Abstract
In addition to the genomes of Arabidopsis (Arabidopsis thaliana) and poplar (Populus trichocarpa), two near-complete rosid genome sequences, grape (Vitis vinifera) and papaya (Carica papaya), have been recently released. The phylogenetic relationship among these four genomes and the placement of their three independent, fractionated tetraploidies sum to a powerful comparative genomic system. CoGe, a platform of multiple whole or near-complete genome sequences, provides an integrative Web-based system to find and align syntenic chromosomal regions and visualize the output in an intuitive and interactive manner. CoGe has been customized to specifically support comparisons among the rosids. Crucial facts and definitions are presented to clearly describe the sorts of biological questions that might be answered in part using CoGe, including patterns of DNA conservation, accuracy of annotation, transposability of individual genes, subfunctionalization and/or fractionation of syntenic gene sets, and conserved noncoding sequence content. This précis of an online tutorial, CoGe with Rosids (http://tinyurl.com/4a23pk), presents sample results graphically.
The strategy of comparing the genome of one organism to that of another organism, with a third genome playing the role of outgroup, is the primary act of comparative or evolutionary genomics (Koonin, 2005). It is always important to know whether the genomes being studied carry the remnants of an ancient polyploidy event because of the known genomic consequences that follow polyploidy (Semon and Wolfe, 2007). One-half of all plant species are recent polyploids and all plant genomes sequenced so far show evidence of one or more ancient polyploidies (Adams and Wendel, 2005). If the genes or genomes being compared have diverged enough to ensure that conserved sequences are functional, then detailed alignments can identify DNA stretches that, rather than encoding a product, must, by default, bind a product encoded by some other sequence (Hardison, 2003; Prakash and Tompa, 2005). Useful divergence has been discussed (Lyons and Freeling, 2008) and is the topic of further discussion in this article. Genomic comparisons are also a quick way to check suspect gene model annotations and to transfer genomic knowledge acquired in one taxon to a less-studied taxon. Evolutionary genomics is always enhanced when tools are available to researchers that help them to easily perform sequence analyses among several genomic regions from multiple organisms and minimize the time it takes to repeat such comparisons. Likewise, visualization software also enriches these analyses by allowing researchers to more easily and better visualize the information contained within genomic sequences and detect patterns of conservation or divergence when comparing multiple genomic regions.
CoGe, our online comparative genomics platform (http://synteny.cnr.berkeley.edu/CoGe), integrates genomic datasets from any organism, DNA alignment and assessment tools, and interactive graphic modules to enable researchers to compare any genomic DNA sequence, feature, or position with any other in any organism (Lyons and Freeling, 2008). In a typical workflow, CoGe allows researchers to begin with a genomic feature of interest (e.g. a gene) in a number of formats (FASTA sequence, GenBank accession, LOC_ number, gene name), find homologous features within the same genome or in different genomes, and compare multiple genomic regions using CoGe's Genome Evolution tool, GEvo. These results can subsequently be refined for further analysis (e.g. change extent and orientation of genomic regions analyzed, change alignment algorithm, or algorithm parameters), raw data can be downloaded, and the analytical configuration saved as a tinyurl for collaborative and future work. Although these types of workflows are useful, having menus of premade GEvo links that anchor syntenic genomic regions from within and among related genomes can save researchers much time. We have made such menus of GEvo links specifically for Arabidopsis (Arabidopsis thaliana) researchers. These links set up comparison of syntenic regions across the currently sequenced rosid genomes Arabidopsis, papaya (Carica papaya), poplar (Populus trichocarpa), and grapevine (Vitis vinifera; Supplemental Information S1 and S2), which is useful because comparing all syntenic chromosomal regions across these genomes would result in an eight-way comparison comprising 28 pairwise sequence alignments. These eight diverged, syntenic rosid chromosomal sequences reflect the results of four sequencing annotation projects and many research papers, as will be reviewed.
Aligning syntenic regions of Arabidopsis chromosomes with their syntenic chromosomal regions in other rosids allows researchers to identify patterns of conservation and divergence in the structure of genomes. By comparing syntenic genomic regions containing several genes, these analyses can determine whether a gene has transposed in the order Brassicales, whether a genomic region is prone to rearrangements, or whether an Arabidopsis (or poplar) gene was retained, fractionated (lost), or tandemly duplicated following a genome duplication event. Analyzing syntenic gene sets (syntelogs, general term that does not distinguish syntenic gene pairs arising by orthology from those derived from intragenomic duplications) at higher resolutions may enable the identification of conserved noncoding sequences (CNSs), possible annotation errors, and the presence of unannotated splice variants. CoGe is designed to support such research.
