Abstract
Light provides crucial positional information in plant development, and the morphogenetic processes that are orchestrated by light signals are triggered by changes of gene expression in response to variations in light parameters. Control of expression of members of the RbcS and Lhc families of photosynthesis-associated nuclear genes by light cues is a paradigm for light-regulated gene transcription, but high-resolution expression profiles for these gene families are lacking. In this study, we have investigated expression patterns of members of the RbcS and Lhc gene families in Arabidopsis (Arabidopsis thaliana) at the cellular level during undisturbed development and upon controlled interference of the light environment. Members of the RbcS and Lhc gene families are expressed in specialized territories, including root tip, leaf adaxial, abaxial, and epidermal domains, and with distinct chronologies, identifying successive stages of leaf mesophyll ontogeny. Defined spatial and temporal overlap of gene expression fields suggest that the light-harvesting and photosynthetic apparatus may have a different polypeptide composition in different cells and that such composition could change over time even within the same cell.
Plants are exquisitely sensitive to their light environment and have evolved sophisticated biochemical systems to perceive intensity, quality, direction, and duration of the light signal (Chen et al., 2004). Information regarding these parameters is used to modulate an amazing variety of developmental processes throughout the plant's life, thus defining light as one of the most important cues in plant development (Fankhauser and Chory, 1997; Franklin et al., 2005). Most of the events choreographed by light require changes in gene expression (Simpson and Herrera-Estrella, 1990; Thompson and White, 1991), and regulation of gene expression by light, particularly that of the genes encoding the small subunits of the Rubisco (RbcS) and the light-harvesting chlorophyll a/b-binding proteins (Lhc; previously known as Cab), has been the subject of extensive investigation (Arguello-Astorga and Herrera-Estrella, 1998).
The enzyme Rubisco is located in the stroma of the chloroplast, where it catalyzes the carboxylation of ribulose bisphosphate in the Calvin cycle and the oxygenation of the same substrate in the photorespiratory pathway (Jensen and Bahr, 1977). Rubisco is a multimeric enzyme consisting of eight small subunits and eight large subunits (Baker et al., 1975), and while the large subunits are encoded by a single chloroplast gene (Bedbrook et al., 1979), the small subunits are encoded by a family of nuclear genes (Dean et al., 1989). In Arabidopsis (Arabidopsis thaliana), the RbcS gene family comprises four members (RbcS1A, RbcS1B, RbcS2B, RbcS3B) that have been divided into two subfamilies, A and B, on the basis of linkage and sequence similarities (Krebbers et al., 1988).
Lhc proteins span the chloroplast thylakoid membrane, coordinate the binding of a number of chlorophylls and carotenoids, and, by assembling with the two photosystems, maximize and regulate light harvesting (Grossman et al., 1995). At present, 21 Lhc genes have been annotated in Arabidopsis (Jansson, 1994; Klimmek et al., 2006). The Lhca1 to Lhca4 genes encode the polypeptides of the light-harvesting complex I (LHCI) associated with the PSI (Jansson, 1994). The LHCI is composed of two heterodimers of Lhca proteins (Lhca1/4 and Lhca2/3) arranged in series (Ben-Shem et al., 2003). Lhca5 monomers or homodimers assemble with the LHCI peripherally at the Lhca2/3 site (Lucinski et al., 2006), while Lhca6 has yet to be characterized. The trimeric LHCII consists of various combinations of three very similar proteins, encoded by the Lhcb1, Lhcb2, and Lhcb3 genes (Jansson, 1994). Five Lhcb1 genes have been characterized (Leutwiler et al., 1986; McGrath et al., 1992), and while the mature proteins encoded by Lhcb1.1, Lhcb1.2, and Lhcb1.3 are identical, the Lhcb1.4 and Lhcb1.5 proteins are slightly divergent (Jansson, 1999). Three Lhcb2 genes have been identified, and the mature proteins encoded by Lhcb2.1 and Lhcb2.2 are identical, while Lhcb2.3 differs by only one amino acid (Jansson, 1999; Klimmek et al., 2006). Lhcb3 is encoded by a single gene (Jansson, 1999). The Lhcb4, Lhcb5, and Lhcb6 proteins are associated with the PSII in monomeric aggregation states (Jansson, 1994), while Lhcb7 and Lhcb8 have not yet been characterized. The two Lhcb4 genes, Lhcb4.1 and Lhcb4.2, encode proteins that display a low level of sequence identity (Jansson, 1999). Single genes encode Lhcb5, Lhcb6, Lhcb7, and Lhcb8 (Jansson, 1999; Klimmek et al., 2006).
The influence of light on regulation of RbcS and Lhc expression is exerted at multiple levels (Tobin and Silverthorne, 1985; Simpson and Herrera-Estrella, 1990; Thompson and White, 1991; Terzaghi and Cashmore, 1995). Light affects transcription of RbcS and Lhc genes, as well as the stability and translation of their transcripts. Furthermore, light has been implicated in the regulation of a variety of posttranslational processes controlling RbcS and Lhc activity, including protein folding, localization, and assembly of polypeptides into protein complexes. Because RbcS and Lhc proteins can interact within each family to form heterodimers, trimers, and higher-order complexes, the activity of the resulting entities could vary depending on their composition, should the different subunits have different biochemical properties. The composition of RbcS and Lhc protein complexes would ultimately depend on which subsets of compatible partners were simultaneously expressed in the same cells and could change under different regimes, were the single constituents differentially regulated by environmental signals. In a wide array of species, individual members of the RbcS and Lhc families have been shown to display a range of expression levels in different organs, with stereotypically strong expression in leaves, significantly lower expression in other green organs, and very low, if any, expression in organs devoid of chloroplasts (Tobin and Silverthorne, 1985; Dean et al., 1989; Simpson and Herrera-Estrella, 1990; Thompson and White, 1991). However, high-resolution expression profiles in the homologous system are available only for selected members of the RbcS and Lhc families, specific organs, and few species (e.g. Jefferson et al., 1987; Langdale et al., 1988; Silverthorne and Tobin, 1990; Bansal et al., 1992; Meier et al., 1995; Fleming et al., 1996), and are lacking for any of the RbcS and Lhc genes in Arabidopsis.
Here, we have characterized expression patterns of Arabidopsis RbcS and Lhc genes at cellular resolution during undisturbed development and upon controlled light context interference. Our data show that despite the large degree of overlap among gene expression patterns, individual members of the RbcS and Lhc families are associated with unique spatio-temporal expression profiles. Furthermore, our results suggest that expression of defined sets of RbcS and Lhc genes identifies distinct stages of leaf tissue ontogeny that cannot be distinguished anatomically. By charting a geography of the possible RbcS and Lhc protein complexes in different cells and tissues under normal and challenged development, we finally supply a platform for future functional studies aimed at dissecting the specialized and redundant roles of the members of these gene families.
RESULTS
Visualization of RbcS and Lhc Expression
Because regulation of RbcS and Lhc expression is accomplished, at least in part, at the transcriptional level (Tobin and Silverthorne, 1985; Simpson and Herrera-Estrella, 1990; Thompson and White, 1991; Terzaghi and Cashmore, 1995), we employed transcriptional reporter gene fusions to visualize RbcS and Lhc expression patterns at high resolution.
While analysis of global gene expression data for the RbcS family is not available, expression profiling has recently shown that the abundantly expressed Lhca1 to Lhca4 and Lhcb1 to Lhcb6 are coregulated in Arabidopsis, although within that cluster, Lhcb6, Lhcb4.1, Lhcb4.2, and Lhcb2.3 are expressed with a partially divergent pattern (Klimmek et al., 2006). Furthermore, the four rarely expressed Lhca5, Lhca6, Lhcb7, and Lhcb8 have been shown to display a distinct regulation profile from that of the abundantly expressed Lhc genes (Klimmek et al., 2006). Therefore, we have here sampled all the RbcS genes, seven representative members of the abundantly expressed Lhc genes, including three of the four genes displaying slightly deviant expression, and three of the four rarely expressed Lhc genes (Supplemental Table S1).
The sequence within 3 kb upstream of the translation start codon is sufficient to recapitulate the endogenous mRNA expression pattern in 80% of the cases for 44 Arabidopsis transcription factors (Lee et al., 2006). Therefore, to construct transcriptional fusions of RbcS and Lhc genes, we used approximately 3 kb of upstream noncoding sequence, whenever primer design made it possible, or the entire upstream noncoding region, whichever was shorter (Supplemental Table S1).
To improve the sensitivity and spatial resolution for detecting gene expression, all the upstream regulatory sequences generated in our study were fused to a nuclear-localized yellow fluorescent protein (YFP) that consists of a translational fusion between the coding region of histone 2A (HTA6; At5g59870) and that of the enhanced YFP (EYFP; Zhang et al., 2005). Because YFP is approximately 50% brighter than GFP and four times brighter than the cyan fluorescent protein (CFP; Dobbie et al., 2008), we reasoned that YFP should allow us to detect fusions at much lower expression levels. Furthermore, because YFP signals can be easily separated from both CFP and GFP fluorescence in plants (e.g. Kato et al., 2002; Sawchuk et al., 2007), YFP fusions should greatly expand the number of possible combinations with preexisting fluorescent marker lines and consequently vastly increase the utility of the resources generated here. Targeting all the YFP produced in each individual cell to the nucleus locally increases the concentration of YFP of 30% to 50%, resulting in enhanced sensitivity of signal detection (Joly, 2007). Furthermore, nuclear localization of YFP allows sufficient spatial separation from coexpressed plasma membrane-targeted fluorescent labels necessary to determine the shape of YFP-expressing cells (see below).
The expression profile conferred by the RbcS and Lhc regulatory sequences was characterized in transgenic Arabidopsis. For each construct, the progeny of 10 to 18 independent transgenic lines were inspected to identify the most representative expression pattern. Detailed expression analysis was performed on the progeny of three homozygous lines per construct. These representative lines were selected based on strong YFP expression, emblematic of the expression profile observed across the entire series of transgenic lines and resulting from single insertion of the transgene. In genetic crosses, the progeny of at least two independent transgenic lines per construct were examined. Attributes used to assess the representative nature of the displayed features and derived reproducibility quotients are reported in Supplemental Table S2. Higher-resolution images are provided in Supplemental Figures S1 to S7.
