Abstract
Pleiotropy plays a central role in theories of adaptation, but little is known about the distribution of pleiotropic effects associated with different adaptive mutations. Previously, we described the phenotypic effects of a collection of independently arising beneficial mutations in Escherichia coli. We quantified their fitness effects in the glucose environment in which they evolved and their pleiotropic effects in five novel resource environments. Here we use a candidate gene approach to associate the phenotypic effects of the mutations with the underlying genetic changes. Among our collection of 27 adaptive mutants, we identified a total of 21 mutations (18 of which were unique) encompassing five different loci or gene regions. There was limited resolution to distinguish among loci based on their fitness effects in the glucose environment, demonstrating widespread parallelism in the direct response to selection. However, substantial heterogeneity in mutant effects was revealed when we examined their pleiotropic effects on fitness in the five novel environments. Substitutions in the same locus clustered together phenotypically, indicating concordance between molecular and phenotypic measures of divergence.
Keywords: parallel evolution, beneficial mutations, pleiotropy, genetics of adaptation
… in dealing with such a complex character as selective value, the essential uniqueness of each allele must never be forgotten.(Sewall Wright 1968, p. 62)
1. Introduction
A major challenge in the study of adaptation is to demonstrate a causal relationship between the action of natural selection on different phenotypes and the underlying genetic changes (Orr & Coyne 1992; Jones 1998; Nachman 2005). This difficulty was aptly described as a ‘chasm’ by Phillips (2005), who emphasized that many studies fall into two broad categories: those that can identify molecular variation, but have limited knowledge of its adaptive significance, and those that can identify ecologically important traits, but have limited knowledge of their genetic bases. Understanding the mapping between genotypes and phenotypes is central to determining why selection leads to divergence and diversification in some instances, and convergence and parallelism in others (Elena & Lenski 2003). Unfortunately, the relationship between genotype, phenotype and adaptation has been demonstrated in only a handful of cases to date (Wichman et al. 1999; Cooper et al. 2001; Bradshaw & Schemske 2003; Nachman et al. 2003; Mundy et al. 2004; Colosimo et al. 2005; Kwiatkowski 2005; Herring et al. 2006; Hoekstra et al. 2006; Knight et al. 2006).
An important question concerns the extent to which similar phenotypic changes are driven by similar changes at the molecular level (Doolittle 1994; Wood et al. 2005; Hegreness & Kishony 2007). Little is known about the distribution of phenotypic effects of different alleles at a given locus (Phillips 2005), and even ‘strict’ definitions of parallelism implicitly assume that different substitutions in the same locus are phenotypically equivalent (Schluter et al. 2004). However, as Wright (1968) cautioned, alleles of even a single locus can differ from one another not just quantitatively, but also qualitatively, owing to their unique pleiotropic effects. Thus, different mutations may be substituted in separate lineages because they provide similar improvements to a selected trait, yet they may cause diverse suites of pleiotropic effects that become important only after an environmental change or some further change in the gene pool. Differences in phenotype that arise from heterogeneous pleiotropic effects can thereby influence the likelihood of subsequent substitutions, leading to divergent evolutionary trajectories over the long term (Mani & Clarke 1990; Travisano et al. 1995a,b; Hodgkin 1998; Bull et al. 2000; MacLean & Bell 2003; MacLean et al. 2004).
Previously, we described the phenotypic effects of a large sample of spontaneous beneficial mutations that arose from a common ancestor in Escherichia coli (Ostrowski et al. 2005). We examined the fitness effects of these mutations in the glucose environment in which they evolved, as well as their pleiotropic (unselected) effects in five novel resource environments. Here we identify the genetic bases of these adaptations by sequencing candidate loci. By associating our earlier phenotypic measures of divergence with the underlying genetic changes, we assess the extent to which parallel and divergent phenotypic evolution resulted from the substitution of unique beneficial mutations, and we disentangle the contributions of directly selected and pleiotropic effects of mutations to that phenotypic evolution.
2. Material and methods
(a) Collection of mutants
Isolation of the mutant clones used in our study has been described in detail elsewhere (Rozen et al. 2002). Briefly, 30 replicate populations were founded with each of two clones that were isogenic except for a single neutral marker, indicating the ability (Ara+) or inability (Ara−) to catabolize arabinose. The marker difference causes clones to form red (Ara−) or white (Ara+) colonies when plated on tetrazolium-arabinose indicator agar, but does not affect fitness in any of the experimental environments considered in this paper (Ostrowski et al. 2005). Populations were founded with Ara+ and Ara− clones in the following ratios: 1 : 100; 1 : 10; 1 : 1; 10 : 1; 100 : 1. Cultures were propagated daily in a glucose minimal medium, according to the protocol given in Lenski et al. (1991). The populations were plated periodically to assess the relative proportions of the Ara+ and Ara− marker states; a sustained increase in the frequency of either state indicated that a beneficial mutation had arisen in this subpopulation, at which point clones of both marker states were isolated and saved. Owing to the stochastic occurrence of beneficial mutations, evolved clones were collected at varying times, but none were collected after 400 generations. The objective was that each isolated clone would contain a single beneficial mutation; our results indicate that this goal was largely, but not perfectly, achieved. For our study, we analysed 27 mutants from the 30 populations; three putative mutants were excluded because they had no significant fitness gain in the environment where they evolved (Ostrowski et al. 2005).
