Abstract
The yucca–yucca moth interaction is one of the most well-known and remarkable obligate pollination mutualisms, and is an important study system for understanding coevolution. Previous research suggests that specialist pollinators can promote rapid diversification in plants, and theoretical work has predicted that obligate pollination mutualism promotes cospeciation between plants and their pollinators, resulting in contemporaneous, parallel diversification. However, a lack of information about the age of Yucca has impeded efforts to test these hypotheses. We used analyses of 4322 AFLP markers and cpDNA sequence data representing six non-protein-coding regions (trnT–trnL, trnL, trnL intron, trnL–trnF, rps16 and clpP intron 2) from all 34 species to recover a consensus organismal phylogeny, and used penalized likelihood to estimate divergence times and speciation rates in Yucca. The results indicate that the pollination mutualism did not accelerate diversification, as Yucca diversity (34 species) is not significantly greater than that of its non-moth-pollinated sister group, Agave sensu latissimus (240 species). The new phylogenetic estimates also corroborate the suggestion that the plant–moth pollination mutualism has at least two origins within the Agavaceae. Finally, age estimates show significant discord between the age of Yucca (ca 6–10 Myr) and the current best estimates for the age of their pollinators (32–40 Myr).
Keywords: adaptive radiation, AFLP, Agave, Agavaceae, Yucca, cpDNA
1. Introduction
Interactions with pollinating insects have played a fundamental role in the evolution and diversification of flowering plants (Grant 1949; Whittall & Hodges 2007), and lineages of plants that have evolved features associated with specialist pollinators are often particularly diverse (Hodges & Arnold 1995; Sargent 2004). Some of the most remarkable adaptations for pollination are found in plants associated with seed-feeding pollinators, such as those of yuccas and yucca moths (Yucca and Hesperoyucca; Tegeticula and Parategeticula, respectively; Pellmyr 2003), figs and fig wasps (Janzen 1979; Machado et al. 2001; Weiblen 2002), Glochidion shrubs and gracillariid (Epicephala) moths (Kato et al. 2003; Kawakita et al. 2004) and the senita cactus and senita moth (Holland & Fleming 1999). In these associations, plants rely almost exclusively on a single pollinator species, whose larvae in turn feed on some of the host's developing seeds.
In the yucca–yucca moth association, which Darwin considered ‘the most remarkable pollination system ever described’ (Darwin 1874), the female moth uses unique tentacular appendages on its mouthparts to gather and manipulate pollen from yucca flowers. The female moth first oviposits into the floral ovary, before actively pollinating the flower by depositing pollen directly onto the stigma in a highly stereotypical manner (Pellmyr 2003). Because the larva requires fertilized seeds to complete its development, and as there are no co-pollinators, active pollination by the female moth is critical for ensuring its reproductive success and the system is an obligate mutualism.
The intimacy and specificity of this pollination system have led to suggestions that pollination by seed feeders presents the opportunity for rapid, simultaneous diversification in both the plants and the pollinators through joint speciation and adaptive radiation (Sanderson & Donoghue 1996; Schluter 2000; Good-Avila et al. 2006). Testing these hypotheses on a macroevolutionary scale requires sufficient species diversity, information about evolutionary patterns and data on the timing of evolutionary events (Kiester et al. 1984; Page 1991). Significant progress has been made in addressing these questions for some sections of the extraordinarily species-rich figs and fig wasps (Weiblen 2004; Machado et al. 2005; Ronsted et al. 2005; Jiang et al. 2006; Marussich & Machado 2007) and for the Glochidion–Epicephala associations (Kawakita et al. 2004), but these hypotheses have yet to be tested for the association between yuccas and yucca moths.
In the past 10 years, the number of recognized pollinating yucca moths has increased to approximately 25 species in all, and a robust phylogeny based on molecular data is now available for the moths (Pellmyr 1999; Althoff et al. 2006; Pellmyr et al. in press). However, information about the timing and rates of speciation in both partners is critical for studying cospeciation (Page 1991) and adaptive radiations (Sanderson & Donoghue 1996; Hodges 1997; Schluter 2000; Pybus et al. 2002), so the lack of robust plant age estimates has prevented the tests of these hypotheses.
