Abstract
Two distinct peroxisomal targeting signals (PTSs), the C-terminal PTS1 and the N-terminal PTS2, are defined. Processing of the PTS2 on protein import is conserved in higher eukaryotes. Recently, candidates for the responsible processing protease were identified from plants (DEG15) and mammals (TYSND1). We demonstrate that plants lacking DEG15 show an expressed phenotype potentially linked to reduced β-oxidation, indicating the impact of protein processing on peroxisomal functions in higher eukaryotes. Mutational analysis of Arabidopsis (Arabidopsis thaliana) DEG15 revealed that conserved histidine, aspartic acid, and serine residues are essential for the proteolytic activity of this enzyme in vitro. This indicates that DEG15 and related enzymes are trypsin-like serine endopeptidases. Deletion of a plant-specific stretch present in the protease domain diminished, but did not abolish, the proteolytic activity of DEG15 against the PTS2-containing glyoxysomal malate dehydrogenase. Fluorescence microscopy showed that a DEG15-green fluorescent protein fusion construct is targeted to peroxisomes in planta. In vivo studies with isolated homozygous deg15 knockout mutants and complemented mutant lines suggest that this enzyme mediates general processing of PTS2-containing proteins.
All peroxisomal proteins are encoded in the nucleus, synthesized on cytosolic ribosomes, and imported as folded proteins with one of two peroxisomal targeting signals (PTSs; Baker and Sparkes, 2005). PTS1 is a C-terminal tripeptide with the consensus sequence Ser-Lys-Leu (SKL; Swinkels et al., 1991). This noncleavable sequence is responsible for the import of the majority of peroxisomal proteins. A smaller number of proteins is imported into peroxisomes by the N-terminal PTS2 with the consensus sequence (R)(L/V/I)-X5-(H)(L/A). It is cleaved in higher eukaryotes, such as mammals and plants, after arrival of the protein in the peroxisome at a conserved Cys cleavage site (Gietl et al., 1994; Kato et al., 1996a; Reumann, 2004). The significance of PTS2 processing is still unknown because the catalytic properties of processed and unprocessed enzymes are similar as reported for glyoxysomal malate dehydrogenase (gMDH; Gietl et al., 1996; Cox et al., 2005) and PTS2 is not processed in lower eukaryotes like yeasts (Table I).
Table I.
Peroxisomal matrix proteins of higher plants, mammals, and yeasts containing the targeting signal PTS2 in the N terminus
TPTS2 is indicated in bold. The conserved Cys near the cleavage site for the peroxisomal processing peptidase in higher eukaryotes is indicated in bold and underlined. (Taken from Helm et al. [2007]. Copyright by The National Academy of Sciences of the USA.)
| Enzyme/Species | Plants and Mammals
|
||
|---|---|---|---|
| N-Terminal Presequence | Cleavage Site* | Mature Subunit | |
| Malate dehydrogenase | |||
| Watermelon | MQPIPDVNQRIARISAHLHPPKSQMEESSALRRANCR*-AKGGAPGFKVAI | ||
| Pumpkin | MKPIPDVNERIARISAHLPPKSQMEEGSVLRRANCR*-AKGGAPGFRVAI | ||
| Alfalfa | MEPNSYANSRITRIASHLNPPNLKMNEHGGSSLTNVHCR*-AKGGTPGFKVAI | ||
| Rice | MEDAAAAARRMERLASHLRPPASQMEESPLLRGSNCR*-AKGAAPGFKVAI | ||
| Rape | MPHKRIAMISAHLQPSFTPQMEAKNSVMGLESCR*-AKGGNPGFKVAI | ||
| Arabidopsis (At5g09660) | MEFRGDANQRIARISAHLTPQMEAKNSVIGRENCR*-AKGGNPGFKVAI | ||
| Arabidopsis (At2g22780) | MPDNQRIARISAHLNPPNLHNQIADGSGLNRVACR*-AKGGSPGFKVAI | ||
| Citrate synthase | |||
| Pumpkin | MPTDMELSPSNVARHRLAVLAAHLSAASLEPPVMASSLEAHCV*-SAQTMVAPP | ||
| Arabidopsis (At2g42790) | MEISERVRARLAVLSGHLSEGKQDSPAIERWCT*-SADTSVAPL | ||
| Arabidopsis (At3g58740) | MEISERARARLAVLNAHLTVSEPNQVLPAIEPWCT*-SAHITAAPH | ||
| Acyl-CoA oxidase | |||
| Pumpkin | MASPGEPNRTAEDESQAAARRIERLSLHLTPIPLDDSQGVEMETC*-AAGKAKA | ||
| Arabidopsis (At5g65110) | MESRREKNPMTEEESDGLIAARRIQRLSLHLSPSLTLSPSLPLVQTETC*-SAR | ||
| 3-Keto-acyl-CoA thiolase | |||
| Cucumber | MEKAINRQSILLHHLRPSSSAYTNESSLSASVC*-AAGDSASY | ||
| Pumpkin | MEKAINRQSILLHHLRPSSSAYSHESSLSASVC*-AAGDSASY | ||
| Mango | MEKAINRQSILLHHLRPSNSSSHNYESALAASVC*-AAGDSAAY | ||
| Rape | MEKAMERQRVLLEHLRPSSSSSHSFEGSLSASAC*-LAGDSAAY | ||
| Arabidopsis (At5g48880) | MEKAIERQRVLLEHLRPSSSSSHNYEASLSASAC*-LAGDSAAY | ||
| Rat | MSESVGRTSAMHRLQVVLGHLAGRPESSSALQAAPC*-SAGFPQAS | ||
| Human | MQRLQVVLGHLRGPADSGWMPQAAPC*-LSGAPQAS | ||
| Phytanoyl-CoA2-hydroxylase | |||
| Rat | MDYTRAGARLQVLLGHLGRPSALQIVAHPVSGPASPANFC*-PEQFQYTL | ||
| Alkyldihydroxyacetone phosphate synthase | |||
| Human | MAEAAAAAGGTGLGAGASYGSAADRDRDPDPDRAGRRLRVLSGHLLGRPREALSTNEC*-KARRA | ||
| Rat | MAEAAGEAGASERDPDAVRARRRLRVLSGHLLGRPQEAPSTNEC*-KARRAASA | ||
| Mouse | MAEAAAGEAGASERDPDAGRARRRLRVLSGHLLGRPQEAPSTNEC*-KARRAASA | ||
| Yeasts | |||
| 3-Keto-acyl-CoA thiolase | |||
| Saccharomyces cerevisiae | MSQRLQSIKDHLVLSAMGLGESKRKNSLLEK | ||
| Candida tropicalis | MDRLNQLSGQLKPNAKQSILQKNPDDVVIV | ||
| Yarrowia lypolytica | MDRLNNLATQLEQNPAKGLDAITSKNPDDV | ||
| Amine oxidase | |||
| H. polymorpha | MERLRQIASQATAASAAPARPAHPLDPLST | ||
Recently, DEG15 and TYSND1 proteases responsible for peroxisomal processing were identified in watermelon (Citrullus vulgaris; Helm et al., 2007) and mammals (Kurochkin et al., 2007). DEG15 belongs to the family of Deg/HtrA proteases, which form trimeric, hexameric, and dodecameric complexes (Clausen et al., 2002; Helm et al., 2007; Krojer et al., 2008) and many of them contain one to three PDZ domains in addition to a trypsin/chymotrypsin-like protease domain. Crystallographic structure analyses indicate that these domains play a role in substrate recognition and activation of the protease domain (Krojer et al., 2002). Arabidopsis (Arabidopsis thaliana) DEG15 (At1g28320), however, is unique because it has a single trypsin-like domain, lacks PDZ domains, and has a PTS1 C-terminal tripeptide, SKL. It further contains a 67-amino acid insertion loop between His and Asp of the His-Asp-Ser catalytic triad (Fig. 1). This loop is conserved in the paralog enzyme of rice (Oryza sativa). The rice DEG15 trypsin-like domain is 45% identical to that of Arabidopsis DEG15. DEG15 from watermelon was suggested to exist as a dimer of 144 kD as well as a monomer of 72 kD with different substrate specificities, the dimer form operating as the peroxisomal processing protease and the monomer as a general degrading protease (Helm et al., 2007). Depending on the Ca2+ concentration, the two forms are interconvertible.