RESULTS
Rosid Genome Facts and Crucial Definitions
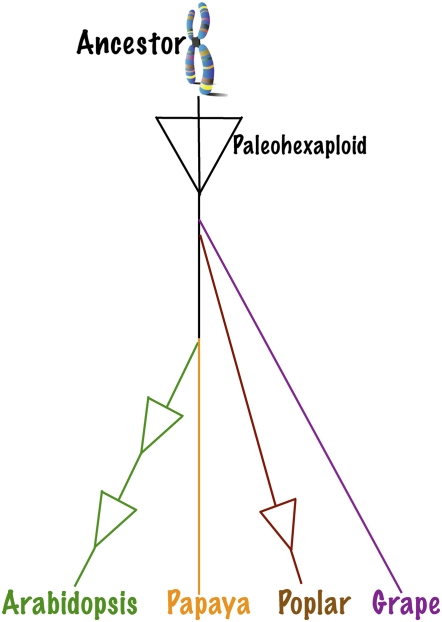
Comparing Arabidopsis, poplar, papaya, and grape genomes is particularly useful because of their phylogenetic relationships and placement of paleotetraploidies relative to speciation events. The phylogenetic tree of Figure 1 is from the Missouri Botanical Garden trees Web site (http://www.mobot.org/MOBOT/research/APweb/welcome.html in March 2008; Rodman et al., 1993; Soltis et al., 1999, 2003; Hall et al., 2002; Davis et al., 2005; Jansen et al., 2007; Zhu et al., 2007) and has been modified to highlight whole genome duplication (WGD) events and their subsequent fractionation back toward the gene content of the diploid ancestor. The WGD events identified in Figure 1 are referenced individually as follows. The Arabidopsis lineage has undergone two sequential WGD events (Bowers et al., 2003) since its divergence from the papaya lineage deep in the order Brassicales. Each Arabidopsis genome duplication event is followed by extensive fractionation (see definitions) that reduced the genome's gene content back toward that of the pretetraploid ancestor. Meanwhile, papaya's lineage has had no such WGD events (Ming et al., 2008).
Figure 1.
Rosid phylogenetic tree from MOBOT decorated with fractionated WGD events. References are in text. If grape is not an official rosid, it is a close outgroup. WGD events are shown as triangles because they are subsequently fractionated so the majority of duplicated genes were removed. The branch lengths that position the tetraploidy events relate to relative divergence times and not to absolute time elapsed, and are drawn to highlight lineage-specific genome duplication events.
Whereas both Arabidopsis and papaya are in the fabids (eurosid I) clade, poplar represents the sister rosid lineage (malvids/eurosid II) whose lineage underwent one minimally fractionated WGD event (Tuskan et al., 2006). Grape (Jaillon et al., 2007; Valesco et al., 2007) is just basal to the rosids or a basal rosid (Soltis et al., 1999, 2003; Jansen et al., 2007; Zhu et al., 2007), and, like papaya, has not had any WGD events in its own lineage since the paleohexaploidy (Jaillon et al., 2007) or gamma polyploidy (Bowers et al., 2003) shared by grape and all rosids. Arabidopsis now has an excellent outgroup in papaya, and all Brassicales have a useful outgroup in grape. Poplar provides an additional outgroup for Brassicales research provided that its own lineage-specific WGD event is reverse fractionated to infer the genomic structure of its preduplicated ancestor. Comparison across multiple outgroups is a powerful tool that, for example, permits unambiguous discrimination between gene gain in Arabidopsis versus gene loss in any one outgroup.
Many eukaryotic gene lineages are characterized by gene family expansions over evolutionary time (Lespinet et al., 2002; Koonin, 2005). Several gene families of Arabidopsis have been shown to have expanded by (1) tandem duplication (Rizzon et al., 2006); (2) retention and/or the avoidance of fractionation following WGD events (see citations below); and (3) transposition (Freeling et al., 2008). Each of these modes of gene family expansion creates paralogs that potentially duplicate the function of the original gene. If retained, this functional duplication sets the stage for biased gene expansion (Blanc and Wolfe, 2004; Seoighe and Gehring, 2004; Maere et al., 2005; Thomas et al., 2006) and subsequent subfunctionalization (Force et al., 1999). Selection against dosage effects (haploinsufficiencies) that might occur due to particular fractionations of a tetraploid (Birchler et al., 2005) is probably a major principle in explaining gene content trends in gene expansion (Freeling, 2008), and probably in increasing morphological complexity as well (Freeling and Thomas, 2006).
Some words have complex or multiple meanings. We use the following definitions and provide examples in the CoGe with Rosids online tutorial and later in this article.
Collinearity
Collinearity is a fact (induction) indicating that a chromosome or segment will tend to have genes in the same order and orientation as they were in an ancestor. As divergence time between any two genomic regions increases, collinearity decreases by virtue of chromosomal or single-gene aberrations, such as inversions, translocations, and deletions.
Synteny
Synteny is an inference (deduction) based on collinearity data that two or more chromosomes or segments are derived from a common ancestor. Genomic regions with collinear features are syntenous, but syntenous segments can have zero shared features (e.g. fully fractionated gene content). If noncollinear gene orders can be explained by chromosomal aberrations, then synteny may often be inferred. Single-gene transpositions obfuscate synteny, as do rearrangement-prone regions (Fortna et al., 2004; Gordon et al., 2007), and can obscure attempts to reconstruct evolutionary history. Genes derived from syntenic regions can be paralogs (e.g. homologs retained from tetraploidy) or orthologs (e.g. from speciation). The term syntelog might be used to identify homologous genes in reference to ancestral chromosomal position only.
Single-Gene Transposition
Single-gene transposition refers to the insertion of one gene into a new location (Freeling et al., 2008). This chromosomal event uniquely destroys collinearity and synteny. Some synonyms: ectopic duplication (Leister, 2004) and ectopic translocation (Ameline-Torregrosa et al., 2007).
