The transfer of sugars is an important process in both biology and biotechnology; the first crystallization of a rhamnosyltransferase is reported.
Keywords: rhamnosyltransferases, WsaF, Geobacillus stearothermophilus
Abstract
The β1,2-rhamnosyltransferase WsaF is involved in the biosynthesis of a polyrhamnan chain which is attached to the surface-layer protein from Geobacillus stearothermophilus NRS 2004/3a. The enzyme belongs to the large retaining GT4 family. To date, no structure of a rhamnosyltransferase has been published. Recombinant purified native WsaF has been crystallized, resulting in crystals that belonged to space group P212121 with unit-cell parameters a = 50.5, b = 56.1, c = 276.8 Å and diffracted to 3.0 Å resolution. Selenomethionine-variant WsaF crystallized in space group P21 with unit-cell parameters a = 75.9, b = 75.5, c = 78.1 Å and diffracted to 2.3 Å resolution.
1. Introduction
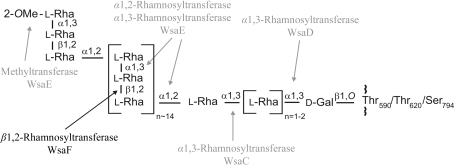
Glycosylation is the most common modification of bacterial surface-layer (S-layer) proteins (Messner et al., 2008 ▶). The intrinsic self-assembly feature of native S-layer (glyco)proteins (Sleytr et al., 2007 ▶) as well as of rationally engineered S-layer neoglycoproteins (Steiner, Hanreich et al., 2008 ▶) into monomolecular arrays with nanometre-scale periodicity makes S-layers ideal matrices for high-density display of tailor-made glycans for interfering with complex biological systems. Envisaged applications of S-layer neoglycoproteins include the fields of receptor mimics, vaccine design and drug delivery using carbohydrate recognition, as well as the fabrication of glycan nanoarrays (Schäffer & Messner, 2004 ▶). WsaF (MW 48.3 kDa; protein accession No. AAR99609) is a β-retaining rhamnosyltransferase which catalyses the formation of a β1,2-linkage in the polyrhamnan biosynthesis of the S-layer protein SgsE in Geobacillus stearothermophilus NRS 2004/3a (Steiner, Novotny et al., 2008 ▶). The enzymes involved in S-layer glycan biosynthesis of G. stearothermophilus NRS 2004/3a are encoded by a polycistronic S-layer glycosylation (slg) gene cluster (GenBank accession No. AF328862; Novotny et al., 2004 ▶). WsaF is one of four rhamnosyltransferases that processively build up the S-layer glycan chain with the structure 2-OMe-α-l-Rhap-(1→3)-β-l-Rhap-(1→2)-α-l-Rhap-(1→[2)-α-l-Rhap-(1→3)-β-l-Rhap-(1→2)-α-l-Rhap-(1→]n=13–182)-α-l-Rhap-(1→[3)-α-l-Rhap]n=1–2-(1→3)-β-d-Galp-(1→, which is O-glycosidically linked to Thr590, Thr620 and Ser794 of the S-layer protein SgsE (Fig. 1 ▶; Schäffer et al., 2002 ▶; Steiner et al., 2006 ▶; Steiner, Novotny et al., 2008 ▶). WsaF transfers a rhamnose residue from dTDP-rhamnose to the α1,2-linked rhamnose in the repeating unit of the glycan chain, as described recently (Steiner, Novotny et al., 2008 ▶). While WsaF belongs to the retaining GT4 family of glycosyltransferases (see http://www.cazy.org/index.html) and thus to the GT-B fold superfamily, the other rhamnosyltransferases of the slg gene cluster are predicted to be members of the inverting GT2 family, which display a GT-A fold. The classification of glycosyltransferases into different families results from overall amino-acid similarities and highly conserved catalytic domains, which reflect the predicted structure of the protein as well as the reaction mechanism (Coutinho et al., 2003 ▶; Lairson et al., 2008 ▶). So far, only six structures of GT4-family members have been published: PimA, a phosphatidyl mannosyltransferase involved in phosphatidyl-myo-inositol mannoside biosynthesis in Mycobacterium smegmatis (Guerin et al., 2007 ▶), WaaG from Escherichia coli, which is an α1,3-glucosyltransferase involved in lipopolysaccharide biosynthesis (Martinez-Fleites et al., 2006 ▶), the avilamycin eurkanate-precursor glycosyltransferase AviT from Streptomyces viridochromogenes (Martinez-Fleites et al., 2006 ▶) and the N-acetylglucosamine transferase MshA from Corynebacterium glutamicum, which is involved in mycothiol biosynthesis (Vetting et al., 2008 ▶), as well as two glycosyltransferases with unknown function, BaGT4 from Bacillus anthracis (Ruane et al., 2008 ▶) and the archaeal WbaZ-1 from Archaeglobus fulgidus (unpublished work; PDB code 2f9f). To date, no structure of a rhamnosyltransferase has been reported. In the present report, we describe the purification, crystallization and preliminary crystallographic analysis of the β1,2-rhamnosyltransferase WsaF from G. stearothermophilus NRS 2004/3a.
