Crystals of the Sox17 HMG domain bound to LAMA1 enhancer DNA-element crystals that diffracted to 2.75 Å resolution were obtained.
Keywords: Sox17, HMG domains, LAMA1
Abstract
Sox17 is a member of the SRY-related high-mobility group (HMG) of transcription factors that have been shown to direct endodermal differentiation in early mammalian development. The LAMA1 gene encoding the α-chain of laminin-1 has been reported to be directly bound and regulated by Sox17. This paper describes the details of initial crystallization attempts with the HMG domain of mouse Sox17 (mSox17-HMG) with a 16-mer DNA element derived from the LAMA1 enhancer and optimization strategies to obtain a better diffracting crystal. The best diffracting crystal was obtained in a condition containing 0.1 M Tris–HCl pH 7.4, 0.2 M MgCl2, 30% PEG 3350 using the hanging-drop vapour-diffusion method. A highly redundant in-house data set was collected to 2.75 Å resolution with 99% completeness. The presence of the mSox17-HMG–DNA complex within the crystals was confirmed and Matthews analysis indicated the presence of one complex per asymmetric unit.
1. Introduction
Sox proteins belong to the HMG box-containing superfamily of proteins. The HMG box constitutes a DNA-binding motif that spans 70–80 amino acids (Wegner, 1999 ▶). In contrast to other non-sequence-specific HMG proteins, Sox proteins are sequence-specific transcription factors that function as key regulators of various developmental pathways (Dragan et al., 2004 ▶; Murphy & Churchill, 2000 ▶). Mammalian genomes encode 20 Sox proteins (Pevny & Lovell-Badge, 1997 ▶; Wegner, 1999 ▶), which can be further subdivided into seven subgroups based on the HMG-box sequences and overall protein architecture (Wegner, 1999 ▶). Sox17, Sox7 and Sox18 belong to subgroup F of this scheme. Sox17 was originally identified as a stage-specific transcription activator during mouse spermatogenesis (Kanai et al., 1996 ▶). Full-length mSox17 consists of 419 amino acids and contains a single HMG box near the N-terminus (Kanai et al., 1996 ▶). Sox17 has been shown to direct endodermal differentiation in early mammalian development (Seguin et al., 2008 ▶). The LAMA1 gene encoding the α-chain of laminin-1 has been reported to be directly bound and regulated by Sox17 in mouse F9 embryonal carcinoma cells (Niimi et al., 2004 ▶). The binding site contains a GACAAT motif, which resembles the consensus sequence bound by most Sox-family members, (A/T)(A/T)CAA(A/T)G (Niimi et al., 2004 ▶).
The Sox2 protein is one of the major players in early development and stem-cell biology. Sox2 is required for the self-renewal of embryonic stem (ES) cells and is a key component of a molecular cocktail that allows the induction of pluripotency from differentiated tissue (Fong et al., 2008 ▶; Takahashi et al., 2007 ▶). The crystal structure (Remenyi et al., 2003 ▶) and NMR structure (Williams et al., 2004 ▶) of the HMG domain of Sox2 in complex with Oct1 and DNA have been reported. The structures revealed that Sox2 consists of three α-helices exhibiting an L-shaped arrangement. Sox2 binds the minor groove of the DNA and bends the DNA towards the major groove with an approximate bend angle of 90° (Remenyi et al., 2003 ▶). The HMG domains of mouse Sox17 and Sox2 exhibit ∼60% sequence identity. However, they are functionally nonredundant and regulate virtually competitive biological processes (Nakagawa et al., 2008 ▶). This study attempts to unravel the biochemical basis of the selective promoter recognition by Sox17. One possibility is that different Sox proteins bend the DNA to different degrees, which may lead to Sox-protein-specific cofactor recruitment (Dragan et al., 2004 ▶; Kamachi et al., 2000 ▶). To study DNA recognition by Sox17 and the bending topology of the complex, we aimed to determine the structure of the HMG domain of mSox17 in complex with DNA. In this report, we describe the protein purification and crystallization of the mouse Sox17 HMG domain with a 16-mer DNA element derived from the LAMA1 enhancer and discuss the effects of different crystallization components in the mother liquor as well as different overhangs on the growth of diffraction-quality crystals.