Fractionation
Fractionation is the loss mechanism by which a duplicated gene, chromosomal segment, or genome tends to return to its preduplication gene content, but not necessarily gene order (Lockton and Gaut, 2005). Fractionation of a tetraploid results in the loss of one of the initial homologs, but not both. A species is polymorphic for duplications arising by local duplication and by single-gene transposition and may be polymorphic for exact patterns of post-tetraploidy fractionation as well. There is no one diploid of a species, so the term diploidizaiton is not always an accurate synonym for fractionation.
Subfunctionalization
Subfunctionalization is the selectively neutral tendency of a duplicated cis-acting unit of function (gene) to lose dispensable sequences (functions) on one, but not both, duplicates, such that the ancestral function is spread over both duplicates (Force et al., 1999). This idea was originally used to explain the over-retention of gene pairs following duplication (Lynch and Force, 2000), but gene content change data following tandem duplication versus WGD events supports dosage effect rather than subfunctionalization as the duplicate retention mechanism (Freeling and Thomas, 2006; Freeling, 2008). In any case, postretention duplicate genes often subfunctionalize (He and Zhang, 2005; Rastogi and Liberles, 2005; Duarte et al., 2006; Ganko et al., 2007; Woolfe and Elgar, 2007).
Plant CNS
Plant CNS is a significantly conserved sequence of nonprotein-coding plant DNA associated with a pair of syntenous (orthologous or homologous/paralogous) genes or segments as evidenced by unfiltered BLASTn hits below the e-value of a 15/15 exact nucleotide match (or equivalent). True CNS identification requires divergence times long enough to ensure that identified regions of sequence similarity are caused by selection for function rather than neutral carryover. Maize (Zea mays)-rice (Oryza sativa; Guo and Moose, 2003; Inada et al., 2003) and Arabidopsis-Arabidopsis α-homologs (Haberer et al., 2004; Thomas et al., 2007) exhibit useful divergence times.
No introduction to comparing homologous DNA sequences is complete without emphasizing the importance of the relative level of sequence divergence. For example, as two homologous noncoding sequences diverge from a common ancestor, at some point in their evolution, nonfunctional regions will be removed or randomized with respect to one another. If two homologous sequences are not sufficiently diverged, then CNSs might be unselected carryover from the ancestor. Such conserved regions cannot be assumed to have function and represent false-positive noise. Detecting functional noncoding sequences based on sequence similarity can yield false positives that may be filtered by employing e-value (or other metrics) cutoff values. However, exact cutoff metrics depend on the alignment algorithm used. Previous work has identified that a BLASTn e-value cutoff for a 15/15 exact nucleotide match or a Lagan window of 21 bp at 70% identity are sufficient criteria when comparisons are under approximately 20 kb (Lyons and Freeling, 2008). When divergence is optimal, CNSs are often more conserved than protein-encoding exons because of wobble in the third codon position. As two sequences diverge beyond the optimum, CNSs become less conserved than exons, and ultimately become undetectable. The degradation of conservation in functional noncoding sequences has been explained by a process known as binding site turnover (Ludwig et al., 1998; Frith et al., 2006; Moses et al., 2006).
Sometimes divergent genomes fractionate to such a large extent that synteny is difficult to detect. For an extreme example, if a post-tetraploid lineage has complete fractionation whereby every duplicated gene is lost from one homologous genomic region or the other, then there is no ability to detect synteny by finding a collinear series of putatively homologous genes. An example of extreme fractionation exists within the genomes of both grape and papaya as evidenced by the three genomes of the ancient pre-rosid hexaploidy located in Figure 1. Two of these three genomes are fractionated so heavily that synteny is barely apparent; the third genome helps to evaluate these data (Lyons et al., 2008). The genomes of any dicot and any monocot are not collinear in any overall way (M. Freeling, unpublished data). If this is due to one or more independent heavily fractionated paleotetraploidies in their lineages, synteny could conceivably be tested if we had a genome of one of the outgroup Anthophyta taxa that predates the monocot-dicot split: Magnoliales, Winteraceae, Laurales, or Illicales. In theory, even fully fractionated tetraploidies can be reconstructed using outgroups.
CoGe contains tools for assessing synteny, divergence levels, and CNS discovery by supporting efficient acquisition, evaluation, and refinement of syntenic alignment analyses. Our platform should prove to be particularly useful for de novo discovery by allowing researchers to quickly specify genomic regions for comparative analyses, adjust alignment algorithms and settings, add or remove genomic regions, extend or contract regions being analyzed, and rerun analyses. Results are generated on the fly and displayed in an interactive system for quick interpretation and subsequent iterative refinement of the analysis.
DISCUSSION
Introducing the CoGe Online Tutorial Titled CoGe with Rosids
The CoGe with Rosids online tutorial (http://tinyurl.com/4a23pk) is a step-by-step walk-through for finding and evaluating syntenic regions among the sequenced rosid genomes and encourages researchers to follow along with their own analyses. At the top of each tutorial page are the links that comprise the titles of the various lessons: Genome Duplications, Gene Lists, GEvo, Finding Synteny, Putting Syntelogs Together, Overview and Analysis, Fractionation, Annotation Errors, Local Duplications and Insertions, CNSs/Subfunctionalization, and Inversions. Each lesson is an entrance to CoGe.