The purpose of this paper is to fill this void by providing information about the age and phylogenetic relationships within Yucca. Here, we have inferred phylogenetic relationships within the genus using relaxed-clock methods to date key nodes in the radiation of the group. A recent analysis of nearly 100 samples using a dataset of 4322 AFLP markers recovered historically recognized sections within the genus and provided evidence for the monophyly of most conservatively delineated taxa (Pellmyr et al. 2007). We combine those AFLP data with approximately 3785 bases of cpDNA sequence data from six non-protein-coding regions to recover a consensus organismal phylogeny for the yuccas, determine the age of the genus Yucca and its phylogenetic placement within the Agavaceae, and estimate absolute and relative speciation rates within Yucca. The results strengthen the case for multiple origins of the mutualism and reject the hypothesis that yucca moth pollination caused accelerated diversification in Yucca. Additionally, the ages inferred here show that diversification of Yucca substantially post-dates the best estimates for the age of their pollinators; we discuss possible implications of this result.
2. Material and methods
(a) Data collection
The genus Yucca L. has not been subject to a comprehensive taxonomic revision, despite considerable interest in the genus, not least from horticulture. Recent studies have cited approximate numbers for the total diversity in the genus, generally ranging from 40 to 50 species. However, a recent AFLP-based analysis that included nearly 100 samples representing the most commonly named taxa (Pellmyr et al. 2007) identified 34 phylogenetically defined species. In the present study, 96 individuals were selected for PCR and sequencing, representing the 34 species identified by Pellmyr et al. (2007) and several putative outgroups, including Agave, Hesperoyucca and Hesperaloe. Whenever possible, samples selected for sequencing were the same individuals included in the AFLP study (Pellmyr et al. 2007).
DNA sequence data were generated from six chloroplast regions (trnT–trnL intergenic spacer, trnL, trnL intron, trnL–trnF intergenic spacer, rps16 intron and clpP intron 2) and were combined with data from homologous regions in Phalaenopsis aphrodite (Orchidaceae) and Acorus calamus (Acoraceae) obtained from GenBank, which served as outgroups. Additionally, data from the trnL gene and trnL–trnF intergenic spacer obtained from GenBank for more than 50 species from across the Asparagales, together with data from A. calamus and Amborella trichopoda (Amborellaceae), were used to infer ages in the Agavaceae and the position of Yucca within the family. The complete list of taxa is available in the electronic supplementary material.
The new data were combined with GenBank sequences to produce four datasets: 1404 bases of cpDNA sequence data from the trnL gene and trnL–trnF intergenic spacer from 153 taxa (including 139 taxa within the Agavaceae sensu lato and 14 outgroups); 3785 bases of cpDNA sequence data from 6 loci from 98 taxa (including 91 Yucca sequences, 3 Hesperoyucca whipplei sequences and 4 outgroups); a combined dataset containing 3366 bases of cpDNA and 4322 AFLP markers from 83 taxa, including 81 Yucca samples and 2 H. whipplei; and a conflict-free dataset (see electronic supplementary material) combining 3366 bases of cpDNA sequences and 4322 AFLP markers from 46 taxa (44 Yucca samples and 2 H. whipplei). Differences in total base counts resulted from the elimination of gap-only characters corresponding to indels in excluded taxa.
Phylogenetic analyses were completed using parsimony and Bayesian inference in PAUP v. 4.0b10 (Swofford 2002) and MrBayes v. 3.1.2 (Huelsenbeck & Ronquist 2001), respectively. Conflict between AFLP and cpDNA data partitions was assessed using an ILD test, partitioned Bremer supports (Baker & DeSalle 1997), and Bayesian analysis, and ParaFit (Legendre et al. 2002) was used to identify a subset of taxa where there was significant (p<0.05) agreement between the data partitions. Divergence times and mutation rates were estimated using penalized likelihood in r8s v. 1.71 (Sanderson 1997, 2002). Relative and absolute speciation rates were compared between Yucca and its sister group (Agave sensu latissimus) using a randomly branching Markovian model (Slowinski & Guyer 1989), a likelihood ratio test (Sanderson & Donoghue 1994), a Yule model (Baldwin & Sanderson 1998), and lineages-through-time plots, calculting Phybus's gamma statistic (Pybus et al. 2002). Detailed descriptions of the laboratory and analytical methods are available as electronic supplementary material.