Figure 1.
Structure of the S-layer glycoprotein glycan of G. stearothermophilus NRS 2004/3a. The site of action of WsaF is indicated with a black arrow. Modified after Steiner, Novotny et al. (2008 ▶).
2. Experimental
2.1. Expression and purification
E. coli BL21 Star cells transformed with pET28a-WsaF (Steiner, Novotny et al., 2008 ▶) were grown in 10 × 1 l Luria–Bertani broth (LB) supplemented with kanamycin (50 µg ml−1) at 310 K. For the expression of selenomethionine-labeled WsaF, an SeMet minimal medium (Guerrero et al., 2001 ▶) containing 50 µg ml−1 l-SeMet was used. Cells of an overnight culture of E. coli BL21 Star transformed with pET28a-WsaF were centrifuged and washed with sterile MilliQ water and the pellet was resuspended in water. This suspension was used as inoculum for the SeMet minimal medium. Expression of WsaF was initiated by addition of isopropyl β-d-1-thiogalactopyranoside to a final concentration of 1 mM to a culture with an OD600 of ∼0.8 and cultivation was continued for an additional 20 h at 293 K. Cells were harvested, washed with cold saline and resuspended in 20 mM cold buffer A [20 mM sodium phosphate buffer pH 7.4 containing 0.5 M NaCl and 20 mM imidazole (10 mM β-mercaptoethanol was added for SeMet-labeled WsaF)]. Cells were disrupted by passage through a cell disrupter (Constant Cell Disruption Systems, Daventry, England) at 207 MPa and the suspension was centrifuged for 20 min at 32 800g. The supernatant was filtered through a 0.45 µm syringe filter and applied onto a 5 ml HisTrap HP prepacked column (GE Healthcare, Uppsala, Sweden) equilibrated in buffer A. Proteins were eluted using a step gradient to 500 mM imidazole (50, 100, 150, 200, 300 and 500 mM imidazole) in buffer A. Fractions were analyzed by SDS–PAGE, pooled and dialyzed overnight against buffer B [25 mM Tris–HCl pH 8.0, 250 mM NaCl, 10% glycerol (10 mM dithiothreitol was added for SeMet-labeled WsaF)]. The dialyzed solution was concentrated to 5 ml using Vivaspin 20 Centrifugal Filter Units (10 000 Da molecular-weight cutoff; Sartorius) and applied onto a Superdex 200 (16/60) column (GE Healthcare) equilibrated in buffer B. WsaF eluted as a single peak and on SDS–PAGE displayed a major band corresponding to the monomer and a faint band at the size of the dimer. The fractions containing WsaF were pooled and concentrated to a protein concentration of 6.6 mg ml−1. Protein concentration was estimated using a NanoDrop 1000 UV–Vis spectrophotometer. Protein identity was confirmed by mass spectrometry. Purified WsaF was flash-frozen in liquid nitrogen and stored at 193 K.
Prior to crystallization of SeMet-labeled WsaF, the N-terminal His tag was cleaved using one unit of thrombin (GE Healthcare) per 100 µg protein for 20 h at 293 K. Completeness of cleavage was analyzed by SDS–PAGE. Thrombin was removed by application of the sample to a 1 ml HiTrap benzamidine column (GE Healthcare). Cleaved SeMet-labeled WsaF was concentrated to 6.5 mg ml−1, flash-frozen in liquid nitrogen and stored at 193 K. Incorporation of SeMet, as well as the cleavage of the His tag, was confirmed by ES–QTOF mass spectrometry.
