Glucosyl-3-phosphoglycerate synthase (GpgS) is a key enzyme that catalyses the first glucosylation step in methylglucose lipopolysaccharide biosynthesis in Mycobacterium spp. Here, the crystallization and preliminary crystallographic analysis of GpgS from M. tuberculosis and of its complex with UDP are reported.
Keywords: glycosyltransferases, methylglucose lipopolysaccharides, Mycobacterium tuberculosis
Abstract
Glucosyl-3-phosphoglycerate synthase (GpgS) is a key enzyme that catalyses the first glucosylation step in methylglucose lipopolysaccharide biosynthesis in mycobacteria. These important molecules are believed to be involved in the regulation of fatty-acid and mycolic acid synthesis. The enzyme belongs to the recently defined GT81 family of retaining glycosyltransferases (CAZy, Carbohydrate-Active Enzymes Database; see http://www.cazy.org). Here, the purification, crystallization and preliminary crystallographic analysis are reported of GpgS from Mycobacterium tuberculosis and of its complex with UDP. GpgS crystals belonged to space group I4, with unit-cell parameters a = 98.85, b = 98.85, c = 127.64 Å, and diffracted to 2.6 Å resolution. GpgS–UDP complex crystals belonged to space group I4, with unit-cell parameters a = 98.32, b = 98.32, c = 127.96 Å, and diffracted to 3.0 Å resolution.
1. Introduction
The 6-O-methylglucosyl-containing lipopolysaccharides (MGLPs; Lee & Ballou, 1964 ▶; Lee, 1966 ▶) and the 3-O-methylmannosyl-containing polysaccharides (MMPs; Gray & Ballou, 1971 ▶; Maitra & Ballou, 1977 ▶) are two unusual polymethylated polysaccharides (PMPS) produced by mycobacteria. Both PMPS localize to the cytoplasm, where they have been proposed to regulate fatty-acid biosynthesis owing to their ability to form stable 1:1 complexes with long-chain fatty acids and acyl-coenzyme A derivatives. In sequestering the products of fatty-acyl synthase I (FAS I), PMPS are thought to facilitate the release of the neo-synthesized chains from the enzyme, thereby not only reopening active sites essential for enzyme turnover but also terminating their elongation (for a review, see Bloch, 1977 ▶). In addition, PMPS have been proposed to serve as general fatty-acyl carriers, the role of which would be to facilitate the further processing of very long and insoluble fatty-acyl CoAs, including mycolic acids, by increasing the tolerance of mycobacteria to high cytoplasmic concentrations of these products while protecting them from degradation (Yabusaki & Ballou, 1979 ▶). The MGLPs of Mycobacterium bovis BCG are composed of ten α-(1→4)-linked 6-O-methylglucosyl residues with a nonreducing end made of the tetrasaccharide 3-O-methyl-d-Glcp-[α-(1→4)-d-Glcp]3-α-(1→, whereas the tetrasaccharide →4)-[α-(1→4)-d-Glcp]3-α(1→6)-d-Glcp-α(1→ linked to position 2 of d-glyceric acid constitutes the reducing end of the molecule. Positions 3 of the second and fourth α-d-Glcp residues (closest to the reducing end) are substituted by single β-d-Glcp residues. The nonreducing end of the polymer can be acylated by a combination of acetate, propionate and isobutyrate. The Glcp residues of the reducing end can be esterified with up to three succinate groups and position 1 of glyceric acid can also be esterified by octanoate. MGLPs occur in the cell as a mixture of four main components that differ in their content of esterified succinic acid (Tuffal et al., 1998 ▶). Although the structures of MGLPs and their fatty-acyl-binding properties have been well established, little is known about the biosynthesis of these important molecules.