A typical question that CoGe aids in answering is starting with one Arabidopsis gene of interest and ending with an analysis of multiple syntenic chromosomes within rosid species. CoGe's workflow for answering this follows.
Find Sequence of Interest
Start with gene name of interest (e.g. At1g07300).
Use FeatView to identify genomic feature of that name and select appropriate feature type (e.g. CDS).
Get sequence of genomic feature.
Find Putative Homologous Sequence in Genomes of Interest
Send sequence to CoGeBlast.
Select organisms of interest whose genomes to search.
Search and evaluate results.
Select genomic regions containing putative homologs.
Evaluate Synteny
Send selected genomic regions to GEvo.
Expand genomic regions (e.g. 100 kb around identified putative homologs).
Compare genomic regions.
Analyze results and evaluate synteny.
Refine Analysis
Orient genomic regions.
Trim or expand genomic regions.
Remove genomic regions without evidence for synteny.
Add additional genomic regions for further analysis.
While it is useful to have such comparative genomic workflows, having premade sets of syntelogous genes can enhance the rate by which genomic regions are analyzed. We have prepared gene sets from syntenic regions in Arabidopsis, papaya, poplar, and grape (Supplemental Information S1 and S2). These lists have links to CoGe's Genome Evolution tool, GEvo, which preloads the genomic regions around each gene using 19 kb on either side of the gene and automatically runs BLASTz (Schwartz et al., 2003) comparisons for all pairwise combinations. Once the initial analysis has been completed, manipulation of the analysis settings is usually needed to orient genomic regions in relation to one another (i.e. reverse complement a region), change the length of the genomic regions analyzed, change the alignment algorithm (e.g. to Lagan [Brudno et al., 2003]), or to change alignment parameters and false-positive noise cutoffs. GEvo's interface provides the means to accomplish such tasks and rerun an analysis quickly. Also, GEvo's results are displayed in an interactive interface that allows researchers to quickly interpret the results and find additional genomic information as needed. For example, clicking on a gene model will display its annotation in an information box with links to additional information and data (e.g. raw sequence). Clicking on regions of sequence similarity (e.g. BLAST hits, which are represented as colored rectangles in GEvo) will get information about that region's similarity and draw a line connecting the aligned pair of sequences. These lines connecting regions of sequence similarity are used to visually evaluate synteny and identify regions of interest. If several regions of sequence similarity are identified that overlap gene models (if available) and are collinear with respect to one another, this is evidence for synteny. However, two regions that have little or no sequence in common can be syntenic if both have collinearity to an outgroup sequence. GEvo also returns all input and output data as downloadable links and provides a tinyurl to regenerate the analysis as last configured for future revisiting or collaboration.
Figure 2 (http://tinyurl.com/4n3npz) shows the results of a GEvo analysis that started with a link provided in the premade gene sets (Supplemental Information S1, row 11228) and was subsequently reoriented and resized to highlight a syntenic comparison between Arabidopsis, papaya, and grape and two regions of poplar derived from its most recent genome duplication event (see Fig. 1). Regions of sequence similarity between papaya and grape have green lines connecting them showing a collinear arrangement. This collinear arrangement is strong evidence for synteny; synteny can be demonstrated for all pairwise comparison of these genomic regions (see tinyurl link and commence research).
Figure 2.
A screenshot of the BLASTz output from a syntenic comparison among chromosomal regions of Arabidopsis, papaya, and grape and two regions of poplar derived from its most recent genome duplication event. Each horizontal image is a visualization of one genomic region. The dashed line in the middle of each region represents the division between the top (5′ on left) and the bottom (5′ on right) strand. Gene models are drawn as colored arrows directly above or below this line, with CDS colored green, RNA blue, and the full gene model as gray to show introns. BLASTz was used to find regions of similarity (i.e. BLAST hits) between all pairwise sequence comparisons and are drawn as colored blocks above or below gene models, with each color representing one pairwise comparison. BLAST hits in the (++) and (+−) orientation are drawn above and below the dashed line, respectively. Green lines are drawn connecting BLAST hits between grape and papaya (second and third genomic regions, respectively). Such collinearity of BLAST hits is evidence for synteny, which is a pattern repeated between all pairwise comparisons of these genomic regions. Gold and orange stars highlight possible annotation errors and missing annotations, respectively, based on discordance of gene models and regions of sequence similarity. Results can be regenerated using http://tinyurl.com/4n3npz and research might be resumed.
Visualizing sequence comparisons in their genomic context allows for the quick identification of possible annotation errors. For example, Figure 2 is labeled with gold stars where gene models are not congruent across syntenic regions and with orange stars where a gene model is missing in a region that is syntenic with other regions with gene models. We will investigate one of these annotation errors further in this section.
Given the phylogenetic tree in Figure 1, we expect there to be up to three additional Arabidopsis regions that are syntenic to the ones shown in Figure 2. There are at least two methods for finding these syntenic regions. The simplest method is to use a premade list of Arabidopsis syntenic regions using papaya as an outgroup (Supplemental Information S2), and the other is to use CoGe to find these regions de novo. The online tutorial shows a step-by-step analysis using the latter method: Briefly, this entails using the CoGe system to find all the coding features in the identified syntenic papaya region, searching the Arabidopsis genome for putative homologs using CoGe's multigenome BLAST tool, sorting the resulting BLAST hits by genomic location, manually identifying a chromosomal series of ordered BLAST hits to identify putative syntenic regions in Arabidopsis, and evaluating those Arabidopsis regions for synteny using GEvo. With experience, this type of workflow takes a few minutes. At the core of this process is CoGe's BLAST tool, CoGeBlast, which has been designed to allow researchers to easily evaluate multiple BLAST hits from multiple query sequences across multiple genomes in their genomic context—an essential feature for manually identifying putative syntenous regions.