RbcS and Lhc Expression in Light-Grown Seedlings
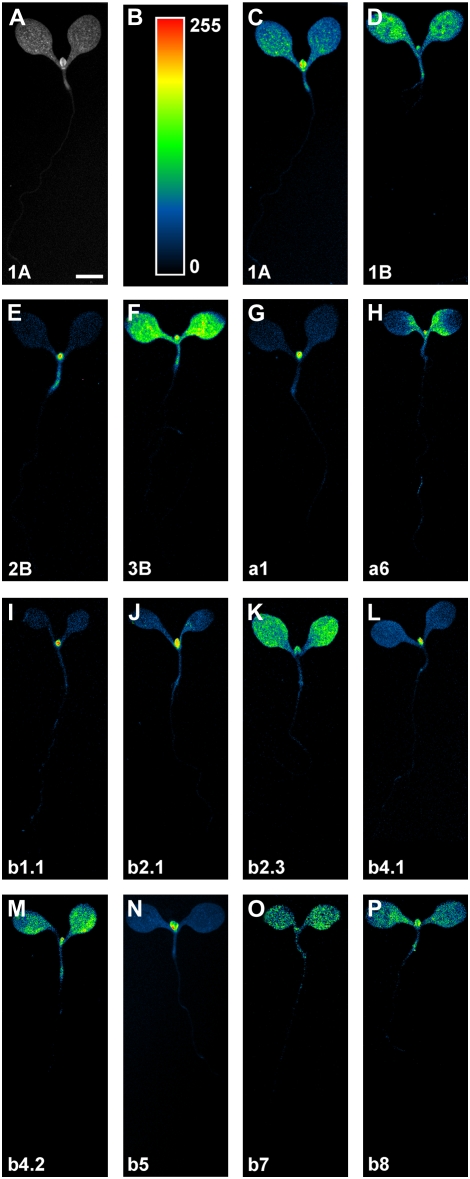
We first asked whether RbcS and Lhc genes were expressed in specific cellular domains within organs of light-grown seedlings. To address this question, we imaged RbcSpro:HTA6:EYFP and Lhcpro:HTA6:EYFP expression in whole seedlings and in cotyledons, hypocotyl, and root of seedlings 4 d after germination (DAG). Detected fluorescence signals were digitized with analog-to-digital converters that support 4096 discrimination levels, and individual images were acquired using the full extent of such resolution (see “Materials and Methods”); however, our eyes can only distinguish few tens of shades of gray (Russ, 2002). Therefore, to fully and readily convey information on fluorescence feature properties, monochrome images were color displayed with a purposely created look-up table (LUT; available as downloadable supplementary file) in which black was used to encode global background, blue to encode local background, and cyan, green, yellow, orange, and red to encode increasing signal intensities (Fig. 1, A–C).
Figure 1.
RbcS and Lhc expression in light-grown seedlings. Bottom left, Gene identity. Seedlings are displayed at identical magnification; therefore, for simplicity, a scale bar is only reported in the first image. A and C to P, Wide-field epifluorescence microscopy. A to C, A LUT in which black was used to encode global background, blue to encode local background, and cyan, green, yellow, orange, and red to encode increasing signal intensities (B) was applied to eight-bit gray-scaled images (A) to generate color-coded images (C). For additional details, see text. 1A, RbcS1A; 1B, RbcS1B; 2B, RbcS2B; 3B, RbcS3B; a1, Lhca1; a6, Lhca6; b1.1, Lhcb1.1; b2.1, Lhcb2.1; b2.3, Lhcb2.3; b4.1, Lhcb4.1; b4.2, Lhcb4.2; b5, Lhcb5; b7, Lhcb7; b8, Lhcb8. Bar: 500 μm.
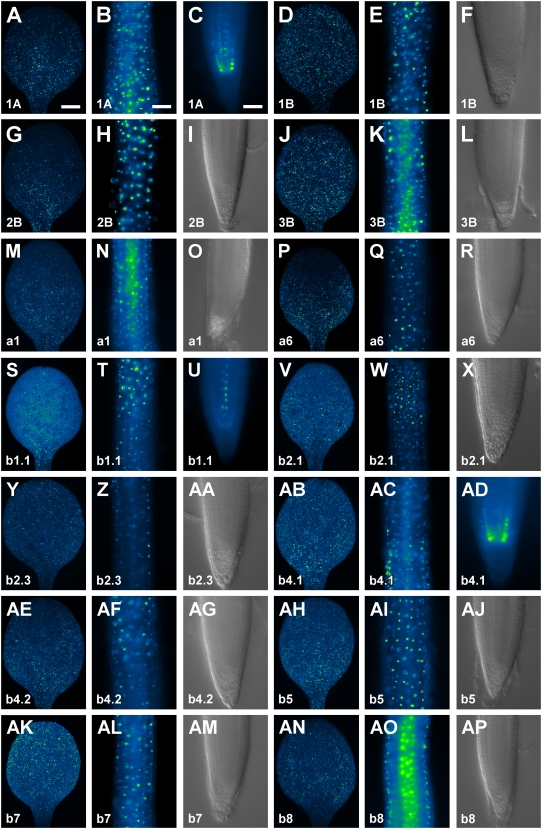
All RbcS and Lhc genes were expressed in cotyledons (Figs. 1, A and C–P, and 2, A, D, G, J, M, P, S, V, Y, AB, AE, AH, AK, and AN). However, while expression of a few genes was uniformly distributed (RbcS1B and RbcS3B) or randomly scattered (Lhcb2.1, Lhcb2.3, Lhcb4.1, and Lhcb8) throughout the entire cotyledon (Fig. 2, D, J, V, Y, AB, and AN), most of the genes showed preferential or exclusive expression to the basal region of the cotyledon (Fig. 2, A, G, M, P, S, AE, and AH), and only Lhcb7 expression was slightly more pronounced in the apical portion of the cotyledon (Fig. 2AK). In general, RbcS and Lhc genes were evenly expressed along the hypocotyl (Figs. 1, A and C–P, and 2, B, E, H, K, N, Q, T, W, Z, AC, AF, AI, AL, and AO). Finally, expression of RbcS and Lhc genes was prevalently absent in roots (Fig. 2, F, I, L, O, R, X, AA, AG, AJ, AM, and AP), but Lhcb1.1 was expressed in vascular cells (Fig. 2U), Lhcb4.1 in the endodermis (Fig. 2AD), and RbcS1A in both vascular and endodermal cells (Fig. 2C).
Figure 2.
RbcS and Lhc expression in seedling organs. In-figure information, color code, and abbreviations are as described in Figure 1. Equivalent organs are displayed at identical magnification; therefore, for simplicity, scale bars are only reported in the first image of the respective organ series. A to E, G, H, J, K, M, N, P, Q, S to W, Y, Z, AB to AF, AH, AI, AK, AL, AN, and AO, Wide-field epifluorescence microscopy. F, I, L, O, R, X, AA, AG, AJ, AM, and AP, Differential interference contrast microscopy. A, D, G, J, M, P, S, V, Y, AB, AE, AH, AK, and AN, Cotyledons, adaxial view. B, E, H, K, N, Q, T, W, Z, AC, AF, AI, AL, and AO, Hypocotyls. C, F, I, L, O, R, U, X, AA, AD, AG, AJ, AM, and AP, Root tips. Bars: A, 150 μm; B, 150 μm; C, 50 μm.
RbcS and Lhc Expression in Mature Plant Organs and Embryos
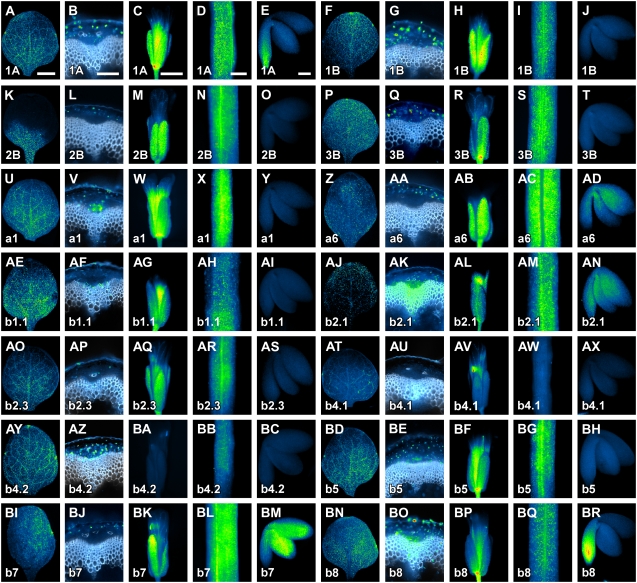
We next asked whether RbcS and Lhc genes were expressed in distinctive cellular territories of mature plant organs and embryos. To address this question, we visualized RbcSpro:HTA6:EYFP and Lhcpro:HTA6:EYFP expression in first leaves of seedlings 14 DAG, in flowers at stages 13 to 15 (Smyth et al., 1990) of plants at the onset of stage 1, at which the first silique has differentiated on the inflorescence (approximately 25 DAG; Altamura et al., 2001), and in stems, siliques, and embryos of plants at the end of stage 2, at which all flowers have differentiated into green siliques (approximately 40 DAG; Altamura et al., 2001).
In mature first leaves, most RbcS and Lhc genes were expressed profusely and without a marked bias (Fig. 3, A, F, P, U, AE, AJ, AO, AY, BD, and BN), except for RbcS2B, which showed predominant expression toward the basal portion of the leaf (Fig. 3K). Only Lhca6, Lhcb4.1, and Lhcb7 showed erratic, inconspicuous expression in mature first leaves (Fig. 3, Z, AT, and BI). With the exception of Lhcb4.1, all RbcS and Lhc genes were expressed in the mature, lowermost region of the stem (Fig. 3, B, G, L, Q, V, AA, AF, AK, AP, AU, AZ, BE, BJ, and BO). Most of the genes were expressed there ubiquitously (Fig. 3, B, G, Q, V, AF, AK, AP, AZ, BE, and BO), but expression of RbcS2B, Lhca6, and Lhcb7 was restricted to the cortex (Fig. 3, L, AA, and BJ). All RbcS and Lhc genes, with the exception of Lhcb4.2, were expressed in mature flowers (Fig. 3, C, H, M, R, W, AB, AG, AL, AQ, AV, BA, BF, BK, and BP). Most of the genes were expressed in both sepals and petals (Fig. 3, C, H, M, R, W, AG, AL, AQ, BF, BK, and BP), while Lhca6 was expressed in the sepals only (Fig. 3AB) and Lhcb4.1 was exclusively expressed in petals (Fig. 3AV). Except for Lhcb4.1, all RbcS and Lhc genes were expressed in mature green siliques (Fig. 3, D, I, N, S, X, AC, AH, AM, AR, AW, BB, BG, BL, and BQ). Finally, expression of RbcS and Lhc genes was typically absent in mature embryos (Fig. 3, J, O, T, Y, AI, AS, AX, BC, and BH), but was detected in the root tip for RbcS1A and Lhcb8 (Fig. 3, E and BR) and in the cotyledons and embryonic axis for Lhca6, Lhcb2.1, and Lhcb7 (Fig. 3, AD, AN, and BM).