Empadinhas and coworkers recently showed that recombinant forms of the GpgS enzymes of M. bovis BCG and M. smegmatis display glucosyl-3-phosphoglycerate activity in vitro, transferring a Glcp residue from UDP-Glc to the 2 position of d-3-phosphoglycerate (PG) to form α(1→2)-d-Glcp-3-phosphoglycerate (GPG; Empadinhas et al., 2008 ▶). Furthermore, recent evidence based on the analysis of an M. smegmatis knockout mutant deficient in the expression of gpgS clearly implicated this enzyme in the transfer of the first Glcp residue of MGLP, generating GPG. This product is subsequently dephosphorylated by an unknown phosphatase to yield the glucosyl-glycerate (GG) subunit found at the reducing end of MGLP (Kaur et al., manuscript in preparation). A second and third Glcp residue are subsequently added by unidentified glucosyltransferase/branching enzymes to form [α-(1→4)-d-Glcp]-α(1→6)-d-Glcp-α(1→2)-d-glyceric acid. Interestingly, a cluster of genes dedicated to the biosynthesis of MGLP has recently been identified in M. smegmatis and M. tuberculosis (Stadthagen et al., 2007 ▶). It was demonstrated that Rv3032 encodes the main α-(1→4)-glucosyltransferase responsible for the elongation of MGLP, whereas Rv3030 encodes the O-methyltransferase likely to be required for the 6-O-methylation of these lipopolysaccharides.
Glycosyltransferases (GTs) can be classified into either ‘inverting’ or ‘retaining’ enzymes according to the anomeric configuration of the reaction substrates and products. A single-displacement mechanism is well established for ‘inverting’ enzymes, whereas the catalytic mechanism for ‘retaining’ enzymes, including GpgS, remains unclear (Empadinhas et al., 2008 ▶; Lairson et al., 2008 ▶). Interestingly, only two protein topologies have been found for the nucleotide sugar-dependent enzymes from the first 29 GT sequence-based families (CAZy, Carbohydrate-Active Enzymes Database; see http://www.cazy.org) for which three-dimensional structures have been reported. These topologies are variations of ‘Rossmann-like’ domains and have been defined as GT-A (Charnock & Davies, 1999 ▶) and GT-B (Vrielink et al., 1994 ▶). Both inverting and retaining enzymes have been found within the GT-A and GT-B fold GTs, indicating that there is no correlation between the overall fold of GTs and their catalytic mechanism. GpgS belongs to the recently defined GT81 family of GTs. The crystallographic characterization of GpgS will shed light on the catalytic mechanism of the GT81 family of GTs and the biosynthesis of MGLP in mycobacteria.
2. Materials and methods
2.1. Cloning, expression and purification
The gpgS gene (Rv1208; http://genolist.pasteur.fr/TubercuList/) was amplified from genomic M. tuberculosis (strain H37Rv) DNA by standard PCR using oligonucleotide primers gpgS_NcoI_Fwd (5′-GGAATTCCATATGACAGCATCGGAGCTGGTCGCC-3′) and gpgS_HindIII_rev (5′-CCCAAGCTTTCAGCGCGGCCGCATCACCTTCATCGG-3′) and Phusion DNA Polymerase (New England Biolabs). The PCR fragment was digested with NdeI and HindIII and ligated to the expression vector pET28a (Novagen), generating pET28a-gpgS. The recombinant GpgS (344 residues) has an additional 20-amino-acid peptide (1-MGSSHHHHHHSSGLVPRGSH-20) at the N-terminus that includes a histidine tag and a thrombin cleavage site.