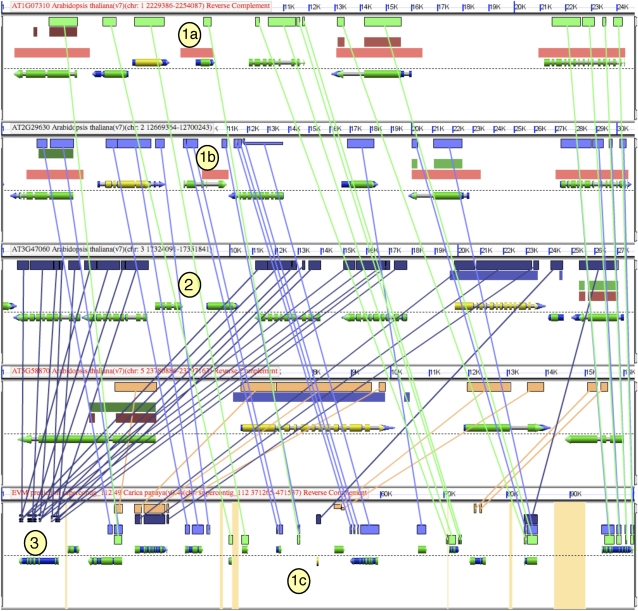
Figure 3 shows the results of finding the three additional regions of Arabidopsis that are syntenic to the previously analyzed regions from Arabidopsis and papaya. This figure reveals the fractionated nature of the Arabidopsis genome (Bowers et al., 2003) resulting from two sequential tetraploidies and fractionation because it diverged from papaya. Each Arabidopsis region shown in Figure 3, except one, has its entire gene content represented and intercalated in papaya, whereas the Arabidopsis regions share only a subset of their combined gene content among one another. This is most easily seen in the papaya image (bottom) in Figure 3, where some papaya subregions have sequence similarity to a single Arabidopsis region, while others match two or three Arabidopsis regions.
Figure 3.
A screenshot of the BLASTz output for a syntenic comparison among four Arabidopsis regions and one papaya region (top to bottom). Figure attributes are as described in the Figure 2 legend. Orange in the background of the papaya genomic region indicates unsequenced regions. Lines connecting regions of sequence similarity have been drawn between each Arabidopsis region and papaya. Circled numbers refer to (1) possible annotation error evidenced by sequence similarity between two regions of Arabidopsis (1a, 1b) and papaya (1c); (2) insertion into a tandem array of locally duplicated genes; and (3) evidence for local duplication in Arabidopsis lineage due to multiple genes mapping to the same region in papaya. This research can be revisited at http://tinyurl.com/2vzcsk.
The graphic representation of Figure 3 enables the identification of a putative annotation error in Arabidopsis denoted by yellow circles 1a and 1b. Here, there is discordance of the gene models with 1a having sequence similarity to 1b that extends approximately 1 kb beyond its 5′ end. By comparison to the Arabidopsis outgroup genome, papaya, more information can be gleaned. There is sequence similarity of 1a to a region in papaya that is without a gene annotation (1c) and probably represents a missed model annotation in papaya. Also, there is sequence similarity to the 5′ region of 1b to a region in papaya with a gene annotation. Using GEvo, this can be further analyzed in more detail by zooming in on just these genomic regions and using a sequence alignment algorithm more sensitive than BLASTz.
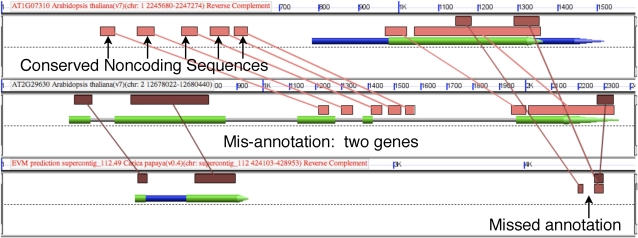
Figure 4 shows the results of an analysis using BLASTn (Altschul et al., 1990). Shorter regions of sequence similarity are detected and a series of five putative CNSs are found 5′ of the gene in the first Arabidopsis region (top). This gene also has annotated untranslated regions (UTRs; blue arrow), which is indicative of supporting experimental evidence for this gene model. The gene model in the second Arabidopsis region (middle) lacks UTRs, which is indicative of automated gene prediction algorithms. Sequence similarity of the Arabidopsis regions to the gene model in papaya strongly suggests that the gene in the second Arabidopsis region is misannotated and is actually two genes. These two putative genes have orthologs in papaya, one of which was annotated and the other missed as well.
Figure 4.
A screenshot of BLASTn output for the comparison among two Arabidopsis regions and one papaya region to identify errors in gene annotations; settings include a false-positive noise cutoff requiring hits to be as or more significant than a 15/15-bp exact nucleotide match (Thomas et al., 2007). Figure attributes are as described in the Figure 2 legend. The research can be revisited at http://tinyurl.com/4lqjdd.