Figure 1.
Comparison of the protease domains of selected Deg/HtrA proteases. Eco, Escherichia coli; Ath, Arabidopsis thaliana; Osa, Oryza sativa; Hsa, Homo sapiens. Reproduced from figure 7 in the supplement of Helm et al. (2007). Copyright by The National Academy of Sciences of the USA.
The human (Homo sapiens), mouse (Mus musculus), and rat (Rattus norvegicus) TYSND1 catalytic protease domain is homologous to that of DEG15, but lacks the 67-amino acid loop domain (Fig. 1). Because TYSND1 activity was inhibited by the Cys protease inhibitor N-methylmaleimide (NEM), it was suggested that the mammalian enzyme is a Cys protease (Kurochkin et al., 2007). We have identified and isolated a deg15 knockout line (SALK line 007184) of Arabidopsis, where processing of pre-gMDH is impaired. With this line, we were able to show that the lack of PTS2 processing results in an increased resistance of the homozygous mutants to the herbicide precursor 4-(2,4-dichlorophenoxy) butyric acid (2,4-DB), which is converted in peroxisomes to the toxic herbicide 2,4-dichlorophenoxyacetic acid (2,4-D) by β-oxidation. Site-directed mutagenesis revealed that the catalytic activity of DEG15 is carried out by the His-Asp-Ser catalytic triad, indicating that this enzyme is a Ser protease. Additionally, we confirmed the peroxisomal localization of DEG15 and investigated the range of proteins, which are processed by this protease.
RESULTS AND DISCUSSION
deg15 Knockout Plants Are Resistant to the Herbicide Precursor 2,4-DB
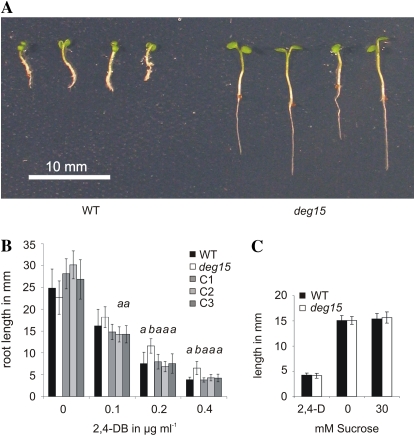
To investigate the impact of PTS2 processing by DEG15 on the function of peroxisomes, we analyzed wild-type and deg15 knockout plants for their ability to grow in the presence of the nontoxic herbicide precursor 2,4-DB. Because 2,4-DB is converted to the toxic herbicide 2,4-D by peroxisomal β-oxidation (Hayashi et al., 2002; Zolman et al., 2007), plants impaired in this function should stay unaffected during such treatment. A clear phenotypic difference was observed between the deg15 knockout mutant and wild-type plants treated with 2,4-DB (Fig. 2A). Mutant plants had significantly longer roots than the wild type when grown on solid medium containing 2,4-DB (Fig. 2, A and B). This phenotype was more pronounced with the increased seedling age (Fig. 2, compare A and B) and at higher concentrations of 2,4-DB and was reversed in deg15 knockout plants complemented with constitutively expressed DEG15 (Fig. 2B, plants C1, C2, and C3). This confirms that the lack of DEG15 is responsible for resistance to 2,4-DB and not to a so-far undetected second mutation in the deg15 knockout line. No difference in root length was observed in wild-type and mutant plants grown in the presence of 2,4-D (Fig. 2C, left), indicating that the increased resistance to 2,4-DB of the mutant line is not due to an altered response to auxin. We further tested the dependence of dark-grown wild-type and deg15 seedlings on Suc. No difference in hypocotyl lengths was observed between etiolated wild-type and mutant seedlings (Fig. 2C, middle and right).
Figure 2.
Resistance of deg15 knockout plants to the herbicide precursor 2,4-DB. A, Photograph of wild-type (WT) and deg15 knockout seedlings grown for 13 d on solid medium containing 0.4 μg mL−1 2,4-DB. B, Analysis of root length of plants grown for 11 d on solid medium without and with different concentrations of 2,4-DB. WT, Wild-type; deg15, deg15 knockout line; C1, C2, and C3, three transformants of the knockout line complemented with DEG15 wild-type cDNA. Significant differences by t tests of root length from deg15 plants at the level of <0.01 are labeled with a; those different from wild type at the same level are labeled with b. The number of plants tested varied from seven to 11 without 2,4-DB, from seven to 19 for 0.1 μg, from 13 to 29 for 0.2 μg, and from 21 to 26 for 0.4 μg mL−1. C, Absence of response of wild-type and deg15 mutant plants to 2,4-D and Suc. At left, root length of plants grown for 11 d in the presence of 0.05 μg mL−1 2,4-D. At middle and right, hypocotyl length of etiolated plants grown for 6 d with or without Suc.
Oilseed plants like Arabidopsis metabolize storage fatty acids during seedling establishment by peroxisomal β-oxidation. If this pathway was impaired in deg15 mutant plants, they would depend on an external energy source, such as sugar, for normal germination. Our result indicates that deg15 mutant plants are able to metabolize enough fatty acids during seedling establishment in darkness to promote normal growth. It is possible that the lack of PTS2 processing does not reduce the efficiency of the enzymatic machinery involved in the β-oxidation of fatty acids and their subsequent conversion to energy. At least for one peroxisomal enzyme, gMDH, it was shown that there is no difference in the catalytic properties of processed and unprocessed enzymes (Gietl et al., 1996; Cox et al., 2005).