Escherichia coli BL21(DE3)pLysS cells transformed with pET28a-gpgS were grown in 3000 ml 2×YT medium supplemented with 25 µg ml−1 kanamycin and 34 µg ml−1 chloramphenicol at 310 K. When the culture reached an OD600 value of 0.6, GpgS expression was induced by adding 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG; MIP). After 4 h at 310 K, cells were harvested and resuspended in 40 ml 50 mM Tris–HCl pH 8.0 (solution A) containing protease inhibitors (Complete EDTA-free; Roche). Cells were then disrupted by sonication (five cycles of 1 min) and the suspension was centrifuged for 20 min at 10 000g. The supernatant was applied onto a HisTrap Chelating column (1 ml; GE Healthcare) equilibrated with 50 mM Tris–HCl pH 8.0, 500 mM NaCl (solution B). The column was washed with solution B until no absorbance at 280 nm was detected. Elution was performed with a linear gradient of 0–500 mM imidazole in 40 ml solution B at 1 ml min−1. Fractions containing GpgS were pooled and diluted 1:5 with solution A. This solution was filtered through a Millipore 0.22 µm filter and applied onto a Mono Q HR 5/5 (GE Healthcare) column equilibrated in solution A. The enzyme was eluted at 300 mM NaCl with a linear gradient of 0–500 mM NaCl in 30 ml solution A at 1 ml min−1. The protein was concentrated using a Vivaspin 20 device (10 000 Da molecular-weight cutoff; Sartorius) and loaded onto a HiLoad 16/60 Superdex 75 (GE Healthcare) equilibrated in 50 mM Tris–HCl pH 8.0, 150 mM NaCl. The resulting preparation displayed a single protein band when run on 10% SDS–PAGE stained with Coomasie brilliant blue (Fig. 1 ▶ a). The purified enzyme (3.6 mg by the Bio-Rad Protein Assay) was concentrated to 10 mg ml−1 in 10 mM Tris–HCl pH 8.0 and stored at 193 K for further use in enzymatic activity assays.
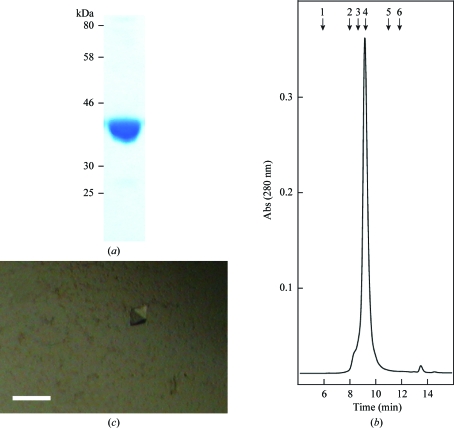
Figure 1.
(a) Purified recombinant GpgS from M. tuberculosis. (b) The dimeric state of pure GpgS in solution was determined by size-exclusion chromatography using a BioSuite 250 5 µm HR SEC column (Waters Corporation). The molecular-weight standards employed were blue dextran (1; 2000 kDa), β-amylase (2; 200 kDa), alcohol dehydrogenase (3; 150 kDa), bovine serum albumin (4; 66 kDa), carbonic anhydrase (5; 29 kDa) and cytochrome c (6; 12.4 kDa). (c) Crystals of GpgS from M. tuberculosis in the presence of UDP. The scale bar is 100 µm in length.
In order to precisely determine the oligomeric state of GpgS in solution, the purified protein was subjected to size-exclusion chromatography using a BioSuite 250 5 µm HR SEC column (Waters Corporation) equilibrated in 50 mM Tris–HCl pH 6.8 and 150 mM NaCl at 1 ml min−1. The column was previously calibrated using gel-filtration standards (Sigma) including β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa) and cytochrome c (12.4 kDa). The results showed that GpgS from M. tuberculosis is a dimer, with no sign of high-order oligomers in the purified preparation (Fig. 1 ▶ b).