3. Results
(a) Phylogeny of Agavaceae
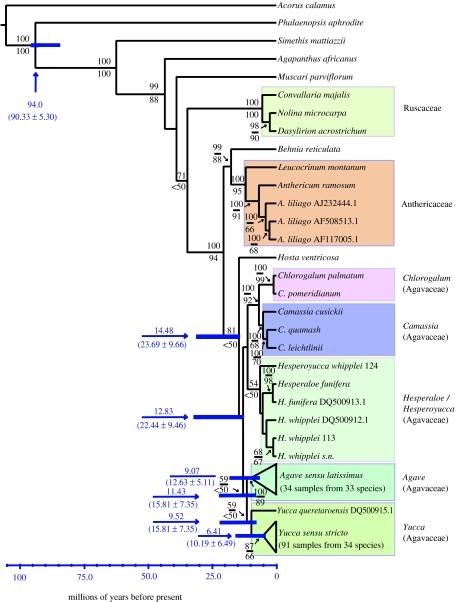
The Bayes consensus tree of the trnL–trnF (153-taxa) dataset (figure 1) showed strong (99–100%) support for the monophyly of the Asparagales, the monophyly of the ‘core’ Asparagales (the Agapanthaceae, Hyacinthaceae, Ruscaceae (=Convallariaceae), Anthericaceae, Behniaceae and Agavaceae) and for a clade containing the Anthericaceae and Agavaceae. There was also moderate support (81%) for the monophyly of the Agavaceae sensu lato (Agavaceae+Hosta). Parsimony bootstrap support was lower but also offered moderate (more than 60%) to strong (100%) support for these same groupings.
Figure 1.
Chronogram of the Agavaceae based on the Bayes consensus tree derived from 153 cpDNA sequences from the trnL gene and the trnL–trnF intergenic spacer. Amborella trichopoda was used to root the topology and was pruned in the r8s analysis. Node labels show Bayesian posterior probabilities (above) and non-parametric bootstrap supports (below). Unlabelled nodes have less than 50% support. Error bars represent the standard deviation of age estimates profiled across post-burn-in trees (below the arrow; values above the arrow represent the node age in Bayes consensus). For the complete consensus tree, see electronic supplementary material.
Within the Agavaceae (figure 1), posterior probabilities offered strong support for the monophyly of the genera Camassia and Chlorogalum, for a clade containing both Camassia and Chlorogalum and for the monophyly of Agave sensu latissimus (Agave, Beschorneria, Furcraea, Manfreda, Polianthes, and Prochnyanthes). Non-parametric bootstrapping also showed moderate support for these groups. Finally, there was weak support (more than 50%) within the Agavaceae for a clade containing Hesperoyucca and Hesperaloe, for the monophyly of Yucca and for a sister-group relationship between Yucca and A. s. latissimus, but support for these relationships was considerably stronger in the analyses of the complete dataset from all the six cpDNA regions (see §3b and figure 2).
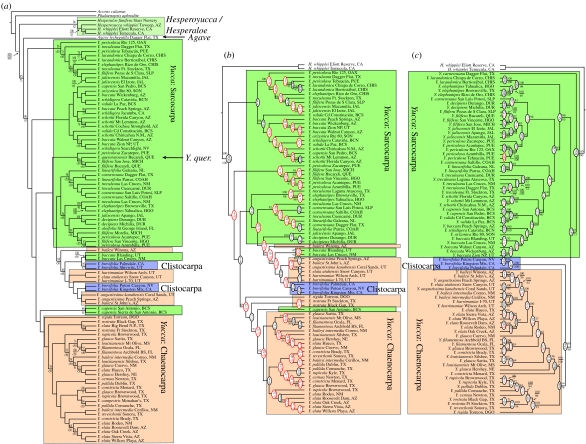
Figure 2.