It is possible that the lack of protein processing specifically affects the β-oxidation of 2,4-DB to 2,4-D. Recently, a protein specific for the β-oxidation of the naturally occurring 2,4-DB analog, indole-3-butyric acid (IBA), was described from Arabidopsis (Zolman et al., 2007), suggesting that an at least partially independent pathway for the β-oxidation of IBA and 2,4-DB may exist in parallel to the β-oxidation of fatty acids.
Other mutant strains that show a response to IBA/2,4-DB treatment, but no sugar dependency for germination in darkness contain a mutated gene for the PEX7 receptor. This protein is a PTS2 recognition factor and is involved in PTS2-dependent peroxisomal import. After translocation, PEX7 is recycled back to the cytoplasm (Baker and Sparkes, 2005). It is tempting to speculate that the lack of PTS2 processing in deg15 plants might interfere with the successful relocation of PTS2 recognition factor PEX7 to the cytoplasm because the machinery involved in this process is not yet well understood (Baker and Sparkes, 2005).
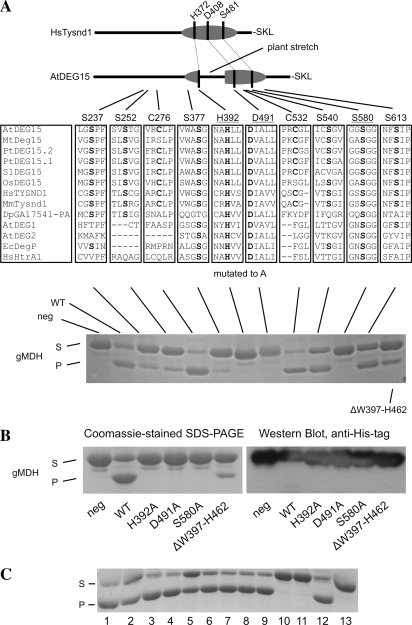
Confirmation That the Proteolytic Activity of DEG15 Is Carried Out by the His-Asp-Ser Catalytic Triad Using Site-Directed Mutants
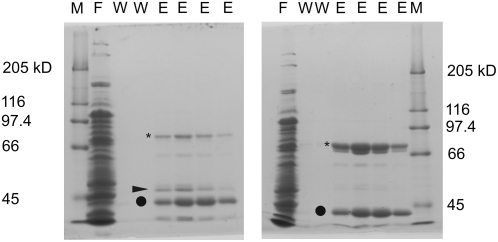
The conserved putative catalytic triad of DEG15 from Arabidopsis (accession no. Q8VZD4, a gene product of At1g28320) is located at positions His-392, Asp-491, and Ser-580 in the amino acid sequence alignment, as compared with DegP of Escherichia coli, DEG15 of rice, human TYSND1, and Arabidopsis DEG2 (Fig. 1). DEG15 proteases from plants contain an additional loop of approximately 60 amino acids between the catalytic residues His-392 and Asp-491. In disagreement with the presence of a conserved putative catalytic triad typical for Ser proteases, the mammalian DEG15 homolog in function TYSND1 was identified as a Cys protease (Kurochkin et al., 2007). To test the involvement of conserved amino acids in the catalytic process, the following residues of DEG15 were mutated to Ala: Ser-237 (DEG15S237A), Ser-252 (DEG15S252A), Cys-276 (DEG15C276A), Ser-377 (DEG15S377A), His-392 (DEG15H392A), Asp-491 (DEG15D491A), Cys-532 (DEG15C532A), Ser-540 (DEG15S540A), Ser-580 (DEG15S580A), and Ser-613 (DEG15S613A; Fig. 3).Arabidopsis DEG15WT, as well as the mutated versions, was expressed in E. coli as recombinant fusion proteins with an N-terminal His6-tag. Additionally, DEG15ΔW397-H462, lacking the plant-specific loop, was expressed under similar conditions to examine the requirement of this region for the activity of plant DEG15. Several protein bands were visible after chromatographic purification of DEG15WT and its mutated versions in Coomassie-stained SDS-polyacrylamide gels (Fig. 4). For DEG15WT (Fig. 4, left), all three major bands were recognized by an anti-His-tag antibody and identified by mass spectrometry as full-length DEG15 (76 kD) or its truncated versions (47 and 40 kD) lacking the protease domain (data not shown). In the eluted fraction of DEG15S580A (Fig. 4, right), a band with the apparent molecular mass of 47 kD was missing, indicating self-cleavage of active DEG15WT (Fig. 4, left, marked by arrow). This band also appeared in the eluted fractions of the other mutated active versions of DEG15 (data not shown).
Figure 3.
Site-directed mutants demonstrate that the His-Asp-Ser catalytic triad carries out the proteolytic activity of Arabidopsis DEG15. A (top), Diagram of Homo sapiens TYSND1 with the location of the His (H372), Asp (D408), and Ser (S481) of the expected catalytic triad and the C-terminal Ser (S), Lys (K), and Leu (L) for the PTS1 signal. It is compared to Arabidopsis DEG15 with its plant-specific stretch between the catalytic His and Asp. Middle, Residues conserved in the majority of DEG15 homologs from Arabidopsis thaliana (At), Medicago truncatula (Mt), Populus tremula (Pt), Solanum lycopersicum (Sl), Oryza sativa (Os), H. sapiens (Hs), Mus musculus (Mm), Drosophila pseudoobscura (Dp), and the respective regions in Arabidopsis DEG1 and DEG2, Escherichia coli (Ec) DegP, and H. sapiens HtrA2. Residues are labeled according to their position in DEG15 from Arabidopsis. Residues forming the potential catalytic triad are underlined and residues mutated to Ala are bold. Bottom, Proteolytic activity of recombinant wild-type and mutated DEG15 toward purified His-6/XpressEpitope-tagged pre-gMDH. S, Substrate; P, processed gMDH. Additionally, a mutant with a deletion of the plant-specific loop (ΔW397-H462) was tested. Only mutation of H392, D491, and S580 to Ala completely eliminated processing. B, N-terminal processing of recombinant His6/XpressEpitope-tagged gMDH by DEG15. Left, Coomassie-stained SDS-PAGE gel. Right, Detection of the N-terminal His-tag by immunoblotting. The tag is absent in the processed substrate. C, Proteolytic activity of DEG15WT against recombinant pre-gMDH in the presence of protease inhibitors. S, Substrate; P, product of proteolysis. Shown are: positive control without inhibitors (line 1), 1 μm pepstatin A (line 2), 2 mm EGTA (line 3), 2 mm EDTA (line 4), 1 mm 1,10-phenanthroline (line 5), 1 mm benzamidine (line 6), 4 μg mL−1 aprotinin (line 7), 10 μm leupeptin (line 8), 20 μm E64 (line 9), 1 mm NEM (line 10), 2 mm NEM (line 11), 1 mm Pefabloc SC (line 12), and control reaction without protease (line 13).