2.2. Crystallization
A broad screening of crystallization conditions using the Classics Suite Screen (Qiagen) was performed with an Oryx 8 workstation using the sitting-drop method (Douglas Instruments Ltd). Crystallization conditions resulting in small crystals were manually reproduced and further optimized in terms of pH, precipitant concentration and drop volume using the hanging-drop vapor-diffusion method. All experiments were carried out at 289 K. The best crystals of GpgS were obtained by mixing 1 µl protein solution (10 mg ml−1) in the presence of 1 mM MgCl2 with 1 µl 100 mM sodium cacodylate pH 6.5, 200 mM trisodium citrate and 30%(v/v) 2-propanol. Crystals appeared after 1 d and grew as rods reaching dimensions of 0.08 × 0.03 × 0.02 mm. Crystals of the GpgS–uridine 5′-diphosphate (UDP; Fluka) complex were obtained by mixing 2 µl protein solution (10 mg ml−1) in the presence of 1 mM MgCl2 and 5 mM UDP with 2 µl 10%(v/v) ethanol and 1.5 M NaCl (Fig. 1 ▶ c). Crystals also appeared after 1 d and grew as rhombuses reaching dimensions of 0.04 × 0.03 × 0.03 mm. In both cases, crystals were transferred to a cryoprotectant solution (25% glycerol in the mother liquor) for 5 min and cryocooled in liquid nitrogen prior to data collection.
2.3. Data collection and processing
The crystals of GpgS belonged to the tetragonal space group I4 and contained two molecules per asymmetric unit, corresponding to a Matthews coefficient of 2.25 Å3 Da−1 and a solvent content of 45.28%. X-ray diffraction data were collected from a single crystal to 2.6 Å resolution on the 4.2.2 beamline (λ = 1.00 Å) at the Advanced Light Source (ALS, Berkeley, USA) equipped with an CCD NOIR-1 MBC system detector and were processed using the program d*TREK v.9.9.1 (Rigaku). The crystals of the GpgS–UDP complex also belonged to the tetragonal space group I4 and diffracted to 3.0 Å resolution (Fig. 2 ▶). In both cases, a total of 360 diffraction images were collected with a 0.5° rotation between images. Data-collection statistics are shown in Table 1 ▶.
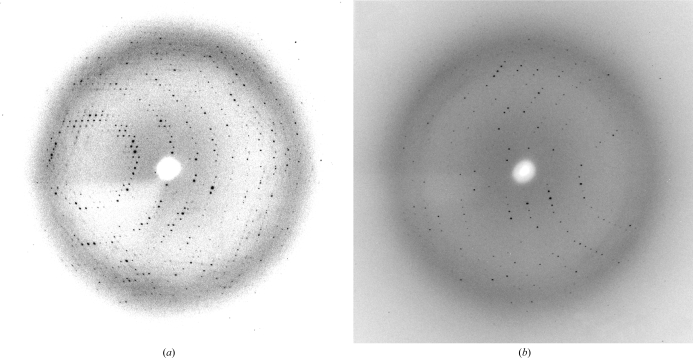
Figure 2.
(a) X-ray diffraction image of an M. tuberculosis GpgS crystal. (b) X-ray diffraction image of an M. tuberculosis GpgS–UDP complex crystal.
Table 1. Data-collection statistics.
Values in parentheses are for the last shell.
| GspS | GspS–UDP | |
|---|---|---|
| Space group | I4 | I4 |
| Crystal system | Tetragonal | Tetragonal |
| Unit-cell parameters (Å) | a = 98.85, b = 98.85, c = 127.64 | a = 98.32, b = 98.32, c = 127.96 |
| Matthews coefficient (Å3 Da−1) | 2.29 | 2.25 |
| Molecules per ASU | 2 | 2 |
| Resolution range (Å) | 2.6 (2.69–2.60) | 3.0 (3.11–3.00) |
| Mosaicity (°) | 1.38 | 0.41 |
| Total observations | 131861 | 88219 |
| Unique reflections | 18835 | 12217 |
| Multiplicity | 7.0 | 7.2 |
| Completeness (%) | 100 (100) | 100 (100) |
| Rmerge† (%) | 10.6 (48.0) | 10.7 (41.3) |
| 〈I/σ(I)〉 | 8.9 (3.0) | 10.3 (3.3) |
R
merge = 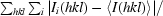
 .
.