Phylogenetic relationships inferred from cpDNA sequence data and AFLPs. Bayes consensus trees of (a) the complete 98-taxa cpDNA dataset, (b) the 83-taxa cpDNA dataset and (c) the 83-taxa AFLP dataset. Nodal indices show Bayesian posterior probabilities (above) and non-parametric bootstrapping support (below). Unlabelled nodes have less than 50% support. Ovals on nodes in (b,c) show partitioned Bremer supports (parsimony) from the cpDNA (top) and AFLPs (bottom). Ovals in red identify conflict between partitions.
(b) Phylogeny of Yucca: 98-taxa dataset
The Bayes consensus tree of the 98-taxa dataset showed strong (96–100%) support for the monophyly of Yucca, for the monophyly of Hesperoyucca and for a sister-group relationship between Hesperoyucca and Hesperaloe (figure 2). There was also moderate support (85% posterior probability) for a clade containing Yucca and Agave. These relationships also received moderate to strong support (73–100%) from non-parametric bootstrapping.
Within Yucca, all three traditionally recognized sections (Sarcocarpa, Clistocarpa and Chaenocarpa) were paraphyletic, contrary to the topology recovered from the AFLP data in previous studies (Pellmyr et al. 2007). However, the relationships recovered here were generally very weakly supported, with most major groups receiving less than 50% posterior probability. One grouping receiving some support (69% posterior probability) was a clade containing Yucca brevifolia (Clistocarpa) and a number of geographically proximate—but morphologically distinct—taxa from the Colorado Plateau region.
(c) Assessment of conflict between partitions
Although the ILD test suggested no statistically significant conflict between the AFLP and cpDNA sequence data in the 83-taxa dataset (p=0.24), the partitioned Bremer supports suggested widespread disagreements between the two data partitions. In a combined parsimony analysis of the two data partitions, 60 of 80 nodes had non-positive Bremer supports from the cpDNA and four nodes received non-positive Bremer supports from the AFLP data. Partitioned Bremer supports for nodes in the Bayes consensus trees also showed widespread disagreements: 61 of the 80 nodes in the topology inferred from the cpDNA sequence data received non-positive supports from the AFLP data, and seven nodes in the topology inferred from the AFLP data received non-positive supports from the cpDNA data (figure 2).
Analysis in a Bayesian context also suggested significant differences between the cpDNA gene tree and the tree inferred from the AFLP data. As in the analysis of the complete 98-taxa cpDNA dataset (§3b), analysis of the smaller 83-taxa cpDNA dataset found sections Chaenocarpa and Sarcocarpa to be polyphyletic, with a clade of plants from the Colorado Plateau being nested within the remaining Sarcocarpa (figure 2). Filtering the post-burn-in trees from the separate analyses suggests that both datasets significantly reject (p≪0.001) the topology favoured by the other.
However, despite the widespread conflicts between partitions, comparison of the topologies and branch lengths using ParaFit revealed significant overall congruence (p<0.001) between the two Bayes consensus trees. Of the 83 taxa for which complete AFLP and cpDNA sequence data were available, 46 contributed significantly (p<0.05) to the overall congruence between the two datasets (see electronic supplementary material). Because the possibility of introgression could be statistically rejected for these taxa, they were included in the combined, conflict-free dataset to compute a phylogeny of Yucca and infer divergence times (§3d and §3e).
(d) Phylogeny of Yucca: 46-taxa dataset
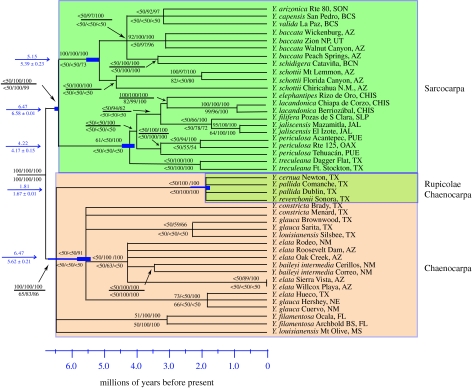
The Bayesian analysis of the conflict-free (46-taxa) dataset found strong support for the monophyly of sections Chaenocarpa and Sarcocarpa and for the series Rupicolae (figure 3). All of these relationships also received moderate (86%) to strong (100%) support from the Bayesian posterior probabilities and non-parametric bootstrapping (figure 3). Separate analyses of the AFLP and cpDNA data for the 46-taxa dataset produced topologies similar to that inferred from the combined data and support indices that were consistent with—although weaker than—those inferred in the combined analysis (figure 3).