Figure 3.
RbcS and Lhc expression in mature plant organs and embryos. In-figure information, color code, and abbreviations are as described in Figure 1. Equivalent organs are displayed at identical magnification; therefore, for simplicity, scale bars are only reported in the first image of the respective organ series. Wide-field epifluorescence microscopy throughout. A, F, K, P, U, Z, AE, AJ, AO, AT, AY, BD, BI, and BN, First leaves 14 DAG, abaxial view. B, G, L, Q, V, Z, AA, AF, AK, AP, AU, AZ, BE, BJ, and BO, Cross sections through the lowermost region of the stem of plants at stage 2 (Altamura et al., 2001); white, lignin autofluorescence. C, H, M, R, W, AB, AG, AL, AQ, AV, BA, BF, BK, and BP, Flowers at stages 13 to 15 (Smyth et al., 1990). D, I, N, S, X, AC, AH, AM, AR, AW, BB, BG, BL, and BQ, Mature siliques of plants at stage 2. E, J, O, T, Y, AD, AI, AN, AS, AX, BC, BH, BM, and BR, Mature embryos of plants at stage 2. Bars: A and D, 1 mm; B and E, 100 μm; C, 250 μm.
RbcS and Lhc Expression during Leaf Development
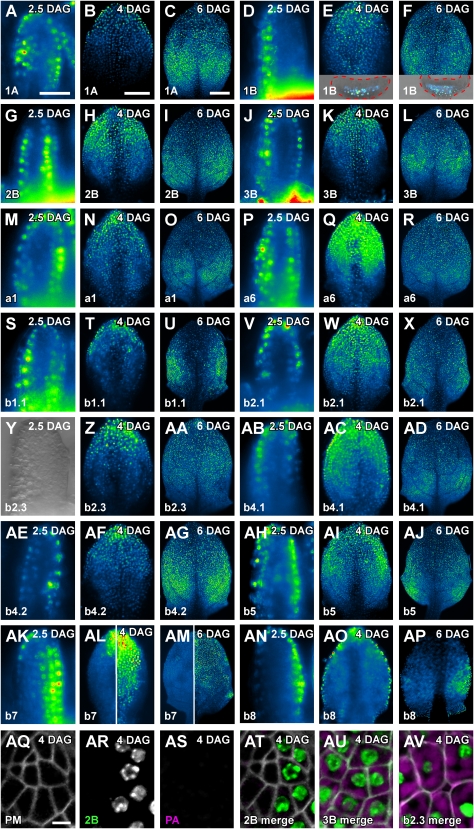
All RbcS and Lhc genes are expressed in mature foliar organs (Figs. 1–3); however, their patterns of initiation, progression, and termination, or persistence, of expression could be remarkably different, even for genes that are expressed similarly at organ maturity. To visualize dynamics of RbcS and Lhc expression over time, we monitored RbcSpro:HTA6:EYFP and Lhcpro:HTA6:EYFP expression in first leaves at 2.5, 4, and 6 DAG.
At 2.5 DAG, most of the RbcS and Lhc genes were expressed throughout the primordia with the exception of its middle region (Fig. 4, A, G, J, M, P, S, V, AB, AE, and AH). At 4 DAG, expression of a subset of those genes (RbcS2B, Lhca6, Lhcb2.1, Lhcb4.1, and Lhcb5) comprised the most apical half of the leaves (Fig. 4, H, Q, W, AC, and AI) and pervaded the entire leaves at 6 DAG (Fig. 4, I, R, X, AD, and AJ). Expression of the complementary subset of genes (RbcS1A, RbcS3B, Lhca1, Lhcb1.1, and Lhcb4.2) was restricted to the most apical one-third of the 4-DAG leaves (Fig. 4, B, K, N, T, and AF), and by 6 DAG, expression encompassed the apical two-thirds of the leaves (Fig. 4, C, L, O, U, and AG). While most of the RbcS and Lhc genes were therefore expressed in largely overlapping spatial domains and with strikingly comparable temporal dynamics, a few RbcS and Lhc genes were expressed with conspicuously distinct patterns. Lhcb2.3 expression was absent in 2.5-DAG primordia, was constrained to the leaf tip at 4 DAG, and embraced the most apical one-half of 6-DAG leaves (Fig. 4, Y–AA). RbcS1B was expressed with an unequivocal preference for the abaxial side of 2.5-DAG primordia, and expression remained confined to the abaxial side of 4- and 6-DAG leaves (Fig. 4, D–F; Supplemental Fig. S8). In contrast, Lhcb7 showed pronounced adaxial expression throughout leaf development, and expression at the abaxial side was largely circumscribed to the leaf tip (Fig. 4, AK–AM). Finally, Lhcb8 was expressed with a prominent epidermal bias at all stages of leaf development (Fig. 4, AN–AP).
Figure 4.
RbcS and Lhc expression during leaf development. In-figure information, color code, and abbreviations are as described in Figure 1. Top right: leaf primordium age in DAG. Leaf primordia at equivalent DAG are displayed at identical magnification; therefore, for simplicity, scale bars are only reported in the first image of the respective DAG series. A to X and Z to AP, Wide-field epifluorescence microscopy. Y, Differential interference contrast microscopy. AQ to AV, Confocal laser scanning microscopy. A, D, G, J, M, P, S, V, Y, AB, AE, AH, AK, and AN, Lateral view (adaxial side of primordia to the right). B, C, E, F, H, I, K, L, N, O, Q, R, T, U, W, X, Z, AA, AC, AD, AF, AG, AI, AJ, AL (left side), AM (left side), and AO to AV, Abaxial view. AL (right side) and AM (right side), Adaxial view. Insets in E and F, Overlay between wide-field epifluorescent and differential interference contrast microscopy images of cross sections through the middle of 4- and 6-DAG leaves, respectively; red dashed line, primordium outline; adaxial side toward top. AQ, Plasma membrane-labeling UBQ10pro:EGFP:LTi6b expression. AR, RbcS2Bpro:HTA6:EYFP expression. AS, Plastid autofluorescence. AT, Overlay of images in AQ-AS; note that cells expressing RbcS2Bpro:HTA6:EYFP cannot be anatomically discriminated from neighboring cells. AU, Overlay of images of UBQ10pro:EGFP:LTi6b and RbcS3Bpro:HTA6:EYFP expression and of plastid autofluorescence. AV, Overlay of images of UBQ10pro:EGFP:LTi6b and Lhcb2.3pro:HTA6:EYFP expression and of plastid autofluorescence. Images of plastid autofluorescence were all acquired at identical settings (see “Materials and Methods” for details). PA, Plastid autofluorescence; PM, Plasma membrane. Bars: A and B, 50 μm; C, 200 μm; AQ, 5 μm.
We next asked whether inception of RbcS and Lhc expression could be assigned to specific stages of leaf subepidermal cell development. We selected expression of RbcS2B, RbcS3B, and Lhcb2.3 as representatives of three different chronologies of gene expression (Fig. 4, A–AP), and adopted the combination of cell morphology, extent of intercellular spaces, and level of plastid intrinsic fluorescence as a stage-specific indicator of leaf subepidermal cell differentiation (Pyke and Page, 1998; Scarpella et al., 2004; Kang et al., 2007). We visualized onset of RbcS2Bpro:HTA6:EYFP, RbcS3Bpro:HTA6:EYFP, and Lhcb2.3pro:HTA6:EYFP expression in 4-DAG leaves of the plasma membrane marker line UBQ10pro:EGFP:LTi6b. RbcS2B and RbcS3B were first expressed in tightly connected polygonal cells (Fig. 4, AQ, AR, AT, and AU); however, unlike RbcS2B, onset of RbcS3B expression was associated with presence of weakly autofluorescent plastids (Fig. 4, AS–AU). Finally, initiation of Lhcb2.3 expression occurred in expanded cells that had acquired a distinctively round shape, that contained large numbers of strongly autofluorescent plastids along the cell surface, and that were separated by conspicuous intercellular spaces (Fig. 4AV).
Cell and Tissue-Specific Expression of RbcS and Lhc Genes in Foliar Organs
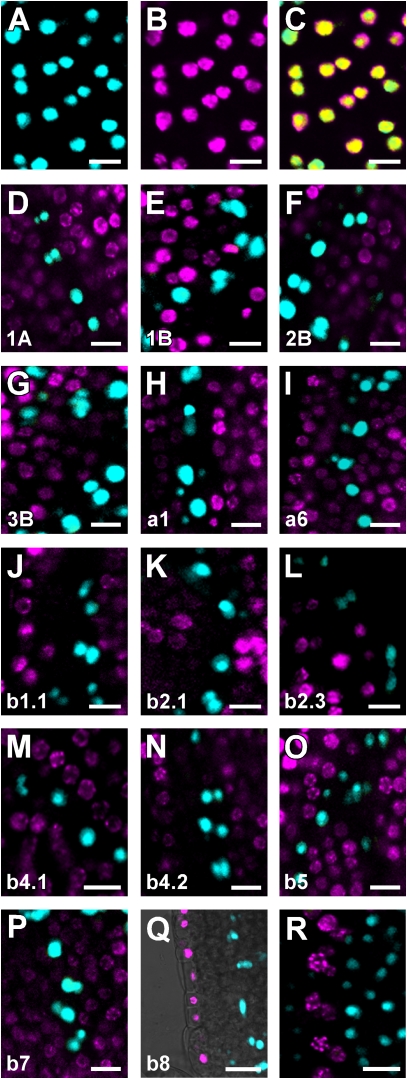
Despite their different dynamics during leaf development, RbcS and Lhc expression seemed to be excluded from leaf veins (Fig. 4). An unambiguous criterion to test such a hypothesis would be to visualize expression of RbcS and Lhc genes relative to that of the early vascular marker gene Athb8 (Baima et al., 1995). Onset of Athb8 gene expression in leaves labels a subset of ground cells that have been specified to vascular cell fate (i.e. preprocambial cells) but that are morphologically indistinguishable from the residual ground cell population (Kang and Dengler, 2004; Scarpella et al., 2004). We therefore rigorously assessed the degree of colocalization between RbcSpro:HTA6:EYFP and Lhcpro:HTA6:EYFP on the one hand and Athb8pro:ECFP:3xNLS on the other. Because of the differential sensitivity of our perception across the visible spectrum (Russ, 2002), to improve discrimination of fluorescence signals in double-labeling images and therefore more accurately visualize signal colocalization, we adopted an extended dual-channel LUT from cyan to magenta through green, yellow, and red (Demandolx and Davoust, 1997). Fluorescence in each detection channel was displayed in either cyan (e.g. Fig. 5A) or magenta (e.g. Fig. 5B). Single-fluorophore images were then merged using a differential operator (see “Materials and Methods”). As a result, preponderance of cyan signal over colocalized magenta signal is encoded in green, opposite in red, and colocalized cyan and magenta signals of equal intensity in yellow (e.g. Fig. 5C). As shown in Figure 5, none of the RbcS and Lhc genes was expressed in Athb8-labeled cells, suggesting that RbcS and Lhc gene expression is excluded from vascular cells, already at the preprocambial stage. To test for possible artifacts induced by fluorophore intrinsic properties (e.g. different maturation time and stability of HTA6:EYFP and ECFP:3xNLS) or detection parameters (e.g. suboptimal excitation wavelength and emission interval), we exchanged fluorophores for the RbcS2B and Athb8 gene expression combination and measured extent of colocalization between RbcS2Bpro:ECFP:3xNLS and Athb8pro:HTA6:EYFP signals (Fig. 5R). The absence of any overlap of fluorescence in reciprocal permutations of RbcS2B and Athb8 regulatory regions with YFP and CFP (compare Fig. 5, F and R) suggests that our colocalization data are fluorophore independent, thus further supporting that RbcS and Lhc genes are not expressed in leaf preprocambial cells.