2. Experimental procedures
2.1. Cloning and expression
The HMG domain of mSox17, spanning amino-acid residues 66–144 of the full-length protein, was PCR-amplified from a cDNA clone (kindly provided by Paul Robson of the Genome Institute of Singapore) using the following DNA primers: 5′-CACCTCTCGCATCCGGCGGCCG-3′ and 5′-CTACTGCTTGCGCCGCCGCGG-3′. The PCR product was introduced into the Gateway entry vector pENTR/TEV/d-TOPO by directional TOPO cloning (Invitrogen). The insert was verified by DNA sequencing and introduced into the Gateway destination pETG-20A by performing a Gateway LR reaction, resulting in the pETG20A-mSox17-HMG expression plasmid. pETG20A-mSox17-HMG was transformed into Escherichia coli BL21 (DE3) cells (Invitrogen) and cells were grown in Luria–Bertani (LB) broth containing 100 µg ml−1 ampicillin and 0.2% glucose at 310 K. When an OD600nm of 0.7 was reached, the temperature was lowered to 303 K and protein expression was induced by adding 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Cells were harvested by centrifugation after 4 h and stored at 193 K.
2.2. Protein purification
Cells were thawed and resuspended in buffer A (50 mM Tris–HCl pH 8.0, 300 mM NaCl, 30 mM imidazole) and disrupted by ultrasonication on ice for 15 min. The lysate was clarified by centrifugation and passed through a 0.22 µm filter. The supernatant was incubated with Ni-Sepharose beads (GE Healthcare) pre-equilibrated with buffer A and Thx-His6-mSox17-HMG was eluted using buffer B (50 mM Tris–HCl pH 8.0, 100 mM NaCl, 300 mM imidazole). The fusion tag and mSox17-HMG were separated by TEV digestion using a substrate:enzyme ratio of 75:1(w:w) at 277 K for approximately 16 h. mSox17-HMG was purified by ion-exchange chromatography using a 6 ml Resource S column (GE Healthcare) pre-equilibrated with buffer C (10 mM Tris–HCl pH 8.0, 100 mM NaCl) and eluted with a salt gradient. Finally, size-exclusion chromatography was performed using a Hi-Prep Superdex-75 16/60 column (GE Healthcare) in buffer C. Fractions containing Sox17 were pooled and concentrated to 5 mg ml−1.
2.3. Crystallization of Sox17 in complex with DNA
PAGE-purified oligonucleotides (Fig. 1 ▶) were purchased at 1 mM concentration (Sigma–Aldrich). Annealing was carried out by combining equimolar amounts of complementary DNA, heating to 368 K for 5 min and gradual cooling to room temperature. The mSox17-HMG–DNA complex was prepared by mixing mSox17-HMG and DNA in a 1:1.2 molar ratio and incubating on ice for 2 h. The complex was concentrated to 5–20 mg ml−1 (as determined using the Bradford method) using a Vivaspin centrifugal filter column with 3000 molecular-weight cutoff (Sartorius BioLab). Crystallization trials were carried out using screens from both Hampton Research and Qiagen dispensed by an Innovadyne robot. Refinements of crystallization hits were conducted manually in a hanging-drop setting.
Figure 1.
16-mer DNA elements derived from the LAMA1 cis-regulatory region used in crystallization trials. (a) CG-overhang DNA. Crystallization of this DNA with mSox17-HMG produced a sharp-edged rhombus-shaped crystal that diffracted to 3.5 Å resolution. (b) TA-overhang DNA. (c) TT-overhang DNA, which did not allow crystal formation. (d) Blunt-ended DNA. Crystallization of protein with this DNA produced two types of crystals: small flat squarish crystals that produced a weak diffraction pattern and elongated composite crystals that diffracted to 2.75 Å resolution.
2.4. X-ray data collection and processing
Crystals for X-ray diffraction studies were subjected to a continuous stream of nitrogen gas without cryoprotective solution. Data were collected using a PLATINUM 135 CCD detector with focused Cu Kα X-rays from an X8 PROTEUM rotating-anode generator (Bruker AXS) controlled by the PROTEUM2 software suite. Processing and scaling were performed using the same PROTEUM2 software (Sheldrick, 2008 ▶). Data were analyzed using XPREP (Sheldrick, 2008 ▶), XTRIAGE (Zwart et al., 2005 ▶) and programs from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶).
3. Results and discussion
3.1. Protein preparation
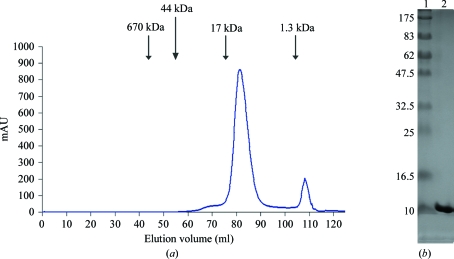
mSox17-HMG was expressed and purified with typical yields of 2.5 mg pure protein per litre of bacterial expression culture. mSox17-HMG eluted from the final Superdex-75 gel-filtration column as a single symmetric peak corresponding to the molecular weight of the monomeric form of the protein (∼10 000 Da; Fig. 2 ▶ a). SDS–PAGE analysis indicates >98% purity after the final purification (Fig. 2 ▶ b).
Figure 2.
