) expressing synthetic miRs (plants are shown below). The TUBULIN (TUB) gene was used as a control. D to F, Phenotypes of a wild-type plant (D), a transgenic line expressing the MYB28-like-miR (E), and a transgenic line expressing the ATR1-like-miR (F). Both miRs were expressed under the control of the 35S CaMV promoter. G, A cross between a plant overexpressing MYB28 and a MYB28-like-miR plant could rescue the MYB28-like-miR phenotype. H, A cross between a plant overexpressing ATR1/MYB34 and a ATR1-like-miR plant could rescue the ATR1-like-miR phenotype. I, Met- and Trp-derived GS accumulation in 35S∷MYB28-like-miR- and 35S∷ATR1-like-miR-expressing plants. The samples were collected from 14-d-old rosette leaves and analyzed by UPLC-qTOF-MS (see “Materials and Methods”). Relative IG levels are shown as means ± se from six independent samples; asterisks indicate values that are statistically significantly different (P < 0.05) compared with wild-type values. The metabolite levels shown are presented as log ratios from the wild type (levels of the latter, therefore, are always zero). For gene names, see Figure 7 legend.
Expressing the MYB28-like-miR resulted in dwarfed plants that produced epinastic leaves (Fig. 2, D and E). The first two leaves lacked trichomes, while older leaves contained trichomes only on their distal side (data not shown). Plants expressing the ATR1-like-miR were also reduced in size, and their rosette leaves were round and light yellow in color (Fig. 2, D and F). To support our observations regarding the activity of the synthetic miR to silence members of both clades, we coexpressed (performed by crossings) either MYB28-like clade members in the background of MYB28-like-miR plants or ATR1-like clade genes in the background of ATR1-like-miR plants. We further showed that the phenotypes of plants expressing either of the synthetic miRs could be rescued (Fig. 2, G and H).
Chemical analysis of plants expressing the MYB28-like-miR and the ATR1-like-miR showed that MYB28-like-miR-expressing plants retained a significant reduction in AG content while the ATR1-like-miR plants exhibited a significant reduction in IG levels (Fig. 2I). Interestingly, in the case of two GSs (3-hydroxypropyl [an AG] and 4MO-I3M [an IG]), we detected a significant decline in plants expressing both MYB28-like-miR and the ATR1-like-miR.
Overexpression of the MYB28-Like Clade and the ATR1-Like Clade Genes Driven by Specific Promoters Results in Severe Morphological Phenotypes and Phenocopies of High-Auxin Mutants, Respectively
To further investigate the functions of the different MYB factors belonging to both clades, we overexpressed all six genes in Arabidopsis. The ASYMMETRIC LEAVES1 (AS1) gene promoter directs relatively early expression in young leaf primordia and vascular tissues but not in the apical meristem (Fig. 3M; Byrne et al., 2000). Overexpression of any of the three MYB28-like clade factors using the AS1 promoter resulted in nearly identical phenotypes: dwarf plants with very short petioles and light green rounded leaves (Fig. 3, A and B). In contrast, plants overexpressing the same set of genes under the control of the late 650 promoter, which drives expression in mature rosette leaves but not in meristems and young primordia (Fig. 3N), showed weak phenotypes (data not shown).
Figure 3.
Phenotypes of plants overexpressing genes of the two clades. A and B, Seedlings (A) and plants (B) overexpressing the MYB28-like clade genes under the control of the early AS1 promoter. C, Overexpression of ATR1-like clade members under the control of the relatively late 650 promoter. D and E, Wild-type (WT) plant (D) and wild-type inflorescence (E). F and G, Plant overexpressing ATR1/MYB34 driven by the AS1 promoter (F) and its inflorescence (G). H and I, Plant overexpressing MYB51 driven by the AS1 promoter (H) and its inflorescence (I). J to L, Expression of a DR5:GUS marker for free auxin production in the wild type (J) and in 650≫ATR1 (K) and 650≫MYB51 (L) backgrounds. M and N, Expression pattern of YFP and GUS reporter genes driven by the AS1 promoter (M) and driven by the 650 promoter (N).
Ectopic expression of the ATR1-like clade members (ATR1/MYB34 and MYB51) under the control of the 650 promoter produced plants with long hypocotyls, epinastic cotyledons, and elongated petioles (Fig. 3C), resembling phenotypes of mutants and transgenic plants overproducing IAA (yucca, sur1, sur2, and CYP79B2 overexpression; Boerjan et al., 1995; Mikkelsen et al., 2000; Zhao et al., 2001). Overexpression of the same genes under the control of the AS1 promoter resulted in plants possessing a pin-like naked stem due to cessation in leaf formation at the vegetative meristem (Fig. 3, D–I) and shoot tips that often developed fused floral organ structures. Due to the sterility of plants expressing these factors early in development (under the control of the AS1 promoter), we generated plants in which expression of the ATR1-like clade members was driven by the 650 promoter that directs expression at later stages of development. The third ATR1-like clade member, MYB122, was also expressed in the same way, but no morphological changes could be detected in transgenic plants (data not shown).
To corroborate our observation on alterations in auxin (IAA) metabolism phenotypes of ATR1/MYB34- and MYB51-overexpressing plants, we introduced the DR5∷GUS construct into the 650≫ATR1/MYB34 and 650≫MYB51 transgenic backgrounds. DR5∷GUS was used as a marker to visualize patterns of free auxin production by analyzing the expression of the GUS gene fused to the highly active synthetic auxin response element, which enables a rapid detection of elevated free auxin concentration, movement, and accumulation (Ulmasov et al., 1997). While expression of DR5∷GUS in wild-type seedlings is restricted to the hydathodes of either cotyledons or true leaves (Aloni et al., 2003; Fig. 3J), in the 650≫ATR1/MYB34 seedlings, DR5∷GUS expression was broadened to the first and secondary veins of cotyledons and did not change in true leaves (Fig. 3K). Expression of DR5∷GUS in 650≫MYB51 was also altered compared with the wild type, and it appeared in petioles (in both cotyledons and leaves) and leaf blades (Fig. 3L). Expression of the DR5∷GUS in MYB28-like-expressing plants was similar to that detected in the wild-type background (data not shown). Altogether, these results suggest that both ATR1-like clade members overproduce IAA.
Transcriptome and Metabolome Changes in Plants Overexpressing the ATR1-Like and MYB28-Like Clade Genes
We used Affymetrix GeneChips and nontargeted metabolomics to carry out a detailed examination of the consequences of overexpressing members of both clades in Arabidopsis. The transcriptomes and metabolomes of lines ectopically expressing MYB76 or MYB29 (MYB28-like clade gene expression driven by the AS1 promoter; see above) and ATR1/MYB34 or MYB51 (ATR1-like clade gene expression driven by the 650 promoter; see above) were compared with those detected in wild-type plants. Two, mass spectrometry-based analytical methods were employed in order to cover a wide range of compound classes present in Arabidopsis. In the first method, ultra-performance liquid chromatography coupled to a quadrupole time-of-flight mass spectrometer (UPLC-qTOF-MS) was used to detect mainly semipolar components (in both electrospray ionization (ESI)-positive and ESI-negative modes). The high resolution and high mass accuracy of the UPLC-qTOF-MS system and tandem mass spectrometry (MS/MS) analysis allows structural elucidation of unknown peaks, although in a large number of cases the identification might be ambiguous (e.g. in the case of isomers). Using this technology for Arabidopsis leaves allowed us to putatively identify and monitor the relative levels of 72 metabolites, mainly secondary metabolites (Supplemental Table S1). In order to profile polar compounds, in particular primary metabolites, we used the previously established gas chromatography-mass spectrometry (GC-MS) analysis of derivatized extracts (Fernie et al., 2004). In Arabidopsis rosette leaves, this technology allowed us to identify and monitor the levels of 63 metabolites, including amino acids, organic acids, sugar alcohols, tricarboxylic acid (TCA) cycle intermediates, soluble sugars, sugar phosphates, and a few secondary metabolites (Supplemental Table S2).
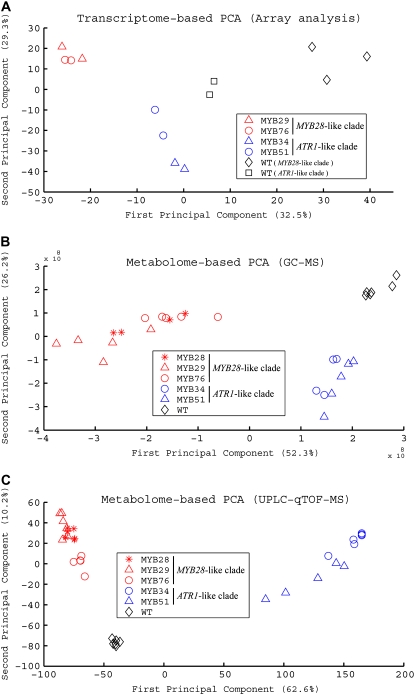
To obtain a broad view of the differences in the transcript and metabolite profiles of the various genotypes, we conducted principal component analysis (PCA) on the data sets derived from the gene expression analysis and metabolite profiling (UPLC-qTOF-MS [operated in the ESI-negative mode] and GC-MS; Fig. 4). Gene and metabolite expression profiles could be clearly distinguished between the genotypes. The transcription profiles of lines ectopically expressing MYB76 or MYB29 (MYB28-like clade) and ATR1/MYB34 or MYB51 (ATR1-like clade) were clearly dissimilar to their corresponding wild-type samples and also showed a clear difference between each other (Fig. 4A). The whole UPLC-qTOF-MS data set was projected on the first two principal components resulting from the PCA. Samples derived from plants overexpressing the ATR1-like clade (MYB51 and ATR1/MYB34) could be clearly differentiated from the profiles of plants overexpressing the MYB28-like factors (MYB28, MYB29, and MYB76) and from the wild type. PCA on the data obtained with GC-MS showed a separation between the two clades of transcription factors and the wild type as good as the separation obtained in the UPLC-qTOF-MS data (Fig. 4, B and C).
Figure 4.
Metabolite and expression profiles of plants overexpressing members of the two GS regulators differ between them and from those of wild-type plants. PCA of data sets obtained using three different technologies: mRNA GeneChip array (A), GC-MS (B), and UPLC-qTOF-MS (C). In all sections, red symbols mark plants overexpressing the MYB28-like clade genes, blue symbols mark plants overexpressing the ATR1-like clade genes, and black symbols mark wild-type (WT) plants.
In the course of this study, we recognized that transcriptional regulators of metabolic pathways might have a broad influence not only on committed steps in a specific metabolic pathway but also on the more central, primary metabolism, which supplies pathways of secondary metabolism its backbone precursors. Therefore, we evaluated the results obtained from the transcriptome and metabolome analyses in the case of both the “proximal” network pathways (defined here as downstream from Trp and Met, including GSs and linked pathways) and the “distal” network pathways (defined here as providing precursors to Trp and Met biosynthesis and additional metabolic pathways currently not known to be directly related to the GS pathway).
The Transcriptomes of Plants Overexpressing Genes of Both Clades
Due to the very high similarity in transcriptome changes between samples derived from overexpression of the same clade members (see “Materials and Methods”; Fig. 4), particularly in genes of the proximal and distal networks, we combined the results of the overexpression of the same clade members in comparison with wild-type plants (ATR1/MYB34 and MYB51 versus the wild type and MYB29 and MYB76 versus the wild type; Figs. 5 and 6).
Figure 6.
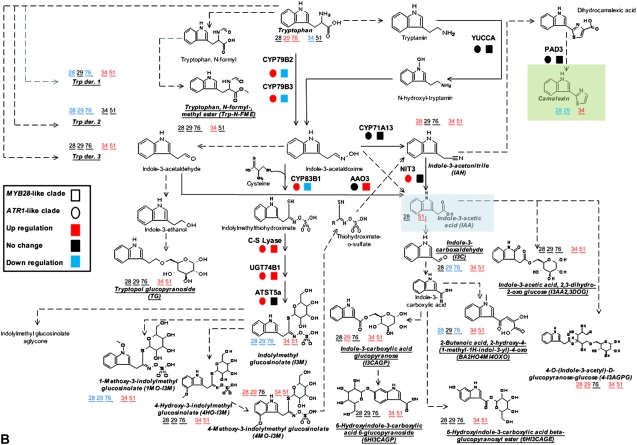
Gene expression and metabolite levels in the distal network pathways related to Met- and Trp-derived GS biosynthesis. Each metabolic pathway is indicated by a different color: Trp and Phe biosynthesis and metabolism in blue, Met biosynthesis and metabolism in pink, TCA cycle in blue, sulfur assimilation and metabolism in yellow, and folate metabolism in green. The mRNA expression analysis was done with joined values obtained in overexpression plants of each clade, while metabolite analysis was done separately for each line (see “Materials and Methods”). All known enzymatic reactions are marked with black arrows, while the predicted or multiple reactions are marked with dotted arrows. Colored squares and circles represent statistically significant changes in gene expression of plants overexpressing the MYB28-like and ATR1-like clade genes, respectively. Putatively identified compounds are marked with squares, and the colored numbers represent statistically significant changes in the corresponding overexpression lines. Detailed information regarding each gene can be found in Supplemental Table S4, and metabolite profiles of the distal network metabolites can be found in Figure 8 (and partially in Supplemental Fig. S2).
Proximal Network Transcriptome Changes Related to the Biosynthesis of GSs
Side Chain Elongation of the Amino Acid
Expression of the METHYLTHIOALKYLMALATE (MAM1 and MAML) synthase genes encoding enzymes catalyzing the first two cycles in the side chain elongation of Met and BRANCHED-CHAIN AMINOTRANSFERASE4 (BCAT4; Schuster et al., 2006) was specifically induced in the MYB28-like clade-overexpressing plants. Two other putative BCAT genes (BCAT3 and BCAT5) were positively regulated by the MYB28-like clade members and may also play a role in this metabolic pathway (Fig. 5A).