Figure 5.
RbcS and Lhc expression in leaf subepidermal cells. In-figure information and abbreviations are as described in Figure 1. A to P and R, Confocal laser scanning microscopy images of first leaves 4 DAG; leaf tips toward top. Q, Overlay of confocal laser scanning microscopy and transmitted light images. A, RbcS2Bpro:ECFP:3xNLS expression. B, RbcS2Bpro:HTA6:EYFP expression. C, Overlay of images in A and B. C to R, Images color-coded with a dual-channel LUT from cyan to magenta through green, yellow, and red (Demandolx and Davoust, 1997). Fluorescence in each detection channel was displayed in either cyan or magenta. Single-fluorophore images were then merged using a differential operator. As a result, preponderance of cyan signal over colocalized magenta signal is encoded in green, opposite in red, and colocalized cyan and magenta signals of equal intensity in yellow. For additional details, see text. D to Q, Cyan, Athb8pro:ECFP:3xNLS; magenta, RbcSpro:HTA6:EYFP or Lhcpro:HTA6:EYFP as indicated by gene identifier at bottom left. R, Cyan, RbcS2Bpro:ECFP:3xNLS; magenta, Athb8pro:HTA6:EYFP. Bars = 10 μm.
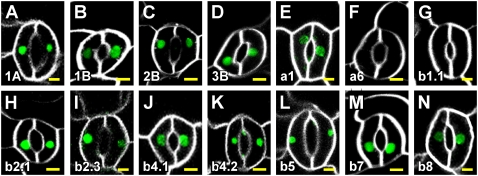
While Lhcb8 was exclusively expressed in epidermal cells of the leaf (Figs. 4, AN–AP, and 5Q), we asked whether expression of RbcS genes and of all other Lhc genes was confined to subepidermal, nonvascular cells. To address this question, we visualized RbcSpro:HTA6:EYFP and Lhcpro:HTA6:EYFP expression in epidermis of 4-DAG cotyledons in which plasma membranes were labeled by the FM 4-64 water-soluble lipophilic dye to expose cell shape. As shown in Figure 6, all genes, with the sole exception of Lhca6 and Lhcb1.1, were expressed in guard cells of stomata.
Figure 6.
RbcS and Lhc expression in cotyledon epidermal cells. In-figure information and abbreviations are as described in Figure 1. Confocal laser scanning microscopy images of adaxial epidermis of cotyledons at 4 DAG. Green, RbcSpro:HTA6:EYFP or Lhcpro:HTA6:EYFP expression as indicated by gene identifier at bottom left; white, FM 4-64-decorated plasma membrane. Bars = 5 μm.
Response of RbcS and Lhc Expression to Different Light Conditions
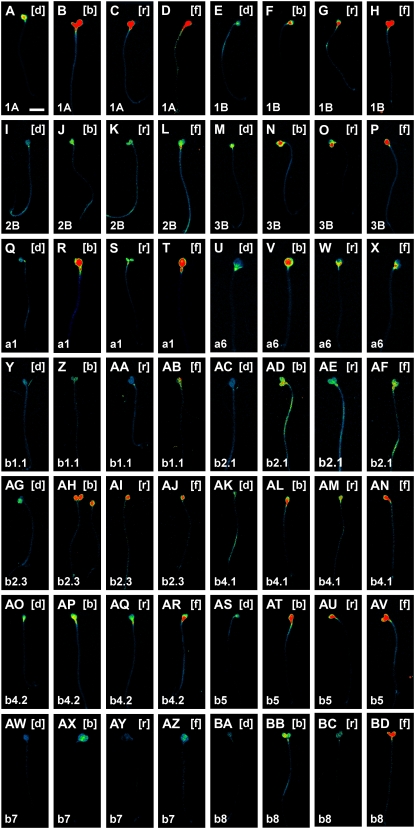
Levels of RbcS and Lhc expression are strongly affected by light signals (Tobin and Silverthorne, 1985; Simpson and Herrera-Estrella, 1990; Thompson and White, 1991; Terzaghi and Cashmore, 1995; Arguello-Astorga and Herrera-Estrella, 1998). We therefore finally asked whether light cues also impinged on the spatial patterns of RbcS and Lhc expression. To address this question, we grew seedlings for 3 d in the dark and either transferred them to blue, red, or far-red light conditions (see “Materials and Methods”) or continued to grow them in the dark. We then visualized RbcSpro:HTA6:EYFP and Lhcpro:HTA6:EYFP expression at 4 DAG.
While elevation of Lhcb7 expression was selectively elicited by blue and far-red light treatments, enhanced expression of all other RbcS and Lhc genes was induced by light, although at different levels, irrespective of its spectrum range (Fig. 7). However, responsiveness of gene expression to light was constrained by organ-specific cues. In fact, while induction of expression for most genes was restricted to cotyledons (Fig. 7, E–L, Q–T, Y–AB, AG–AN, and AW–AZ), a few genes (RbcS1A, Lhcb2.1, and Lhcb4.2) displayed light-stimulated enhancement of expression in both cotyledons and hypocotyls (Fig. 7, A–D, AC–AF, and AO–AR). Furthermore, expression of RbcS3B, Lhca6, Lhcb5, and Lhcb8 was induced in both cotyledons and hypocotyls in blue and far-red light but solely in the cotyledons in red light (Fig. 7, M–P, U–X, AS–AV, and BA–BD). Finally, up-regulation of RbcS and Lhc gene expression was not observed in the root (not shown).
Figure 7.
Response of RbcS and Lhc expression to different light conditions. In-figure information, color code, and abbreviations are as described in Figure 1. Top right, Light treatment. Seedlings are displayed at identical magnification; therefore, for simplicity, scale bars are only reported in the first image. Wide-field epifluorescence microscopy throughout. [b], Seedlings grown for 1 d in blue light following 3 d of growth in the dark; [d], seedlings grown for 4 d in the dark; [f], seedlings grown for 1 d in far-red light following 3 d of growth in the dark; [r], seedlings grown for 1 d in red light following 3 d of growth in the dark. For additional details, see text. Bar = 2 mm.
DISCUSSION
Global gene expression profiling provides invaluable information on gene expression levels and their regulation at whole-genome scale, but is typically characterized by a limited degree of discrimination in the spatial dimension (Kehr, 2003; Brandt, 2005; Lange, 2005; Lee et al., 2005). High-resolution gene expression analysis is therefore the perfect complement to ongoing genome-wide approaches to the study of transcript accumulation patterns. In this study, we have investigated expression patterns of members of the RbcS and Lhc photosynthesis-associated nuclear gene families in Arabidopsis at the cellular level during undisturbed development and upon controlled light context interference. Transcription of RbcS and Lhc genes could be modulated by regions other than the upstream regulatory sequences used in our study (e.g. Ali and Taylor, 2001). Furthermore, abundance of RbcS and Lhc transcripts can be regulated at the posttranscriptional level (Tobin and Silverthorne, 1985; Simpson and Herrera-Estrella, 1990; Thompson and White, 1991). However, our results are in good agreement with Arabidopsis RbcS and Lhc expression profiles reported previously (Dedonder et al., 1993; Klimmek et al., 2006) or extracted from publicly accessible large-scale microarray data sets (Birnbaum et al., 2003; Leonhardt et al., 2004; Schmid et al., 2005; Suh et al., 2005; Winter et al., 2007; Supplemental Table S3; Supplemental Fig. S9; and below), suggesting that, at least in Arabidopsis, cellular dynamics of RbcS and Lhc expression are largely determined by transcription patterns. On the other hand, growth conditions, developmental stages, and spatial resolution in available microarray experiments were often very different from those in our study. Therefore, similarity in expression data should still be interpreted with caution. Because RbcS and Lhc expression is additionally subject to extensive translational and posttranslational regulation (Tobin and Silverthorne, 1985; Thompson and White, 1991), patterns of gene transcript abundance alone may not necessarily reflect levels of active RbcS and Lhc proteins. However, to our knowledge, none of the reported translational or posttranslational control mechanisms has been shown to impinge on spatio-temporal profiles of RbcS and Lhc expression, suggesting that cellular patterns of their expression in Arabidopsis development may be accurately visualized by transcriptional fusions. While no expression profiling, at any level or type of resolution, can ever provide direct evidence of gene function, our topography of RbcS and Lhc expression nonetheless allows us to infer some of their less prominent biological properties.