Figure 3.
Chronogram of Yucca based on the Bayes consensus of the 43-taxa conflict-free dataset. Node labels show Bayesian posterior probabilities (above) and non-parametric bootstrap supports (below) from the cpDNA, AFLP and combined datasets, respectively. Unlabelled nodes have less than 50% support. Error bars on nodes represent the standard deviation of age estimates profiled across post-burn-in trees (below the arrow; values above the arrow represent the node age in Bayes consensus). The two Hesperoyucca samples were used to root the tree and have been pruned for the r8s analysis.
(e) Divergence time estimates and rates of evolution
The age of Agavaceae including Hosta, estimated from the Bayes consensus, was approximately 14.5±0.94 Myr (figure 1). Ages estimated from the Bayes consensus tree were consistently lower than the mean age across post-burn-in trees (table 1). The largest disparity in this regard is in the age of Yucca, perhaps owing to alternate placements for the basal taxon Y. queretaroensis (see §4).
Table 1.
Ages within the Asparagales estimated using penalized likelihood from the Bayes consensus tree of the trnL–trnF (153-taxa) dataset and profiled across post-burn-in trees.
| taxon | Bayes consensus tree | across post-burn-in trees | ||||
|---|---|---|---|---|---|---|
| age (Myr) | maximum (95% CI) | minimum (95% CI) | mean | s.d. | Good-Avila et al. | |
| Asparagales | 94 | 94 | 93.91 | 90.33 | 5.3 | 60 |
| Agavaceae+Hosta | 14.48 | 15.14 | 13.26 | 23.69 | 9.66 | 25.8 |
| Agavaceae sensu stricto | 12.83 | 13.35 | 12.34 | 22.44 | 9.46 | — |
| Yucca+Agave | 11.43 | 12.04 | 10.99 | 15.81 | 7.35 | — |
| Agave s. l. | 9.07 | 9.46 | 8.69 | 12.63 | 5.11 | 9.8±3.3 |
| Yucca | 9.52 | 10.15 | 9.05 | 15.81 | 7.35 | 17.2±2.3 |
| ‘crown’ Yucca | 6.41 | 6.81 | 6.06 | 10.19 | 6.49 | — |
Mutation rates in the trnL–trnF dataset across the Agavaceae showed stark differences between substitution rates and insertion/deletion rates. Substitution rates in the trnL gene and the trnL–trnF intergenic spacer, profiled across post-burn-in trees, were 0.0006±0.000021 substitutions per site per Myr, while insertions and deletions occurred at a rate of 0.06±0.05 insertions per deletions per locus per Myr. In addition, there was considerable variation in mutation rates between loci (table 2), with the trnT–trnL region showing the highest substitution rate (0.004 substitutions per site per Myr) and the rps16 intron showing the highest rate of insertion/deletion events (0.25 mutations per locus per Myr). Across all loci, observed mutation rates were at least an order of magnitude higher for indels than for substitutions.
Table 2.
Substitution and insertion/deletion rates within Yucca.