Elution profile of mSox17-HMG run on a Superdex 75 gel-filtration column calibrated with molecular-weight standards. (a) mSox17-HMG elutes as a single symmetric peak corresponding to a molecular weight of ∼10 kDa. (b) 12% SDS–PAGE analysis showing 1 µg purified mSox17-HMG (lane 2) and molecular-weight markers (lane 1; kDa).
3.2. Crystallization
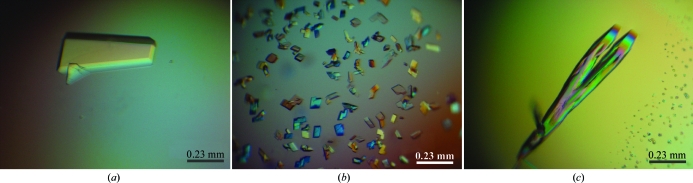
Initial crystal trials were carried out using a 16-mer element derived from the LAMA1 cis-regulatory region containing CG sticky ends (Fig. 1 ▶ a). Although the mSox17-HMG–DNA complex was initially purified by an additional gel-filtration step following complex formation, crystals could readily be obtained by directly setting up crystallization trials. Repeated freeze–thaw cycles do not seem to affect the crystal quality. However, extensive efforts in optimizing the crystallization conditions and cryobuffers did not yield data beyond 3.5 Å resolution (Fig. 3 ▶ a). In addition, data processing was hampered by overlapping lattices and high mosaicity.
Figure 3.
mSox17-HMG–DNA complex crystals. (a) A crystal of mSox17-HMG–CG-overhang DNA that diffracted to 3.5 Å resolution. (b) Small flat squarish crystals of an mSox17-HMG–blunt-ended DNA complex grown in a buffer containing 0.1 M MgCl2 that diffracted poorly. (c) Large composite crystals of an mSox17-HMG–blunt-ended DNA complex that diffracted to 2.75 Å resolution.
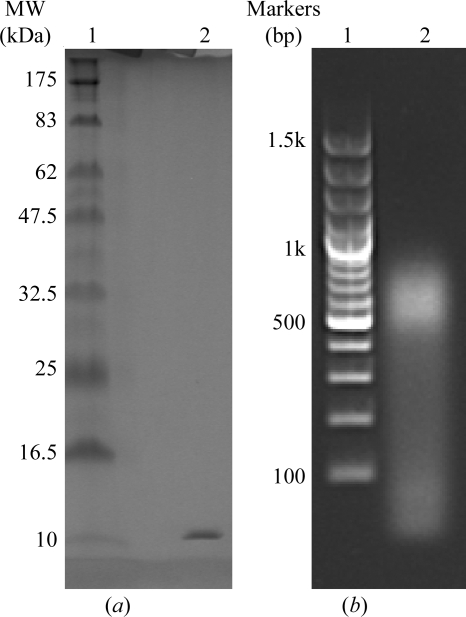
Subsequently, we assessed the effect of various DNA ends on crystal formation. To this end, we performed further screenings with three additional variants of DNA derived from the LAMA1 enhancer (Figs. 1 ▶ b, 1 ▶ c and 1 ▶ d). We found that neither the TA-overhang nor the TT-overhang enabled crystal formation. However, using blunt-ended DNA we observed rapid crystal growth under a variety of conditions. Crystals started to appear after 30 min in 0.1 M Tris–HCl pH 8.0, 30% PEG 4000, 0.2 M MgCl2 with 20 mg ml−1 complex concentration at room temperature. The presence of both protein and DNA was confirmed by washing and dissolving several crystals in fresh mother liquor followed by SDS–PAGE and agarose-gel electrophoresis (Figs. 4 ▶ a and 4 ▶ b). Refinement of the initial hit condition revealed that reservoirs containing at least 50 mM MgCl2 are necessary for crystal formation. Interestingly, macroscopically different crystal forms were observed at different concentrations of MgCl2. Flat plates with defined edges grew in buffers containing 100 mM MgCl2 but diffracted poorly (6.6 Å resolution; Fig. 3 ▶ b). The best-diffracting crystal was grown in a reservoir solution containing 200 mM MgCl2.
Figure 4.
Several crystals of Sox17–blunt-ended DNA complex were dissolved and run on (a) 12% SDS–PAGE stained with SimplyBlue Stain (Invitrogen), showing a band corresponding to the mSox17-HMG protein, and (b) 1% agarose gel stained with ethidium bromide (the sample was not boiled to denature the protein, therefore free and protein-bound DNA can be seen).
Large crystals formed in the pH range 7.4–8.6. pH values greater than 8.6 facilitated nucleation, while pH values lower than 7.4 produced composite crystal plates with undefined edges that showed weak birefringence under polarizing light.