Development of the Core Structure
The next phase in GS biosynthesis, glucone formation, involves five major consecutive reactions (oxidation, oxidation and conjugation, C-S cleavage, glucosylation, and sulfation) catalyzed by enzymes that are specific to either Met- or Trp-derived GSs (Halkier and Gershenzon, 2006). Different members of the CYP79 family of cytochrome P450 enzymes are responsible for catalyzing the conversion to aldoximes in either pathway. Both CYP79B2 and CYP79B3, which encode enzymes catalyzing the conversion of Trp to IAOx (Hull et al., 2000), were positively regulated by the ATR1-like clade factors and showed down-regulation in plants overexpressing the MYB28-like clade genes (Fig. 5B). On the contrary, the MYB28-like clade genes positively regulated CYP79F1 and CYP79F2, which encode enzymes metabolizing the short-chain Met derivatives to aliphatic aldoximes (Reintanz et al., 2001), while the same genes were not changed in expression in the ATR1-like clade-expressing plants (Fig. 5A). The next step in the pathway is mediated by the CYP83 enzymes, which produce an activated, oxidized form of the aldoxime (Halkier and Gershenzon, 2006). While expression of CYP83B1 associated with Trp GSs was induced by the ATR1-like clade genes and repressed by the MYB28-like clade genes (Fig. 5B), expression of CYP83A1, mediating Met oxidation, was up-regulated by MYB28-like clade genes but remained the same in plants overexpressing the ATR1-like clade genes (Fig. 5A).
Following the second oxidation step, two reactions catalyzed by a C-S lyase (Mikkelsen et al., 2004) and a glycosyltransferase (UGT74B1) are believed to be executed by enzymes common to both Met- and Trp-derived GSs (Grubb et al., 2004). Indeed, genes corresponding to enzymes catalyzing these two steps showed induction in plants expressing genes of both clades (Fig. 5). A family of sulfotransferases (AtST) catalyzes the last step in glucone formation. In accordance with the preferred activity of the AtST5a recombinant enzymes toward Trp-derived desulfoglucosinolates (Piotrowski et al., 2004), its corresponding gene was positively induced in the ATR1-like clade-overexpressing plants (Fig. 5B). On the other hand, expression of AtST5b, in which the enzyme preference was reported to be higher toward Met-derived desulfoglucosinolates, was increased in MYB28-like-overexpressing plants (Fig. 5A). Expression of the third sulfotransferase gene (AtST5c) that is also highly active with Met-derived desulfoglucosinolates was positively regulated in plants overexpressing genes of both clades (Fig. 5A; Piotrowski et al., 2004).
Secondary Side Chain Modifications
The initially formed parent GS structure is subjected to a wide range of secondary modifications. The AOP3 gene (2-oxoglutarate-dependant dioxygenase) encodes the enzyme catalyzing the oxidation of the side chain. It converts methylsulfinylalkyl to hydroxyl GSs. Our gene expression analysis points to induction of AOP3 by the MYB28-like clade genes (Fig. 5A). Thus, the expression analysis using arrays clearly suggests that members of the MYB28-like clade and ATR1-like clade factors activate the Met- and Trp-derived GS pathways, respectively.
The Trp-derived aldoxime is an intermediate branch point in the synthesis of IGs, the auxin IAA, and the phytoalexin camalexin. As shown above, the ATR1-like clade members positively regulated the Trp and aldoxime biosynthetic genes. However, in the case of camalexin, both CYP71A13 (Nafisi et al., 2007), which catalyzes the biosynthesis of IAN from IOAx, and PHYTOALEXIN DEFICIENT3 (PAD3), which catalyzes the conversion of dihydrocamalexic acid to camalexin (Schuhegger et al., 2006), were not effected by members of either clade (Fig. 5B). In the case of auxin, IAA is produced by both the IAOx and YUCCA pathways and the disassembly of IGs (Fig. 5). The array experiments showed that the YUCCA pathway was not transcriptionaly affected. IGs could be degraded into IAN, which in turn can be hydrolyzed by nitrilases into IAA. We indeed detected induction of the NITRILASE3 (NIT3) and NIT4 genes (Fig. 5B), although the latter is not associated with IAN metabolism but rather with cyanide detoxification (Kutz et al., 2002).
Transcriptome Changes in the Distal Network of Plants Overexpressing the ATR1-Like and MYB28-Like Clade Genes
We next evaluated the influence of overexpressing members of both clades on the distal network of the transcriptome related to GS biosynthesis (i.e. those providing precursors to Trp and Met biosynthesis) and additional metabolic pathways currently not known to be directly related to the GS pathway (Fig. 6). GS skeletons are sulfur rich, and their precursors are derived from the sulfur assimilation pathway (Fahey et al., 2001). The results from GeneChips analysis clearly pointed to a strong induction in gene expression in both (or either) clades of the pathways involving sulfate assimilation and its metabolism, Ser and Cys metabolism (Fig. 6). Transcripts associated with sulfur transport and adenosine-5′-phosphosulfate (APS; Fig. 6) metabolism were induced in plants overexpressing members of both clades (Fig. 6). ATP sulfurylase catalyzes the formation of the APS branch point metabolite from sulfate. In one branch, APS is further metabolized by APS kinase to 3′-phosphoadenosyl-5′-phosphosulfate (PAPS), the substrate for the AtST sulfotransferases that catalyze the last step in GS (AG and IG) glucone formation (Fig. 5). In a second branch, APS is also metabolized to sulfite, and a dual induction (in plants overexpressing genes of both clades) in transcript levels was also observed for the gene encoding the enzyme carrying out this reaction (APS reductase [APR]). The induced activity of APR might reduce the availability of APS for PAPS, and this might be compensated by the recycling of excess sulfite to sulfate in the precursor of APS, via sulfite oxidase, so that its corresponding transcript was induced only in the MYB28-like-overexpressing plants (Fig. 6). Downstream of APR (Fig. 6), sulfite is reduced by sulfite reductase to H2S, which is incorporated together with O-acetyl-l-Ser to form Cys through Cys synthase. Transcripts corresponding to sulfite reductase and Cys synthase were induced in plants overexpressing both clades (Fig. 6). Cys also serves as an important cofactor for reactions catalyzed by CYP83B1 and CYP83B1 in both pathways of GS biosynthesis, but is also further metabolized through cystathionine and homo-Cys to Met. In accordance with the latter metabolic pathway, expression of the genes encoding cystathionine γ-synthase, cystathionine β-lyase, and homo-Cys S-methyltransferase (HMT) were induced just in the MYB28-like-overexpressing lines. Interestingly, catabolism of Met through the conversion to S-adenosyl-Met for its recycling to homo-Cys (and back to Cys) is possibly induced only in the ATR-like clade by the increased transcript levels of S-adenosyl Met synthase in plants overexpressing this clade member. Met synthase (Ravanel et al., 2004; Rébeillé et al., 2006) carries out the methylation of homo-Cys to Met, using a methyl group from 5-methyltetrahydrofolate (5-methyl-THF). Similar to HMT, Met synthase transcript levels were induced only in the MYB28-like clade-overexpressing lines.
Starting from the citric acid cycle, through Asp biosynthesis and catabolism, down to homo-Cys is a major metabolic route to Met biosynthesis (Fig. 6). A dramatic coexpression of transcripts was evident along this route in plants overexpressing the MYB28-like clade members but not in the case of ATR1-like-overexpressing plants. All genes corresponding to the 11 enzymatic steps starting from oxaloacetate in the citric acid cycle and up to Met were induced in the MYB28-like-overexpressing plants (Fig. 6; Supplemental Table S4, asterisk).
The conversion of Gln to Glu that serves as a precursor for Asp formation is mediated by Gln-oxoglutarate aminotransferase, and its corresponding transcript was also induced only in plants overexpressing the MYB28-like clade genes. Another two metabolic routes that were mainly induced at the transcript level in the MYB28-like-overexpressing plants were the folates and biotin pathways (Fig. 6). The role of tetrahydrofolate derivatives is to transport and donate one-carbon (C1) units, and the 5-methyl-THF intermediate is the methyl donor for the formation of Met through the Met synthase-catalyzed reaction (Fig. 6).
The aromatic amino acids Trp and Phe serve as precursors for the formation of GSs in Arabidopsis. Plants overexpressing the ATR1-like clade genes showed induced expression of genes catalyzing the reaction in the shikimate pathways, leading to the formation of chorismate (DHS, DDS, DDDH, S5D, EPSPS, and CHS). In the same plants, genes acting downstream of chorismate, that are part of the Trp and Phe biosynthetic pathways, were also induced (PD, AATA, AS, PAT, PAI, IGPS, TSA, and TSB). Phe serves as the precursor for benzylglucosinolate (BG) formation, and in the Arabidopsis ecotype Landsberg erecta (Ler), this type of GS accumulates in seeds.
Metabolomics of Plants Overexpressing Genes of the Two Clades
As described above, the information obtained from gene expression analysis was complemented by metabolic profiling using UPLC-qTOF-MS and GC-MS (using extracts derived from plants overexpressing the MYB28-like clade [MYB28, MYB29, and MYB 76] and the ATR1-like clade [MYB51 and ATR1/MYB34] genes). Nontargeted metabolite analysis performed by UPLC-qTOF-MS resulted in the detection of 15,943 and 13,473 mass signals (using the MarkerLynx program) in the positive and negative ionization modes, respectively. Identification of the putative metabolites was initially performed by a “mass-to-mass” search, in which mass signals extracted by MarkerLynx were compared with accurate masses of previously reported Arabidopsis metabolites (Supplemental Table S6). Overall, we were able to assign the putative identities of 53 metabolites in the Arabidopsis rosette leaves based on the mass-to-mass analysis and additional approaches, which included (1) the use of accurate mass for assignment of a possible empirical formula followed by a search in metabolite databases (e.g. KNApSAcK [http://prime.psc.riken.jp/KNApSAcK], Database of Natural Products [Chapman & Hall/CRC], and the MOTO database [http://appliedbioinformatics.wur.nl/moto]); (2) dual-energy measurements (Supplemental Table S1); (3) using DLEMMA, a novel approach for metabolite identification (see explanation of the DLEMMA approach in Supplemental Data Set S1); and (4) MS/MS analysis. We also putatively identified 19 additional metabolites only according to their accurate mass; these are presented in Supplemental Table S1, but they were not considered for biological interpretation in this study.
In order to estimate the number of differential metabolites between any of the transgenic lines and the wild type, statistical filtering was applied to the mass signals data. A total of 2,815 and 2,929 mass signals in the positive and negative ionization modes (Supplemental Table S7), respectively, were significantly different in one of the genotypes versus the wild type (assessed by Kruskal-Wallis nonparametric one-way ANOVA; see “Materials and Methods”; Supplemental Table S3). This set of differential mass signals was consequently analyzed in order to cluster together masses belonging to the same metabolite (see “Materials and Methods”). After clustering of differential masses (combining both ionization modes), 1,812 groups were formed (1,400 of which were singletons), and these groups provided an estimation of the total number of differential metabolites we detected in this study. Of the 53 putatively identified metabolites, 34 were also significantly different between at least one of the transgenic plants and the wild type (Table I; Supplemental Table S1).
Table I.