Uniqueness and Redundancy among RbcS and Lhc Expression Patterns: Structural and Functional Implications
A major discovery of the Arabidopsis genome sequencing project was the finding that a surprisingly large number of genes encode isoforms of the same polypeptide (The Arabidopsis Genome Initiative, 2000). Large-scale genome duplication events are a common feature of plant genomes and are mainly responsible for the large number of duplicated individual loci (Bowers et al., 2003; Langham et al., 2004; Paterson et al., 2004; Maere et al., 2005). Duplicate genes may acquire advantageous mutations that become subject to selection and lead to a new function; alternatively, both the ancestral and duplicated gene can accumulate mutations that may lead to the subdivision of the functions of the ancestral gene. These processes of neo- and subfunctionalization occur mainly through mutations in regulatory sequences, rather than mutations in the coding sequence (Blanc and Wolfe, 2004; Haberer et al., 2004; Langham et al., 2004; Wang et al., 2004; Casneuf et al., 2006). If each individual member of a given gene family supplies a set of nonredundant functions, as genetic evidence (Briggs et al., 2006) and evolutionary considerations (Nowak et al., 1997) seem to suggest, diversified expression of gene family members would then provide metabolic diversity and flexibility to individual cells. The physiological fine-tuning of the cell would be further enhanced if protein family members cooperated in multimeric complexes of variable composition. RbcS and Lhc proteins can interact with members of the same family to form heterodimers, trimers, and higher-order complexes (Baker et al., 1975; Jansson, 1994; Ben-Shem et al., 2003; Lucinski et al., 2006). However, the existence of overlapping expression among members of these gene families is a prerequisite for such type of interaction to occur in vivo. Our findings show that, in Arabidopsis, members of the RbcS and Lhc families are expressed in distinct, yet redundant, spatio-temporal domains, suggesting that the light-harvesting and photosynthetic apparatus may have a different polypeptide composition in different cells and that such composition could change over time even within the same cell. Aggregation status of Lhca6, Lhcb7, and Lhcb8 within the LHCs is yet to be determined, but our results suggest that samples other than mature leaves, the materials typically used in biochemical studies of photosystems, will have to be used to address such a question. In fact, Lhca6 is expressed only transiently in leaf development and displays inconspicuous, erratic expression in mature leaves, while expression of Lhcb7 and Lhcb8 is restricted to a small subpopulation of cells throughout leaf development.
The precise biological function of each member of the RbcS and Lhc families will eventually have to be defined by single or higher-order loss-of-function mutations, but our results can already serve as a baseline to explore the biological consequences of mutations in these genes. RbcS1A, but none of the other RbcS genes, is expressed in root tips. Because the RbcL gene, which encodes the large, catalytic subunit of Rubisco, is also expressed in root tips (Isono et al., 1997), a functional isoform of the multimeric enzyme may be present in these tissues. Two additional observations in our study support that root tip expression of RbcS1A (and RbcL) do not represent spurious gene activities, but may be related to yet-to-be-uncovered functions for photosynthetic genes in the root. First, two members of the Lhc family, Lhcb1.1 and Lhcb4.1, are also expressed in the root tip. Second, not only are Lhcb1.1 and Lhcb4.1 expressed in these tissues, but their cumulative root tip expression domain surprisingly matches that of RbcS1A. Global gene expression profiling revealed low levels of expression of Lhcb4.1 in the root tip (Birnbaum et al., 2003; Supplemental Table S3). Presence of RbcS and Lhcb1 transcripts was also detected (Supplemental Table S3), but their identity could not unambiguously be ascertained because, in the ATH1 GeneChip, the same Affymetrix locus identifier is assigned to all the RbcS genes on the one hand and to Lhcb1.1, Lhcb1.2, and Lhcb1.3, collectively, on the other. Finally, more recent proteomics efforts have identified RbcL, RbcS1A, and Lhcb4.1 polypeptides in the Arabidopsis root (Baerenfaller et al., 2008), suggesting that in Arabidopsis Rbc and Lhc gene expression data largely mirror protein accumulation profiles. Because RbcS and Lhc activity is subject to extensive control at the posttranslational level (Tobin and Silverthorne, 1985; Thompson and White, 1991), even patterns of protein abundance alone may, however, not necessarily reflect levels of functional RbcS and Lhc proteins. While the significance of the expression of photosynthesis-related genes and proteins in what are generally perceived as photosyntetically inactive tissues still eludes us, our findings suggest a means to uncover the unique biological roles of RbcS1A, Lhcb1.1, and Lhcb4.1. Extensive overlap of gene expression in most plant tissues may preclude dissecting the function of these genes in single mutants, but focusing phenotypic analysis on the root tip could provide the possibility to unequivocally assign a nonredundant function.
Uniqueness and Redundancy among RbcS and Lhc Expression Patterns: Gene Expression and Developmental Ramifications
Transcriptomes from different plants or organs at different physiological states have generated a vast amount of information that can be monitored and compared. However, data from gene expression profiling of whole plants or entire organs have to be interpreted with caution, as genes that appear to be sporadically expressed at the organ level may be expressed at high levels in very small cellular fields (Kehr, 2003; Brandt, 2005; Lange, 2005; Lee et al., 2005). Furthermore, if only mature stages of organ development are sampled for transcript analysis, ephemeral gene expression may be underestimated or entirely missed. Previous studies identified Lhca6, Lhcb7, and Lhcb8 as being rarely expressed in Arabidopsis (Jansson, 1999; Klimmek et al., 2006), a finding that is supported by the extremely low hybridization signal associated with these genes in available microarray data sets (Supplemental Table S3); however, in our study, Lhca6, Lhcb7, and Lhcb8 were expressed at levels comparable to those of the abundantly expressed genes Lhca1 and Lhcb1.1 (Figs. 1–7; Klimmek et al., 2006). The apparent discrepancy vanishes if one considers that Lhca6, Lhcb7, and Lhcb8 are always expressed in a subset of cells in each individual organ. Furthermore, the proportion of cells expressing these genes decreases during organ development, such that at organ maturity, the stage typically sampled in microarray experiments (Supplemental Table S3), expression of Lhca6, Lhcb7, and Lhcb8 is maintained in very small cellular domains.
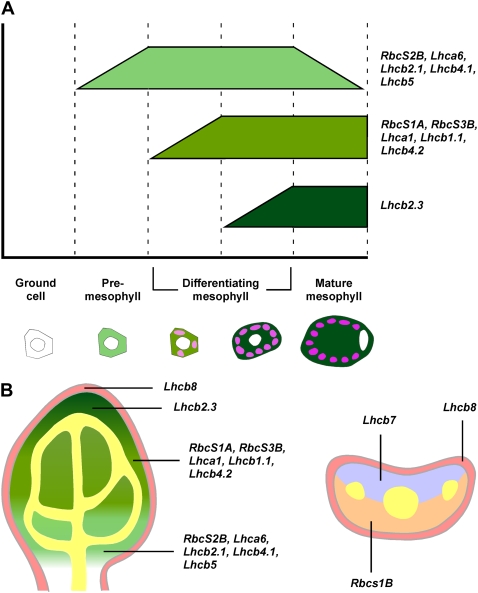
Technologies aimed at characterizing the population of genes that are transcribed in individual cell types, such as fluorescence-activated cell sorting (Birnbaum et al., 2005), are enormously facilitated by, or completely rely on, the availability of cell type-specific markers. Gene expression profiles selectively labeling distinct cell types are already available and have been indispensable to the identification of the gene expression complement of a variety of cells and tissues through single cell-type sampling (Birnbaum et al., 2003; Nawy et al., 2005). However, lack of markers for mesophyll has hampered the extension of those approaches to the characterization of the cohort of genes that molecularly define this tissue. Further, cell state-specific markers have been crucial for the characterization of tissue ontogeny, especially of steps preceding acquisition of morphological conspicuity (e.g. Malamy and Benfey, 1997; Scarpella et al., 2004; Gordon et al., 2007). While the physiological and biochemical properties of mesophyll cells have been the focus of investigation for the past 300 years (for review, see Govindjee et al., 2006), details of their development are largely unexplored, especially in C3 plants such as Arabidopsis (Pyke and Lopez-Juez, 1999). During leaf development, most of the RbcS and Lhc genes were expressed in essentially overlapping subepidermal nonvascular domains and with amazingly comparable dynamics, which only seemed to differ for the temporal aspects of the initiation and termination of their expression (Fig. 8). We identified a collection of genes whose onset of expression labeled initial steps of subepidermal tissue ontogeny (RbcS2B, Lhca6, Lhcb2.1, Lhcb4.1, and Lhcb5), a set of genes that became expressed at early stages of mesophyll development (RbcS1A, RbcS3B, Lhca1, Lhcb1.1, and Lhcb4.2), and a late marker of mesophyll differentiation (Lhcb2.3). Subepidermal cells that initiate expression of genes belonging to the two latter groups (RbcS1A, RbcS3B, Lhca1, Lhcb1.1, Lhcb4.2, and Lhcb2.3) can be discriminated because of accumulation of autofluorescent plastids, acquisition of round cell shape, presence of intercellular spaces, or a combination of those features. However, onset of expression of genes of the first cluster (RbcS2B, Lhca6, Lhcb2.1, Lhcb4.1, and Lhcb5) identifies a distinct cell state within a population of anatomically indistinguishable ground cells. Sustained levels of expression of this set of genes overlap with initiation of expression of RbcS and Lhc genes marking onset of mesophyll anatomical differentiation (Fig. 8A). Further, cells that express RbcS and Lhc genes fail to express the preprocambial marker gene Athb8, which labels ground cells specified to a vascular fate. Therefore, we conclude that expression of RbcS and Lhc genes of the first group identifies a “premesophyll” cell state that cannot be distinguished anatomically. Mutual exclusivity of premesophyll and preprocambial states supports the view that mesophyll and vascular cell identity acquisition represent antagonistic pathways in leaf subepidermal tissue ontogeny (Scarpella et al., 2004; Kang et al., 2007; Sawchuk et al., 2008). While no single regulatory element is unique to the upstream noncoding regions of the genes belonging to each of the three groups of mesophyll developmental stage-specific markers, each of these cohorts of genes is characterized by the presence of a distinct array of overrepresented elements in their upstream sequences (Supplemental Table S4). Regulatory regions of premesophyll genes are characterized by overrepresentation of the Tbox required for promoter activity of genes that are exclusively expressed in green organs (Yang et al., 1993; Kwon et al., 1994; Chan et al., 2001). Further, overrepresentation of the I-box, which has been suggested to mediate photosynthetic tissue-specific expression (Martinez-Hernandez et al., 2002; Maclean et al., 2008), is unique to upstream sequences of genes labeling intermediate stages of leaf mesophyll development. Finally, the Lhcb2.3 upstream noncoding region is distinguished by overrepresentation of the Box II necessary for mesophyll-specific expression (Nishiuchi et al., 1995). Although each of these regulatory elements has been implicated in expression in green tissues, their impact on stage-specific expression in leaf mesophyll ontogeny remains to be addressed experimentally.