(b) Competition assays
The competition assays are described in detail in Ostrowski et al. (2005). All clones competed against their common ancestor of the opposite marker state in six carbon sources: glucose; mannitol; maltose; N-acetylglucosamine (NAG); galactose; and melibiose. For each assay, the mutant was mixed in equal proportions with its ancestor and the two strains were allowed to grow and compete for 24 hours in each type of medium. Mixed cultures were plated at the start and the end of the assay to assess the changes in abundance. Relative fitness was calculated as the ratio of the two competitors' realized Malthusian parameters (Lenski et al. 1991), which are their net rates of growth in competition. Each fitness assay was replicated three-fold in each novel resource and 15-fold in glucose, the selected environment.
(c) DNA sequencing
Previous analyses of a long-term evolution experiment, initiated from the same ancestor and employing the same glucose-limited environment, identified five genes or gene regions as candidate loci for adaptation: pykF; nadR; hokB/sokB; spoT; and an upstream non-coding region of pbpA-rodA (Schneider et al. 2000; Cooper et al. 2001, 2003; Woods et al. 2006). Primers were designed to cover the overlapping regions of these genes or gene regions. PCR products were purified on a GFX column and the sequencing was done using an ABI automated sequencer. The genes or gene regions of interest were sequenced in their entirety for all clones at least once. All sequences were compared with that of the ancestor, and any conflicts that could not be resolved by eye were re-sequenced. Candidate mutations were confirmed only when they could be detected on both strands and in sequences arising from a minimum of two independent PCRs.
(d) Identical mutations
Of the 21 mutations found in our study (see below), three pairs were identical at the nucleotide sequence level. These identical mutations raise two methodological issues. First, are they truly independent, or might they have resulted from cross-contamination between evolving populations? Second, should they be included or excluded from the statistical analyses? Regarding the possibility of contamination, two lines of evidence support their independence. (i) The observed pairwise proportion of substitutions that were identical can be calculated for the 20 clones in which mutations were found in sequenced genes. The resulting value of 1.6% (three pairs identical out of 190 possible pairs) is similar to the value of 2.1% obtained for the long-term evolving populations (Woods et al. 2006), where markers embedded in that experiment, along with multiple different mutations found in each population, absolutely exclude any cross-contamination. (ii) One spoT mutation found in our study is identical to a mutation previously found in one of the long-term populations (Cooper et al. 2003); that previous clone has several other mutations that are not present in our clone, and thus we are certain that the exact same spoT mutation arose independently in these two populations. Given that these few identical mutations evolved independently, we retained them in most of our statistical analyses. However, we also repeated all our analyses on reduced datasets where we excluded one member of each pair; the exclusion of these genotypes did not alter any of our conclusions.
(e) Statistical analyses
First, to assess the phenotypic similarity of genotypes with mutations in the same locus, we performed an analysis to cluster the genotypes according to their fitness effects in the six resources. The distance metric used for the analysis was a normalized Euclidean distance. Clustering proceeded by an algorithm that iteratively joined the closest genotype to a given cluster, where the location of the cluster is based on the average distance among points in the cluster (SYSTAT 2002); other methods produced similar results. Second, to determine whether there was a significant association between the phenotypic effects of mutations and the underlying genetic changes, we performed an analysis of similarity (ANOSIM) using the PRIMER-E software package (Clarke 1993; Clarke & Gorley 2001). The analysis uses a distance matrix to rank all pairs of genotypes from most similar (lowest rank) to least similar (highest rank), and then calculates the difference in the mean rank of the between-group comparisons to that of the within-group comparisons. Statistical significance is determined by a permutation test, where gene names are assigned at random to genotypes, and the analysis is repeated. Where feasible, all possible permutations of the dataset were performed; otherwise, 1000 permutations were performed. Finally, one-way and two-way analyses of variance (ANOVA) tests were performed in SAS (SAS Institute 1999). For the nested two-way ANOVA, F-tests were constructed in a manner analogous to that described in Goldberg & Scheiner (2001), except that a Satterthwaite correction was used where the data were unbalanced. For simplicity, we round the d.f. to the nearest whole number when we present such F-tests in the text.
3. Results
(a) Molecular basis of adaptation
Sequencing the five genes or gene regions in 27 clones uncovered 21 mutations: 13 in spoT; 5 in nadR; and one each in pbpA-rodA, pykF and hokB/sokB. Eighteen of the mutations were unique, while three were found in two different clones. A complete list of the identified mutations is provided in the electronic supplementary material. We detected only a single mutation in each clone, with one exception (a pykF-spoT double mutant in genotype 9990; see electronic supplementary material), consistent with the expectation that these clones should typically harbour single beneficial mutations (Rozen et al. 2002; Ostrowski et al. 2005).
In addition to sequencing candidate genes, we performed growth rate assays in ribose; deletion mutations in the ribose operon (rbs) that result in loss of growth on that resource are common in populations evolved under the same conditions. These mutations occur at unusually high frequency owing to an adjacent mobile genetic element. They confer a slight beneficial effect and they reached high frequency by 500 generations in 7 out of 12 long-term populations, with their rapid spread promoted by hitch-hiking with other beneficial mutations of large effect (Cooper et al. 2001). Thus, rbs deletion mutations are not only candidates for adaptive mutations in that operon, but also should serve as sentinels for detecting secondary beneficial mutations. However, all 27 evolved clones grew well on ribose, indicating that their ribose operons remained intact (data not shown).