3. Results and discussion
Recombinant GpgS from M. tuberculosis (Rv1208) with an N-terminal histidine tag was expressed in E. coli BL21(DE3)pLysS cells as a soluble protein. The enzyme was purified to apparent homogeneity by a combination of metal-ion affinity, anion-exchange and gel-filtration chromatography steps. The final yield was 1.2 mg of GpgS per litre of 2×YT medium culture. Interestingly, when run on polyacrylamide gel electrophoresis under denaturing conditions GpgS proved to be a monomer, in good agreement with the theoretical molecular-weight value of 36.54 kDa (Fig. 1 ▶ a). However, when submitted to gel filtration the native enzyme appeared to have a molecular weight of 69.03 kDa, strongly suggesting that GpgS is a homodimeric protein in solution (Fig. 1 ▶ b).
Crystals of GpgS were found in a number of conditions from the Classics Suite Screen (Qiagen) in which salts and/or organic solvents were general precipitants. As depicted in Fig. 2 ▶, complete data sets were collected for GpgS and the GpgS–UDP complex at 2.6 and 3.0 Å resolution, respectively, using synchrotron radiation. Molecular-replacement methods will be carried out using the X-ray structure of the M. avium ssp. paratuberculosis orthologue (PDB code 1ckj; 83% identity), which was reported to be a putative glycosyltransferase of unknown function (Fulton et al., 2008 ▶), as a search model.
Acknowledgments
We thank Dr Karolin Luger (Department of Biochemistry, Colorado State University) for providing us with full access to her crystallography facilities and Dr Jay C. Nix (4.2.2 beamline, ALS, Berkeley, USA) for help with data collection. This work was supported by NIAID, National Institutes of Health, grant 5R01AI064798-3.
References
- Charnock, S. J. & Davies, G. J. (1999). Biochemistry, 38, 6380–6385. [DOI] [PubMed]
- Bloch, K. (1977). Adv. Enzymol. Relat. Areas Mol. Biol.45, 1–84. [DOI] [PubMed]
- Empadinhas, N., Albuquerque, L., Mendes, V., Macedo-Ribeiro, S. & da Costa, M. S. (2008). FEMS Microbiol. Lett.280, 195–202. [DOI] [PubMed]
- Fulton, Z., Crellin, P. K., Brammananth, R., Zaker-Tabrizi, L., Coppel, R. L., Rossjohn, J. & Beddoe, T. (2008). Acta Cryst. F64, 428–431. [DOI] [PMC free article] [PubMed]
- Gray, G. R. & Ballou, C. E. (1971). J. Biol. Chem.246, 6835–6842. [PubMed]
- Lairson, L. L., Henrissat, B., Davies, G. J. & Withers, S. G. (2008). Annu. Rev. Biochem.77, 521–555. [DOI] [PubMed]
- Lee, Y. C. (1966). J. Biol. Chem.241, 1899–1908. [PubMed]
- Lee, Y. C. & Ballou, C. E. (1964). J. Biol. Chem.239, PC3602–PC3603. [PubMed]
- Maitra, S. K. & Ballou, C. E. (1977). J. Biol. Chem.252, 2459–2469. [PubMed]
- Stadthagen, G., Sambou, T., Guerin, M., Barilone, N., Boudou, F., Korduláková, J., Charles, P., Alzari, P. M., Lemassu, A., Daffé, M., Puzo, G., Gicquel, B., Rivière, M. & Jackson, M. (2007). J. Biol. Chem.282, 27270–27276. [DOI] [PubMed]
- Tuffal, G., Albigot, R., Rivière, M. & Puzo, G. (1998). Glycobiology, 8, 675–684. [DOI] [PubMed]
- Vrielink, A., Ruger, W., Driessen, H. P. & Freemont, P. S. (1994). EMBO J.13, 3413–3422. [DOI] [PMC free article] [PubMed]
- Yabusaki, K. K. & Ballou, C. E. (1979). J. Biol. Chem.254, 12314–12317. [PubMed]