Differential metabolites putatively identified by UPLC-qTOF-MS in Arabidopsis rosette leaves
| No. | Putative Metabolite in Arabidopsis Leaves | Short Name | Metabolite Classa | Retention Time | Ionization Mode | Molecular Weight (m/z Detected; M + H) | Molecular Weight (m/z Detected; M − H) | Molecular Weight (Theoretical Mass) | m/z Errorb | Molecular Formula |
|---|---|---|---|---|---|---|---|---|---|---|
| min | ppm | |||||||||
| 1 Ac | Anthranilic acidd | AntA | Sh. der. | 6.40 | Positive | 138.0554 | 138.0555 | −0.72 | C7H7NO2 | |
| 2 B | 12-Hydroxy jasmonic acid glucoside | 12HJAG | Jasmon der. | 14.73 | Negative | 387.1635 | 387.1655 | 5.20 | C18H28O9 | |
| 3 A | p-Coumaric acidd | p-CA | Phenyl. | 7.71 | Negative | 163.0390 | 163.0395 | −3.07 | C9H8O3 | |
| 4 B | Synapoyl-Glciso1 | SG | Phenyl. | 6.27 | Negative | 385.1114 | 385.1135 | 5.4 | C17H22O10 | |
| 5 B | Synapoyl-Glciso2 | SG | Phenyl. | 6.70 | Negative | 385.1114 | 385.1135 | 5.4 | C17H22O10 | |
| 6 C | Sinapoyltartronate | ST | Phenyl. | 7.70 | Negative | 325.0564 | 325.0564 | 0.00 | C14H14O9 | |
| 7 B | Feruloyl tartaric acid | FTA | Phenyl. | 5.70 | Negative | 325.0547 | 325.0560 | 4.00 | C14H14O19 | |
| 8 B | Kaempferol 3-O-6-O-rhamnosyl-glucoside 7-O-rhamnoside | K3-O-RG,7-O-R | Phenyl. | 7.27 | Negative | 739.2074 | 739.2086 | −1.62 | C33H40O19 | |
| 9 B | 3-Hydroxypropyl glucosinolate | 3OHP | AGs | 1.20 | Negative | 376.0380 | 376.0372 | 2.13 | C10H19NO10S2 | |
| 10 B | 8-Methylthiooctyl glucosinolate | 8MTO | AGs | 15.40 | Negative | 476.1090 | 476.1083 | 1.47 | C16H31NO9S3 | |
| 11 B | 7-Methylthioheptyl glucosinolate | 7MTH | AGs | 11.87 | Negative | 462.0930 | 462.0926 | 0.87 | C15H29NO9S3 | |
| 12 B | 8-Methylsulfinyloctyl glucosinolate | 8MSOO | AGs | 4.52 | Negative | 492.1040 | 492.1032 | 1.63 | C16H31NO10S3 | |
| 13 B | 7-Methylsulfinylheptyl glucosinolate | 7MSOH | AGs | 3.03 | Negative | 478.0880 | 478.0875 | 1.05 | C15H29NO10S3 | |
| 14 B | Methylsulfonyloctyl glucosinolate | MSO | AGs | 5.72 | Negative | 508.0988 | 508.0981 | 1.40 | C16H31NO11S3 | |
| 15 B | 9-Methylsulfinylnonyl glucosinolate | 9MSN | AGs | 7.16 | Negative | 506.1188 | 506.1188 | 0.00 | C17H33NO10S3 | |
| 16 C | 3-Benzoyloxypropyl glucosinolate | 3BOP | BGs | 8.30 | Negative | 480.0642 | 480.0634 | 1.70 | C17H23NO11S2 | |
| 17 B | 4-Hydroxyindol-3-ylmethyl glucosinolate | 4HO-I3M | IGs | 3.23 | Negative | 463.0480 | 463.0481 | −0.22 | C16H20N2O10S2 | |
| 18 B | 1-Methoxyindole glucosinolate | 1MO-I3M | IGs | 6.09 | Negative | 477.0640 | 477.0638 | 0.42 | C17H22N2O10S2 | |
| 19 B | 4-Methoxyindole glucosinolate | 4MO-I3M | IGs | 8.65 | Negative | 477.0640 | 477.0638 | 0.42 | C17H22N2O10S2 | |
| 20 B | Indol-3-yl-methyl glucosinolate | I3M | IGs | 4.27 | Negative | 447.0540 | 447.0532 | 1.79 | C16H20N2O9S2 | |
| 21 A | Tryptophan | Trp | aaa | 3.73 | Positive | 205.0985 | 205.0977 | 3.90 | C11H12N2O2 | |
| 22 A | Camalexin | Camalexin | Trp. der | 17.23 | Positive | 201.0492 | 201.0486 | 2.98 | C11H8N2S | |
| 23 B | 1-(1H-indole-3-carboxylate), glucopyranose | I3CAGP | Trp. der | 7.30 | Negative | 322.0930 | 322.0927 | 0.93 | C15 H17 N O7 | |
| 24 B | 6-Hydroxyindole-3-carboxylic acid 6-O-β-d-glucopyranoside | 6HI3CAGP | Trp. der | 2.20 | Negative | 338.0873 | 338.0876 | 0.90 | C15H17NO8 | |
| 25 C | 6-Hydroxyindole-3-carboxylic acid β-d-glucopyranosyl ester | 6HI3CAGE | Trp. der | 2.70 | Negative | 338.0873 | 338.0876 | 0.90 | C15H17NO8 | |
| 26 B | Tryptophan, N-formyl-, methyl ester | Trp-N-FME | Trp. der | 9.89 | Negative | 245.0915 | 245.0926 | −4.49 | C13H14N2O3 | |
| 27 B | 1H-Indole-3-carboxaldehyde | I3C | Trp. der | 10.30 | Negative | 144.0447 | 144.0449 | −1.39 | C9H7NO | |
| 28 B | 2-Butenoic acid, 2-hydroxy-4-(1-methyl-1H-indol-3-yl)-4-oxod | BA2HO4MI4OXO | Trp. der | 8.47 | Negative | 244.0606 | 244.0610 | −1.64 | C13H11NO4 | |
| 29 B | Tryptopol-glucopyranosided | TG | Trp. der | 9.50 | Negative | 322.1302 | 322.1291 | 2.79 | C16H21NO6 | |
| 30 C | 1H-Indole-3-acetic acid, 2,3-dihydro-2-oxo Glc | I3AA2,3DOG | Trp. der | 5.49 | Negative | 352.1034 | 352.1032 | 0.57 | C16H19NO8 | |
| 31 C | 4-O-(Indole-3-acetyl)-d-glucopyranose Glc | 4-I3AGPG | Trp. der | 11.40 | Positive | 500.1767 | 500.1768 | −0.20 | C22H29NO12 | |
| 32 | Tryptophan derivative (1)d | Trp. Der-1 | Trp. der | 15.95 | Negative | 606.1200 | ||||
| 33 | Tryptophan derivative (2)d | Trp. Der-2 | Trp. der | 18.40 | Negative | 606.1200 | ||||
| 34 | Tryptophan derivative (3)d | Trp. Der-3 | Trp. der | 18.61 | Negative | 606.1200 |
Compounds identified by DLEMMA (L. Feldberg and A. Aharoni, unpublished data). Phenyl., Phenylpropanoid; Trp. der, Trp derivative; Sh. der., shikimate derivative; aaa, aromatic amino acid; Jasmon der., jasmonic acid derivative.
m/z Error, Difference between theoretical and found m/z values in ppm.
Confidence levels in the metabolite identification: A, metabolites identified by a standard; B, metabolites identified by dual-energy injection; C, metabolites identified by MS/MS fragments obtained from the positive or negative ionization mode.
Metabolites identified by standard injection.
The UPLC-qTOF-MS analysis of mostly secondary metabolites was complemented by GC-MS metabolite profiling of derivatized extracts. Following a statistical test (see “Materials and Methods”), 48 of the 63 detected metabolites, mostly organic acids and sugars, were found to be significantly different in at least a single genotype compared with the wild type (Supplemental Table S2).
The Proximal Network Metabolome: Changes in Levels of Both Classes of GSs
The levels of AGs and IGs and other metabolites of the defined proximal network (metabolites downstream to either Trp or Met) were compared between rosette leaves derived from the transgenic plants overexpressing the various MYB factors and wild-type plants (Figs. 6 and 7). MYB28-like clade-overexpressing plants showed increased levels of seven Met-derived GSs (Fig. 5A): 3-hydroxypropyl, 8-methylsulfinyloctyl, 7-methylthioheptyl, 8-methylthiooctyl, 9-methylsulfinylnonyl, methylsulfonyloctyl, and 7-methylsulfinylheptyl. Most intriguing was the finding that levels of the same seven GSs were reduced, in the majority of cases, in leaves derived from the ATR1-like clade-overexpressing plants (Fig. 7). On the other hand, the ATR1-like clade-overexpressing plants exhibited significant accumulation of four IGs (Fig. 5B), 4-methyoxy-3-indolylmethyl (4MO-I3M), 1-methoxy-3-indolylmethyl (1MO-I3M), indolylmethyl (I3M), and 4-hydroxy-3-indolylmethyl (4HO-I3M), compared with wild-type plants. Also in this case, I3M and 1MO-I3M showed reduced levels in plants overexpressing the other clade members (i.e. the MYB28-like clade; Fig. 7). The third IG, 4HO-I3M, was not detected in MYB28-like clade-overexpressing plants, while levels of the fourth IG, 4MO-I3M, were also induced in the MYB28-like clade-overexpressing plants. Taken together, these results demonstrated that the increased accumulation of one type of GSs in transgenic plants overexpressing members of either clade resulted in decreased levels of the other type of GSs (Fig. 7). This cross talk was also evident in the case of the transcriptome analysis (Fig. 5).
Figure 7.
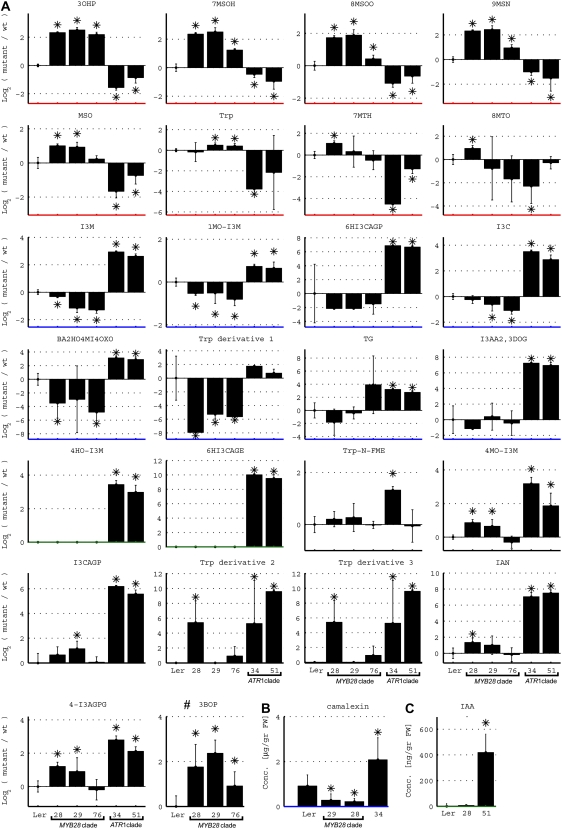
Accumulation of proximal network metabolites related to Met- and Trp-derived GS pathways. A, Levels in wild-type (wt) plants (Ler) and plants overexpressing the MYB28-like clade (MYB28, MYB29, and MYB76) and the ATR1-like clade (ATR1/MYB34 and MYB51) genes. The metabolite levels in these charts are presented as log ratios from the wild type (levels of the latter, therefore, are always zero). B and C, Camalexin concentrations in wild-type and MYB29-, MYB28-, and ATR1/MYB34-overexpressing plants (leaf tissue) after treatment with AgNO3 (B) and levels of IAA in wild-type plants and plants overexpressing MYB28 and MYB51 (leaf tissue; C). Metabolite levels are shown as means ± se from six (UPLC-qTOF-MS analysis) or five (GC-MS analysis) independent samples; asterisks indicate values that are significantly different (P < 0.05) in comparison with the wild type. The different metabolites are ordered according to the different behaviors, indicated with different colors of the x axes, as follows: red, increase in the MYB28-like clade and decrease in the ATR1-like clade; blue, decrease in the MYB28-like clade and increase in the ATR1-like clade; green, increase in the ATR1-like clade; black, increase in both the MYB28-like clade and the ATR1-like clade. The full names of the detected compounds in this analysis are as follows: Trp indole-3-carboxylate glucopyranose (I3CAGP), 9-methylsulfinylnonyl glucosinolate (9MSN), methylsulfonyloctyl glucosinolate (MSO), 6-hydroxyindole-3-carboxylic acid 6-O-β-glucopyranoside (6HI3CAGP), 6-hydroxyindole-3-carboxylic acid β-glucopyranosyl ester (6HI3CAGE), 3-benzoyloxypropyl glucosinolate (3BOP), Trp N-formyl methyl ester (Trp-N-FME), 1H-indole-3-carboxaldehyde (I3C), 2-butenoic acid, 2-hydroxy-4-(1-methyl-1H-indole-3-yl)-4-oxo (BA2HO4MI4OXO), tryptopol glucopyranoside (TG), 1H-indole-3-acetic acid, 2,3-dihydro-2-oxo Glc (I3AA2,3DOG), 4-O-(indole-3-acetyl)-glucopyranose Glc (4-I3AGPG), 3-hydroxypropyl glucosinolates (3OHP), 7-methylsulfinylheptyl glucosinolates (7MSOH), 8-methylthiooctyl glucosinolates (8MTO), 8-methylsulfinyloctyl glucosinolates (8MSOO), 7-methylthioheptyl glucosinolates (7MTH), 1-methoxyindole glucosinolates (1MO-I3M), indole-3-yl-methyl glucosinolates (I3M), 4-methoxyindole glucosinolates (4MO-I3M), and 4-hydroxyindole-3-yl-methyl glucosinolates (4HO-I3M).
The Proximal Network Metabolome: Changes Related to the Biosynthesis of Metabolites Downstream of Trp
Accumulation of the phytoalexin camalexin cannot be detected in wild-type plants without being induced by a variety of microorganisms, such as Pseudomonas syringae and Alternaria brassisicola, and by abiotic factors, such as AgNO3 (Glawischnig et al., 2004). After AgNO3 treatment, plants overexpressing the ATR1-like clade members exhibited 3- to 4-fold higher levels of camalexin compared with wild-type plants (Fig. 7). Plants overexpressing the MYB28-like genes, on the other hand, produced lower levels of camalexin upon AgNO3 induction (3- to 4-fold) relative to AgNO3-treated wild-type plants (Fig. 7). These results demonstrate once more the reciprocal negative feedback regulation between the two GS pathways. Such a negative correlation in levels between plants overexpressing genes of the two clades was not detected in the case of a different proximal network metabolite, auxin. We analyzed the amount of free IAA in rosette leaves expressing the MYB51 (under the control of the 650 promoter) and MYB28 (under the control of the AS1 promoter) factors (Fig. 7) and revealed that IAA levels were increased 10-fold in MYB51-overexpressing plants compared with their levels in wild-type plants but were not changed in the MYB28-overexpressing plants.
The extensive metabolic analyses using GC-MS and UPLC-qTOF-MS allowed us to putatively identify and monitor the levels of additional proximal network components. Although a clear reciprocal decline in IAA levels in MYB28-overexpressing plants was not detected, levels of one of its derivatives, indole-3-carboxyladehyde (I3C), was induced in the ATR1-like gene-overexpressing plants and reduced in the MYB76- and MYB29-overexpressing plants (Figs. 5B and 7). The level of IAN, the substrate for IAA formation and the precursor for camalexin biosynthesis, was very dramatically increased in the ATR1-like clade-overexpressing plants. This metabolite could be formed both from IAOx and as a product of IG hydrolysis (Fig. 7). IAN was also slightly increased in the MYB28-overexpressing plants.