Figure 8.
Schematic summary of RbcS and Lhc expression in leaf development. A, Graphic representation of temporal expression profiles and associated stages of leaf mesophyll cell development. Green bars indicate the duration of maximum expression of each cohort of genes; ramped termini denote gradual initiation or decline of gene expression. B, Tissue map of a paradermal (left) or a transverse (right) median section through a developing leaf illustrating fields of gene expression. Different shades of green indicate consecutive onset of gene expression and corresponding stages of mesophyll ontogeny as defined in A. Orange, abaxial side; pink, epidermis; purple, adaxial side; yellow, preprocambial and procambial cells as visualized through Athb8 expression (Baima et al., 1995; Kang and Dengler, 2004; Scarpella et al., 2004).
In our study, adaxial expression of Lhcb7 and abaxial expression of RbcS1B label cells in the leaf that will give rise to palisade and spongy mesophyll, respectively, before any overt anatomical differentiation has taken place (Pyke et al., 1991). Because the adaxial side of the leaf is that which is exposed to the highest light intensity, it is not immediate to reconcile the expression pattern of Lhcb7 in the leaf with the protein's predicted light-harvesting function in light-limiting conditions. Alternatively, Lhcb7 could function in nonphotochemical quenching to dissipate energy under conditions where the absorbed light exceeds the electron transfer capacities of the thylakoid complexes contributing to primary photochemistry (Szabo et al., 2005), and a similar function has indeed already been suggested for Lhcb4, Lhcb5, and Lhcb6 (Farber et al., 1997; Dall'Osto et al., 2005). Carbon dioxide fixation is proportional to light intensity, but Rubisco content and carbon fixation at the abaxial and adaxial surfaces of the leaf are comparable (Nishio et al., 1993; Sun and Nishio, 2001). This suggests the presence of Rubisco isoforms at the abaxial side that are more efficient in carbon fixation at light-limiting conditions than the isoforms existing at the adaxial side. RbcS1B is the only RbcS gene to be exclusively expressed at the abaxial side of the leaf, which suggests that RbcS1B could modify the catalytic properties of Rubisco and increases its enzymatic efficiency in light-limiting conditions. Alternatively, because cells of the spongy mesophyll are separated by extensive intercellular spaces, comparable levels of Rubisco at the abaxial and adaxial surfaces could reflect a higher content of Rubisco per cell in the spongy mesophyll. Because the amount of small Rubisco subunits regulates large subunit expression (Khrebtukova and Spreitzer, 1996; Rodermel et al., 1996), additional RbcS1B at the abaxial side of the leaf could simply be part of a mechanism for regulating Rubisco abundance. The precise assignment of a function to RbcS1B, and to RbcS genes in general, will, however, have to await the development of methods to quantify the presence or proportion of different small Rubisco subunits in the holoenzyme. Irrespective of the specific role of RbcS1B, its upstream noncoding region, but not that of any of the other genes in our study, contains the AACGGGTGAA sequence required for suppression of expression in the adaxial domain of the leaf primordium (Supplemental Table S4; Watanabe and Okada, 2003).
Except for RbcS2B and Lhca6, whose expression across the entire plant is predominantly or exclusively associated with subepidermal, photosynthetic cells, each of the other genes has additional, conspicuous expression domains in nonmesophyll cells in organs other than the leaf. While it will be interesting to understand the molecular basis of the nonmesophyll expression domains, all the markers can already begin to assist in the spatial and temporal dissection of leaf mesophyll ontogeny, and expression of RbcS2B and Lhca6 can be used to readily visualize and molecularly characterize mesophyll cells and their development throughout the plant.
MATERIALS AND METHODS
Vector Construction
The origins of the Athb8pro:ECFP:3xNLS and the Athb8pro:HTA6:EYFP lines have been previously described (Sawchuk et al., 2007). To generate the RbcSpro:HTA6:EYFP and Lhcpro:HTA6:EYFP lines, sequences upstream of RbcS or Lhc coding regions, respectively, were amplified from Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) genomic DNA using Finnzymes Phusion high-fidelity DNA polymerase (New England BioLabs) and gene-specific primers (Supplemental Table S1), integrated into pDONR221(Invitrogen) with BP clonase II (Invitrogen), sequence-checked, and recombined into the Gateway-adapted pFYTAG binary vector (Zhang et al., 2005) using LR clonase II (Invitrogen). The RbcS2Bpro:ECFP:3xNLS line was generated by recombining the pDONR221-integrated RbcS2B upstream sequences into the Gateway-adapted pBGCN binary vector (Kubo et al., 2005) with LR clonase II (Invitrogen). To generate the UBQ10pro:EGFP:LTi6b line, the EGFP:LTi6b sequence was amplified from genomic DNA of a Ubi3pro:EGFP:LTi6b line (Vidaurre et al., 2007; a generous gift of M. Aida and B. Scheres) with the EYFP KpnI Forw (TATGGTACCATGGTGAGCAAGGGCGAG) and pEGAD SacI Rev (CCCGAGCTCAATAAATTCCTCACATAAACCAACG) primers using Finnzymes Phusion high-fidelity DNA polymerase (New England BioLabs), subcloned, sequence-checked, and cloned into a derivative of the pC1300-PolyA binary vector (a kind gift of J. Mathur) to give rise to pC1300-EGFP:LTi6b. Approximately 1.5 kb of sequences upstream of the UBQ10 (At4g05320) coding sequence were amplified from Arabidopsis (ecotype Col-0) genomic DNA with the UBQ10 HindIII Forw (CTCAAGCTTTCCCATGTTTCTCGTCTGTC) and the UBQ10 SmaI Rev (CGACCCGGGCTGTTAATCAGAAAAACTCAG) primers using Finnzymes Phusion high-fidelity DNA polymerase (New England BioLabs), subcloned, sequence-checked, and cloned into pC1300-EGFP:LTi6b to give rise to UBQ10pro:EGFP:LTi6b.
Plant Material, Transformation, and Growth Conditions
In all experiments, seeds were surface sterilized, synchronized, and germinated on growth medium (half-strength Murashige and Skoog salts [Sigma-Aldrich], 15 g L−1 Suc [Fisher Scientific], 0.5 g L−1 MES [Sigma Chemical], 0.8% [w/v] agar [Bioshop Canada], pH 5.7) at a density of 1 seed cm−2 as previously described (Scarpella et al., 2004). Sealed plates were incubated at 25°C under continuous fluorescent light (blue450:red633:far-red740 12:22:3 μmol m−2 s−1). We define DAG as days following exposure of imbibed seeds to light. For analysis of gene expression under controlled light conditions, germination was induced by incubating sealed plates at 25°C under fluorescent light (blue450:red633:far-red740 12:22:3 μmol m−2 s−1) for 2 h. Plates were then wrapped in aluminum foil and incubated at a 70° angle at 25°C in the dark. At 3 DAG, plates were either incubated at 25°C in the dark for an additional 24-h period or unwrapped and incubated at 25°C for 24 h under blue (450 nm: 19 μmol m−2 s−1), red (633 nm: 11 μmol m−2 s−1), or far-red (740 nm: 7 μmol m−2 s−1) light provided by 470-nm, 633-nm, or 731-nm A19 DecorLED 270° solid-state lamps (Ledtronics), respectively. Seedlings were transferred at 5 DAG to Promix BX soil (Evergro/Westgro) in 7- × 7- × 8-cm pots at a density of 0.1 seedlings cm−2 and grown at 22°C under fluorescent light (blue450:red633:far-red740 14:88:6 μmol m−2 s−1) in a 16-h-light/8-h-dark cycle. Arabidopsis (ecotype Col-0) was transformed with Agrobacterium tumefaciens strain GV3101∷MP90 (Koncz and Schell, 1986) harboring the RbcSpro:HTA6:EYFP, Lhcpro:HTA6:EYFP, or RbcS2Bpro:ECFP:3xNLS constructs by the floral dip method (Clough and Bent, 1998). Primary transformants were selected on growth medium supplemented with 200 μg mL−1 carbenicillin (Bioshop Canada), 10 μg mL−1 glufosinate ammonium (Crescent Chemical), and 50 μg mL−1 nystatin (Bioshop Canada).