(b) Concordance between measures of genotypic and phenotypic similarity
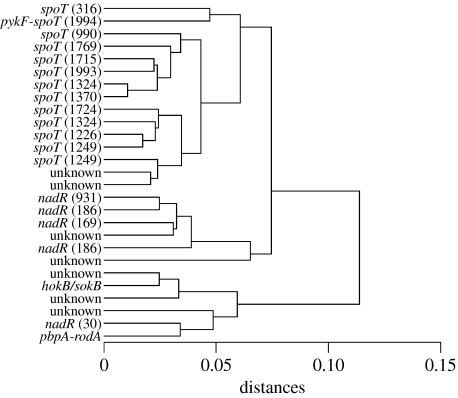
We used hierarchical cluster analysis to group mutants according to their fitness effects on six different resources and then overlaid the cluster diagram with the locus in which mutations were subsequently identified (figure 1). All the 27 clones included in this analysis had significant fitness gains relative to their ancestor in glucose (Ostrowski et al. 2005). In seven clones, we found no mutations in any of the five candidate genes that we sequenced; however, all of these ‘unknowns’ must carry beneficial mutations in one or more other unidentified loci, given their increased fitness in the glucose environment.
Figure 1.
Hierarchical cluster analysis based on estimates of relative fitness of 27 mutant clones on six different resources. The phenotypic distance metric is a normalized Euclidean distance. The identities of the loci where mutations were found in candidate genes are shown on the left. Numbers in parentheses indicate the location of the mutation within the corresponding gene, from the first position of the start codon.
A visual inspection of the cluster diagram suggests that mutations in the same locus tend to cluster together, which indicates that they produce similar suites of phenotypic effects (figure 1). To quantify the statistical support for this association, we performed an ANOSIM, as summarized in table 1. The ANOSIM tests specifically whether clones with mutations in the same locus are phenotypically more similar to one another than are clones with mutations in different loci. The global test shows an R-statistic equal to 0.800, which is highly significant (p≤0.001). The ANOSIM on individual pairs of loci also confirms that there are significant phenotypic differences between those mutations in nadR and spoT. Other pairwise comparisons between loci were not significant, but the number of mutations was always very small for one of the loci. In all comparisons involving the spoT locus, the observed value of the R-statistic was the most extreme of all possible permutations (table 1), indicating that spoT is highly divergent in its fitness effects compared with the other loci.
Table 1.
ANOSIM testing for phenotypic similarity among mutations at the same versus different loci.
| comparisons | R-statistic | significance level | possible permutations | permutations performed | permutations≥observed R |
|---|---|---|---|---|---|
| global test | 0.800 | <0.001 | 7 054 320 | 1000 | 0 |
| pairwise tests | |||||
| spoT versus nadR | 0.821 | <0.001 | 6188 | 1000 | 0 |
| spoT versus spoT and pykF | 0.833 | 0.077 | 13 | 13 | 1 |
| spoT versus hokB/sokB | 0.902 | 0.077 | 13 | 13 | 1 |
| spoT versus pbpA-rodA | 0.942 | 0.077 | 13 | 13 | 1 |
| nadR versus spoT and pykF | 0.920 | 0.167 | 6 | 6 | 1 |
| nadR versus hokB/sokB | −0.160 | 0.667 | 6 | 6 | 4 |
| nadR versus pbpA-rodA | 0.320 | 0.333 | 6 | 6 | 2 |
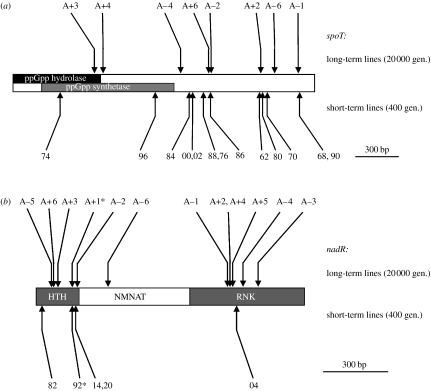
While it is the concordance between phenotypic and molecular patterns that is most noticeable, the exceptions are also interesting, especially in the light of additional information. For example, two of the identified spoT mutations are phenotypically divergent from all other mutants with substitutions in this locus (figure 1). One of these genotypes carries a second mutation at the pykF locus, which may account for its outlier status. However, the other genotype, spoT(316), is the only spoT mutation in the region of sequence overlap between the ppGpp synthetase and hydrolase functional domains of the gene (figure 2a), although the boundaries of these regions are not known precisely (Gentry & Cashel 1996). One possibility, therefore, is that the fitness effects of mutations in this region differ from those in other regions of the gene. More generally, the relative scarcity of beneficial mutations in the synthetase and hydrolase regions, as opposed to the C-terminal regulatory domain (figure 2a), may reflect functional constraints on the types of mutations that can produce the beneficial effect. This concentration of beneficial mutations in the C-terminal domain is seen not only among the mutations found in these single mutation clones (11 out of 13 spoT mutations in this domain) but also in the lines that evolved for 20 000 generations during a long-term experiment in the same environment (6 out of 8 mutations in this domain; Cooper et al. 2003).
Figure 2.