The levels of additional proximal network metabolites downstream of Trp were strongly and specifically induced in plants overexpressing ATR1-like clade genes but not in plants overexpressing the MYB28-like clade members (Figs. 5B and 7). These included (1) the Trp derivative Trp-N-formyl methyl ester; (2) the IAOx derivative tryptol glucopyranoside; (3) three derivatives of I3C, 2-butenoic acid, 2-hydroxy-4-(1-methyl-1H-indole-3-yl)-4-oxo, 6-hydroxyindole-3-carboxylic acid glucopyranosyl ester, and 6-hydroxyindole-3-carboxylic acid 6-glucopyranoside; and (4) the IAA derivate 1H-indole-3-acetic acid, 2,3-dihydro-2-oxo hexose (Figs. 5B and 7). Another derivative of I3C, indole-3-carboxylic acid 6-glucopyranose, the putative precursor of 6-hydroxyindole-3-carboxylic acid 6-glucopyranoside, was induced in plants overexpressing both ATR1-like members but was also increased in MYB29-overexpressing plants (Figs. 5B and 7).
Two unknown Trp derivatives (Trp derivatives 2 and 3; Fig. 7) were induced in the ATR1-like-overexpressing plants and in the MYB28-expressing plants. A third Trp derivative (Trp derivative 1) was reduced in levels in all three MYB28-like clade-overexpressing plants but did not change in the ATR1-clade-overexpressing plants. Finally, levels of Trp itself were strongly down-regulated in the ATR1/MYB34-overexpressing plants, while they were slightly increased in plants overexpressing the MYB29 and MYB76 members of the MYB28-like clade (Figs. 5B and 7). Another indolic compound, a derivative of IAA, 4-O-(indole-3-acetyl)-d-glucopyranose Glc, showed a significant increase in levels in plants overexpressing the two ATR1-like clade members but also, albeit more moderately, in plants overexpressing MYB28 and MYB29 (Fig. 7).
Metabolome Changes in the Distal Network of Plants Overexpressing the ATR1-Like and MYB28-Like Clade Genes
We next evaluated the influence of overexpressing both clade members on the distal network of the metabolome related to GS biosynthesis. Overall, a total of 53 distal network metabolites were detected as differentially produced in at least a single transgenic plant compared with the wild type (from both GC-MS and LC-MS analyses; Fig. 8; Supplemental Fig. S2). The levels of 40 of these metabolites in the different genotypes are depicted in Figure 8. A relatively large set of metabolites were increased in levels in all five overexpression lines. They included several acids (glycolic acid, propanoic acid, 2,4-dihydroxybutanoic acid, glyceric acid, Hyp, and threonic acid), sugars (Xyl, 1,6-anhydroglucose, and trehalose), and O-acetyl-Ser. The latter metabolite is a major precursor for Cys that is utilized in the biosynthesis of both classes of GSs (Fig. 6). On the other hand, Ser, the precursor for O-acetyl-Ser, was down-regulated in the ATR1-like clade-overexpressing plants but up-regulated in plants overexpressing MYB28 and MYB76 from the MYB28-like clade (Fig. 8).
Figure 8.
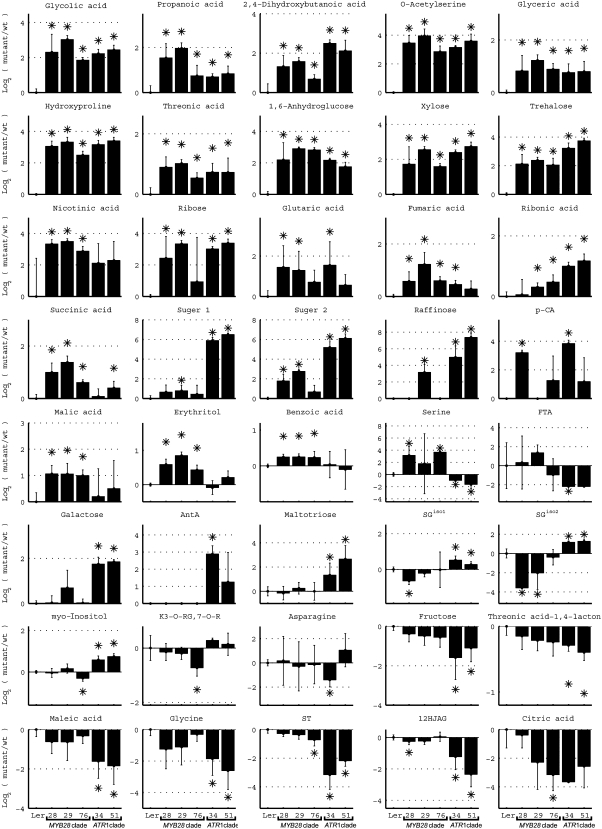
Accumulation of representative distal network metabolites related to Met- and Trp-derived GS pathways. Levels in wild-type plants (Ler) and plants overexpressing the MYB28-like clade (MYB28, MYB29, and MYB76) and the ATR1-like clade (ATR1/MYB34 and MYB51) genes. Metabolite levels are shown as means ± se from six (UPLC-qTOF-MS analysis) or five (GC-MS analysis) independent samples; asterisks indicate values that are significantly different (P < 0.05) in comparison with the wild type. The metabolite levels in these charts are presented as log ratios from the wild type (levels of the latter, therefore, are always zero). The full names of the detected compounds in this analysis are as follows: anthranilic acid (AntA), p-coumaric acid (p-CA), synapoyl-Glc (SG; isomers [iso] 1 and 2), 12-hydroxyjasmonic acid 12-O glucoside (12HJAG), feruloyl tartaric acid (FTA), kaempferol 3-O[6-O-(rhamnosyl)glucoside] 7-O-rhamnoside (K3-O-RG,7-O-R).
The TCA cycle supplies precursors to Met biosynthesis, and components of this pathway were induced in plants overexpressing genes of both clades (but not all), including fumaric acid, glutaric acid, and succinic acid (Figs. 6 and 8). Malic acid, another component of the TCA cycle, as well as nicotinic acid, benzoic acid, and erythritol were only induced in the MYB28-like clade-overexpressing plants. The phenylpropanoid derivative benzoic acid is a precursor for the formation of BGs (produced typically by Arabidopsis seeds; Fig. 6), among them 3-benzoyloxypropyl glucosinolate (3BOP), which was induced in leaves of plants overexpressing the MYB28-like clade members (Figs. 5A and 7).
Interestingly, metabolites derived from Phe were differentially expressed in the transgenic lines, albeit not in the same manner among the various genotypes (Figs. 6 and 8). These included p-coumaric acid, sinapic acid derivatives (two sinapoyl Glc isomers [SGiso1 and SGiso2] and sinapoyl tartronate), feruloyl tartaric acid, and a derivative of the flavonol kaempferol. Among the metabolites that were specifically induced in plants overexpressing the ATR1-like clade members (and not changed in the overexpression of the other clade), we detected several sugars (maltotriose, galactinol, and d-Gal) and anthranilic acid, which is the precursor for Trp biosynthesis (Figs. 6 and 8; Supplemental Fig. S2).
The compound PAPS is required not only for GS biosynthesis but also for various other sulfation reactions. In one such reaction, the jasmonic acid derivative 12-hydroxyjasmonic acid is either sulfonated (Gidda et al., 2003) by a sulfotransferase (and PAPS) or glycosylated (Liechti and Farmer, 2006). Supporting the competition for PAPS between glycosylation and sulfonation of 12-hydroxyjasmonic acid in Arabidopsis leaves, we detected a decrease in the levels of 12-hydroxyjasmonic acid 12-O-β-glucoside in plants overexpressing members of the two clades, particularly those of the ATR1-like clade (Figs. 6 and 8).
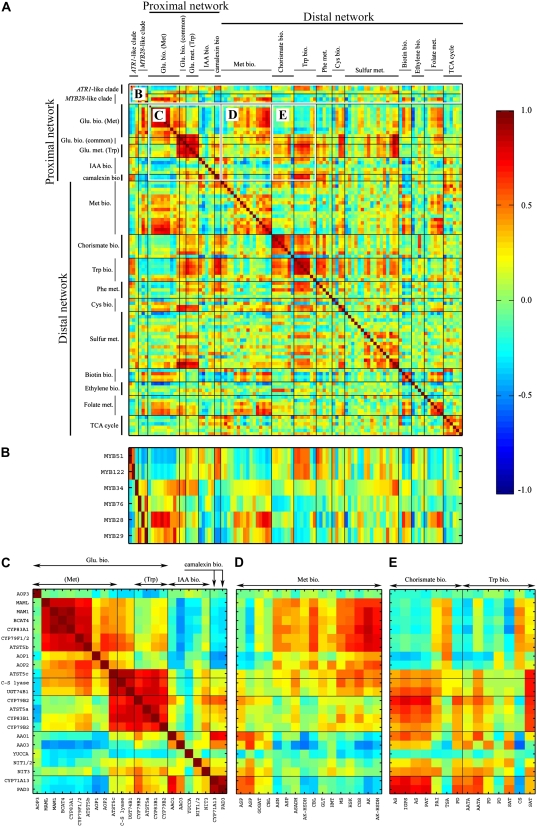
Expression Correlation Analysis between the Two Clade Members and the Proximal and Distal Network Genes in Response to Various Biological Perturbations
From the results obtained above, it was evident that the five GS regulators we examined act directly or indirectly to activate or repress multiple branches of the metabolome while controlling the balance between them. In order to examine how the expression of the six GS regulators is coordinated with the various metabolic processes and how these processes are coordinated with each other, we collected publicly available Arabidopsis gene expression data that are derived from hundreds of experiments and that represent 211 different biological perturbations. Next, we calculated the correlation matrix for the six MYB transcription factors and the entire set of proximal network and distal network genes that showed a significant change in the overexpressing plants (Fig. 9A; Supplemental Table S9). Figure 9B shows an enlargement of the correlation matrix between the six MYB transcription factors and the proximal and distal network genes. The results showed that MYB29 and MYB76 exhibit a positive correlation (r = 0.63) between them and that MYB29 is also positively correlated (although less) to MYB28 (r = 0.29), which is negatively correlated to both MYB51 and MYB122 (r = −0.44 and r = −0.37, respectively), which are positively correlated (0.55) between themselves. Surprisingly, ATR1/MYB34, which is part of the ATR1-like clade, is negatively correlated with its clade members (MYB51 and MYB122) and positively correlated with the MYB28-like clade members.
Figure 9.
Correlation in expression of members of the two clades and the proximal and distal network genes in response to various biological perturbations. The correlation matrix is for six members of the two clades, all of the proximal and distal network genes that exhibited a statistically significant expression change in one of the overexpression plants. The correlation matrix was calculated using hundreds of publicly available GeneChip experiments (see “Materials and Methods”) representing 211 different biological perturbations. Interesting correlation regions from the complete correlation matrix (A) are indicated in B to E.
MYB51 and MYB122 showed a positive correlation to the proximal network genes that are involved in Trp-derived GS biosynthesis, camalexin, and IAA biosynthesis but a negative correlation to the proximal network genes that are involved in Met-derived GS biosynthesis. In contrast, the MYB28-like clade showed a positive correlation to the proximal network genes that are involved in Met-derived GS biosynthesis. While MYB28 showed positive (albeit relatively weak) correlation to the proximal network genes that are involved in Trp-derived GS biosynthesis, MYB29 and MYB76 were not correlated to the same genes. All three members of the MYB28-like clade showed negative correlation to genes that are involved in IAA and camalexin biosynthesis. ATR1/MYB34, which as mentioned above is correlated with the MYB28-like clade members, showed similar correlations to MYB28, as it was positively correlated with the proximal network genes that are involved in Met-derived GS biosynthesis, showed positive correlation to the proximal network genes that are involved in Trp-derived GS biosynthesis and negative correlation to genes that are involved in IAA and camalexin biosynthesis.
The correlation matrix of the proximal network genes (Fig. 9C) showed that each group of genes (Met- versus Trp-derived GS biosynthesis) presents a very high positive correlation within it, but there is also a positive correlation between the two groups, meaning that genes from both groups are frequently active simultaneously. In addition, strong negative correlation is detected between the Met-derived GS genes and the IAA and camalexin biosynthesis genes, while the Trp-derived GS genes exhibit positive correlation with these genes.
Of the entire set of proximal network genes, the Met biosynthesis distal network genes showed the strongest positive correlation with those involved in Met-derived GS biosynthesis, but it was also positive to the common GS biosynthesis genes and Trp-derived GS biosynthesis genes (Fig. 9D). It showed a negative correlation to the genes involved in IAA and camalexin biosynthesis (Fig. 9D). On the other hand, the Trp biosynthesis distal network genes exhibited a strong and positive correlation with all of the proximal network genes, excluding the Met-derived GS biosynthesis genes, to which they had a strong negative correlation (Fig. 9E).
Plants Overexpressing Both Clades Deter Oviposition of the Whitefly B. tabaci
GSs are known to function as a defense mechanism against insect herbivores in members of the Brassicaceae family (Rask et al., 2000). Here, we examined the effect of GS accumulation detected in transgenic plants overexpressing the different members of the MYB28-like and ATR1-like clades on the plant's ability to influence insect behavior. The experiments studied the oviposition decisions of the phloem-feeding insect B. tabaci, a major agricultural pest of field and horticultural crops worldwide (Brown et al., 1995). In most flying insects with offspring of limited mobility, the ability of adult females to distinguish and avoid poor-quality hosts for oviposition should strongly influence reproductive success and lifetime fitness through enhanced juvenile growth and survival.