| Yucca TMRCA=6.41 | |||||
| category | clpP intron 2 substitutions per site per Myr | rps16 substitutions per site per Myr | trnL−trnF substitutions per site per Myr | trnL+trnL intron substitutions per site per Myr | trnT−trnL substitutions per site per Myr |
| mean | 7.637×10−5±0.0001386 | 7.844×10−5±0.0001867 | 0.0001252±0.0001055 | 0.0002571±0.0003492 | 0.004088±0.007401 |
| category | clpP intron 2 indels per locus per Myr | rps16 indels per locus per Myr | trnL−trnF indels per locus per Myr | trnL+trnL intron indels per locus per Myr | trnT−trnL indels per locus per Myr |
| mean | 0.0077688±0.0132624 | 0.2498±0.2227 | 0.09190±0.1219 | 0.003824±0.02233 | 0.04534±0.07893 |
| Yucca TMRCA=10.14 | |||||
| category | clpP intron 2 substitutions per site per Myr | rps16 substitutions per site per Myr | trnL−trnF substitutions per site per Myr | trnL+trnL intron substitutions per site per Myr | trnT−trnL substitutions per site per Myr |
| mean | 6.349×10−5±0.000119 | 5.732×10−5±0.0001453 | 8.773×10−5±7.392×10−5 | 0.0001875±0.0002609 | 0.0029812±0.006704 |
| category | clpP intron 2 indels per locus per Myr | rps16 indels per locus per Myr | trnL–trnF indels per locus per Myr | trnL+trnL intron indels per locus per Myr | trnT–trnL indels per locus per Myr |
| mean | 0.006974±0.01226 | 0.1771±0.1591 | 0.07270±0.09689 | 0.003792±0.003197 | 0.03578±0.06328 |
(f) Comparisons of diversification rates
The comparison of species numbers and the likelihood ratio test indicates no significant difference in speciation rates between Yucca s. s. and Agave s. latissimus (p=0.25 and 0.20, respectively). Likewise, the tests that explicitly considered topology and branch lengths suggest that the two groups do not differ in their tempo of diversification. The rate of speciation in A. s. latissimus under the Yule model was estimated to be 0.21±0.001 species per lineage per Myr; within Yucca, the speciation rate was estimated to be 0.33±0.06. These estimates are separated by slightly less than two standard deviations.
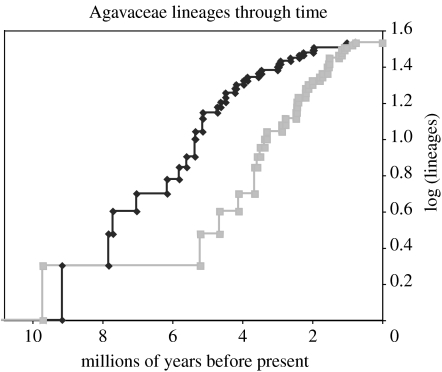
Although the lineages-through-time plots (figure 4) indicate that Agave appears to have diversified earlier than Yucca, the gamma statistics (−4.56 and −3.23, respectively) suggested that the rates of speciation in both groups have declined significantly over time (p=0.026 for Agave s.l.; p<0.001 for Yucca).
Figure 4.
Lineages-through-time plot for Agave. s. latissimus (black diamonds) and Yucca (grey squares). Both groups show significant declines in speciation rates over time. Values for Pybus's gamma are −4.56 and −3.23, respectively.
4. Discussion
The phylogenies inferred here show strong support for the Agavaceae s. lato as currently recognized (Bogler et al. 2006), including Hosta as a basal taxon, sister to the remainder of the Agavaceae. Our results also offer strong support (99% posterior probability) for a clade containing the Anthericaceae and Behniaceae, sister to the Agavaceae s. lato.
Within the Agavaceae, there was strong support (100% posterior probability) for the monophyly of Camassia and Chlorogalum, and these genera are strongly supported as sister taxa. In the combined (98 taxa) cpDNA dataset, there was relatively strong support (85% posterior probability) for the monophyly of a group containing Agave s. latissimus and Yucca, to the exclusion of Hesperoyucca. This result corroborates previous studies arguing that Hesperoyucca did not belong within Yucca s. s. and that there may therefore have been more than one origin of the yucca–yucca moth mutualism (Bogler & Simpson 1995; Bogler et al. 1995; Clary & Simpson 1995).
There is evidence for considerable conflict between the AFLP and cpDNA datasets, particularly in the placement of Clistocarpa. The AFLP data place Clistocarpa as a sister group to the Chaenocarpa. In the cpDNA data, the Clistocarpa were nested within the Sarcocarpa, along with a number of other species from the Colorado Plateau region. This conflict most likely represents introgression by the plastid genome. All of the taxa in this group were from the Mojave Desert and Colorado Plateau region of Arizona, California, and Utah, suggesting a strong biogeographic signature that would be consistent with chloroplast introgression, and this relationship has also been recovered in previous studies that relied on chloroplast data (Hanson 1993). Together, these results suggest that introgression may not be uncommon within Yucca.