Microtechniques and Microscopy
Whole seedlings, dissected seedling organs, embryos, and stem hand-sections were mounted in water with a 0.17-mm coverslip (Fisher Scientific; VWR). Dissected silique and flower samples were imaged in plastic dishes with water-submerged pedicels. Seedlings, flowers, siliques, and embryos were viewed with a 1× Planapochromat (NA, 0.041; WD, 55 mm) objective of a Leica MZ 16FA stereomicroscope (Leica Microsystems) equipped with an HBO103 mercury vapor short-arc lamp (Osram), and images were captured with an Andor iXonEM+ camera (Andor Technology). YFP was visualized using a 500/20 excitation filter and a 535/30 emission filter (Leica Microsystems). Seedling organs and stem sections were viewed with a 5× Fluar (NA, 0.25; WD, 12.5 mm), 10× Planapochromat (NA, 0.45; WD, 2.0 mm), 20× Planapochromat (NA, 0.8; WD, 0.55 mm), or 40× Planapochromat (NA, 0.95; WD, 0.25 mm) objective of an Axio Imager.M1 microscope (Carl Zeiss) equipped with a Hamamatsu ORCA-AG camera (Hamamatsu Photonics). YFP was visualized with a 50%-attenuated HBO103 mercury vapor short-arc lamp (Osram) using a BP 500/20 excitation filter, an FT515 beam splitter, and a BP 535/30 emission filter (Carl Zeiss). Lignin autofluorescence was visualized using a BP 390/22 excitation filter, an FT420 beam splitter, and a BP 460/50 emission filter (Carl Zeiss). Samples were exposed for 0.05 to 1 s during image acquisition, because longer exposure times induce physiological damage, mitotic arrest, and cell death (Dixit and Cyr, 2003). Electron multiplication gain or output amplifier gain values were set to match the incoming signal with the input range of the camera analog-to-digital converter (Nordberg and Sluder, 2007). For visualization of light responsiveness of gene expression, imaging parameters were optimized for dark-grown samples, and all other samples were imaged at identical settings. Because high resolution is required to achieve visible separation of fluorescence signals that are in close proximity but are not spatially overlapping (Smallcombe, 2001), for colocalization analysis, leaves were observed with a 63× Planaprochromat oil (NA, 1.4; WD, 0.19 mm) objective of a Zeiss Axiovert 100M microscope equipped with a Zeiss LSM 510 laser module confocal unit (Carl Zeiss). For visualization of GFP and YFP, or of CFP and YFP, GFP or CFP was excited with the 458-nm line of an argon laser at 55% of output (equivalent to approximately 6 A) and 85% to 100% transmission and emission detected with a BP480-520 filter, while YFP was excited with the 514-nm line of an argon laser at 1% to 45% transmission and emission detected with a BP565-615 filter. For visualization of plastid autofluorescence, chlorophyll was excited with the 543-nm line of a helium-neon laser at 3% transmission and emission detected with a BP650-710 filter. For plasma membrane stainings, seedlings were incubated in 10 μg mL−1 FM 4-64 (Invitrogen) for 2 min immediately prior to imaging with a 63× Planaprochromat oil (NA, 1.4; WD, 0.19 mm) objective of a Zeiss Axiovert 100M LSM 510 laser scanning confocal microscope (Carl Zeiss). For visualization of YFP and FM 4-64, YFP was excited with the 488-nm line of an argon laser at 5% to 30% transmission and emission detected with a BP500-550 filter, while FM 4-64 was excited with the 543-nm line of a helium neon laser at 5% to 10% transmission and emission detected with a BP565-615 filter. Optical slices of identical thickness were used to collect fluorophore emission signals in multiple labeling experiments, and identical settings were adopted to detect chlorophyll autofluorescence in all samples. Sequential excitation and collection of emission from individual fluorophores were performed in high-speed channel switching (multitrack) line scanning mode. Under these conditions, signal bleed-through of the different fluorophores across different photomultiplier channels was never observed. Signal-to-noise ratio was increased during image acquisition by four-frame temporal averaging (Russ, 2002).
Image Analysis and Processing
All images were cropped using Adobe Photoshop 7.0 (Adobe Systems) and turned into 8-bit images using ImageJ (National Institutes of Health, http://rsb.info.nih.gov/ij). Brightness and contrast were not altered for images of gene expression in different light environments. For all other images, brightness and contrast were adjusted through linear stretching of the histogram in ImageJ (National Institutes of Health). Images were color-displayed by applying in ImageJ (National Institutes of Health) a purposely created LUT in which black was used to encode global background, blue to encode local background, and cyan, green, yellow, orange, and red to encode increasing signal intensities (available as downloadable supplemental file). Signal colocalization was visualized with an extended dual-channel LUT from cyan to magenta through green, yellow, and red (Demandolx and Davoust, 1997). Fluorescence in each detection channel was displayed in either cyan or magenta and then merged using the differential operator in Adobe Photoshop 7.0 (Adobe Systems). As a result, a preponderance of cyan signal over colocalized magenta signal is encoded in green, opposite in red, and colocalized cyan and magenta signals of equal intensity in yellow. All images were assembled into figures and labeled in Canvas 8 (ACD Systems International).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. RbcS and Lhc expression in light-grown seedlings.
Supplemental Figure S2. RbcS and Lhc expression in seedling organs.
Supplemental Figure S3. RbcS and Lhc expression in mature plant organs and embryos.
Supplemental Figure S4. RbcS and Lhc expression during leaf development.
Supplemental Figure S5. RbcS and Lhc expression in leaf subepidermal cells.
Supplemental Figure S6. RbcS and Lhc expression in cotyledon epidermal cells.
Supplemental Figure S7. Response of RbcS and Lhc expression to different light conditions.
Supplemental Figure S8. RbcS1B expression in 4- and 6-DAG leaves.
Supplemental Figure S9. Genevestigator-generated organ-specific and developmental RbcS and Lhc expression chart.
Supplemental Table S1. Genes, loci, and primers.
Supplemental Table S2. Reproducibility criteria and indices.
Supplemental Table S3. RbcS and Lhc expression levels from publicly available microarray data sets.
Supplemental Table S4. Regulatory elements associated with specific RbcS and Lhc gene expression patterns.
Supplemental File S1. Additional literature cited.
Supplemental File S2. LUT used in this study.
Supplementary Material
Acknowledgments
We thank Mitsuhiro Aida, Taku Demura, David Galbraith, Jaideep Mathur, and Ben Scheres for generously providing plasmids and seeds, Mike Deyholos for help with microarray data set analysis, and Thomas Berleth, Luis Herrera-Estrella, and June Simpson for invaluable comments on the manuscript.
This work was supported by a Discovery Grant of the Natural Sciences and Engineering Research Council of Canada (NSERC), by an Alberta Ingenuity (AI) New Faculty Grant, and by the Canada Research Chairs Program. T.J.D. was supported by an NSERC CGS-M Scholarship and an AI Student Scholarship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Enrico Scarpella (enrico.scarpella@ualberta.ca).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ali S, Taylor WC (2001) The 3′ non-coding region of a C4 photosynthesis gene increases transgene expression when combined with heterologous promoters. Plant Mol Biol 46 325–333 [DOI] [PubMed] [Google Scholar]
- Altamura MM, Possenti M, Matteucci A, Baima S, Ruberti I, Morelli G (2001) Development of the vascular system in the inflorescence stem of Arabidopsis. New Phytol 151 381–389 [Google Scholar]
- Arguello-Astorga G, Herrera-Estrella L (1998) Evolution of light-regulated plant promoters. Annu Rev Plant Physiol Plant Mol Biol 49 525–555 [DOI] [PubMed] [Google Scholar]
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320 938–941 [DOI] [PubMed] [Google Scholar]
- Baima S, Nobili F, Sessa G, Lucchetti S, Ruberti I, Morelli G (1995) The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121 4171–4182 [DOI] [PubMed] [Google Scholar]
- Baker TS, Eisenberg D, Eiserling FA, Weissman L (1975) The structure of form I crystals of D-ribulose-1,5-diphosphate carboxylase. J Mol Biol 91 391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal KC, Viret JF, Haley J, Khan BM, Schantz R, Bogorad L (1992) Transient expression from cab-m1 and rbcS-m3 promoter sequences is different in mesophyll and bundle sheath cells in maize leaves. Proc Natl Acad Sci USA 89 3654–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedbrook JR, Coen DM, Beaton AR, Bogorad L, Rich A (1979) Location of the single gene for the large subunit of ribulosebisphosphate carboxylase on the maize chloroplast chromosome. J Biol Chem 254 905–910 [PubMed] [Google Scholar]
- Ben-Shem A, Frolow F, Nelson N (2003) Crystal structure of plant photosystem I. Nature 426 630–635 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Jung JW, Wang JY, Lambert GM, Hirst JA, Galbraith DW, Benfey PN (2005) Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat Methods 2 615–619 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302 1956–1960 [DOI] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422 433–438 [DOI] [PubMed] [Google Scholar]
- Brandt SP (2005) Microgenomics: gene expression analysis at the tissue-specific and single-cell levels. J Exp Bot 56 495–505 [DOI] [PubMed] [Google Scholar]
- Briggs GC, Osmont KS, Shindo C, Sibout R, Hardtke CS (2006) Unequal genetic redundancies in Arabidopsis: a neglected phenomenon? Trends Plant Sci 11 492–498 [DOI] [PubMed] [Google Scholar]
- Casneuf T, De Bodt S, Raes J, Maere S, Van de Peer Y (2006) Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biol 7 R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, Guo L, Shih MC (2001) Promoter analysis of the nuclear gene encoding the chloroplast glyceraldehyde-3-phosphate dehydrogenase B subunit of Arabidopsis thaliana. Plant Mol Biol 46 131–141 [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher plants. Annu Rev Genet 38 87–117 [DOI] [PubMed] [Google Scholar]
- Clough S, Bent A (1998) Floral dip: a simplified method for transformation of Arabidopsis. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Dall'Osto L, Caffarri S, Bassi R (2005) A mechanism of nonphotochemical energy dissipation, independent from PsbS, revealed by a conformational change in the antenna protein CP26. Plant Cell 17 1217–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean C, Pichersky E, Dunsmuir P (1989) Structure, evolution, and regulation of RbcS genes in higher plants. Annu Rev Plant Physiol Plant Mol Biol 40 415–439 [Google Scholar]
- Dedonder A, Rethy R, Fredericq H, Van Montagu M, Krebbers E (1993) Arabidopsis rbcS genes are differentially regulated by light. Plant Physiol 101 801–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demandolx D, Davoust J (1997) Multicolour analysis and local image correlation in confocal microscopy. J Microsc 185 21–36 [Google Scholar]
- Dixit R, Cyr R (2003) Cell damage and reactive oxygen species production induced by fluorescence microscopy: effect on mitosis and guidelines for non-invasive fluorescence microscopy. Plant J 36 280–290 [DOI] [PubMed] [Google Scholar]
- Dobbie IM, Lowndes NF, Sullivan KF (2008) Autofluorescent proteins. In KF Sullivan, ed, Fluorescent Proteins, Ed 2, Vol 85. Academic Press, London, pp 1–22 [DOI] [PubMed]
- Fankhauser C, Chory J (1997) Light control of plant development. Annu Rev Cell Dev Biol 13 203–229 [DOI] [PubMed] [Google Scholar]