(a) Location of mutations found in the spoT locus after 20 000 generations (top; Cooper et al. 2003) and associated with the first selective sweep within 400 generations (bottom; this study). Clone identification numbers listed in table S1 in the electronic supplementary material are abbreviated here to include only their last two digits, which are unique to each clone (e.g. 9990 is listed as 90). The two shaded regions of the gene correspond to the ppGpp hydrolase and synthetase domains (Gentry & Cashel 1996). Arrows indicate locations of mutations, from the N-terminus (left) to the C-terminus (right); mutations that affect the same codon are indicated by a single arrow with multiple clone labels. (b) Location of mutations found in the nadR locus after 20 000 generations (top; Woods et al. 2006) and associated with the first selective sweep within 400 generations (bottom; this study). The approximate domain boundaries are indicated, as described in Kurnasov et al. (2003): HTH, helix-turn-helix required for the repressor function; NMNAT, NMN adenylyltransferase; RNK, ribosylnicotinamide kinase. Asterisks indicate mutations caused by the insertion of IS150 elements.
Beneficial mutations are also confined to particular regions of the nadR gene (figure 2b). Studies in E. coli and Salmonella enterica have shown that NadR is a protein that both regulates nicotinamide adenine dinucleotide (NAD) biosynthesis and is involved in the transport of NAD precursors into the cell (Penfound & Foster 1996; Raffaelli et al. 1999; Kurnasov et al. 2003; Grose et al. 2005). These different functions appear to be mediated through three different domains of the NadR protein. The N-terminal domain forms a DNA-binding helix-turn-helix motif that regulates NAD biosynthetic genes by binding to downstream operator sequences. The central portion of the gene confers weak adenylyltransferase activity, catalysing the conversion of the NAD precursor nicotinamide mononucleotide (NMN) to NAD. Finally, comparative studies suggest that the C-terminal region confers ribosylnicotinamide kinase activity, mediating the phosphorylation of N-ribosylnicotinamide to produce NMN, which can be transported across the inner membrane of the cell (Kurnasov et al. 2003).
For the single mutation lines analysed here and from a previous analysis of this gene in the long-term lines (Woods et al. 2006), all but one of the 17 nadR mutations arose in either the N- or C-terminal regions (figure 2b). It is also interesting that the nadR(186) and nadR(931) mutations, which impact the N- and C-terminal domains, respectively, nonetheless cluster together based on their phenotypic effects (figure 1). Although the domains have distinct biochemical functions, this clustering implies that these mutations have similar consequences across the set of resource environments where fitness effects were assayed. On the other hand, the nadR(30) mutation does not cluster phenotypically with the four other nadR mutations in our study. This particular mutation is a single base-pair deletion, near the start of the gene, which causes a translational stop at codon 14. This premature stop codon might account for its phenotypic difference, although several other insertion and deletion mutations in the N-terminal region of nadR (genotypes 14, 20 and 92; figure 2) cluster phenotypically with the nadR mutation in the C-terminal region (figure 1). Notably, no mutations in the short-term lines (and only one in the long-term lines) arose in the central portion of the nadR gene, which encodes the adenylyltransferase function. The regulatory function of this domain has not been studied in E. coli, but in S. enterica some mutations have been shown to cause a ‘super-repressor’ phenotype and loss of transport function (Penfound & Foster 1996; Grose et al. 2005). In any case, the clustering of the mutations in certain domains is again consistent with constraints on the types of mutations that are beneficial under the particular environmental conditions where they evolved. In this instance, most beneficial mutations are found in those domains where mutations are likely to reduce, rather than enhance, the function of the NAD repressor.
It is also very interesting that many of the phenotypic clusters encompass mutations at multiple loci (including ‘unknown’), even the well-defined cluster that contains 11 of the 13 spoT mutations found here (figure 1). Although the identity of the gene in which some mutation produces these similar phenotypic effects may be unknown, the fact that a mutation truly exists in each clone is known because all of the clones included in this study showed significant fitness improvement relative to their ancestor in the glucose environment where they evolved (Ostrowski et al. 2005). The two unknown mutations that are similar in their phenotypic effects to mutations in spoT are especially interesting because previous research on the long-term lines also demonstrated the existence of a mutation at some unknown locus that produced similar effects on global gene expression profiles to those caused by a beneficial spoT mutation (Cooper et al. 2003). More generally, our data also indicate that beneficial mutations at different loci can sometimes have similar phenotypic effects, despite the overall concordance between the affected locus and phenotypic effects demonstrated by the ANOSIM (table 1).
One final point to note in this section is that the three clone pairs having identical mutations—nadR(186), spoT(1324) and spoT(1249)—cluster with other clones that carry mutations in the same gene (figure 1). However, the clones comprising each pair are not each other's closest neighbours. The observation that two clones with the exact same mutation are not always more similar than the clones with different mutations in the same locus indicates that some of the observed variability simply reflects measurement error in the fitness assays. As noted in §2, we have repeated the ANOSIM and other analyses on reduced datasets from which we excluded one member of each pair that shared identical mutations, and the overall concordance between genotype and phenotype remained highly significant.
(c) Targets of selection
To develop an understanding of how the six different resources individually and collectively contribute to the observed phenotypic clustering, we performed a principal components analysis (PCA). The first and second principal components explain 46.0% and 24.3% of the variance, respectively. The factor loadings are shown in figure 3. Three of the resources—glucose, NAG and mannitol—all share a common uptake mechanism, which is the phosphotransferase system (PTS). It is striking that the vectors for these resources are virtually indistinguishable in their contributions to the first two principal components. By contrast, the three non-PTS resources—maltose, galactose and melibiose—are divergent from both the PTS resources and one another, consistent with previous work suggesting that the PTS is an important target of selection in the glucose-limited evolution environment (Travisano & Lenski 1996). Previous work on these mutations identified strong positive correlations between fitness in glucose and fitness in NAG, mannitol and maltose (Ostrowski et al. 2005). However, the correlation between fitness in glucose and maltose was puzzling in the light of its classification as a non-PTS resource. The PCA thus suggests that there is some underlying heterogeneity in the correlated response of these mutants to maltose when compared with the PTS resources, which was not evident based solely on the individual correlations (figures 3 and 4).