Lastly, the identity and phylogenetic affinities of Y. queretaroensis remain enigmatic. The sequence generated for this project is nested deep within Yucca in the section Sarcocarpa, whereas the sequence obtained from GenBank falls as sister to all other yuccas. There is reason for scepticism about both of these placements. The former may be the result of introgression, as this sequence is strongly supported (96% posterior probability) as sister to two samples of Yucca filifera, including one collected at the same site as our sample of Y. queretaroensis. The latter result—placing Y. queretaroensis as sister to all other yuccas—is consistent with the previously published AFLP phylogeny (Pellmyr et al. 2007), but this relationship received only weak support (59% posterior probability) and was extremely unstable throughout the analysis. Minority topologies from the post-burn-in trees place this sample as sister to Agave s. latissimus, or sister to Hesperoyucca or sister to the clade containing Yucca and Agave. s. latissimus.
(a) Age estimates
Overall, the results suggest a surprisingly recent origin of Yucca. Whereas previous work on yucca moths suggested that the plant–moth mutualism may be of Eocene age (Pellmyr & Leebens-Mack 1999), these results suggest that the genus is 9–16 Myr old, with the Yucca crown group (i.e. excluding Y. queretaroensis) having diversified 6–10 Myr ago. Although these estimates are quite low, they are in line with the results from other recent studies: Good-Avila et al. (2006) estimated the age of Yucca to be 13–17 Myr and Eguiarte et al. (2000) estimated the common ancestor of the Sarcocarpa and Chaenocarpa to be ca 6 Myr old. The oldest fossil from the lineage is estimated to be 14 Myr old (Tidwell & Parker 1990), although it is unclear whether this specimen belongs within Yucca or it may represent an extinct stem group.
The sections within the Yucca crown group differentiated rapidly 4–6 Myr ago, with the highest rates of lineage formation occurring 3–4 Myr ago (figure 4). This radiation appears to have occurred more recently and somewhat more rapidly than in Agave s. latissimus, but there was no statistically significant difference in mean speciation rates between the genera under any analytical approach (see §4c).
(b) Rates of evolution in the chloroplast genome
The mutation rate estimates showed that rates of evolution in the chloroplast genome are generally slow, with all regions showing a substitution rate of less than 0.005 substitutions per site per Myr. Rates of evolution may be particularly slow within the yuccas; substitution rates in the trnL gene and the trnL–trnF intergenic spacer within Yucca were roughly half of the average rate found here for the Asparagales as a whole. However, these low mutation and substitution rates may not be unusual for the chloroplast genome generally; whereas we found sequence variation at approximately 5% of sites in the cpDNA dataset, a recent survey of these same gene regions found that sequence variation within genera averages approximately 3% (Shaw et al. 2005).
There was considerable variation in substitution rates between loci. Consistent with the findings of Shaw et al. (2005), the trnT–trnL intergenic spacer showed substitution rates roughly an order of magnitude greater than in the more commonly studied trnL–trnF region, but indels proved to be a surprisingly useful source of phylogenetically informative data, with considerably higher overall mutation rates. Indels and substitutions in the trnT–trnL intergenic spacer may therefore have greater usefulness in future phylogenetic work.
(c) Diversification of the yucca–yucca moth pollination mutualism
Two long-standing hypotheses about the evolution of obligate pollination mutualism are that they may spur rapid diversification and that the plants and their specialist pollinators may tend to speciate in parallel (Grant 1949; Kiester et al. 1984; Hodges & Arnold 1995; Sargent 2004). Our results provide evidence against elevated rates of diversification in Yucca and do not support strict-sense cospeciation in terms of the contemporaneity of diversification.
We find no support for the idea that speciation rates have been accelerated in Yucca, as no analysis found significant differences in speciation rates between Yucca and its sister group. With more than seven times as many species as in Yucca, the diversity of Agave s. latissimus alone should argue against the hypothesis that yucca moth pollination promotes accelerated speciation. Because we find that Agave s. latissimus is the sister group to Yucca, their relative diversity is a fair comparison of diversification rates. Although the lineages-through-time plot (figure 4) indicates that Yucca diversified more recently, and although absolute speciation rates per lineage are somewhat higher in Yucca than in its sister group, this difference was not statistically significant.
It is possible that incomplete sampling of species diversity in Agave s. latissimus could have biased our estimates of speciation rates, or that the apparent difference in diversity could be due to uneven taxonomic effort, but we found no significant difference in raw number of species either, even using a cautious lower estimate for Yucca. Indeed, these considerations suggest that our tests of the key innovation hypothesis are conservative.