- Farber A, Young AJ, Ruban AV, Horton P, Jahns P (1997) Dynamics of xanthophyll-cycle activity in different antenna subcomplexes in the photosynthetic membranes of higher plants (the relationship between zeaxanthin conversion and nonphotochemical fluorescence quenching). Plant Physiol 115 1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AJ, Manzara T, Gruissem W, Kuhlemeier C (1996) Fluorescent imaging of GUS activity and RT-PCR analysis of gene expression in the shoot apical meristem. Plant J 10 745–754 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Larner VS, Whitelam GC (2005) The signal transducing photoreceptors of plants. Int J Dev Biol 49 653–664 [DOI] [PubMed] [Google Scholar]
- Gordon SP, Heisler MG, Reddy GV, Ohno C, Das P, Meyerowitz EM (2007) Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134 3539–3548 [DOI] [PubMed] [Google Scholar]
- Govindjee, Beatty JT, Gest H, Allen JF, editors (2006) Discoveries in Photosynthesis, Vol 20. Springer, Dordrecht, The Netherlands
- Grossman AR, Bhaya D, Apt KE, Kehoe DM (1995) Light-harvesting complexes in oxygenic photosynthesis: diversity, control, and evolution. Annu Rev Genet 29 231–288 [DOI] [PubMed] [Google Scholar]
- Haberer G, Hindemitt T, Meyers BC, Mayer KF (2004) Transcriptional similarities, dissimilarities, and conservation of cis-elements in duplicated genes of Arabidopsis. Plant Physiol 136 3009–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono K, Niwa Y, Satoh K, Kobayashi H (1997) Evidence for transcriptional regulation of plastid photosynthesis genes in Arabidopsis thaliana roots. Plant Physiol 114 623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson S (1994) The light-harvesting chlorophyll a/b-binding proteins. Biochim Biophys Acta 1184 1–19 [DOI] [PubMed] [Google Scholar]
- Jansson S (1999) A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci 4 236–240 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RG, Bahr JT (1977) Ribulose 1,5-bisphosphate carboxylase-oxygenase. Annu Rev Plant Physiol 28 379–400 [Google Scholar]
- Joly E (2007) Optimising blue fluorescent protein (BFP) for use as a mammalian reporter gene in parallel with Green Fluorescent Protein (GFP). Nature Precedings. http://hdl.handle.net/10101/npre.2007.1259.1 (May 15, 2008)
- Kang J, Dengler N (2004) Vein pattern development in adult leaves of Arabidopsis thaliana. Int J Plant Sci 165 231–242 [Google Scholar]
- Kang J, Mizukami Y, Wang H, Fowke L, Dengler NG (2007) Modification of cell proliferation patterns alters leaf vein architecture in Arabidopsis thaliana. Planta 226 1207–1218 [DOI] [PubMed] [Google Scholar]
- Kato N, Pontier D, Lam E (2002) Spectral profiling for the simultaneous observation of four distinct fluorescent proteins and detection of protein-protein interaction via fluorescence resonance energy transfer in tobacco leaf nuclei. Plant Physiol 129 931–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehr J (2003) Single cell technology. Curr Opin Plant Biol 6 617–621 [DOI] [PubMed] [Google Scholar]
- Khrebtukova I, Spreitzer RJ (1996) Elimination of the Chlamydomonas gene family that encodes the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Proc Natl Acad Sci USA 93 13689–13693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimmek F, Sjodin A, Noutsos C, Leister D, Jansson S (2006) Abundantly and rarely expressed Lhc protein genes exhibit distinct regulation patterns in plants. Plant Physiol 140 793–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of the TL-DNA gene 5 controls the tissue specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204 383–396 [Google Scholar]
- Krebbers E, Seurinck J, Herdies L, Cashmore AR, Timko MP (1988) 4 Genes in 2 Diverged Subfamilies Encode the Ribulose-1,5-Bisphosphate Carboxylase Small Subunit Polypeptides of Arabidopsis-Thaliana. Plant Mol Biol 11 745–759 [DOI] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HB, Park SC, Peng HP, Goodman HM, Dewdney J, Shih MC (1994) Identification of a light-responsive region of the nuclear gene encoding the B subunit of chloroplast glyceraldehyde 3-phosphate dehydrogenase from Arabidopsis thaliana. Plant Physiol 105 357–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale JA, Rothermel BA, Nelson T (1988) Cellular pattern of photosynthetic gene expression in developing maize leaves. Genes Dev 2 106–115 [DOI] [PubMed] [Google Scholar]
- Lange BM (2005) Single-cell genomics. Curr Opin Plant Biol 8 236–241 [DOI] [PubMed] [Google Scholar]
- Langham RJ, Walsh J, Dunn M, Ko C, Goff SA, Freeling M (2004) Genomic duplication, fractionation and the origin of regulatory novelty. Genetics 166 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN (2006) Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci USA 103 6055–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Levesque M, Benfey PN (2005) High-throughput RNA isolation technologies. New tools for high-resolution gene expression profiling in plant systems. Plant Physiol 138 585–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutwiler LS, Meyerowitz EM, Tobin EM (1986) Structure and expression of three light-harvesting chlorophyll a/b-binding protein genes in Arabidopsis thaliana. Nucleic Acids Res 14 4051–4064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucinski R, Schmid VH, Jansson S, Klimmek F (2006) Lhca5 interaction with plant photosystem I. FEBS Lett 580 6485–6488 [DOI] [PubMed] [Google Scholar]
- Maclean D, Jerome CA, Brown AP, Gray JC (2008) Co-regulation of nuclear genes encoding plastid ribosomal proteins by light and plastid signals during seedling development in tobacco and Arabidopsis. Plant Mol Biol 66 475–490 [DOI] [PubMed] [Google Scholar]
- Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y (2005) Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA 102 5454–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124 33–44 [DOI] [PubMed] [Google Scholar]
- Martinez-Hernandez A, Lopez-Ochoa L, Arguello-Astorga G, Herrera-Estrella L (2002) Functional properties and regulatory complexity of a minimal RBCS light-responsive unit activated by phytochrome, cryptochrome, and plastid signals. Plant Physiol 128 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JM, Terzaghi WB, Sridhar P, Cashmore AR, Pichersky E (1992) Sequence of the fourth and fifth Photosystem II type I chlorophyll a/b-binding protein genes of Arabidopsis thaliana and evidence for the presence of a full complement of the extended CAB gene family. Plant Mol Biol 19 725–733 [DOI] [PubMed] [Google Scholar]
- Meier I, Callan KL, Fleming AJ, Gruissem W (1995) Organ-specific differential regulation of a promoter subfamily for the ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit genes in tomato. Plant Physiol 107 1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T, Lee JY, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN (2005) Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17 1908–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio JN, Sun J, Vogelmann TC (1993) Carbon fixation gradients across spinach leaves do not follow internal light gradients. Plant Cell 5 953–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiuchi T, Nakamura T, Abe T, Kodama H, Nishimura M, Iba K (1995) Tissue-specific and light-responsive regulation of the promoter region of the Arabidopsis thaliana chloroplast omega-3 fatty acid desaturase gene (FAD7). Plant Mol Biol 29 599–609 [DOI] [PubMed] [Google Scholar]
- Nordberg JJ, Sluder G (2007) Practical aspects of adjusting digital cameras. In G Sluder, DE Wolf, eds, Digital Microscopy, Ed 3, Vol 81. Academic Press, San Diego, pp 159–169 [DOI] [PubMed]
- Nowak MA, Boerlijst MC, Cooke J, Smith JM (1997) Evolution of genetic redundancy. Nature 388 167–171 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA 101 9903–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K, Lopez-Juez E (1999) Cellular differentiation and leaf morphogenesis in Arabidopsis. Crit Rev Plant Sci 18 527–546 [Google Scholar]
- Pyke KA, Marrison JL, Leech RM (1991) Temporal and spatial development of the cells of the expanding 1st leaf of Arabidopsis thaliana (L) Heynh. J Exp Bot 42 1407–1416 [Google Scholar]
- Pyke KA, Page AM (1998) Plastid ontogeny during petal development in Arabidopsis. Plant Physiol 116 797–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodermel S, Haley J, Jiang CZ, Tsai CH, Bogorad L (1996) A mechanism for intergenomic integration: abundance of ribulose bisphosphate carboxylase small-subunit protein influences the translation of the large-subunit mRNA. Proc Natl Acad Sci USA 93 3881–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ JC (2002) The Image Processing Handbook, Ed 4. CRC, Boca Raton, FL
- Sawchuk MG, Donner TJ, Scarpella E (2008) Auxin transport-dependent, stage-specific dynamics of leaf vein formation. Plant Signal Behav 3 286–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawchuk MG, Head P, Donner TJ, Scarpella E (2007) Time-lapse imaging of Arabidopsis leaf development shows dynamic patterns of procambium formation. New Phytol 176 560–571 [DOI] [PubMed] [Google Scholar]
- Scarpella E, Francis P, Berleth T (2004) Stage-specific markers define early steps of procambium development in Arabidopsis leaves and correlate termination of vein formation with mesophyll differentiation. Development 131 3445–3455 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Silverthorne J, Tobin EM (1990) Post-transcriptional regulation of organ-specific expression of individual rbcS mRNAs in Lemna gibba. Plant Cell 2 1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J, Herrera-Estrella L (1990) Light-regulated gene expression. Crit Rev Plant Sci 9 95–109 [Google Scholar]
- Smallcombe A (2001) Multicolor imaging: the important question of co-localization. Biotechniques 30 1240–1242, 1244–1246 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MC, Samuels AL, Jetter R, Kunst L, Pollard M, Ohlrogge J, Beisson F (2005) Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol 139 1649–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Nishio J (2001) Why abaxial illumination limits photosynthetic carbon fixation in spinach leaves. Plant Cell Physiol 42 1–8 [DOI] [PubMed] [Google Scholar]
- Szabo I, Bergantino E, Giacometti GM (2005) Light and oxygenic photosynthesis: energy dissipation as a protection mechanism against photo-oxidation. EMBO Rep 6 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR (1995) Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol 46 445–474 [Google Scholar]
- The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Thompson WF, White MJ (1991) Physiological and molecular studies of light-regulated nuclear genes in higher plants. Annu Rev Plant Physiol Plant Mol Biol 42 423–466 [Google Scholar]
- Tobin EM, Silverthorne J (1985) Light regulation of gene expression in higher plants. Annu Rev Plant Physiol 36 569–593 [Google Scholar]
- Vidaurre DP, Ploense S, Krogan NT, Berleth T (2007) AMP1 and MP antagonistically regulate embryo and meristem development in Arabidopsis. Development 134 2561–2567 [DOI] [PubMed] [Google Scholar]
- Wang J, Tian L, Madlung A, Lee HS, Chen M, Lee JJ, Watson B, Kagochi T, Comai L, Chen ZJ (2004) Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics 167 1961–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Okada K (2003) Two discrete cis elements control the Abaxial side-specific expression of the FILAMENTOUS FLOWER gene in Arabidopsis. Plant Cell 15 2592–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2 e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Kwon HB, Peng HP, Shih MC (1993) Stress responses and metabolic regulation of glyceraldehyde-3-phosphate dehydrogenase genes in Arabidopsis. Plant Physiol 101 209–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Gong FC, Lambert GM, Galbraith DW (2005) Cell type-specific characterization of nuclear DNA contents within complex tissues and organs. Plant Methods 1 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.