Figure 3.
Factor loadings plot for the PCA. Variables included in the analysis were relative fitness in six resources: GLU, glucose; NAG, N-acetylglucosamine; MAN, mannitol; MAL, maltose; GAL, galactose; and MEL, melibiose. PTS resources share a common mechanism for transport across the inner membrane of the cell, whereas non-PTS resources use a variety of different mechanisms.
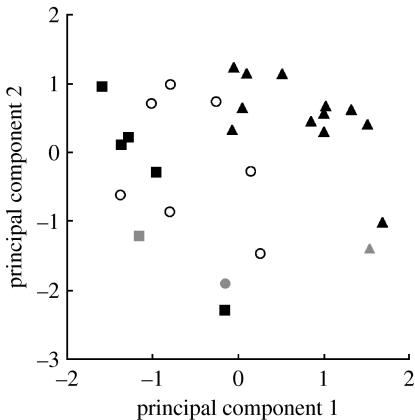
Figure 4.
Plot of the genotypes according to their factor scores for the first two principal components. The identity of the genes where mutations were found has been overlaid onto the plot: black triangles, spoT; black squares, nadR; grey circles, pbprod A; grey squares, hokB/sokB; grey triangles, spoT and pykF; open circles, unknown.
(d) Selected versus pleiotropic effects of beneficial mutations
To examine variation in the directly selected effects of the 19 single mutations that were mapped to known genes, we performed an ANOVA on the mutations' fitness effects in glucose, where the independent mutations were nested within their corresponding locus. As shown in table 2, neither the affected locus nor the mutation within a locus had significant effects on performance in glucose. However, the picture was more complex when we ran one-way ANOVAs separately for the spoT and nadR mutants. These subsidiary analyses indicated that the five mutations in nadR were variable in their fitness effects in glucose (F4,69=3.82, p=0.007), while the 12 single mutations in spoT were not (F11,167=1.28, p=0.239). The overall pattern is thus one of strong but not perfect parallelism, both within and across loci, when the focal trait is the direct response to selection (i.e. fitness in the glucose medium).
Table 2.
ANOVA on fitness effects of 19 independent beneficial mutations, each mapped to a known locus, in the glucose medium where they evolved.
| source | d.f. | MS | F | p |
|---|---|---|---|---|
| locus | 3 | 0.0125 | 2.49 | 0.100 |
| mutation (locus) | 15 | 0.0050 | 1.30 | 0.201 |
| error | 265 | 0.0039 |
In contrast to the near-uniformity among mutations in their selected effects in the glucose medium, we see substantial variation in their pleiotropic effects as determined by correlated responses to the novel resource environments. As summarized in table 3, there are significant interactions for both resource×locus and resource×mutation (locus), in addition to significant main effects of locus and resource. The significant resource×locus interaction indicates that the different genes harbouring beneficial mutations, while similar in the benefits they confer in the selected environment, entail different suites of pleiotropic effects across the set of unselected resources. By extension, the resource×mutation (locus) interaction indicates variability even among different alleles of the same locus. This latter point, however, is sensitive to the inclusion of a particular nadR mutant, the same one that fails to cluster with the other nadR mutants (figures 1 and 4) and also underlies the significant variation among nadR mutants in glucose. When this mutation was dropped from the analysis, the resource×mutation (locus) interaction is no longer significant (p=0.718). This dependence could indicate that this nadR mutation is qualitatively different from the others. However, an alternative possibility is that this particular clone bears some unknown secondary mutation—in which case, we cannot be confident that the phenotypes we measure result solely from different substitutions in the nadR locus. In any case, this dependence does not impact our main conclusion that mutations in different loci have different phenotypic effects, because excluding this atypical nadR mutant would strengthen that result. Taken together, our results demonstrate that different beneficial mutations, which provide a similar selective benefit in the environment where they arose, nonetheless entail divergent suites of pleiotropic effects in novel environments.
Table 3.
ANOVA on fitness effects of 19 independent beneficial mutations in five novel resource environments.
| source | d.f. | MS | F | p |
|---|---|---|---|---|
| resource | 4 | 0.3336 | 10.54 | 0.0006 |
| locus | 3 | 0.1402 | 4.11 | 0.036 |
| mutation (locus) | 15 | 0.0029 | 0.92 | 0.540 |
| resource×locus | 12 | 0.0358 | 7.67 | <0.0001 |
| resource×mutation (locus) | 60 | 0.0047 | 1.48 | 0.0253 |
| error | 189 | 0.0032 |
4. Discussion
Evolutionary biology suffers from a paucity of examples where both the genetic basis of an adaptive change and its phenotypic consequences are known. Large samples of such well-characterized beneficial mutations are necessary for identifying the multiple paths available to adaptive evolution, the similarities and differences between the paths and the consequences that each step along a given path entails for subsequent evolution (Travisano et al. 1995a; Lenski et al. 2003; Weinreich et al. 2006; Ostrowski et al. 2007; Poelwijk et al. 2007).