Figure 4.
Purification of DEG15wt (left) and DEG15S580A (right) by affinity chromatography with Ni2+-nitrilotriacetic acid. Asterisk indicates 76-kD full-length protein, arrow indicates 47-kD product of putative self-cleavage, and circle marks 40-kD truncated DEG15. Purification of DEGH392A and DEG15D491A was similar to DEG15S580A; all other purifications were similar to DEG15WT. The major fragment band at 40 kD was absent in DEG15S237A. M, Marker; F, flow through; W, wash fraction; E, elution fraction.
The full-length and truncated DEG15 spontaneously formed oligomers with different subunit combinations that could not be removed by standard chromatographic procedures (data not shown). Since pre-gMDH is processed by DEG15 in vivo (Helm et al., 2007), we used purified recombinant Arabidopsis pre-gMDH, expressed with an N-terminal His6/XpressEpitope-tag in E. coli, as substrate for recombinant DEG15WT and its mutated versions in vitro (Fig. 3A, bottom). A mock assay was performed in the absence of added DEG15 as a negative control. DEG15WT was processed upon incubation of full-length His/XpressEpitope-tagged recombinant pre-gMDH (41.4 kD) to an approximately 8-kD smaller fragment as visualized by Coomassie-stained SDS-polyacrylamide gels (Fig. 3A, bottom). Immunoblotting with anti-His-tag antibody confirmed that processing of recombinant pre-gMDH occurs at the N terminus because the resulting large fragment of approximately 33 kD no longer contained the His-tag (Fig. 3B). The processed gMDH fragment was analyzed by N-terminal Edman sequencing (data not shown), indicating that cleavage in the in vitro assay occurs at the same site as in vivo (CR*-AKGGN; Table I). DEG15H392A, DEG15D491A, and DEG15S580A showed no proteolytic activity against pre-gMDH (Fig. 3A, bottom). Mutation of the residues Ser-540 and Ser-613 to Ala did not alter the activity of the protease, whereas the activity was reduced after mutation of Ser-237 and Ser-252 (75% less substrate was degraded as compared to DEG15WT).
Mutation of Ser-377 drastically reduced proteolytic activity of this construct and 90% less substrate was degraded as compared to DEG15WT (Fig. 3A, bottom). The deletion of the plant-specific stretch (amino acid residues Trp-397 to His-462) led to reduced substrate processing (25% as compared to DEG15WT). Interestingly, the activity of recombinant DEG15 was slightly enhanced by the exchange of Cys residues Cys-276 and Cys-532 to Ala (approximately 120% of the activity of DEG15WT was measured). These data show that His-392, Asp-491, and Ser-580, but neither the plant-specific stretch nor any of the other investigated residues, are essential for proteolytic activity of Arabidopsis DEG15.
Inhibition of TYSND1 by NEM, but not by E64, both universal inhibitors of Cys proteases, led to the conclusion that TYSND1 represents a novel class among Cys proteases (Kurochkin et al., 2007) Therefore, we tested the effect of various protease inhibitors on recombinant Arabidopsis DEG15WT using pre-gMDH as a substrate (Fig. 3C). Similar to TYSND1, the proteolytic activity of DEG15 in our protease assays was abolished by the Cys protease inhibitor NEM (Fig. 3C, lines 10 and 11), but not by E64 (Fig. 3C, line 9). However, one should mention here that NEM is not a specific Cys protease inhibitor, but a broad-acting agent that may be able to inhibit every enzyme containing free thiol groups due to its alkylation activity. Lack of inhibition of Arabidopsis DEG15 by general Ser protease inhibitors, such as benzamidine (Fig. 3C, line 6), aprotinin (Fig. 3C, line 7), leupeptin (Fig. 3C, line 8), or Pefabloc (Fig. 3C, line 12), can be explained by the observation that some Ser protease subclasses are resistant to the action of broad-range inhibitors. For example, the only known inhibitor for E. coli DegP is diisopropylfluorophosphate (Swamy et al., 1983; Lipinska et al., 1990). No inhibition of DEG15 was observed in the presence of pepstatin A, an inhibitor of aspartyl proteases (Fig. 3C, line 2) or metalloprotease inhibitors, such as EGTA (Fig. 3C, line 3), EDTA (Fig. 3C, line 4), or 1,10-phenanthroline (Fig. 3C, line 5).
Complexity of inhibitor studies is further supported by the fact that DEG15 from watermelon was inhibited by general Ser/Cys protease inhibitors, but not by the widely used Ser protease inhibitor phenylmethylsulfonyl fluoride (PMSF; Helm et al., 2007). Enzymatic activity of recombinant Arabidopsis DEG15 was only slightly reduced by PMSF (data not shown). Because NEM is a reagent for the covalent modification of Cys residues, the conclusion from the inhibitor studies is that either 1 of 9 or 18 Cys residues present in DEG15 or TYSND1, respectively, is important for enzymatic activity. Alternatively, a covalent modification of a Cys in the substrate may abolish cleavage.
DEG15 from watermelon was reported to occur either as a monomer or as a dimer (Helm et al., 2007), with the monomer acting as a general protease and the dimer as the peroxisomal processing protease. Size-exclusion chromatography of DEG15S580A showed two distinct peaks, suggesting the presence of at least two oligomeric states formed by full-length 76-kD and truncated 40-kD DEG15S580A (data not shown). Preliminary cross-linking studies using glutaraldehyde (Azem et al., 1998) revealed that 76- and 40-kD DEG15S580A formed 290-, 212-, and 97-kD complexes that might represent 4-mers to 7-mers, 3-mers to 5-mers, and 2-mers, respectively, depending on the stoichiometry of the full-length and truncated forms (data not shown). Because we were not able to separate both DEG15S580A forms, it is not possible to precisely identify the oligomeric state of the protease that is responsible for proteolytic activity in the assays presented here. In summary, our data show that Arabidopsis DEG15 is a Ser protease with the catalytic triad formed by residues His-392, Asp-491, and Ser-580, and the plant-specific stretch is not directly involved in proteolysis. Concerning the high similarity on amino acid level between DEG15 and TYSND1 and the absence of a conserved Cys, we propose that also the mammalian protease is Ser type.