Also seen in Figure 3 are two other patterns frequently encountered among comparisons of this type: (1) the expansion by local duplication of a gene into a tandem array as seen in the third Arabidopsis region; and (2) an insertion into that array that is not syntenic with any other Arabidopsis or papaya region. The tandem array is evidenced by comparison to papaya (yellow circle 3) where four genes (all annotated as being similar to glycosyl hydrolase family 3 proteins) from the third Arabidopsis region have sequence similarity to a single gene in papaya (dark blue boxes and lines). In the middle of this array are two additional genes. Because these genes are not present in papaya, and papaya has a single copy of the gene present in the Arabidopsis tandem array, it is likely that this tandem array was formed after the divergence of Arabidopsis and papaya. Subsequent to the formation of the tandem array, there were one or more transposition events that inserted the additional two genes. The two transposed genes encode F-box family proteins; this occurrence is consistent with the types of genes known to have transposed in Arabidopsis (Freeling et al., 2008).
The possibility exists that the F-box genes could have been lost in the papaya lineage rather than being more recent insertions into Arabidopsis. We can discriminate between loss and gain events through comparison to additional outgroup syntenic regions. Following the logic inherent in the rosid phylogeny shown in Figure 1, examination of the one grape and/or two poplar syntenic regions should answer this question. This experiment is easily accomplished either using CoGeBlast, as described, or using the appropriate premade menus of GEvo links. When this is done, the grape and two poplar chromosomal regions show a similar gene content as papaya (there are no F-box genes; data not shown; please see http://tinyurl.com/2q7uzs), except that one poplar region has a truncated version of the duplicated Arabidopsis gene.
CONCLUSION
Here we have shown how using our comparative genomics system, CoGe, allows for the comparison, visualization, and subsequent manipulation of multiple chromosomal DNA sequences from multiple organisms (http://synteny.cnr.berkeley.edu/CoGe). Over the past year, two more genomes in the rosid clade have been sequenced so that four different families are now represented. The botanical model, Arabidopsis, is composed of four fractionated subgenomes that are distinct to its lineage. Arabidopsis is the best annotated eudicot genome by far, but many of its genes encode entirely unknown functions and some have annotation errors. Papaya serves as a much-needed primary outgroup for Arabidopsis, and both grape and poplar comprise useful secondary outgroups. Here, we presented data using our system to perform DNA sequence alignments that validate synteny, document fractionation, uncover misannotated gene models, identify missed gene models, discover CNSs, and show the alignment signatures of local gene duplications and gene transpositions.
Whereas comparative genomics systems, such as CoGe, are essential for analyzing the ever-increasing number of new genomes being sequenced, CoGe itself is evolving to better assist new research questions. GEvo, for example, is a useful multigenomic region analysis tool, but has several limitations. First is the physical limit to the extent of genomic space analyzed. While useful for analyzing regions of up to a couple of megabases, GEvo is not useful for analyses on a larger scale (e.g. whole chromosomes). The graphic results may take hours to process due to many thousands of identified regions of sequence similarity and genes, and their display is often too dense for researchers to visually identify interesting patterns of genome evolution. Likewise, when analyzing more than 15 genomic regions at once, the results returned to a researcher's computer often overload their system (although this problem is mitigated by newer computers). Also, a researcher's ability to interact with the results in a meaningful way is often limited by the size of their monitor.
Fortunately, CoGe's design allows new tools to be developed in a flexible way and, when built, they have immediate access to CoGe's genome database and its preexisting toolset. We are currently developing a new genome-wide comparison tool for identifying syntenic regions. When complete, this tool will allow researchers the ability to select any two genomes for comparison and generate syntenic dot-plots for quick identification of syntenic regions. These dot-plots, in turn, will have links that will take researchers to GEvo to evaluate syntenic regions of interest in more detail. We have recently released a beta version of this tool, called SynMap, which can be accessed from CoGe's homepage.
Rosid researchers would greatly benefit by having an outgroup to the rosid paleohexaploidy. Although the asterid clade, sister to the rosid clade, looks promising because genome sequences are available from the orders Lamiales and Solanales, analysis of several unannotated short contigs of Mimulus guttatus available from the Joint Genome Institute (ftp://ftp.jgi-psf.org/pub/JGI_data/Mimulus_guttatus) that are approximately 100 kb in length show extensive synteny with grape. This one-to-one mapping of genomic regions (e.g. http://tinyurl.com/6hssn7, http://tinyurl.com/5pcjuz, http://tinyurl.com/6h6673) is highly indicative that the paleohexaploidy predates the divergence of the rosid and asterid clades. It may be some time before researchers have an appropriate paleohexaploid outgroup genome sequence.
MATERIALS AND METHODS
Method for Generating Arabidopsis-Papaya-Grape Two Poplar Gene List with GEvo Links
Arabidopsis (Arabidopsis thaliana; The Arabidopsis Information Resource [TAIR], version 7) was blasted against papaya (Carica papaya; EVM, 1.0 release) and grape (Vitis vinifera; genoscope version 1) using BLASTn using default settings and an e-value filter of 0.001. The top hit to each was taken as a potential syntelog. To generate syntelogs to poplar (Populus trichocarpa), which has had a relatively recent genome duplication event, DAGchainer (Haas et al., 2004) was used. If a single matching syntenic region was found, a single poplar gene was included (if one matched the Arabidopsis query sequence). If two matching syntenic regions were found, a gene from each region was used (if there were matches to the Arabidopsis query sequence). These sets of genes were then used to generate links to GEvo with appropriate anchors for each matching genome.