Here we used a candidate gene approach to identify the molecular basis of a collection of 27 beneficial mutations in E. coli. We found many instances of parallelism at the gene level, with 13 beneficial mutations in one gene and five in another gene. We even found a few mutations that were identical at the nucleotide level, as well as some other mutations in the same or nearby codons to those found previously in analyses of lines that evolved for 20 000 generations under the same conditions (Cooper et al. 2003; Woods et al. 2006). The concentration of beneficial mutations in these few genes, and even specific sites within these genes, implies that phenotypic parallelism may result from the finite number of possible mutations that can produce the most beneficial phenotypic effects appropriate to a particular selective environment, thus supporting an ‘oligogenic’ model of adaptive evolution (Tanksley 1993; Bradshaw et al. 1998; Orr 2005; Wood et al. 2005).
We demonstrated significant associations between genotype and phenotype, with different beneficial mutations in the same locus having similar phenotypes. However, there was little or no variation among different mutations, even across several loci, based on their fitness effects in the glucose environment in which they were selected. Rather, it was the pleiotropic effects of different substitutions—that is, their correlated effects on fitness in several novel resources—that revealed latent heterogeneity among these beneficial mutations.
These patterns of striking parallelism in selected responses, coupled with substantial divergence in correlated responses, derive from the same set of genetic changes. The contrast between the uniformity of direct responses and the diversity of correlated responses vividly demonstrates the importance of multiple genetic paths in evolution. It also helps to reconcile the diversification so evident in nature with the parallelism more typically observed in laboratory-based studies of evolution (Wichman et al. 1999; Cooper et al. 2003). Consistent with this interpretation are other laboratory studies in which subtle changes in the environment substantially altered the evolutionary responses. For example, Travisano (1997) showed very different responses for E. coli that evolved in otherwise identical environments containing either glucose or maltose, even though maltose is simply a glucose dimer. Similarly, Mongold et al. (1999) demonstrated that mutations which allowed E. coli to grow at otherwise lethally high temperatures were distinct from changes that allowed E. coli to thrive at only slightly lower temperatures. Here we demonstrate not only a multiplicity of mutational paths allowing adaptation to any particular environment, but also that heterogeneity in the correlated responses among these genetic paths contributes to the evolution of biological diversity.
Acknowledgments
We thank D. Rozen for providing the strains, N. Hajela for technical assistance and T. Cooper for comments on the manuscript. This research has been supported by grants to R.E.L. from the National Science Foundation and the DARPA ‘FunBio’ programme and a training fellowship to E.A.O. from the Keck Center for Interdisciplinary Bioscience Training of the Gulf Coast Consortia (NLM grant no. 5T15LM07093).
Footnotes
Electronic supplementary material is available at http://dx.doi.org/10.1098/rspb.2007.1244 or via http://journals.royalsociety.org.
Supplementary Material
The position for each mutation is indicated relative to the first basepair of the corresponding gene's start codon. One clone (show in boldface) was a double mutant and is therefore listed twice
References
- Bradshaw H.D, Schemske D.W. Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature. 2003;426:176–178. doi: 10.1038/nature02106. doi:10.1038/nature02106 [DOI] [PubMed] [Google Scholar]
- Bradshaw H.D, Otto K.G, Frewen B.E, McKay J.K, Schemske D.W. Quantitative trait loci affecting differences in floral morphology between two species of monkeyflower (Mimulus) Genetics. 1998;149:367–382. doi: 10.1093/genetics/149.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull J.J, Badgett M.R, Wichman H.A. Big-benefit mutations in a bacteriophage inhibited with heat. Mol. Biol. Evol. 2000;17:942–950. doi: 10.1093/oxfordjournals.molbev.a026375. [DOI] [PubMed] [Google Scholar]
- Clarke K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993;18:117–143. doi:10.1111/j.1442-9993.1993.tb00438.x [Google Scholar]
- Clarke K.R, Gorley R.N. PRIMER-E; Plymouth, UK: 2001. PRIMER v.5: user manual/tutorial. [Google Scholar]
- Colosimo P.F, et al. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. doi:10.1126/science.1107239 [DOI] [PubMed] [Google Scholar]
- Cooper V.S, Schneider D, Blot M, Lenski R.E. Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B. J. Bacteriol. 2001;183:2834–2841. doi: 10.1128/JB.183.9.2834-2841.2001. doi:10.1128/JB.183.9.2834-2841.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T.F, Rozen D.E, Lenski R.E. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc. Natl Acad. Sci. USA. 2003;100:1072–1077. doi: 10.1073/pnas.0334340100. doi:10.1073/pnas.0334340100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle R.F. Convergent evolution: the need to be explicit. Trends Biochem. Sci. 1994;19:15–18. doi: 10.1016/0968-0004(94)90167-8. doi:10.1016/0968-0004(94)90167-8 [DOI] [PubMed] [Google Scholar]
- Elena S.F, Lenski R.E. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 2003;4:457–469. doi: 10.1038/nrg1088. doi:10.1038/nrg1088 [DOI] [PubMed] [Google Scholar]