DEG15 Colocalizes with gMDH in the Peroxisomes of Nicotiana benthamiana
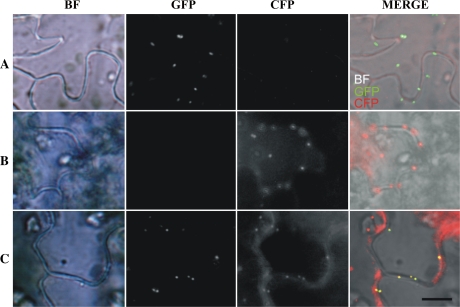
To determine the location of DEG15 in the leaf peroxisomes we constructed a plasmid encoding a GFP fused to the N terminus of DEG15S580A. An inactive version of DEG15 was chosen to prevent its autodegradation in planta. A second plasmid encoding a cyan fluorescent protein (CFP) fused to the C terminus of pre-gMDH was constructed as a control. Both fusion constructs were placed under the control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter. N. benthamiana leaves were transformed transiently by Agrobacterium (Agrobacterium tumefaciens)-mediated leaf infection (Wydro et al., 2006) with the individual constructs and cotransformed with both plasmids (Fig. 5). The micrographs in Figure 5, row A, show a leaf segment from a transformant expressing GFP-DEG15S580A under bright-field (BF) illumination, with a filter passing only the green fluorescence wavelength (GFP) and a filter passing the CFP. The merged BF, GFP, and CFP image identifies the DEG15S580A protein distinct spot within the cell. In row B, corresponding series of a leaf from a transformant expressing the CFP-tagged gMDH identifies it in the peroxisomes (image CFP and MERGE). A leaf segment expressing both the DEG15S580A and the CFP-tagged gMDH in row C demonstrates the colocalization of the two fluorescing proteins in the peroxisomes (GFP, CFP, MERGE). This is a first clear-cut demonstration of a colocalization of DEG15 together with a marker enzyme in the peroxisome.
Figure 5.
Colocalization of DEG15 and gMDH in peroxisomes in transiently transformed N. benthamiana leaves. BF, GFP fluorescence, CFP fluorescence. Row A, Transformation with plasmid-encoding GFP-DEG15. Row B, Transformation with plasmid encoding gMDH-CFP. Row C, Cotransformation with plasmids encoding GFP-DEG15 and gMDH-CFP. Scale bar = 10 μm.
The Range of Proteins Processed by DEG15
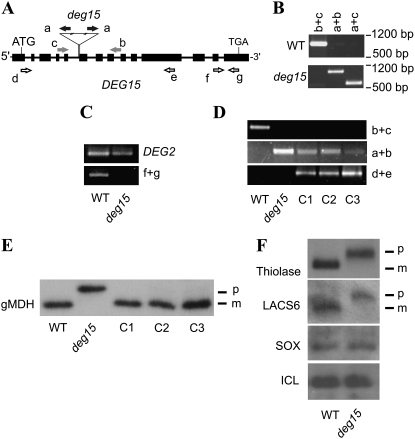
Because DEG15 processed pre-gMDH in vitro (Fig. 3A, bottom) and colocalized with gMDH in planta (Fig. 5), we examined whether DEG15 processes PTS2-containing proteins in vivo. The deg15 knockout mutant Salk_007184 carrying a tandem T-DNA insertion within the fifth intron (Fig. 6A) was obtained from the Salk Institute Arabidopsis insertion line mutant collection (Alonso et al., 2003). A homozygous line of the mutant was isolated (Fig. 6B) that was unable to transcribe the DEG15 gene as shown by reverse transcription (RT)-PCR using the DEG2 gene, encoding a chloroplast DEG2 protein (Haussuhl et al., 2001; Huesgen et al., 2006), as a control (Fig. 6C). To validate that DEG15 processes the PTS2 peptide of pre-gMDH, we complemented the deg15 mutant line with DEG15 cDNA under the control of the constitutively expressed CaMV 35S promoter. Three potential transformant lines (C1-3) were identified and confirmed for the presence of the DEG15 cDNA in the deg15 knockout background (Fig. 6D). Immunoblot analysis revealed PTS2 processing of pre-gMDH in the three deg15 knockout lines complemented with DEG15 cDNA (Fig. 6E). This confirms unambiguously that DEG15 is carrying out processing of the pre-gMDH in planta.
Figure 6.
DEG15 processes PTS2-containing proteins in vivo. A, Schematic representation of the DEG15 gene. Exons are represented by black boxes, introns as black lines. Positions of start and stop codons and the position and orientation of the tandem T-DNA insertion are labeled. Positions of the binding sites of the primers used (Table II) are indicated by arrows. T-DNA insertion primer (black), line-specific screening primers (gray), primers for screening the cDNA insertions in complemented mutant lines (white). B, Genetic analysis of the insertion line by PCR with primers as indicated in A. C, RT-PCR using wild-type and deg15 mutant plants to confirm the absence of the DEG15 transcript with the primers indicated in A. Transcript of DEG2 was used as a positive control. D, Genetic analysis of complementation lines C1, C2, and C3 by PCR with primers as indicated in A to confirm the DEG15 cDNA insertion into the deg15 knockout mutant background. E, PTS2 processing of pre-gMDH (p) to mature gMDH (m) in planta assayed by immunoblotting. Leaf extracts from wild type, the deg15 knockout mutant, and the three deg15 lines complemented with DEG15 cDNA (C1-3) were analyzed. F, PTS2 processing of thiolase and LACS6 assayed by immunoblotting as in E. No processing of the PTS1-containing peroxisomal proteins, such as SOX and ICL, was observed. p, Precursor; m, mature protein.
We analyzed processing of other PTS2-containing proteins, such as 3-keto-acyl-CoA thiolase (thiolase; Kato et al., 1996b) and long-chain acyl-CoA synthetase 6 (LACS6; Hayashi et al., 2002) or PTS1-containing proteins, such as sulfite oxidase (SOX; Hansch et al., 2006) and isocitrate lyase (ICL; Charlton et al., 2005) by DEG15. Whereas thiolase and LACS6 were not processed in the deg15 mutant, no difference between wild-type and mutant line was observed for SOX and ICL, as proven by immunoblotting (Fig. 6F). These data indicate that DEG15 is the processing protease for PTS2 in higher eukaryotes.
CONCLUSION
In the work presented here, we were able to show an expressed phenotype of a higher eukaryotic organism lacking peroxisomal processing. With the available Arabidopsis loss-of-function mutant and its complementation strains, it will be interesting to further evaluate the impact and benefit of this process, especially on a molecular basis. Additionally, we could solve the discrepancy in classification of plant DEG15 and its mammalian paralog TYSND1 concerning the catalytic mechanism. We showed that DEG15, and most probably also TYSND1, are Ser proteases. Furthermore, we could unambiguously demonstrate that DEG15 is a peroxisomal protease that processes PTS2-containing proteins in vivo.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) and mutant plants were cultivated on soil at a photon flux density of 150 μmol photons m−2 s−1 under short-day conditions (8-h light, 21°C/16-h dark, 19°C). For 2,4-DB treatment, seeds were surface sterilized and sown on agar plates containing 2.2 g L−1 Murashige and Skoog salts, 30 mm Suc, and 8 g L−1 agar, supplemented with 2,4-DB or 2,4-D at indicated concentrations. After 11 d, plants were analyzed for expressed phenotypes. For seedling-establishing assays, seeds were surface sterilized and sown on agar plates containing 2.2 g L−1 Murashige and Skoog salts, 8 g L−1 agar, and 0 and 30 mm Suc, respectively. After exposure to light for 7 h, plants were grown in darkness for 6 d.