Method for Generating Four Arabidopsis-Papaya Gene List with GEvo Links
We analyzed the collinearity between the longest 200 papaya scaffolds and the five Arabidopsis chromosomes. We used BLASTP (E-value < 1e-5, top five hits) to search for similarities among complete sets of annotated papaya (EVM, 1.0 release) and Arabidopsis genes (TAIR, version 7). For simplicity, the papaya genes were renamed according to their positions on the scaffolds. We applied a multiple genome and subgenome alignment algorithm MCscan (Tang et al., 2008) to identify and display collinearity. Briefly, the program first enumerates pairwise collinearity between all pairs of papaya scaffolds and Arabidopsis chromosomes through a dynamic programming procedure very similar to DAGchainer. The default scoring scheme (configurable) is min (log10 e-value, 50) match score for one gene pair, and −1 gap penalty for each 10-kb distance between any two consecutive gene pairs. All tandem matches are collapsed using one representative gene pair that has the lowest BLASTP e-value. This is followed by a statistical test used in Wang et al. (2006), and an e-value is calculated for each block. The syntenic blocks that exceed score 300 and e-value <1e-10 are retained. These pairwise collinear segments are then grouped into blocks and progressively realigned, using papaya scaffolds as reference. This creates multiple alignment views of several genomic regions that show similar gene orders. Finally, for each syntenic block, we manually removed regions that are derived from a more ancient duplication event (γ). This pipeline generates a gene list that contains many multialigned blocks that show one papaya region usually matching to up four Arabidopsis regions.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Information S1. At-Cp-Vv-2Pt.xls.
Supplemental Information S2. At4-Cp.xls.
Supplementary Material
Acknowledgments
We thank the funding agencies and sequencing teams worldwide for the public genome databases analyzed by CoGe. E.L. thanks B.C.T. for system guidance as a BOFH.
This work was supported by the National Science Foundation (NSF; grant nos. DBI 034937 and DBI 0701871 to M.F. [CoGe] and grant nos. MCB–0450260 and DBI–0553417 to A.P. [Cp-4At list of five syntenic genes/positions]); the University of Hawaii and the U.S. Department of Defense (grant no. W81XWH0520013 to M.A. [papaya sequence]); Maui High Performance Computing Center (grant to M.A.); and the Hawaii Agriculture Research Center (grant to R.M. and coworkers).
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Eric Lyons (elyons@nature.berkeley.edu).
The online version of this article contains Web-only data.
References
- Adams KL, Wendel JF (2005) Polyploidy and genome evolution in plants. Curr Opin Plant Biol 8 135–141 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215 403–410 [DOI] [PubMed] [Google Scholar]
- Ameline-Torregrosa C, Wang BB, O'Bleness MS, Deshpande S, Zhu H, Roe B, Young ND, Cannon SB (2007) Identification and characterization of NBS-LRR genes in the model plant Medicago truncatula. Plant Physiol 146 5–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Riddle NC, Auger DL, Veitia RA (2005) Dosage balance in gene regulation: biological implications. Trends Genet 21 219–226 [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422 433–438 [DOI] [PubMed] [Google Scholar]
- Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S (2003) LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res 13 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CC, Webb CO, Wurdack KJ, Jaramillo CA, Donoghue MJ (2005) Explosive radiation of Malpighiales supports a mid-cretaceous origin of modern tropical rain forests. Am Nat 165 E36–E65 [DOI] [PubMed] [Google Scholar]
- Duarte JM, Cui L, Wall PK, Zhang Q, Zhang X, Leebens-Mack J, Ma H, Altman N, dePamphilis CW (2006) Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol Biol Evol 23 469–478 [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortna A, Kim Y, MacLaren E, Marshall K, Hahn G, Meltesen L, Brenton M, Hink R, Burgers S, Hernandez-Boussard T, et al (2004) Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol 2 E207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M (2008) The evolutionary position of subfunctionalization, downgraded. Genome Dynamics 4 25–40 [DOI] [PubMed] [Google Scholar]
- Freeling M, Lyons E, Pedersen B, Alam M, Ming R, Lisch D (2008) Many or most genes in Arabidopsis transposed after the origin of the order Brassicales. Genome Res (in press) [DOI] [PMC free article] [PubMed]
- Freeling M, Thomas BC (2006) Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res 16 805–814 [DOI] [PubMed] [Google Scholar]
- Frith MC, Ponjavic J, Fredman D, Kai C, Kaweai J, Carninci P, Hayshizaki Y, Sandelin A (2006) Evolutionary turnover of mammalian transcription start sites. Genome Res 16 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganko EW, Meyers BC, Vision TJ (2007) Divergence in expression between duplicated genes in Arabidopsis. Mol Biol Evol 24 2298–2309 [DOI] [PubMed] [Google Scholar]
- Gordon L, Yang S, Tran-Gyamfi M, Baggott D, Christensen M, Hamilton A, Crooijmans R, Groenen M, Lucas S, Ovcharenko I, et al (2007) Comparative analysis of chicken chromosome 28 provides new clues to the evolutionary fragility of gene-rich vertebrate regions. Genome Res 17 1603–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Moose SP (2003) Conserved noncoding sequences among cultivated cereal genomes identify candidate regulatory sequence elements and patterns of promoter evolution. Plant Cell 15 1143–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberer G, Hindemitt T, Meyers BC, Mayer KF (2004) Transcriptional similarities, dissimilarities, and conservation of cis-elements in duplicated genes of Arabidopsis. Plant Physiol 136 3009–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Wortman JR, Salzberg SL (2004) DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics 20 3643–3646 [DOI] [PubMed] [Google Scholar]