- Gentry D.R, Cashel M. Mutational analysis of the Escherichia coli spoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol. Microbiol. 1996;19:1373–1384. doi: 10.1111/j.1365-2958.1996.tb02480.x. doi:10.1111/j.1365-2958.1996.tb02480.x [DOI] [PubMed] [Google Scholar]
- Goldberg D.E, Scheiner S.M. ANOVA and ANCOVA: field competition experiments. In: Scheiner S.M, Gurevitch J, editors. Design and analysis of ecological experiments. Oxford University Press; New York, NY: 2001. pp. 77–98. [Google Scholar]
- Grose J.H, Bergthorsson U, Roth J.R. Regulation of NAD synthesis by the trifunctional NadR protein of Salmonella enterica. J. Bacteriol. 2005;187:2774–2782. doi: 10.1128/JB.187.8.2774-2782.2005. doi:10.1128/JB.187.8.2774-2782.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegreness M, Kishony R. Analysis of genetic systems using experimental evolution and whole-genome sequencing. Genome Biol. 2007;8:201–204. doi: 10.1186/gb-2007-8-1-201. doi:10.1186/gb-2007-8-9-r201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring C.D, et al. Comparative genome resequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat. Genet. 2006;38:1406–1412. doi: 10.1038/ng1906. doi:10.1038/ng1906 [DOI] [PubMed] [Google Scholar]
- Hodgkin J. Seven types of pleiotropy. Int. J. Dev. Biol. 1998;42:501–505. [PubMed] [Google Scholar]
- Hoekstra H.E, Hirschmann R.J, Bundey R.A, Insel P.A, Crossland J.P. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science. 2006;313:101–104. doi: 10.1126/science.1126121. doi:10.1126/science.1126121 [DOI] [PubMed] [Google Scholar]
- Jones C.D. The genetic basis of Drosophila sechellia's resistance to a host plant toxin. Genetics. 1998;149:1899–1908. doi: 10.1093/genetics/149.4.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight C.G, Zitzmann N, Prabhakar S, Antrobus R, Dwek R, Hebestreit H, Rainey P.B. Unraveling adaptive evolution: how a single point mutation affects the protein coregulation network. Nat. Genet. 2006;38:1015–1022. doi: 10.1038/ng1867. doi:10.1038/ng1867 [DOI] [PubMed] [Google Scholar]
- Kurnasov O.V, Polanuyer B.M, Ananta S, Sloutsky R, Tam A, Gerdes S.Y, Osterman A.L. Ribosylnicotinamide kinase domain of NadR protein: identification and implications of NAD biosynthesis. J. Bacteriol. 2003;184:6906–6917. doi: 10.1128/JB.184.24.6906-6917.2002. doi:10.1128/JB.184.24.6906-6917.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski D.P. How malaria has affected the human genome and what human genetics can teach us about malaria. Am. J. Hum. Genet. 2005;77:171–190. doi: 10.1086/432519. doi:10.1086/432519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski R.E, Rose M.R, Simpson S.C, Tadler S.C. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 1991;138:1315–1341. doi:10.1086/285289 [Google Scholar]
- Lenski R.E, Ofria C, Pennock R.T, Adami C. The evolutionary origin of complex features. Nature. 2003;423:139–144. doi: 10.1038/nature01568. doi:10.1038/nature01568 [DOI] [PubMed] [Google Scholar]
- MacLean R.C, Bell G. Divergent evolution during an experimental adaptive radiation. Proc. R. Soc. B. 2003;270:1645–1650. doi: 10.1098/rspb.2003.2408. doi:10.1098/rspb.2003.2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean R.C, Bell G, Rainey P.B. The evolution of a pleiotropic fitness tradeoff in Pseudomonas fluorescens. Proc. Natl Acad. Sci. USA. 2004;101:8072–8077. doi: 10.1073/pnas.0307195101. doi:10.1073/pnas.0307195101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani G.S, Clarke B.C. Mutational order: a major stochastic process in evolution. Proc. R. Soc. B. 1990;240:29–37. doi: 10.1098/rspb.1990.0025. doi:10.1098/rspb.1990.0025 [DOI] [PubMed] [Google Scholar]
- Mongold J.A, Bennett A.F, Lenski R.E. Evolutionary adaptation to temperature. VII. Extension of the upper thermal limit of Escherichia coli. Evolution. 1999;53:386–394. doi: 10.1111/j.1558-5646.1999.tb03774.x. doi:10.2307/2640775 [DOI] [PubMed] [Google Scholar]
- Mundy N.I, Badcock N.S, Hart T, Scribner K, Janssen K, Nadeau N.J. Conserved genetic basis of a quantitative plumage trait involved in mate choice. Science. 2004;303:1870–1873. doi: 10.1126/science.1093834. doi:10.1126/science.1093834 [DOI] [PubMed] [Google Scholar]
- Nachman M.W. The genetic basis of adaptation: lessons from concealing coloration in pocket mice. Genetica. 2005;123:125–136. doi: 10.1007/s10709-004-2723-y. doi:10.1007/s10709-004-2723-y [DOI] [PubMed] [Google Scholar]
- Nachman M.W, Hoekstra H.E, D'Agostino S.L. The genetic basis of adaptive melanism in pocket mice. Proc. Natl Acad. Sci. USA. 2003;100:5268–5273. doi: 10.1073/pnas.0431157100. doi:10.1073/pnas.0431157100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H.A. The probability of parallel evolution. Evolution. 2005;59:216–220. doi:10.1111/j.0014-3820.2005.tb00907.x [PubMed] [Google Scholar]
- Orr H.A, Coyne J.A. The genetics of adaptation: a reassessment. Am. Nat. 1992;140:725–742. doi: 10.1086/285437. doi:10.1086/285437 [DOI] [PubMed] [Google Scholar]
- Ostrowski E.A, Rozen D.E, Lenski R.E. Pleiotropic effects of beneficial mutations in Escherichia coli. Evolution. 2005;59:2343–2352. [PubMed] [Google Scholar]
- Ostrowski E.A, Ofria C, Lenski R.E. Ecological specialization and adaptive decay in digital organisms. Am. Nat. 2007;73:E1–E20. doi: 10.1086/510211. doi:10.1086/510211 [DOI] [PubMed] [Google Scholar]
- Penfound, T. & Foster, J. W. 1996 Biosynthesis and recycling of NAD. In Escherichia coli and Salmonella: cellular and molecular biology, vol. 1 (ed. F. C. Neidhardt), pp. 721–730. Washington, DC: ASM Press.