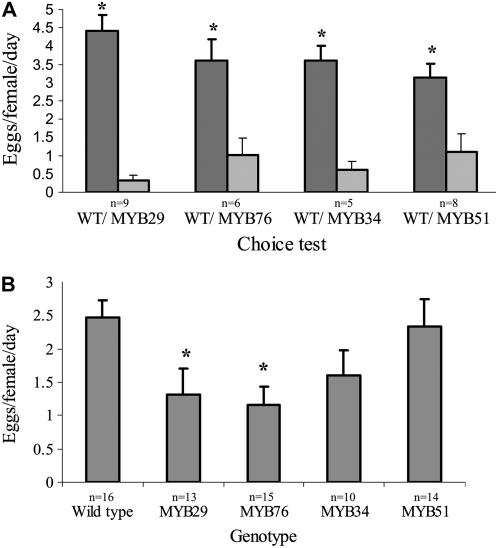
We first conducted choice experiments. Relative to wild-type plants, B. tabaci females preferred to lay significantly fewer eggs on plants overexpressing the MYB28-like clade members MYB29 (paired Student's t test; P < 0.0001) and MYB76 (paired Student's t test; P = 0.048) and those overexpressing the ATR1-like clade members ATR1/MYB34 (paired Student's t test; P = 0.002) and MYB51 (paired Student's t test; P = 0.027; Fig. 10A).
Figure 10.
Oviposition of B. tabaci females (Biotype Q) on five Arabidopsis genotypes: wild type, plants overexpressing MYB29 and MYB76 (both with expression driven by the AS1 promoter), and plants overexpressing ATR1/MYB34 and MYB51 (both with expression driven by the 650 promoter). A, Choice experiments. Vertical bars represent means and se. Asterisks denote significant differences compared with the wild type (paired Student's t test; P < 0.05). Dark and light gray bars represent wild-type plants (WT) and MYB-overexpressing plants, respectively. B, No-choice experiments. Asterisks denote significant differences compared with the wild type (Dunnett's test; P < 0.05). n, Number of biological replicates.
To further test the effect of AGs and IGs on B. tabaci, we conducted no-choice experiments. The number of eggs oviposited per female on wild-type plants was significantly higher than the number of eggs oviposited on MYB29 (ANOVA: F4,63 = 3.43, P = 0.013; Dunnett's test [wild type versus MYB29]: P = 0.049]) and MYB76 (Dunnett's test [wild type versus MYB76]: P = 0.016) plants that accumulate AGs (Fig. 10B). There was no significant difference in oviposition on wild-type plants and ATR1/MYB34 (Dunnett's test [wild type versus ATR1/MYB34]: P = 0.254) and MYB51 (Dunnett's test [wild type versus MYB51]: P = 0.995) plants that accumulate IGs (Fig. 10B).
DISCUSSION
Secondary or specialized plant metabolites such as GSs obtain their basic carbon skeletons from precursor pathways that are represented by primary metabolites, including amino acids, lipids, and carbohydrates. Biosynthesis of secondary metabolites is a costly process, since they are often produced to very high levels and could not be recycled to their basic source. Consequently, switching on a pathway involved in synthesizing such compounds is likely to be a well-coordinated process in which activation occurs at multiple points, from the very primary pathways forming the basic structures to the last committed step in the formation of a specialized metabolite. Despite the issue raised above, only a limited set of reports described detailed and parallel analyses of primary and secondary metabolism and the interface between them.
Here, transgenic plants overexpressing the two clades of R2R3-MYB transcription factors described recently by several groups (Gigolashvili et al., 2007a, 2007b; Hirai et al., 2007; Sonderby et al., 2007) served as an excellent tool for such a study, given that they control secondary metabolic pathways that retain similar primary precursors (amino acids) and that their secondary products share the need for sulfur in their basic skeleton. Transcriptome analysis using Affymetrix GeneChips and metabolomics by the use of mass spectrometry-based technologies were employed in order to obtain the broadest coverage of gene and metabolite expression. Using these approaches, we followed transcripts and metabolites belonging to either the proximal network of GSs and related structures or the distal network of metabolic pathways, generating precursors for their biosynthesis or additional distinct pathways. Although only a relatively small portion of the metabolome could be identified by metabolomics in its current state, such technologies allowed us to putatively identify and monitor the relative levels of more than 130 primary and secondary metabolites in Arabidopsis leaves. Our metabolite measurements represent steady-state analysis of pathway intermediates and end products. While such analysis provides new information with respect to metabolic activity of the plant or the tissue and response to perturbations, it does not represent the dynamics of the metabolic network. Steady-state analysis could be misleading, as the flux through a pathway could change without elevation in pool sizes or the end product, which might have an increased turnover (Fernie et al., 2005). Thus, measurements of flux, as by stable isotope labeling of metabolites, can be most useful to follow the dynamics of metabolic changes that occur in the transgenic plants overexpressing the various MYB regulators, which were investigated here for steady-state metabolite levels.
The Cross Talk between the Trp and Met Branching Pathways Is Not Restricted to AGs and IGs But Includes Camalexin and the IAA Derivative Indole-3-Carboxyladehyde
In view of the fact that GSs are sulfur-containing compounds, it is likely that they are not only important for plant defense but also play a role in the mechanism of controlling the total sulfur pool in the cell. In fact, since GSs contain sulfur atoms, they could be an efficient form of storage compared with free sulfur and possibly even less toxic. Thus, a change in one GS pathway would require balancing of the sulfur pool by modulating the total GS levels, and this could be achieved through an opposite change in levels of a second GS biosynthesis pathway. Previous reports have also described this phenomenon for GS biosynthesis (Hemm et al., 2003; Gigolashvili et al., 2007a, 2007b). In a different report (Gigolashvili et al., 2008), the authors described a transient transactivation assay in Arabidopsis cells that showed that MYB factors controlling AG biosynthesis transcriptionaly repress the IG regulators by directly acting on their promoters. They suggested that this may lead to the metabolic balance between the two GS pathways. However, our array data and those of Sonderby et al. (2007) did not show such a transcriptional repression in plants overexpressing the AG regulators, as expression of the IG regulatory factors was not reduced. With regard to changes in transcripts detected in this study, plants overexpressing the ATR1-like clade showed an increase in the levels of CYP79B2, CYP79B3, and CYP83B1 (IG biosynthesis), while the same transcripts were down-regulated in the MYB28-like-overexpressing plants. Curiously, we could not detect reciprocal changes in transcripts corresponding to the AG biosynthesis pathway, although all five AGs detected did show a reciprocal negative feedback behavior.
The combination of extensive metabolome and transcriptome analyses carried out in this work pointed to other cases of reciprocal negative regulation between the Trp and Met branching pathways. We discovered that the cross talk between the pathways starting from Trp and those of Met is not restricted to AGs and IGs, as detected here and by others, but also includes camalexin (after AgNO3 treatment), the IAA derivative indole-3-carboxyladehyde, and an unknown Trp derivative. Grubb and Abel (2006) suggested two alternative explanations for these compensatory mechanisms between the two biosynthesis pathways. As one possibility, they proposed that it is achieved by pathway intermediates or end products that serve as inhibitors of enzymes; a second explanation was the competition between the cytochrome P450 monooxygenases of the core pathway for electrons needed for their activity. The electrons are provided by NADPH, and a block or activation in one pathway would result in decreased or increased availability of NADPH for the other pathway. Our data point to some transcriptional changes that take part in the cross talk between the two GS pathways. However, as the genes showing reciprocal behavior are limited to part of the IG pathway and to only two IGs (I3M and 1MO-13M), it is likely a combination of changes in transcription and enzyme activities, possibly through inhibitors or reduced cofactors, that mediate this cross talk.
Nontargeted Metabolite Analysis Revealed That Overexpression of the ATR1-Like Clade Genes Has a Much Broader Effect on the Metabolism of Indolic Compounds Than Described Previously
Previous reports of plants overexpressing the ATR1-like clade members showed that they exhibited increased IGs and free IAA levels (Zhao et al., 2002). Nontargeted metabolite profiling conducted in the course of this study showed that the effect on indolic compounds is much broader and is not confined to IGs and free IAA. It included increased production of various Trp derivatives, IAN, derivatives of IAA and indole carboxylic acid, and finally of the phytoalexin camalexin. A simultaneous increase in levels of both IAN and IAA was also detected in plants overexpressing CYP79B2 (Zhao et al., 2002). Thus, the question regarding the regulation of IG biosynthesis in concert with the maintenance of homeostasis of multiple metabolites produced by side branches that share the same basic precursors is most complex. Although we used overexpression to demonstrate the effect on metabolism, the results with transgenic plants might be a good indicator for the surrounding metabolic pathways that have to be balanced when IG production is induced, such as in response to herbivory.
Gigolashvili et al. (2007a) suggested that in spite of the common precursors between IAA and IG biosynthetic pathways, they could be specifically regulated by the different activities of MYB122, ATR1/MYB34, and MYB51. This was based on the observations that the MYB51 overexpression and the dominant mutant (HIG1-1D) lines did not display altered morphology and that the levels of IAA were only moderately increased in these plants. In contrast, overexpression of MYB51 in our study (under the control of a different, specific promoter) resulted in severe morphological phenotypes that strongly resembled those observed by Gigolashvili et al. (2007a) in plants overexpressing MYB122 and ATR1/MYB34. Moreover, measurement of free auxin in the MYB51-overexpressing plants compared with wild-type plants showed a strong increase (up to 10-fold relative to the wild type) in MYB51 plants. The conclusion that overexpression of MYB51, as detected previously for ATR1 overexpression lines (Celenza et al., 2005), results in high levels of free IAA was also supported by the observation that an auxin reporter (DR5∷GUS) was much more active in seedlings overexpressing the ATR1-like clade members than in wild-type seedlings (including in MYB51-overexpressing plants). In fact, the increase in IAA levels detected in MYB51-overexpressing plants was at least five times higher than that detected in the atr1D and 35S∷ATR1 lines (Celenza et al., 2005) and that detected in CYP79B2-overexpressing plants (Zhao et al., 2002). Thus, when overexpressed, MYB51, like ATR1/MYB34, also could influence the homeostasis of auxin.
While elongated hypocotyls and petioles and epinastic cotyledon phenotypes have been observed previously in mutants exhibiting inhibition of IAOx reactions and overexpression of the prealdoxime gene CYP79B2 (Grubb and Abel, 2006), the phenotypes obtained by overexpression of MYB51 and ATR1/MYB34 driven by the AS1 and 650 promoters were much more severe. In particular, expression under the control of the AS1 promoter resulted in plants possessing a pin-like naked stem due to cessation in organ formation at the vegetative meristem and fusion of floral organs. These phenotypes closely resembled the pin1 mutant phenotype, in which polar auxin transport is impaired (Okada et al., 1991). It suggests that the normal levels of free IAA in the inflorescence axes required in early developmental stages of floral bud formation in Arabidopsis were dramatically increased and resulted in the pin-like phenotypes.
Earlier work suggested that there are multiple Trp-dependent and possibly even independent IAA biosynthesis pathways (Delker et al., 2008). The accumulation of free auxin in plants overexpressing the ATR1-like clade may possibly aid in understanding the biosynthesis of this important phytohormone. The increase in IAN levels and the induction of NIT3 in the ATR1-like clade-overexpressing plants might imply that the induced IAA formation is a result of the activation of a metabolic pathway that involves IAN. This is supported by the increased production of camalexin in the same plants, since it was recently suggested that CYP71A13 converts IAOx to IAN and furthermore by yet unknown enzymes to dihydrocamalexic acid, which is subsequently converted to camalexin by PAD3 (or CYP71B15; Zhou et al., 1999). Nevertheless, we did not detect an increase in either CYP71A13 or PAD3 gene expression or in the YUCCA gene, which encodes a monooxygenase catalyzing the N-hydroxylation of tryptamine, the precursor for the formation of IAOx. Thus, it might be that the increased production of IAA and camalexin in the ATR1-like clade-overexpressing plants is carried out by yet undescribed genes in one of the pathways utilizing IAN or through posttranscriptional regulation. Yet, it could not be ruled out that an IAN-independent pathway is activated for both IAA and camalexin biosynthesis in these plants.
Transcriptional and Metabolic Hubs in the Distal Network of Metabolic Pathways Supplying Precursors to GS Biosynthesis
It is clear that precursor feeding pathways need to be activated in order to support the production of GSs, although an open question remaining is what are the direct targets of the different factors studied here in the underlying metabolic pathways. The recently described SULFUR LIMITATION1 (SLIM1) gene adequately exemplifies this regulatory mechanism, in which the same transcription factor possesses gene targets in both primary and secondary metabolism. The SLIM1 transcription factor regulates sulfur assimilation in a sulfur-deficient environment in Arabidopsis roots by inducing the expression of genes associated with sulfate transport (Maruyama-Nakashita et al., 2006). Simultaneously, the same factor enhanced GS degradation by activating the expression of a thioglucosidase (myrosinase) gene (At2g44460) that releases the aglycons of GSs. Sulfate can further be released from the aglycons and recycled by its use in primary metabolism. The slim1 mutation affected the expression of genes related to GS metabolism, including CYP79B2/B3, MAM1, MAML, CYP79F1/F2, BCAT, CYP83B1, and, interestingly, also of the ATR1/MYB34 gene.