Plasmid Constructions
A DNA fragment encoding Arabidopsis DEG15 (At1g28320) was generated by PCR with RAFL04-20-D23 clone (Sakurai et al., 2005) as a template, using primers 1501 and 1504 (Table II). The resulting DNA fragment was cloned blunt ended into the Ecl136II-site of the pBAD/HisA plasmid (Invitrogen), resulting in the plasmid pHS18 for overexpression of DEG15 as a fusion protein. The RAFL04-20-D23 clone was also used as a template for site-directed mutagenesis with primers 1517 and 1518 generating a plasmid containing a sequence coding for DEG15S580A, called pHS19. The plasmid for overexpression of DEG15S580A as a fusion protein (pHS26) was generated like pHS18, with pHS19 as template. The plasmids for overexpression of DEG15S237A (pHS113), DEG15S252A (pHS129), DEG15C276A (pHS124), DEG15S377A (pHS117), DEG15H392A (pHS42), DEG15D491A (pHS43), DEG15C532A (pHS115), DEG15S540A (pHS116), and DEG15S613A (pHS114) were generated by site-directed mutagenesis of pHS18 with the primers 1573 and 1574 (pHS113), 1589 and 1590 (pHS129), 1585 and 1586 (pHS124), 1581 and 1582 (pHS117), 1553 and 1554 (pHS42), 1551 and 1552 (pHS43), 1577 and 1578 (pHS115), 1579 and 1580 (pHS116), and 1575 and 1576 (pHS114), respectively. For the plasmid containing the DNA encoding the DEG15 version lacking the plant-specific stretch W397-H462 (DEG15ΔW397-H462), two fragments were generated by PCR using RALF clone R21163 as a template. Primers 1501 and 1566 or 1565 and 1519 were used, respectively. The two DNA fragments were ligated by PCR and cloned blunt ended into the Ecl136II-site of the pBAD/HisA plasmid resulting in the plasmid pHS50.
Table II.
Primers and their sequences used in this study
| Name | 5′-Sequence-3′ |
|---|---|
| 1501 | ATGGATGTGTCTAAAGTTGTCAG |
| 1504 | TCATAACTTGCTAGGGATCACAT |
| 1517 | GCATCCTGGTGGCGCTGGTGGTGCTGTTC |
| 1518 | GAACAGCACCACCAGCGCCACCAGGATGC |
| 1553 | CTAACAAATGCTGCCCTGCTCGAGCCGTGGAGGTATG |
| 1554 | CACGGCTCGAGCAGGGCAGCATTTGTTAGTATGAGACC |
| 1551 | GGAACAATTAGCAATTGCCTTACTGCAGCTAGAATATGTCCC |
| 1552 | GCTGCAGTAAGGCAATTGCTAATTGTTCCTTGCAAATATAGAC |
| 1501 | ATGGATGTGTCTAAAGTTGTCAG |
| 1566 | CACACGTATGTCTCTCGGCTCAAGCAGGTGAGCATTTG |
| 1565 | CACCTGCTTGAGCCGAGAGACATACGTGTGCGTTTG |
| 1519 | TATGGTACCTCATAACTTGCTAGG |
| 1541 | CACCATGGATTACAAGGATGACGACGATAAGGATGTGTCTAAAGTTGTC |
| 1532 | GCGTGTACATCATAACTTGCTAGGGATCACATC |
| 1573 | CACTAGGCGCTCCCTTTGGAATCCTTTCACC |
| 1574 | CAAAGGGAGCGCCTAGTGCTACAAGTGTATC |
| 1575 | CTCAACTTCGCGATCCCATGTGCAGTCTTGGC |
| 1576 | CATGGGATCGCGAAGTTGAGATGCGGTATAA |
| 1577 | CCAAGAGCCGGCCTTTCTCCTTCTATTTGTTC |
| 1578 | GAGAAAGGCCGGCTCTTGGTCCGAAGAGTCC |
| 1579 | CTATTTGTGCCGGCGTTGTAGCAAAGGTAG |
| 1580 | CTACAACGCCGGCACAAATAGAAGGAGAAAG |
| 1581 | GTTTGGGCTGCAGGTATTATTCTTAACGAAC |
| 1582 | GAATAATACCTGCAGCCCAAACACCATCATTG |
| 1585 | GTTCGAGCTCTCCCTGGAATGGAAGGGGCTCC |
| 1586 | GGGAGAGCTCGAACATCAGCTATCATCAGTGAC |
| 1589 | CAGCGTAGCAACTGGATCCATTGCGAATAG |
| 1590 | CAATGGATCCAGTTGCTACGCTGTTAAAAAAG |
| gMDH1 | GAGTTTCGTGGAGATGCCAACC |
| gMDH2 | TCATTTTCTGATGAATTCAACACC |
| gMDH5 | CACCATGGAGTTTCGTGGAGATGCCAACC |
| gMDH6 | TTTTCTGATGAATTCAACACCTTTC |
| a | TGGTTCACGTAGTGGGCCATCG |
| b | GGAAATCATGATGACCTCTAGTCG |
| c | GCGTATTCTTTTCAGGGTCAGC |
| d | ATGGATGTGTCTAAAGTTGTCAG |
| e | CTTAGTGGTCTAATCAAAATGCC |
| f | CATCTCAACTTCAGCATCCCATG |
| g | TCATAACTTGCTAGGGATCACAT |
For construction of a sequence coding for N-terminally GFP-tagged DEG15S580A under control of the constitutive CaMV 35S promoter, pHS19 was used as a template for PCR using the same primers as for the construction of pHS18. The fragment was cloned blunt ended into the SmaI site of the pEZT-CL plasmid (provided by D. Erhardt, Carnegie Institution), generating pHS41.