- Phillips P.C. Testing hypotheses regarding the genetics of adaptation. Genetica. 2005;123:15–24. doi: 10.1007/s10709-004-2704-1. doi:10.1007/s10709-004-2704-1 [DOI] [PubMed] [Google Scholar]
- Poelwijk F.J, Kiviet D.J, Weinreich D.M, Tans S.J. Empirical fitness landscapes reveal accessible evolutionary paths. Nature. 2007;445:383–386. doi: 10.1038/nature05451. doi:10.1038/nature05451 [DOI] [PubMed] [Google Scholar]
- Raffaelli N, Lorenzi T, Mariani P.L, Emanuelli M, Amici A, Ruggieri S, Magni G. The Escherichia coli NadR regulator is endowed with nicotinamide mononucleotide adenylyltransferase activity. J. Bacteriol. 1999;181:5509–5511. doi: 10.1128/jb.181.17.5509-5511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen D.E, de Visser J.A.G.M, Gerrish P.J. Fitness effects of fixed beneficial mutations in microbial populations. Curr. Biol. 2002;12:1040–1045. doi: 10.1016/s0960-9822(02)00896-5. doi:10.1016/S0960-9822(02)00896-5 [DOI] [PubMed] [Google Scholar]
- SAS Institute 1999 SAS, v. 8. Cary, NC: SAS Institute.
- Schluter D, Clifford E.A, Nemethy M, McKinnon J.S. Parallel evolution and inheritance of quantitative traits. Am. Nat. 2004;163:809–822. doi: 10.1086/383621. doi:10.1086/383621 [DOI] [PubMed] [Google Scholar]
- Schneider D, Duperchy E, Coursange E, Lenski R.E, Blot M. Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence-mediated mutations and rearrangements. Genetics. 2000;156:477–488. doi: 10.1093/genetics/156.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SYSTAT 2002 SYSTAT, v. 10.2. Richmond, CA: SYSTAT.
- Tanksley S.D. Mapping polygenes. Annu. Rev. Genet. 1993;27:205–233. doi: 10.1146/annurev.ge.27.120193.001225. doi:10.1146/annurev.ge.27.120193.001225 [DOI] [PubMed] [Google Scholar]
- Travisano M. Long-term experimental evolution in Escherichia coli. V. Environmental constraints on adaptation and divergence. Genetics. 1997;146:471–479. doi: 10.1093/genetics/146.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travisano M, Lenski R.E. Long-term experimental evolution in Escherichia coli. IV. Targets of selection and the specificity of adaptation. Genetics. 1996;143:15–26. doi: 10.1093/genetics/143.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travisano M, Mongold J.A, Bennett A.F, Lenski R.E. Experimental tests of the roles of adaptation, chance, and history in evolution. Science. 1995a;267:87–90. doi: 10.1126/science.7809610. doi:10.1126/science.7809610 [DOI] [PubMed] [Google Scholar]
- Travisano M, Vasi F, Lenski R.E. Long-term experimental evolution in Escherichia coli. III. Variation among replicate populations in correlated responses to novel environments. Evolution. 1995b;49:189–200. doi: 10.1111/j.1558-5646.1995.tb05970.x. doi:10.2307/2410304 [DOI] [PubMed] [Google Scholar]
- Weinreich D.M, Delaney N.F, DePristo M.A, Hartl D.L. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. doi:10.1126/science.1123539 [DOI] [PubMed] [Google Scholar]
- Wichman H.A, Badgett M.R, Scott L.A, Boulianne C.M, Bull J.J. Different trajectories of parallel evolution during viral adaptation. Science. 1999;285:422–424. doi: 10.1126/science.285.5426.422. doi:10.1126/science.285.5426.422 [DOI] [PubMed] [Google Scholar]
- Wood T.E, Burke J.M, Rieseberg L.H. Parallel genotypic adaptation: when evolution repeats itself. Genetica. 2005;123:157–170. doi: 10.1007/s10709-003-2738-9. doi:10.1007/s10709-003-2738-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods R, Schneider D, Winkworth C.L, Riley M.A, Lenski R.E. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc. Natl Acad. Sci. USA. 2006;103:9107–9112. doi: 10.1073/pnas.0602917103. doi:10.1073/pnas.0602917103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Evolution and the genetics of populations. Vol 1: genetic and biometric foundations. University of Chicago Press; Chicago, IL: 1968. pp. 61–62. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The position for each mutation is indicated relative to the first basepair of the corresponding gene's start codon. One clone (show in boldface) was a double mutant and is therefore listed twice