Indeed, both our microarray and metabolic profiling data point to a clear activation of specific metabolic pathways of the distal network in plants overexpressing members of the two clades. Parts of these pathways serve as carbon skeletons, sulfur and methyl group donors for the biosynthesis of Met and Trp. Expression of genes involved in the sulfur assimilation pathway, the formation of Cys and PAPS, which are important precursors for both IG and AG biosynthesis, were induced in plants overexpressing genes of either clade. At the metabolite level, O-acetyl-l-Ser, the precursor for Cys biosynthesis, was also induced in plants overexpressing members of both clades. Plants overexpressing the MYB28-like clade members (producing AGs) showed induction of genes in the TCA cycle and further downstream through Asp and Gln up to Met. This was supported by an increase in malate and succinate detected in plants expressing one of the three MYB28-like genes.
In the case of the ATR1-like clade gene-overexpressing plants (producing IGs), activation of genes along the route leading from the shikimate pathway to the formation of chorismate and the synthesis of indole and subsequently Trp was evident. In accordance, Celenza et al. (2005) described up-regulation of three IG-related CYP genes but also two Trp biosynthesis-related genes (ASA1 and TSB1) in the dominant atr1D mutant and 35S∷ATR1 plants. Gigolashvili et al. (2007a) showed that MYB51 could activate reporter expression driven by the DHS1 and TSB1 upstream regions but not of the ASA1 gene. This might be related to the increase in anthranilic acid (the product of the ASA1 reaction) observed here in plants overexpressing the ATR1/MYB34 gene and not in the case of MYB51 overexpression. The extensive utilization of Trp in the proximal network pathway of IG biosynthesis could explain the decrease in levels of this amino acid in ATR1/MYB34-overexpressing plants and its increase in MYB29- and MYB76-overexpressing plants.
Overexpression of Genes of the Two Clades Underlines Novel Links to Additional Metabolic Pathways, Including Those of Jasmonic Acid, Folate, Benzoic Acid, and Various Phenylpropanoids
The results obtained by transcriptome and metabolome analyses provided us with new insight into different metabolic pathways that are linked to GS metabolism. It is possible that the various MYB regulators retain direct targets among genes of these pathways. Genes taking part in folate metabolism were induced in plants overexpressing the MYB28-like clade genes. The compound 5-methyl-THF produced in the folate pathway serves as the methyl donor for the biosynthesis of Met from homo-Cys in the reaction catalyzed by Met synthase (Rébeillé et al., 2006).
The reduction in levels of 12-hydroxyjasmonic acid 12-O-β-glucoside in plants overexpressing genes of both clades provided strong evidence for the competition between the utilization of PAPS for sulfunization of GSs and sulfunization and/or glycosylation of 12-hydroxyjasmonic acid. Genes involved in PAPS formation are induced in transgenic plants overexpressing genes of both clades; therefore, PAPS levels increase. Once more PAPS availability is utilized for the production of 12-hydroxyjasmonate sulfate on behalf of 12-hydroxyjasmonate-glucoside, which, therefore, is reduced in levels.
Another interesting observation was the accumulation of benzoic acid, p-coumaric acid, and several other Phe derivatives of sinapate, ferulate and kaempferol. Benzoic acid levels increased only in the MYB28-like clade-overexpressing plants. While the biosynthetic route leading to benzoic acid is not completely resolved, it serves among other reactions for the esterification of BGs. The alkyl portion of the side chain in BGs was suggested to derive from the chain elongation of Met (Graser et al., 2001). The seeds and siliques of Arabidopsis accumulate a series of BGs, the most abundant of them are 3BOP and 4-benzoyloxybutyl glucosinolate. Since plants overexpressing the MYB28-like clade genes accumulated 3BOP in their leaves, it might suggest that the same factors are involved in the regulation of BG biosynthesis in Arabidopsis seeds. The significant decrease in seed 3BOP levels detected by Sonderby et al. (2007) in T-DNA insertion mutants of MYB28 and MYB29 supports this idea. The same authors also showed that seeds of plants overexpressing the MYB76 gene produce more 4-benzoyloxybutyl glucosinolate, while levels of 3BOP remain the same as in wild-type seeds.
Various classes of metabolites produced through the phenylpropanoid pathway are produced in Arabidopsis, acting as, for example, UV-B radiation protectants, phytoalexins, allelopathic compounds, mediators of plant hormone signaling, and facilitators of pollen tube growth (Tohge et al., 2005; Yu and Jez, 2008). Genes that are specific for different branches of phenylpropanoid metabolism are typically transcriptionally regulated by a combination of different classes of regulatory proteins (Skirycz et al., 2007). With similarity to GS metabolism, members of the R2R3-MYB large family of transcription factors are largely involved in the control of various branches of the phenylpropanoid pathway (Stracke et al., 2001). Three genes (MYB11, MYB12, and MYB111), which share significant structural similarity and form subgroup 7 in the Arabidopsis R2R3-MYB family, were shown to control the production of flavonol glycosides (flavonoids), acting in seedlings in an additive manner due to their differential spatial activity (Stracke et al., 2007). In an analogous manner, it appears that the regulation of GS biosynthesis is also orchestrated (but not only) by two related clades of MYB factors that together create a unique subgroup in the Arabidopsis MYB family phylogeny.
Sonderby et al. (2007) showed that overexpression of MYB28, MYB29, and MYB76 led to decreased transcript levels of a MYB transcription factor (MYB4) that suppresses the accumulation of sinapate esters. In addition, transcript levels of SNG1, responsible for the conversion of sinapoyl Glc to sinapoyl malate, were increased in all three genotypes, while expression of BRT1, responsible for the conversion of sinapate to sinapoyl Glc, was reduced in all three. The same authors suggested that the MYB factors may be involved in cross talk between sinapate and aliphatic GS metabolism. Here, we detected changes in the levels of sinapate and ferulate derivatives that generate sunscreen esters and lignin precursors and a derivative of the flavonol kaempferol. In addition, the level of p-coumaric acid was increased in MYB28- and ATR1/MYB34-overexpressing plants. The accumulation of p-coumaric acid synthesized upstream in the general phenylpropanoid pathway supports the explanation provided by Hemm et al. (2003) that the phenylpropanoid O-methylation steps catalyzed by caffeoyl-CoA O-methyltransferase and caffeic acid O-methyltransferase, required for the synthesis of ferulic acid and sinapic acids, respectively, could be perturbed by intermediates of the GS pathway.
How Is the Metabolic Balance Maintained by the Two Clades of Arabidopsis GS Biosynthesis Regulators?
The questions remain regarding how the myriad metabolic pathways and their intermediates that are linked to GS biosynthesis are kept in balance upon changes in GS metabolism and the different roles of the GS MYB regulators in these homeostatic mechanisms. The combined results from previous work and our work in this study using overexpression plants and loss-of-function lines and the correlation analysis based on large gene expression data sets suggest two possible scenarios occurring in Arabidopsis plants. The basic assumption for this model is that the MYB28-like clade and ATR1-like clade genes activate the AG and IG biosynthetic pathway genes, respectively. In the first situation, both Trp-derived GSs, camalexin and IAA, are produced and these activities are mediated by the MYB51 and MYB122 genes. In the second scenario, both Trp- and Met-derived GSs are produced simultaneously, but IAA and camalexin biosynthesis are repressed. This is mediated by the activity of the three MYB28-like clade members and ATR1/MYB34. Thus, the ATR1/MYB34 is a major factor that determines which of the two scenarios will take place, since it is coregulated with the MYB28-like clade genes.
Aliphatic GSs Are More Potent Deterrents to B. tabaci Than Indole GSs
Behavioral experiments on oviposition decisions showed that B. tabaci females preferred to lay eggs on wild-type plants when allowed to choose between wild-type plants and plants overexpressing MYB28- and ATR1-like clade genes. This suggests that GS accumulation may enhance the ability of the plant to protect itself from B. tabaci attack. Furthermore, we showed that AGs exhibited more potent deterrent properties than IGs, as B. tabaci females laid more eggs on transgenic plants ectopically expressing the ATR1-like clade genes (accumulating IGs) than on plants ectopically expressing the MYB28-like genes (accumulating AGs). Mewis et al. (2005) reported that plants respond to insect damage by systemically accumulating higher levels of GSs, which presumably increases their resistance to subsequent attacks. Moreover, recent studies have shown that overexpression of MYB51 in Arabidopsis plants reduces the leaf area consumed by fourth instar larvae of Spodoptera exigua in dual-choice assays (Gigolashvili et al., 2007a) and that overexpression of MYB28 in Arabidopsis plants impaired S. exigua larval development (Gigolashvili et al., 2007b). Due to the large impact that generalist herbivores such as B. tabaci and S. exigua have on agriculture, identification of plant factors that regulate resistance to these insects can significantly contribute to the development of novel control strategies for pest control.
In this study, we identified the MYB28-like clade genes by carrying out a genetic dissection of the abaxial and adaxial leaf domains followed by an examination of abaxial-enriched transcripts. In preliminary experiments to corroborate the array data, the expression pattern of the MYB28-like clade member genes in young leaves of 2-weeks-old seedlings was examined in transgenic plants expressing the YFP reporter gene driven by their upstream region. The MYB28-like clade member genes showed that they exhibit prominent abaxial-localized expression, in particularly MYB76 (data not shown).
In most angiosperms, the polarity of leaves on the adaxial-abaxial sides could be associated with differences in morphology and anatomy (Bowman et al., 2002). These differences correspond to the function of surfaces on either side, for example, light capture on the adaxial side of leaves. Many aphids and whiteflies feed primarily on the abaxial surface of leaves, and several theories have been proposed to explain this behavior of insects: the phloem tissue is more accessible from the underside of the leaf, the possibility of a thinner abaxial cuticle, the ease of stylet penetration through spongy mesophyll, which is less dense than palisade mesophyll, protection from elements such as rain and high solar energy, reduced predation, and reduced accumulation of excreta (Chu, 1995; Powell et al., 2006). In addition, synthesis of anti-herbivore compounds in the abaxial leaf domain can provide better protection to the meristem against phloem-feeding insects, since the abaxial side of young leaves covers the meristem from the outside and therefore serves as a physical barrier as well. Consequently, it will be highly beneficial for the plant to protect itself from herbivory by activating the GS biosynthetic pathway through abaxial expression of the MYB regulatory genes in young leaves. Since dissecting the two sides of a leaf is technically difficult, examples of metabolic pathways activated on one side of leaves and not on the other are scarce. Future work investigating the complex regulation of GS biosynthesis should aim at a higher resolution analysis of transcripts and metabolites in unique cell layers and single cells.
MATERIALS AND METHODS
Plant Lines and Growth Conditions
Unless stated differently, all Arabidopsis (Arabidopsis thaliana) plants described were in the Ler background and were grown under cool-white fluorescent light in long-day conditions (18 h of light/6 h of dark, 18°C). The Arabidopsis lines containing a T-DNA insertion (ecotype Columbia) in MYB28 (SALK_136312) and MYB76 (SALK_055242) were obtained from the SIGnAL project collection (Alonso et al., 2003), and the insertion lines of the MYB29 and MYB51 genes (SM_3_34316 and SM_3_16332) were obtained from the Nottingham Arabidopsis Stock Centre (Scholl et al., 2000). Multiple mutant plants were generated by cross-fertilizing homozygous mutants. The mutant phb-1d was described previously (McConnell et al., 2001). In cases in which the transactivation system was used (Alvarez et al., 2006), the vectors are described by “≫” across the text. Vectors in which the promoter was fused directly to the gene are marked by double asterisks.
Cloning Procedures and Plant Transformation
Transactivation lines were generated by transcriptional fusion of promoters to chimeric LhG4 and cDNAs subcloned behind an operator array (Moore et al., 1998) in the BJ36 vector (oligonucleotides used for cloning cDNAs or promoters are described in Supplemental Table S5). The precursors for the MYB28-like-miR and ATR1-like-miR synthetic miRs were synthesized as described previously (Alvarez et al., 2006), and after sequence verification, the pre-miR was cloned downstream of the 35S CaMV promoter (in pART7). Constructs were subcloned into the pMLBART binary vector and introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. Transgenic plants were generated by the floral dipping method (Clough and Bent, 1998), and transformants were selected in soil on the basis of resistance to the herbicide BASTA. The ANT∷LhG4 line was provided by Michael Lehnard (Schoof et al., 2000); 10OP∷KAN2 and AS1∷LhG4 were described earlier (Eshed et al., 2001).
Analysis of Gene Expression with RT-PCR
For RT-PCR, total RNA was extracted with the RNeasy RNA isolation kit (Qiagen) from 2-week-old seedlings. Approximately 1 μg of DNaseI-treated RNA was primed with oligo(dT) and converted to complementary DNA using SuperScript II reverse transcriptase (Invitrogen). The cDNA concentrations in different tissue samples were equalized semiquantitatively based on the amplification of the α-TUBULIN gene. Subsequent PCRs were carried out using standard protocols (gene-specific primers are listed in Supplemental Table S5).
Microarray Hybridization
Microscissors were used to collect leaves from 14-d-old seedlings whose cotyledons and hypocotyls were removed. For each sample, we pooled leaves of approximately 30 seedlings that showed a clear morphological phenotype (progeny of a single transformation event line). Total RNA (15 μg) was extracted with the RNeasy RNA isolation kit (Qiagen). Labeled copy RNA was prepared and hybridized to Affymetrix ATH1 GeneChips, according to the manufacturer's guidelines. All experiments used two independent biological replicates.