2.2. Crystallization
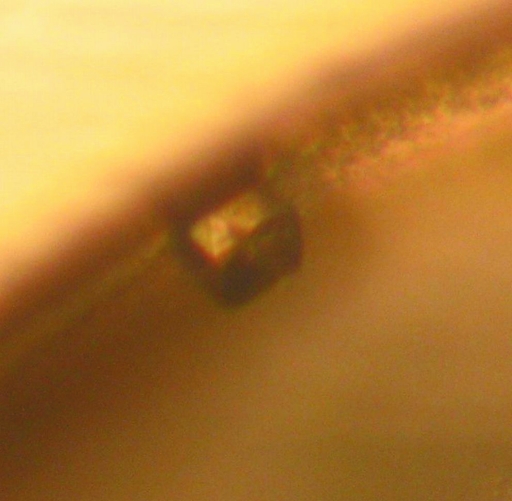
For native WsaF, broad screening of crystallization conditions using Emerald Wizard I and II Screens (Emerald Biosystems) was performed by the hanging-drop vapour-diffusion method (protein concentration 6.6 mg ml−1; drops were formed by mixing 1 µl protein solution with 1 µl well solution). Crystallization conditions resulting in small crystals were further optimized by varying the buffer, pH, salt, PEG concentration and drop volume. All experiments were carried out at 293 K. The best crystals were obtained by mixing 1 µl protein solution (6.6 mg ml−1) with 1 µl well solution consisting of 25%(w/v) PEG 1000, 200 mM NaCl and 100 mM Tris–HCl pH 8.0. Crystals appeared after 4 d at 293 K (Fig. 2 ▶) and grew to 50 µm in size. Prior to data collection, crystals were transferred to a cryoprotectant solution (well solution) and flash-frozen in liquid nitrogen.
Figure 2.
Crystal of native WsaF from G. stearothermophilus NRS 2004/3a grown in 25%(w/v) PEG 1000, 200 mM NaCl and 100 mM Tris–HCl pH 8.0.
However, this crystallization condition was not optimal for SeMet-labeled WsaF containing DTT. Thus, screening of crystallization conditions using Nextal PEGs, pHClear, JCSG and Classics was performed using a Cartesian Honeybee 963 dispensing system (Genomic Solutions, Ann Arbor, Michigan, USA) with the sitting-drop vapour-diffusion method (two protein concentrations were used, 5.7 and 3.6 mg ml−1; the drops consisted of 150 nl protein and 150 nl well solution). Crystal conditions were further optimized manually using the hanging-drop vapour-diffusion method by mixing 1 µl protein solution (4 mg ml−1) and 1 µl well solution. The best crystals by visual examination were obtained using 18%(w/v) PEG 3350 and 0.22 M ammonium chloride. The crystals were cryoprotected by addition of 20% glycerol to the well solution and flash-frozen in liquid nitrogen.
3. Results and discussion
3.1. Preliminary crystallographic characterization
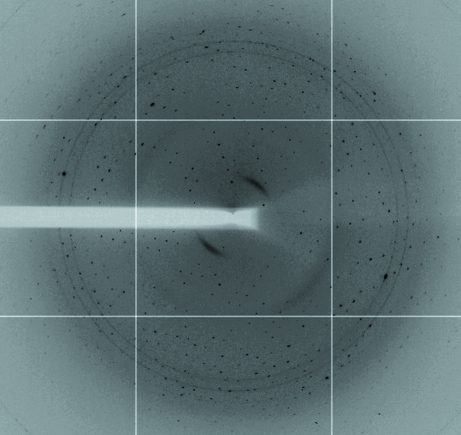
Crystals of native WsaF belonged to the orthorhombic space group P212121 and contained two molecules per asymmetric unit, corresponding to a Matthews coefficient of 2.0 Å3 Da−1 and a solvent content of 38%. X-ray diffraction data were collected from a single crystal to 3.0 Å resolution on the ID14-4 beamline (λ = 0.979) at the ESRF (Grenoble, France) equipped with an ADSC Q315 detector. The data were collected over a range of 140° with 0.2° steps. The data were processed with HKL-2000 (Otwinowski & Minor, 1997 ▶). Data-collection statistics are shown in Table 1 ▶. MOLREP from the CCP4 program suite (Collaborative Computational Project, Number 4, 1994 ▶) was used to calculate a self-rotation function. The χ = 180° section of the self-rotation function revealed a twofold noncrystallographic axis intersecting the crystallographic a and b axes at 45°. This indicates a possible noncrystallographic dimer. The use of Phaser (Collaborative Computational Project, Number 4, 1994 ▶) for molecular replacement with other GT4-family glycosyltransferases was unsuccessful. Thus, SeMet-labeled WsaF was produced for structural determination using single-wavelength anomalous diffraction (SAD). The crystals of SeMet-labeled WsaF belonged to the primitive monoclinic space group P21 and contained two molecules per asymmetric unit (Matthews coefficient of 2 Å3 Da−1 and solvent content of 38%). X-ray diffraction data were collected from a single crystal to 2.28 Å resolution on the I03 beamline (λ = 0.9764 Å) at Diamond (Didcot, England) equipped with an ADSC Q315 detector (Fig. 3 ▶). Four data sets were combined, resulting in an overall 1260° of data. The data were processed with HKL-2000. The self-rotation function showed no clear twofold axis. The data indicated the presence of anomalous scattering. SHELXD (Sheldrick, 2008 ▶) indicated that the anomalous signal extends to 3.2 Å based on a ΔF/σ(ΔF) cutoff of 0.8. Thus far, SHELXD has not located the substructure and our efforts continue.