- Hall JC, Sytsma KJ, Iltis HH (2002) Phylogeny of Capparaceae and Brassicaceae based on chloroplast sequence data. Am J Bot 89 1826–1842 [DOI] [PubMed] [Google Scholar]
- Hardison RC (2003) Comparative genomics. PLoS Biol 1 E58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang J (2005) Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169 1157–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada DC, Bashir A, Lee C, Thomas BC, Ko C, Goff SA, Freeling M (2003) Conserved noncoding sequences in the grasses. Genome Res 13 2030–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449 463–467 [DOI] [PubMed] [Google Scholar]
- Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Muller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, et al (2007) Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA 104 19369–19374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV (2005) Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet 39 309–338 [DOI] [PubMed] [Google Scholar]
- Leister D (2004) Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet 20 116–122 [DOI] [PubMed] [Google Scholar]
- Lespinet O, Wolf YI, Koonin EV, Aravind L (2002) The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res 12 1048–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockton S, Gaut BS (2005) Plant conserved non-coding sequences and paralogue evolution. Trends Genet 21 60–65 [DOI] [PubMed] [Google Scholar]
- Ludwig MZ, Patel NH, Kreitman M (1998) Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change. Development 125 949–958 [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A (2000) The probability of duplicate gene preservation by subfunctionalization. Genetics 154 459–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E, Freeling M (2008) How to usefully compare homologous plant genes and chromosomes as DNA sequence. Plant J 53 661–673 [DOI] [PubMed] [Google Scholar]
- Lyons E, Pedersen B, Kane J, Freeling M (2008) The value of nonmodel genomes and an example using SynMap within CoGe to dissect the hexaploidy that predates rosids. Tropical Plant Biol (in press)
- Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y (2005) Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA 102 5454–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Saw JH, Senin P, Wang W, Ly BV, Lewis KL, et al (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452 991–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses AM, Pollard DA, Nix DA, Iyer VN, Li XY, Biggin MD, Eisen MB (2006) Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput Biol 2 e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash A, Tompa M (2005) Discovery of regulatory elements in vertebrates through comparative genomics. Nat Biotechnol 23 1249–1256 [DOI] [PubMed] [Google Scholar]
- Rastogi S, Liberles DA (2005) Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol Biol 5 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzon C, Ponger L, Gaut BS (2006) Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comput Biol 2: e115 [DOI] [PMC free article] [PubMed]
- Rodman J, Price RA, Karol K, Conti E, Systma KJ, Palmer JD (1993) Nucleotide sequences of the rbcL gene indicate monophyly of mustard oil plants. Ann Mo Bot Gard 80 686–699 [Google Scholar]
- Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W (2003) Human-mouse alignments with BLASTZ. Genome Res 13 103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semon M, Wolfe KH (2007) Consequences of genome duplication. Curr Opin Genet Dev 17 505–512 [DOI] [PubMed] [Google Scholar]
- Seoighe C, Gehring C (2004) Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet 20 461–464 [DOI] [PubMed] [Google Scholar]
- Soltis DE, Senters AE, Zanis MJ, Kim S, Thompson JD, Soltis PS, Ronse De Craene LP, Endress PK, Farris JS (2003) Gunnerales are sister to other core eudicots: implications for the evolution of pentamery. Am J Bot 90 461–470 [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE, Chase MW (1999) Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature 402 402–404 [DOI] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH (2008) Synteny and collinearity in plant genomes. Science 320 486–488 [DOI] [PubMed] [Google Scholar]
- Thomas BC, Pedersen B, Freeling M (2006) Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res 16 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BC, Rapaka L, Lyons E, Pedersen B, Freeling M (2007) Intragenomic conserved noncoding sequences in Arabidopsis. Proc Natl Acad Sci USA 104 3348–3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604 [DOI] [PubMed] [Google Scholar]
- Valesco R, Zharkikh A, Troggio M, Cartwright DA, al Cestaro A e (2007) A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS One 2: e1326 [DOI] [PMC free article] [PubMed]
- Wang X, Shi X, Li Z, Zhu Q, Kong L, Tang W, Ge S, Luo J (2006) Statistical inference of chromosomal homology based on gene collinearity and applications to Arabidopsis and rice. BMC Bioinformatics 7 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, Elgar G (2007) Comparative genomics using Fugu reveals insights into regulatory subfunctionalization. Genome Biol 8 R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XY, Chase MW, Qiu YL, Kong HZ, Dilcher DL, Li JH, Chen ZD (2007) Mitochondrial matR sequences help to resolve deep phylogenetic relationships in rosids. BMC Evol Biol 7 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.