Microarray Data Analysis
Signal values were obtained using the RMA algorithm (Irizarry et al., 2003) implemented using the R programming language, which is currently the gold standard technique for this purpose. Using t test statistics, it was established (data not shown) that there are little or no significant differences between the ATR1 (MYB34)- and MYB51-overexpressing lines and between the MYB29- and MYB76-overexpressing lines; therefore, we joined each group of arrays in the comparison of those lines with the wild-type plants (ATR1/MYB34 and MYB51 versus the wild type and MYB29 and MYB76 versus the wild type). In these comparisons, we used the following criteria to identify significant differences in gene expression levels: genes with log ratio above 0.5 and t test P value below 0.01, and genes with log ratio above 1 and t test P value below 0.05. These criteria were tested on the expression levels of all genes that are marked as enzymes using the AraCyc metabolic database (ftp://ftp.arabidopsis.org/home/tair/Pathways/).
Nontargeted Metabolic Analysis of Semipolar Compounds by UPLC-qTOF-MS
Nontargeted metabolic analysis was performed on leaves of plants overexpressing the ATR1-like clade genes (ATR1/MYB34, n = 6; MYB51, n = 5), MYB28-like clade genes (MYB28, n = 6; MYB29, n = 6; MYB76, n = 5), and wild-type genes (n = 5). For each sample, we pooled leaves of approximately six plants that showed a clear morphological phenotype (progeny of a single transformation event line). Leaves of 14-d-old plants (100 mg) were harvested, frozen, ground, and extracted as described by Glawischnig et al. (2004), with modifications. Extraction was performed for 40 min in 450 μL of MeOH:H2O (80:20) at 60°C. After centrifugation, the supernatant was filtered through a Millex-GV MF (PDV) 0.22-μm filter, and the filtrate was analyzed by LC-MS. The sample (5 μL) was applied to an Acquity UPLC system (Waters) and separated on a BEH C18 Acquity column (100 × 2.1 mm, 1.7 μm; Waters) under a linear gradient elution program with solvent A (0.1% formic acid in 5% acetonitrile-95% water) and solvent B (0.1% formic acid in acetonitrile): 0% to 28% solvent B (22 min), 28% to 40% solvent B (to 22.5 min), 40% to 100% solvent B (to 23 min), 100% solvent B (to 24.5 min), and 100% solvent A (to 26 min). Elution was performed at 0.3 mL min−1 flow, and the column temperature was 35°C. Metabolites were detected by UV absorption (318 nm), and [M − H]− ions for specific GSs were detected in a q-TOF Premier mass analyzer (Waters). The electrospray probe was operated at 3 kV. The source and desolvation temperatures were 125°C and 275°C, respectively. Mass spectra analyses were carried out by the UPLC-qTOF instrument (Waters Premier QTOF), with the UPLC column connected online to a UV light detector (measuring at 318 nm; Waters Acquity) and then to the MS detector. A mixture of 15 standard compounds, injected after each 10 samples, was used as the quality-control sample. MassLynx software version 4.1 (Waters) was used to control the instrument and calculate accurate masses.
Analysis of LC-qTOF-MS Metabolomics Data
The markers (mass signals) obtained from the MarkerLynx software were processed using a custom-made filtering statistical script written in MATLAB 7.0.4 (MathWorks). MarkerLynx often misses the true value of mass signals in the data and marks them as zeros. Therefore, the first stage of the analysis was to distinguish between erroneously marked zero values and true “absent” calls. Two scenarios were considered when examining the replicates for each of the markers. In the first, the mean intensity of the marker is in the highest 90% of the overall data and there is one zero value out of five replicates. The zero value is removed from further analysis. In the second scenario, the intensity is in the highest 90% and there are two or more zero values. A confident assignment of the marker levels for that group cannot be made. Therefore, the marker is excluded completely from the analysis; the mean intensity of nonzero values for the marker in the group is low. Therefore, the zeros are true calls and the zero values are replaced by the detection threshold of the instrument calculated from the overall distribution of the lowest values in the data. To assess whether the different genotypes in the analysis vary in the composition of metabolites, Kruskal-Wallis nonparametric one-way ANOVA was performed on each of the markers. The resulting P values were controlled for multiple hypotheses testing using a 5% false discovery rate cutoff (Benjamini and Hochberg, 1995). For each of the significantly different markers, a series of Mann-Whitney rank sum tests were carried out to find which of the overexpression lines differs from the wild type in the marker's abundance. To control for multiple hypotheses testing (five genotypes versus wild-type tests), once again a 5% false discovery rate cutoff was taken for each of the markers. Statistically different markers were clustered according to the similarity in their abundance profiles across different samples and according to the proximity in their retention time as described by Mintz-Oron et al. (2008). For metabolite identification, we continued only with clusters containing more than one mass signal. Metabolites were identified using standard compounds by comparison of their retention times, UV spectra, MS/MS fragments, and dual-energy fragments. Identification of metabolites for which standards were not available was carried out as described by Mintz-Oron et al. (2008). Chemical structures of the Met and Trp derivatives are presented in Supplemental Table S8.
GC-MS Profiling of Derivatized Extracts
The GC-MS analysis was performed on leaves of plants overexpressing the ATR1-like clade genes (ATR1/MYB34, n = 4; MYB51, n = 5), MYB28-like clade genes (MYB28, n = 5; MYB29, n = 5; MYB76, n = 5), and wild-type genes (n = 4). For every sample, we pooled leaves of approximately six plants that showed a clear morphological phenotype (progeny of a single transformation event line). Analysis of polar compound extracts was performed following the protocol described by Mintz-Oron et al. (2008). Briefly, frozen ground tissue powder (100 mg) was extracted in 700 μL of methanol with 30 μL of internal standard (ribitol; 0.2 mg in 1 mL of water). After mixing vigorously, the extract was sonicated in a bath sonicator for 20 min and centrifuged at 20,000g. Chloroform (375 μL) and water (750 μL) were added to the supernatant, and the mixture was vortexed and centrifuged. Aliquots of the upper methanol-water phase (500 μL) were taken and lyophilized. The derivatization method of the lyophilized sample was as described by Mintz-Oron et al. (2008). Sample volumes of 1 μL were injected into the GC column. A retention time standard mixture (14 μg mL−1 in pyridine:n-dodecane, n-pentadecane, n-nonadecane, n-docosane, n-octacosane, n-dotracontane, and n-hexatriacontane) was injected after each set of six samples. The GC-MS system was composed of a COMBI PAL autosampler (CTC Analytics), a Trace GC Ultra gas chromatograph equipped with a PTV injector, and a DSQ quadrupole mass spectrometer (ThermoElectron). GC was performed on a 30-m × 0.25-mm × 0.25-μm Zebron ZB-5ms MS column (Phenomenex). The PTV split technique was carried out as follows. Samples were analyzed in the PTV solvent split mode. PTV inlet temperature was set at 45°C, followed by the following temperature program: hold at 45°C for 0.05 min, raise to 70°C with a ramp rate of 10°C s−1, hold at this temperature for 0.25 min, transfer to column stage (raising to 270°C with a ramp rate of 14.5°C s−1 and hold at 270°C for 0.8 min), and finish by a cleaning stage (raising to 330°C with a ramp rate of 10°C s−1 and hold at 330°C for 10 min). For separation of the metabolites, we used the chromatographic GC conditions described by Mintz-Oron et al. (2008).
Analysis of GC-MS Data
The reconstructed ion chromatograms and mass spectra were evaluated using Xcalibur software version 1.4 (ThermoFinnigan). Compounds were identified by comparison of their retention index and mass spectrum with those generated for authentic standards analyzed on our instrument. When the corresponding standards were not available, compounds were putatively identified by comparison of their retention index and mass spectrum with those present in the mass spectra library of the Max-Planck-Institute for Plant Physiology (Q_MSRI_ID; http://csbdb.mpimp-golm.mpg.de/csbdb/gmd/msri/gmd_msri.html) and the commercial mass spectra library NIST (www.nist.gov). The response values for metabolites resulting from the Xcalibur processing method were normalized to the ribitol internal standard. This was carried out by dividing the peak area of the metabolite by the peak area of ribitol.
Statistical Analysis of GC-MS Data
In order to test if the level of each metabolite in the transgenic overexpression line was significantly different from its levels in the wild-type plants, we used a standard t test. In cases in which the metabolite was above the detection level in both lines (transgenic and wild type), a two-sample version of the t test was used; in cases in which the metabolite was above the detection level in only one of the lines (transgenic or wild type), a one-sample version of the t test was used. In order to remove outliers in cases in which some of the replicates had detectable levels of the metabolite and some did not, we excluded the outlier replicates only in cases of clear majority, as in cases in which four replicates had detectable levels and two replicates did not.
Analysis of GSs and Camalexin by UPLC-qTOF-MS
For GSs and camalexin analyses, we used the same chromatographic conditions and instrument parameters as described for the nontargeted profiling by UPLC-qTOF-MS (see above). Methylsulfinylalkyl- and methylthioalkyl-type GSs and IGs were identified by their m/z values and mass fragmentation patterns. Camalexin was analyzed using the same extraction and chromatographic conditions and quantified against a calibration curve prepared from a camalexin standard (a kind gift from Jane Glazebrook).
Analysis of Free Auxin (IAA)
Analysis of free auxin (IAA) was performed on a data set derived from plants overexpressing MYB51 (n = 4), MYB28 (n = 3), and the wild type (n = 4). For every sample, we pooled leaves of approximately six plants that showed a clear morphological phenotype (progeny of a single transformation event line). We followed a protocol kindly provided by Dr. Jennifer Normanly that was partially described earlier (Normanly et al., 1993). In brief, frozen plant tissue (100 mg of 14-d-old leaves) was extracted with a solution containing 35% 0.2 m imidazole at pH 7.0 and 65% isopropanol. The 13C-labeled IAA (Cambridge Isotope Laboratories) was added as an internal standard (40 ng g−1 fresh weight of tissue), and the samples were equilibrated for 1 h in the dark at 4°C. Subsequently, 50,000 dpm of 3H-labeled IAA (Amersham) was added as a radiotracer. After centrifugation, the extracted solution was diluted 10-fold with water and loaded on the preequilibrated amino anion-exchange SPE cartridge. After washing, the samples were eluted using five portions of 600 μL of 0.25% phosphoric acid. Most radioactive fractions were combined and passed through the SPE cartridge, then loaded with 200 mg of epoxide resin (Bio-Rad 156-0000 Macroprep Epoxide support). Free IAA was eluted with five portions of 300 μL of methanol, and radioactive fractions were combined. For GC-MS analysis, a 900-μL aliquot of the sample was methylated with 1.5 mL of ethereal diazomethane. Solvents were evaporated under an N2 stream, and the residue was resuspended in 50 μL of ethyl acetate for injection to the GC-MS system. Auxin analysis was performed with a Trace GC Ultra system coupled to a DSQ mass spectrometer (ThermoFinnigan), used in the electron ionization mode. The analytes were separated on a Phenomenex Zebron ZB-5MS capillary column (30 m × 0.25 mm i.d.; film thickness, 0.25 μm). Samples were injected in the PTV splitless mode, and the oven temperature program was as follows. Initial temperature of 40°C was increased at a rate of 15°C min−1 to 300°C (maintained for 4.5 min). Helium was used as carrier gas, and the flow rate was 1.2 mL min−1, the interface temperature was 250°C, and the source temperature was 280°C. Ions with m/z 130, 136, 189, and 195 were monitored. The analytes were quantified by measuring the area ratios of analyte to the internal standard and comparing these ratios with the ratios of the calibration curve of IAA standards that underwent the same process as the samples.
AgNO3 Induction of Camalexin
For camalexin induction, a thin film of 5 mm AgNO3 and 0.02% Silwet L-77 was created on 2-week-old rosette leaves by spraying. The tissue was harvested 12 h after spraying for LC-MS analysis.
Whitefly Oviposition Experiments
A whitefly (Bemisia tabaci) colony (Q biotype) was collected from the Arava Valley in 2003. The colony was maintained on cotton (Gossypium hirsutum ‘Acala’) under standard conditions of 27°C ± 2°C and 14-h-light/10-h-dark photoperiod. Arabidopsis plants (Ler background) were grown under cool-white fluorescent light in long-day conditions (18 h of light/6 h of dark, 18°C) for 5 weeks. All experiments used 4-cm-diameter round pots. Choice and no-choice experiments were conducted in a temperature-controlled room with an approximately 20°C/26°C night/day cycle and a 14-h-light/10-h-dark photoperiod cycle. In the choice experiments, for each replicate three virgin females were isolated at the late fourth instar stage and placed in 3-L jar cages. After emerging, these females were allowed to choose between plants overexpressing one of the MYB factors (MYB29, MYB76, ATR1/MYB34, or MYB51) and wild-type plants (after bolting). Females were allowed to oviposit for 12 to 14 d in each experiment. At the end of each trial, all of the eggs on the abaxial surface of the cauline leaves (the preferred ovipositing and feeding site of B. tabaci) were counted with a stereoscope and the daily number of eggs per emerged female was determined. Student's t test for paired comparisons was carried out for each wild-type and transgenic plant combination (JMP statistical software version 6.0.0; SAS Institute). In the no-choice experiments, for each replicate three virgin females were isolated at the late fourth instar stage and placed in 0.7-L jar cages containing two plants from one of the five plant genotypes (wild type, MYB29, MYB76, ATR1/MYB34, or MYB51). After approximately 13 d, eggs were counted as in the choice experiments (see above) and the daily number of eggs per emerged female was determined. One-way ANOVA was carried out to compare daily oviposition per female on the five plant genotypes. Dunnett's post hoc test was used to compare oviposition on wild-type plants with oviposition on plants overexpressing the MYB factors (JMP statistical software version 6.0.0).
Microscopy
Tissue preparation, histological analyses, tissue clearing, and GUS staining were as described (Eshed et al., 1999).