The crystal quality was improved by replacing PEG 4000 with PEG 3350; PEG 3350 in the range 26–30% produced diffraction-quality crystals. At PEG 3350 concentrations below 30%, crystals either redissolved or disintegrated into crystalline precipitate. At 30% PEG 3350 crystals remained stable for several weeks. The final optimized condition consisted of 0.1 M Tris–HCl pH 7.4, 30% PEG 4000, 0.2 M MgCl2 and 10 mg ml−1 mSox17-HMG–DNA complex (Fig. 3 ▶ c).
3.3. Data collection
The best crystal diffracted to 2.75 Å resolution using an in-house data-collection facility equipped with a CCD detector. Because no obvious ice rings were observed, cryoprotectant was not added. XPREP assigned the enantiomorphic pair P3121/P3221 as the most likely space group, but P31/P32 should also be considered if trials to solve and refine the structure fail. Unit-cell parameters and data-collection statistics are given in Table 1 ▶. The data set is highly redundant and complete, with good merging statistics. The value of the Matthews coefficient (Matthews, 1968 ▶) is 2.3 Å3 Da−1 for one molecule in the asymmetric unit and the estimated solvent content is 45%. Attempts to determine the structure were made using the Sox2–DNA complex structures as a search model with Phaser (McCoy et al., 2005 ▶). Structure solution will also be attempted using selenomethionine derivatization.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| Detector | PLATINUM 135 CCD |
| Space group | P3121 or P3221 |
| Unit-cell parameters | |
| a (Å) | 68.75 |
| b (Å) | 68.75 |
| c (Å) | 66.34 |
| α (°) | 90 |
| β (°) | 90 |
| γ (°) | 120 |
| Resolution of data set (Å) | 66.33–2.75 (2.85–2.75) |
| No. of observed reflections | 49201 |
| No. of unique reflections | 4970 (490) |
| Redundancy | 9.86 (7.60) |
| Completeness (%) | 99 (100) |
| Mean I/σ(I) | 22.75 (3.75) |
| Rint† (%) | 6.9 (39) |
R
int = 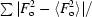
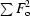 .
.
Acknowledgments
We are grateful to Robert Robinson for facilitating data collection at the Institute of Molecular and Cell Biology (IMCB), Singapore. We also wish to thank Raymond C. Stevens and Kumar Singh Saikatendu from the Scripps Research Institute for their help with the initial data-collection attempts. This work was supported by the Agency for Science, Technology and Research (A*STAR) in Singapore.
References
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Dragan, A. I., Read, C. M., Makeyeva, E. N., Milgotina, E. I., Churchill, M. E., Crane-Robinson, C. & Privalov, P. L. (2004). J. Mol. Biol.343, 371–393. [DOI] [PubMed]
- Fong, H., Hohenstein, K. A. & Donovan, P. J. (2008). Stem Cells, 26, 1931–1938. [DOI] [PubMed]
- Kamachi, Y., Uchikawa, M. & Kondoh, H. (2000). Trends Genet.16, 182–187. [DOI] [PubMed]
- Kanai, Y., Kanai-Azuma, M., Noce, T., Saido, T. C., Shiroishi, T., Hayashi, Y. & Yazaki, K. (1996). J. Cell Biol.133, 667–681. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed]
- Murphy, F. V. IV & Churchill, M. E. (2000). Structure, 8, R83–R89. [DOI] [PubMed]
- Nakagawa, M., Koyanagi, M., Tanabe, K., Takahashi, K., Ichisaka, T., Aoi, T., Okita, K., Mochiduki, Y., Takizawa, N. & Yamanaka, S. (2008). Nature Biotechnol.26, 101–106. [DOI] [PubMed]
- Niimi, T., Hayashi, Y., Futaki, S. & Sekiguchi, K. (2004). J. Biol. Chem.279, 38055–38061. [DOI] [PubMed]
- Pevny, L. H. & Lovell-Badge, R. (1997). Curr. Opin. Genet. Dev.7, 338–344. [DOI] [PubMed]
- Remenyi, A., Lins, K., Nissen, L. J., Reinbold, R., Scholer, H. R. & Wilmanns, M. (2003). Genes Dev.17, 2048–2059. [DOI] [PMC free article] [PubMed]
- Seguin, C. A., Draper, J. S., Nagy, A. & Rossant, J. (2008). Cell Stem Cell, 3, 182–195. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K. & Yamanaka, S. (2007). Cell, 131, 861–872. [DOI] [PubMed]
- Wegner, M. (1999). Nucleic Acids Res.27, 1409–1420. [DOI] [PMC free article] [PubMed]
- Williams, D. C. Jr, Cai, M. & Clore, G. M. (2004). J. Biol. Chem.279, 1449–1457. [DOI] [PubMed]
- Zwart, P. H., Grosse-Kunstleve, R. W. & Adams, P. D. (2005). CCP4 Newsl.42