To create a rescue construct, At1g28320 was amplified from RAFL clone R21163 by PCR using the primers 1541 and 1532 and cloned into pENTR/D-TOPO (Invitrogen) according to the manufacturer's instructions. The resulting plasmid pHS38 served as an entry vector in a gateway reaction with pEarleygate100 (Earley et al., 2006) creating the rescue vector pHS58.
cDNA encoding the open reading frame of gMDH (At5g09660) was generated by PCR from the CD4-34 library (Arabidopsis Biological Resource Center [ABRC]), using primers gMDH1 and gMDH2. The DNA fragment was subcloned into the vector pCRII-TOPO (Invitrogen) according to manufacturer's instructions, resulting in the plasmid pHS48. The coding sequence was reamplified from pHS48 by using the same primers as stated before and cloned blunt ended into the Ecl136II-site of the pBAD/HisA plasmid, resulting in the plasmid pHS51. Using pHS48 as template and primers gMDH5 and gMDH6, a DNA fragment was created and cloned into the pENTR/D-TOPO vector (Invitrogen) according to the manufacturer's instructions, resulting in plasmid pHS63. This served as an entry vector in a gateway reaction (Invitrogen) with pEarleygate102 (Earley et al., 2006). This plasmid, named pHS64, encodes a C-terminally CFP-tagged gMDH under the control of the CaMV 35S promoter.
All primers were obtained from Operon Biotechnologies and were analyzed by sequencing (GATC Biotech AG) for orientation and integrity of the insert. The position of the T-DNA was confirmed by sequencing of PCR products (GATC Biotech AG). Suppression of gene expression was confirmed by transcribing isolated RNA (Rneasy mini kit; Qiagen) from wild-type and insertion line mutants to cDNA (QuantiTect reverse transcription kit; Qiagen) and amplification by PCR with primers f and g (for sequences, see Table II). For complementation studies, homozygous mutant line Salk_007184 was transformed with Agrobacterium (Agrobacterium tumefaciens) carrying the rescue vector pHS58 by the floral-dip method according to Weigel and Glazebrook (2002). Potential transformants were screened for the insertion of the cDNA by PCR using primers d and e. Agrobacterium strains GV2606 and GV3101 were transformed with plasmids pHS41, pHS64, or pHS58, respectively, by the freeze-thaw method according to Weigel and Glazebrook (2002).
Isolation of Homozygous deg15 Deletion Mutant, Its Complementation, and Nicotiana benthamiana Infiltration
T-DNA insertion line mutants (Salk_007184) were obtained from the Nottingham Arabidopsis Stock Centre (NASC; Alonso et al., 2003). Homozygous deg15 mutant plants were identified by PCR using T-DNA (primer a) and gene-specific (primers b und c) primers (for sequences, see Table II). Primers were designed with the online primer design tool provided by SALK (http://signal.salk.edu/tdnaprimers.2.html). PCR elongation times were chosen to allow only products ≤ 1,500 bp to be amplified.
The position of the T-DNA was confirmed by sequencing of PCR products (GATC Biotech AG). Suppression of gene expression was confirmed by transcribing isolated RNA (Rneasy mini kit; Qiagen) from wild-type and insertion line mutants to cDNA (QuantiTect reverse transcription kit; Qiagen) and amplification by PCR with primers f and g. For complementation studies, homozygous mutant line Salk_007184 was transformed with Agrobacterium carrying the rescue vector pHS58 by the floral-dip method according to Weigel and Glazebrook (2002). Potential transformants were screened for the insertion of the cDNA by PCR using primers d and e. PCR elongation times were chosen to allow only products ≤1,500 bp to be amplified. Agrobacterium strains GV2606 and GV3101 were transformed with plasmids pHS41, pHS64, or pHS58, respectively, by the freeze-thaw method according to Weigel and Glazebrook (2002).
Leaves of N. benthamiana were infiltrated with Agrobacterium carrying the appropriate vector as described in Wydro et al. (2006).
Overexpression and Purification
To obtain the N-terminally His6-tagged recombinant protein, Escherichia coli strain Top10F (Invitrogen) harboring the appropriate plasmid (pHS18, pHS26, pHS42, pHS43, pHS50, or pHS51, respectively) was grown to an OD600 of 0.5 and the expression of recombinant proteins was induced by adding l-Ara to a final concentration of 0.02% (w/v). After 3 h at 37°C, cells were harvested by centrifugation at 5,000g and resuspended in lysis buffer (50 mm HEPES, pH 7.8, 300 mm NaCl, and 0.5 mm dithiothreitol). In the case of DEG15 and its variants, additional dithiothreitol was added to a final concentration of 3 mm to prevent oxidation of free Cys. In the case of gMDH, Complete Protease Inhibitor Mix minus EDTA (Roche Diagnostics GmbH) was added according to the manufacturer's instructions. Cells were lysed by ultrasonification (30 s, followed by incubation on ice for 60 s, repeated 10×) and centrifuged at 26,500g for 90 min at 4°C. The supernatant was applied to a Ni2+- nitrilotriacetic column using an FPLC system (GE Healthcare Europe GmbH), washed with lysis buffer (see above), supplemented with 50 mm imidazole, and eluted with a linear gradient from 50 to 300 mm imidazole. Proteins were eluted at approximately 150 mm imidazole (DEG15 and its variants) or 125 mm imidazole (gMDH), respectively. Elution fractions of DEG15 and its variants were directly used for protease activity assays. Elution fractions containing gMDH were analyzed by SDS-PAGE and assayed for their ability to reduce oxaloacetate.
Protease Activity Assays
All protease assays were performed with 5 μL of elution fraction from the protease purification (containing approximately 7.5–10 μg of total protein) in 50 μL total volume at 30°C in 50 mm HEPES, pH 7.8, and 300 mm NaCl. Time-course experiments indicated that no proteolytic activity was assayed after 12 h of incubation (data not shown); therefore, all protease assays were performed overnight to ensure maximal degradation of the substrate. Elution buffer (see Overexpression and Purification) containing 250 mm imidazole was used as a mock control. gMDH was used as a substrate at a final concentration of 1.6 μg μL−1. Protease inhibitors were used in concentrations as indicated in Figure 3. N-terminal Edman sequencing of in vitro-processed gMDH was performed by Proteome Factory AG.
SDS-PAGE and Immunoblotting
SDS-PAGE and immunoblotting were conducted according to Haussuhl et al. (2001). For the detection of gMDH, leaf extracts from 3-week-old plants were used. Extracts from 4- or 7-d-old etiolated Arabidopsis seedlings were used for the detection of ICL or thiolase and SOX, respectively. Antibodies against gMDH were raised against purified gMDH from watermelon (Citrullus vulgaris; Gietl et al., 1996) and used at a 1:5,000 dilution. Antibody dilutions of 1:1,000 were used for thiolase and LACS6 (Hayashi et al., 2002), 1:500 for SOX (Hansch et al., 2006), and 1:5,000 for ICL (Charlton et al., 2005). All antibodies were applied for 14 h at room temperature. A horseradish peroxidase-conjugated antibody against His5 (Qiagen) was used according to the manufacturer's instructions.
Fluorescence Microscopy
GFP and CFP fluorescence was observed under an Olympus BX51 epifluorescence microscope equipped with a Nikon DXM1200 digital camera system (Olympus Europe). For transmitted light viewing, Koehler's illumination was used. Fluorescence of GFP and CFP has been dissected using the filter sets 41020 and 31045 (Chroma Technology), respectively. Multichannel fluorescence pictures were taken and composed with the help of the software LUCIA (Nikon GmbH). Exposure times were 7 s for GFP and 2 s for CFP, respectively.