Gene Expression Correlation Analysis
Publicly available expression data were obtained from the Nottingham Arabidopsis Stock Centre (http://affymetrix.arabidopsis.info/AffyWatch.html), which contains hundreds of publicly available expression profiles. In this study, we focused on several hundred experiments that represent 211 different short-term (ranging from minutes to a few days) biological perturbations. Signal values were obtained using the RMA algorithm (Irizarry et al., 2003) implemented using the R programming language, which is currently the gold standard technique for this purpose. Next, we calculated the correlation matrix between a selected set of genes that showed a significant change in the overexpression lines.
The Arabidopsis Genome Initiative (http://www.arabidopsis.org) identifiers for the genes and gene products in this study are as follows: MYB28, AT5G61420; MYB29, AT5G07690; MYB76, AT5G07700; ATR1/MYB34, AT5G60890; MYB51, AT1G18570; and MYB122, AT1G74080. Microarray data from this article have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus data repository (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE7570.
Supplemental Data
The following materials are available in the online version of this article:
Supplemental Figure S1. Expression patterns of the different MYB cluster members in later stages of development.
Supplemental Figure S2. Additional differential distal network compounds analyzed and detected by GC-MS.
Supplemental Table S1. List of putative metabolites identified in Arabidopsis rosette leaves (ecotype Ler) by UPLC-qTOF-MS and MS/MS analyses.
Supplemental Table S2. List of putative metabolites identified in Arabidopsis rosette leaves (ecotype Ler) by GC-MS analysis.
Supplemental Table S3. Differential mass signals obtained by UPLC-qTOF-MS analysis of Arabidopsis rosette leaves in negative and positive ionization mode.
Supplemental Table S4. EC reaction number, ATG, short names, and long names of enzymes depicted in Figures 5, 6, and 9, along with their differential expression levels (and statistical significance) in the different overexpression lines/clades.
Supplemental Table S5. Oligonucleotides used in this study for cloning full-length cDNAs and promoter regions of genes belonging to one of the ATR1-like or MYB28-like clades.
Supplemental Table S6. List of putative metabolites identified in Arabidopsis rosette leaves collected from publicly available sources.
Supplemental Table S7. Metabolomics data obtained from UPLC-qTOF-MS and GC-MS instruments.
Supplemental Table S8. Structures of Trp- and Met-derived metabolites presented in Figure 5.
Supplemental Table S9. Correlation matrix used for the heat map in Figure 9A.
Supplemental Data Set S1. DLEMMA additional information.
Supplementary Material
Acknowledgments
We thank Jennifer Normanly for providing us the protocol for auxin analysis and for sending the epoxide resin, Jane Glazebrook for the camalexin standard, Maggie Levy for help with GS analysis, Ilana Rogachev for help in the setup of auxin analysis, Alex Brandis for diazomethane preparation, and Michael Lehnard for the ANT∷LhG4 line. We thank Arye Tishbee for the LC-MS analysis, Shirley Horn-Saban for GeneChip hybridizations, Stan Alvarez for the synthetic miR design, Evyatar Steiner for technical assistance, and Gad Galili for his comments on the manuscript.
This work was supported by the Israel Ministry of Science (project no. 3–2552), the European Union project META-PHOR (contract no. FOODCT–2006–036220), Mr. and Mrs. Mordechai Segal, the Henry S. and Anne Reich Family Foundation, and the Israel Science Foundation (grant no. 764/07 to H.L. and grant no. 971/04 to S.M.)
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Asaph Aharoni (asaph.aharoni@weizmann.ac.il).
The online version of this article contains Web-only data.
References
- Aloni R, Schwalm K, Langhans M, Ullrich CI (2003) Gradual shifts in sites of free-auxin production during leaf-primordium development and their role in vascular differentiation and leaf morphogenesis in Arabidopsis. Planta 216 841–853 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Alvarez JP, Pekker I, Goldshmidt A, Blum E, Amsellem Z, Eshed Y (2006) Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J, Fink GR (1998) A Myb homologue, ATR1, activates tryptophan gene expression in Arabidopsis. Proc Natl Acad Sci USA 95 5655–5660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 7 289–300 [Google Scholar]
- Bennett RN, Wenke T, Freudenberg B, Mellon FA, Ludwig-Muller J (2005) The tu8 mutation of Arabidopsis thaliana encoding a heterochromatin protein 1 homolog causes defects in the induction of secondary metabolite biosynthesis. Plant Biol 7 348–357 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Cervera MT, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Van Onckelen H, Van Montagu M, Inze D (1995) Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7 1405–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Eshed Y, Baum SF (2002) Establishment of polarity in angiosperm lateral organs. Trends Genet 18 134–141 [DOI] [PubMed] [Google Scholar]
- Brown JK, Frohlich DR, Rosell RC (1995) The sweet potato or silver leaf whiteflies: biotypes of Bemisia tabaci or a species complex? Annu Rev Entomol 40 511–534 [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA (2000) Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971 [DOI] [PubMed] [Google Scholar]
- Celenza JL, Quiel JA, Smolen GA, Merrikh H, Silvestro AR, Normanly J, Bender J (2005) The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol 137 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu NS (1995) Sympathetic response to betel chewing. J Psychoactive Drugs 27 183–186 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Delker C, Raschke A, Quint M (2008) Auxin dynamics: the dazzling complexity of a small molecule's message. Planta 227 929–941 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Bowman JL (1999) Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99 199–209 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL (2001) Establishment of polarity in lateral organs of plants. Curr Biol 11 1251–1260 [DOI] [PubMed] [Google Scholar]
- Fahey JW, Zalcmann AT, Talalay P (2001) The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56 5–51 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Geigenberger P, Stitt M (2005) Flux an important, but neglected, component of functional genomics. Curr Opin Plant Biol 8 174–182 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L (2004) Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol 5 763–769 [DOI] [PubMed] [Google Scholar]
- Gidda SK, Miersch O, Levitin A, Schimdt J, Wasternack C, Varin L (2003) Biochemical and molecular characterization of a hydroxyjasmonate sulfotransferase from Arabidopsis thaliana. J Biol Chem 278 17895–17900 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Berger B, Mock HP, Müller C, Weisshaar B, Flügge UI (2007. a) The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J 50 886–901 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Engqvist M, Yatusevich R, Müller C, Flügge UI (2008) HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol 177 627–642 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Yatusevich R, Berger B, Müller C, Flügge UI (2007. b) The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J 51 247–261 [DOI] [PubMed] [Google Scholar]
- Glawischnig E, Hansen BG, Olsen CE, Halkier BA (2004) Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc Natl Acad Sci USA 101 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser G, Oldham NJ, Brown PD, Temp U, Gershenzon J (2001) The biosynthesis of benzoic acid glucosinolate esters in Arabidopsis thaliana. Phytochemistry 57 23–32 [DOI] [PubMed] [Google Scholar]
- Grubb CD, Abel S (2006) Glucosinolate metabolism and its control. Trends Plant Sci 11 89–100 [DOI] [PubMed] [Google Scholar]
- Grubb CD, Zipp BJ, Ludwig-Muller J, Masuno MN, Molinski TF, Abel S (2004) Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J 40 893–908 [DOI] [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J (2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57 303–333 [DOI] [PubMed] [Google Scholar]
- Hemm MR, Ruegger MO, Chapple C (2003) The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell 15 179–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, Araki R, Sakurai N, Suzuki H, Aoki K, et al (2007) Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci USA 104 6478–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull AK, Vij R, Celenza JL (2000) Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci USA 97 2379–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 249–264 [DOI] [PubMed] [Google Scholar]
- Kim JH, Durrett TP, Last RL, Jander G (2004) Characterization of the Arabidopsis TU8 glucosinolate mutation, an allele of TERMINAL FLOWER2. Plant Mol Biol 54 671–682 [DOI] [PubMed] [Google Scholar]
- Kutz A, Muller A, Hennig P, Kaiser WM, Piotrowski M, Weiler EW (2002) A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana. Plant J 30 95–106 [DOI] [PubMed] [Google Scholar]
- Levy M, Wang Q, Kaspi R, Parrella MP, Abel S (2005) Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J 43 79–96 [DOI] [PubMed] [Google Scholar]
- Liechti R, Farmer EE (2006) Jasmonate biochemical pathway. Sci STKE 322 cm3. [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H (2006) Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18 3235–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411 709–713 [DOI] [PubMed] [Google Scholar]
- Mewis I, Appel HM, Hom A, Raina R, Schultz JC (2005) Major signaling pathways modulate Arabidopsis glucosinolate accumulation and response to both phloem-feeding and chewing insects. Plant Physiol 138 1149–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Hansen CH, Wittstock U, Halkier BA (2000) Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J Biol Chem 275 33712–33717 [DOI] [PubMed] [Google Scholar]
- Mikkelsen MD, Naur P, Halkier BA (2004) Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J 37 770–777 [DOI] [PubMed] [Google Scholar]
- Mintz-Oron S, Mandel T, Rogachev I, Feldberg L, Lotan O, Yativ M, Wang Z, Jetter R, Venger I, Adato A, et al (2008) Gene expression and metabolism in tomato fruit surface tissues. Plant Physiol 147 823–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore I, Galweiler L, Grosskopf D, Schell J, Palme K (1998) A transcription activation system for regulated gene expression in transgenic plants. Proc Natl Acad Sci USA 95 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nafisi M, Goregaoker S, Botanga CJ, Glawischnig E, Olsen CE, Halkier BA, Glazebrook J (2007) Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19 2039–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J, Cohen JD, Fink GR (1993) Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc Natl Acad Sci USA 90 10355–10359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y (1991) Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3 677–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski M, Schemenewitz A, Lopukhina A, Muller A, Janowitz T, Weiler EW, Oecking C (2004) Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. J Biol Chem 279 50717–50725 [DOI] [PubMed] [Google Scholar]
- Powell G, Tosh CR, Hardie J (2006) Host plant selection by aphids: behavioral, evolutionary, and applied perspectives. Annu Rev Entomol 51 309–330 [DOI] [PubMed] [Google Scholar]
- Rask L, Andreasson E, Ekbom B, Eriksson S, Pontoppidan B, Meijer J (2000) Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol Biol 42 93–113 [PubMed] [Google Scholar]
- Ravanel S, Block MA, Rippert P, Jabrin S, Curien G, Rébeillé F, Douce R (2004) Methionine metabolism in plants: Chloroplasts are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the cytosol. J Biol Chem 279 22548–22557 [DOI] [PubMed] [Google Scholar]
- Raybould AF, Moyes CL (2001) The ecological genetics of aliphatic glucosinolates. Heredity 87 383–391 [DOI] [PubMed] [Google Scholar]
- Rébeillé F, Jabrin S, Bligny R, Loizeau K, Gambonnet B, Van Wilder V, Douce R, Ravanel S (2006) Methionine catabolism in Arabidopsis cells is initiated by a γ-cleavage process and leads to S-methylcysteine and isoleucine syntheses. Proc Natl Acad Sci USA 103 15687–15692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt M, Brown PD, Schneider B, Oldham NJ, Stauber E, Tokuhisa J, Kliebenstein DJ, Mitchell-Olds T, Gershenzon J (2002) Benzoic acid glucosinolate esters and other glucosinolates from Arabidopsis thaliana. Phytochemistry 59 663–671 [DOI] [PubMed] [Google Scholar]
- Reintanz B, Lehnen M, Reichelt M, Gershenzon J, Kowalczyk M, Sandberg G, Godde M, Uhl R, Palme K (2001) Bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell 13 351–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH (2000) Seed and molecular resources for Arabidopsis. Plant Physiol 124 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T (2000) The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100 635–644 [DOI] [PubMed] [Google Scholar]
- Schuhegger R, Nafisi M, Mansourova M, Petersen BL, Olsen CE, Svatos A, Halkier BA, Glawischnig E (2006) CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol 141 1248–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster J, Knill T, Reichelt M, Gershenzon J, Binder S (2006) BRANCHED-CHAIN AMINOTRANSFERASE4 is part of the chain elongation pathway in the biosynthesis of methionine-derived glucosinolates in Arabidopsis. Plant Cell 18 2664–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, Jozefczuk S, Stobiecki M, Muth D, Zanor MI, Witt I, Mueller-Roeber B (2007) Transcription factor AtDOF4;2 affects phenylpropanoid metabolism in Arabidopsis thaliana. New Phytol 175 425–438 [DOI] [PubMed] [Google Scholar]
- Skirycz A, Reichelt M, Burow M, Birkemeyer C, Rolcik J, Kopka J, Zanor MI, Gershenzon J, Strnad M, Szopa J, et al (2006) DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J 47 10–24 [DOI] [PubMed] [Google Scholar]
- Smolen G, Bender J (2002) Arabidopsis cytochrome P450 cyp83B1 mutations activate the tryptophan biosynthetic pathway. Genetics 160 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolen GA, Pawlowski L, Wilensky SE, Bender J (2002) Dominant alleles of the basic helix-loop-helix transcription factor ATR2 activate stress-responsive genes in Arabidopsis. Genetics 161 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonderby IE, Hansen BG, Bjarnholt N, Ticconi C, Halkier BA, Kliebenstein DJ (2007) A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLoS ONE 2 e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50 660–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4 447–456 [DOI] [PubMed] [Google Scholar]
- Tantikanjana T, Yong JW, Letham DS, Griffith M, Hussain M, Ljung K, Sandberg G, Sundaresan V (2001) Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes Dev 15 1577–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai Y, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe D, Kitayama M, et al (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42 218–235 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu O, Jez JM (2008) Nature's assembly line: biosynthesis of simple phenylpropanoids and polyketides. Plant J 54 750–762 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291 306–309 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J, Ecker JR, Normanly J, Chory J, Celenza JL (2002) Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev 16 3100–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Glazebrook J (1999) Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11 2419–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.