Table 1. Data-collection statistics.
Values in parentheses are for the last resolution shell
| Apo WsaF | SeMet-labeled WsaF | |
|---|---|---|
| Space group | P212121 | P21 |
| Crystal system | Orthorhombic | Monoclinic |
| Unit-cell parameters (Å, °) | a = 50.5, b = 56.1, c = 276.8, α = β = γ = 90 | a = 75.9, b = 75.5, c = 78.1, α = γ = 90, β = 108.3 |
| Matthews coefficient (Å3 Da−1) | 2 | 2 |
| Molecules per ASU | 2 | 2 |
| Resolution range (Å) | 47.5–3.0 | 47.3–2.28 |
| Mosaicity (°) | 0.5 | 0.8 |
| Total observations | 573232 | 1764457 |
| Unique reflections | 15120 | 39051 |
| Completeness (%) | 90.9 (56.1) | 99.9 (98.9) |
| Rmerge (%) | 8.5 (32.7) | 15.8 (58) |
| Multiplicity | 4.8 | 14.4 |
| I/σ(I) | 15.7 (1.9) | 14.1 (3.3) |
Figure 3.
X-ray diffraction image of a crystal of SeMet-labeled G. stearothermophilus NRS 2004/3a WsaF.
Acknowledgments
X-ray diffraction data were collected at the ESRF (Grenoble, France) and at Diamond (Didcot, England). This work was supported by the Austrian Science Fund Project J2746 (to KS). The Wellcome Trust supported this science through a program grant to JHN.
References
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Coutinho, P. M., Deleury, E., Davies, G. J. & Henrissat, B. (2003). J. Mol. Biol.328, 307–317. [DOI] [PubMed]
- Guerin, M. E., Kordulakova, J., Schaeffer, F., Svetlikova, Z., Buschiazzo, A., Giganti, D., Gicquel, B., Mikusova, K., Jackson, M. & Alzari, P. M. (2007). J. Biol. Chem.282, 20705–20714. [DOI] [PubMed]
- Guerrero, S. A., Hecht, H.-J., Hofmann, B., Biebl, H. & Singh, M. (2001). Appl. Microbiol. Biotechnol.56, 718–723. [DOI] [PubMed]
- Lairson, L. L., Henrissat, B., Davies, G. J. & Withers, S. G. (2008). Annu. Rev. Biochem.77, 521–555. [DOI] [PubMed]
- Martinez-Fleites, C., Proctor, M., Roberts, S., Bolam, D. N., Gilbert, H. J. & Davies, G. J. (2006). Chem. Biol.13, 1143–1152. [DOI] [PubMed]
- Messner, P., Steiner, K., Zarschler, K. & Schäffer, C. (2008). Carbohydr. Res.343, 1934–1951. [DOI] [PMC free article] [PubMed]
- Novotny, R., Schäffer, C., Strauss, J. & Messner, P. (2004). Microbiology, 150, 953–965. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Ruane, K. M., Davies, G. J. & Martinez-Fleites, C. (2008). Proteins, 73, 784–787. [DOI] [PubMed]
- Schäffer, C. & Messner, P. (2004). Glycobiology, 14, 31R–42R. [DOI] [PubMed]
- Schäffer, C., Wugeditsch, T., Kählig, H., Scheberl, A., Zayni, S. & Messner, P. (2002). J. Biol. Chem.277, 6230–6239. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sleytr, U. B., Huber, C., Ilk, N., Pum, D., Schuster, B. & Egelseer, E. M. (2007). FEMS Microbiol. Lett.267, 131–144. [DOI] [PubMed]
- Steiner, K., Hanreich, A., Kainz, B., Hitchen, P. G., Dell, A., Messner, P. & Schäffer, C. (2008). Small, 4, 1728–1749. [DOI] [PMC free article] [PubMed]
- Steiner, K., Novotny, R., Werz, D. B., Zarschler, K., Seeberger, P. H., Hofinger, A., Kosma, P., Schäffer, C. & Messner, P. (2008). J. Biol. Chem.283, 21120–21133. [DOI] [PMC free article] [PubMed]
- Steiner, K., Pohlentz, G., Dreisewerd, K., Berkenkamp, S., Messner, P., Peter-Katalinić, J. & Schäffer, C. (2006). J. Bacteriol.188, 7914–7921. [DOI] [PMC free article] [PubMed]
- Vetting, M. W., Frantom, P. A. & Blanchard, J. S. (2008). J. Biol. Chem.283, 15834–15844. [DOI] [PMC free article] [PubMed]