Acknowledgments
We thank RIKEN for providing the cDNA clone RAFL04-20-D23 (RIKEN Tsukuba Institute, Japan) and the ABRC for the SALK line (SALK_007184) and the cDNA library CD4-34 (ABRC Stock Center, Ohio State University). We also thank Dr. Diter von Wettstein (Washington State University) for valuable comments on this manuscript, Dr. Ansgar Gruber for help with the microscopy, Silvia Kuhn for technical assistance, and Dr. Dietmar Funck for helpful advice. We are especially grateful to Mikio Nishimura (Okazaki, Japan) for providing us with the antibodies against thiolase and LACS6, Robert Hänsch (Braunschweig, Germany) for providing us with the antibody against SOX, and Alison Baker (Leeds, UK) for providing us with the antibody against ICL.
This work was supported by the Deutsche Forschungsgemeinschaft (grant nos. AD92/8–2 and AD92/8–3 to I.A. and GI154/9–4 and GI154/9–5 to C.G.) and the Konstanz University (grant to I.A.).
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Iwona Adamska (iwona.adamska@uni-konstanz.de).
Open access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Azem A, Weiss C, Goloubinoff P (1998) Structural analysis of GroE chaperonin complexes using chemical cross-linking. Methods Enzymol 290 253–268 [DOI] [PubMed] [Google Scholar]
- Baker A, Sparkes IA (2005) Peroxisome protein import: some answers, more questions. Curr Opin Plant Biol 8 640–647 [DOI] [PubMed] [Google Scholar]
- Charlton WL, Johnson B, Graham IA, Baker A (2005) Non-coordinate expression of peroxisome biogenesis, beta-oxidation and glyoxylate cycle genes in mature Arabidopsis plants. Plant Cell Rep 23 647–653 [DOI] [PubMed] [Google Scholar]
- Clausen T, Southan C, Ehrmann M (2002) The HtrA family of proteases: implications for protein composition and cell fate. Mol Cell 10 443–455 [DOI] [PubMed] [Google Scholar]
- Cox B, Chit MM, Weaver T, Gietl C, Bailey J, Bell E, Banaszak L (2005) Organelle and translocatable forms of glyoxysomal malate dehydrogenase. The effect of the N-terminal presequence. FEBS J 272 643–654 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45 616–629 [DOI] [PubMed] [Google Scholar]
- Gietl C, Faber KN, van der Klei IJ, Veenhuis M (1994) Mutational analysis of the N-terminal topogenic signal of watermelon glyoxysomal malate dehydrogenase using the heterologous host Hansenula polymorpha. Proc Natl Acad Sci USA 91 3151–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietl C, Seidel C, Svendsen I (1996) Plant glyoxysomal but not mitochondrial malate dehydrogenase can fold without chaperone assistance. Biochim Biophys Acta 1274 48–58 [DOI] [PubMed] [Google Scholar]
- Hansch R, Lang C, Riebeseel E, Lindigkeit R, Gessler A, Rennenberg H, Mendel RR (2006) Plant sulfite oxidase as novel producer of H2O2: combination of enzyme catalysis with a subsequent non-enzymatic reaction step. J Biol Chem 281 6884–6888 [DOI] [PubMed] [Google Scholar]
- Haussuhl K, Andersson B, Adamska I (2001) A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO J 20 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, De Bellis L, Hayashi Y, Nito K, Kato A, Hayashi M, Hara-Nishimura I, Nishimura M (2002) Molecular characterization of an Arabidopsis acyl-coenzyme a synthetase localized on glyoxysomal membranes. Plant Physiol 130 2019–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Luck C, Prestele J, Hierl G, Huesgen PF, Frohlich T, Arnold GJ, Adamska I, Gorg A, Lottspeich F, et al (2007) Dual specificities of the glyoxysomal/peroxisomal processing protease Deg15 in higher plants. Proc Natl Acad Sci USA 104 11501–11506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huesgen PF, Schuhmann H, Adamska I (2006) Photodamaged D1 protein is degraded in Arabidopsis mutants lacking the Deg2 protease. FEBS Lett 580 6929–6932 [DOI] [PubMed] [Google Scholar]
- Kato A, Hayashi M, Kondo M, Nishimura M (1996. a) Targeting and processing of a chimeric protein with the N-terminal presequence of the precursor to glyoxysomal citrate synthase. Plant Cell 8 1601–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Hayashi M, Takeuchi Y, Nishimura M (1996. b) cDNA cloning and expression of a gene for 3-ketoacyl-CoA thiolase in pumpkin cotyledons. Plant Mol Biol 31 843–852 [DOI] [PubMed] [Google Scholar]
- Krojer T, Garrido-Franco M, Huber R, Ehrmann M, Clausen T (2002) Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416 455–459 [DOI] [PubMed] [Google Scholar]
- Krojer T, Sawa J, Schafer E, Saibil HR, Ehrmann M, Clausen T (2008) Structural basis for the regulated protease and chaperone function of DegP. Nature 453 885–890 [DOI] [PubMed] [Google Scholar]
- Kurochkin IV, Mizuno Y, Konagaya A, Sakaki Y, Schonbach C, Okazaki Y (2007) Novel peroxisomal protease Tysnd1 processes PTS1- and PTS2-containing enzymes involved in beta-oxidation of fatty acids. EMBO J 26 835–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinska B, Zylicz M, Georgopoulos C (1990) The HtrA (DegP) protein, essential for Escherichia coli survival at high temperatures, is an endopeptidase. J Bacteriol 172 1791–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S (2004) Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol 135 783–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Satou M, Akiyama K, Iida K, Seki M, Kuromori T, Ito T, Konagaya A, Toyoda T, Shinozaki K (2005) RARGE: a large-scale database of RIKEN Arabidopsis resources ranging from transcriptome to phenome. Nucleic Acids Res 33 D647–D650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swamy KH, Chung CH, Goldberg AL (1983) Isolation and characterization of protease Do from Escherichia coli, a large serine protease containing multiple subunits. Arch Biochem Biophys 224 543–554 [DOI] [PubMed] [Google Scholar]
- Swinkels BW, Gould SJ, Bodnar AG, Rachubinski RA, Subramani S (1991) A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J 10 3255–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Wydro M, Kozubek E, Lehmann P (2006) Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotiana benthamiana. Acta Biochim Pol 53 289–298 [PubMed] [Google Scholar]
- Zolman BK, Nyberg M, Bartel B (2007) IBR3, a novel peroxisomal acyl-CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol Biol 64 59–72 [DOI] [PubMed] [Google Scholar]