It is possible that both yuccas and their sister group have experienced rapid radiations, as previously postulated by Good-Avila et al. (2006). However, the combined Yucca+Agave s. latissimus clade is not significantly more diverse than its sister group (Camassia (six species)+Chlorogalum (five species)+Hesperaloe (five species)+Hesperoyucca (one species)) (p=0.12). Furthermore, the speciation rates inferred here are not particularly high in an absolute sense; whereas we estimated speciation rates of 0.21 and 0.33 species per lineage per Myr in Agave s. latissimus and Yucca, respectively, speciation rates in recognized adaptive radiations such as the Hawaiian silverswords or the South African Cape flora are as high as 0.56 and 4.18, respectively (Baldwin & Sanderson 1998; Verboom et al. 2003). That said, Good-Avila et al. (2006) inferred much higher speciation rates (0.32 species per lineage per Myr) within the more narrowly circumscribed Agave sensu lato (Agave+Manfreda+Polianthes+Prochnyanthes), so it is possible that some lineages within the Agavaceae have undergone adaptive radiations.
The age estimates also find no support for contemporaneous diversification of the plants and their pollinators. Whereas the diversification of the Yucca crown group began 6.41–10.19 Myr ago, previous estimates indicate that yucca moths began to diversify ca 40±11.1 Myr (ago; Pellmyr & Leebens-Mack 1999). Although the age estimates for the pollinating moths have recently been revised downwards to 32 Myr (Gaunt & Miles 2002), these are still several million years prior to our very oldest estimates for the common ancestor yuccas and are tens of millions of years older than the maximum-likelihood estimate.
Though one or both of the molecular clock estimates could be in error, if we accept both of these results, we must postulate at least one of two possible historical scenarios: either the earliest pollinating moths fed on some other group of plants or there have been multiple origins of Yucca association within the Prodoxidae. Although it is conceivable that yucca moths initially diversified on a different group of plants—say, for example, an ancient radiation of yuccas that is now largely extinct—this scenario would require extensive host switching by the moths and the concerted extinction of many former plant species. The other alternative that there were multiple origins of pollination behaviour within the Prodoxidae would seem unparsimonious in the extreme; the two genera that comprise the pollinating yucca moths (Tegeticula and Parategeticula) form a monophyletic group, and both partners in pollination mutualism show remarkable adaptations associated with the interaction. A complete cospeciation study, incorporating information about ages, phylogenetic congruence and correlation in branch lengths between the insects and their hosts, might help to select among these alternatives.
Although such an analysis is beyond the scope of the present paper, at the broadest level, the phylogeny estimated here strongly suggests a history of host switching. Previous work by Bogler et al. (1995) found that the anomalous species Hesperoyucca whipplei (previously Yucca whipplei) was the sister group to the genus Hesperaloe, implying that pollination by yucca moths might either have arisen independently in Hesperoyucca or have been lost in Hesperaloe. The new data presented here suggest that Hesperoyucca is only distantly related to Yucca, and that the common ancestor of these groups is therefore unlikely to have been pollinated by yucca moths. If so, then Hesperoyucca would have been colonized by Tegeticula maculata, forming a second origin of the yucca–yucca moth mutualism.
The phylogenetic relationships in both partners are now well resolved and the next important step in uncovering the evolution of host use and issues of cospeciation will be a formal and comprehensive analysis of codiversification, considering all of the Agavaceae-feeding lineages within the Prodoxidae.
Acknowledgments
We thank Karen Clary, Eric Keith, Abisai García-Mendoza, Carlos Beutelspacher and Rogers McVaugh for site information and Paul Wilson for Y. brevifolia tissue. Big Bend N.P., Black Gap Wildlife Management Area, Laguna Atascosa NWR, Ocala NF, Torrey Pines State Park, the Nature Conservancy Larsen Reserve, Mojave National Preserve, Sabal Palm Grove Sanctuary, Snow Canyon State Park, Walnut Canyon NM, the Elliott Chaparral Reserve and Coronado N. F. gave permission to gather plant tissue. The Mexican Secretary of the Environment and Natural Resources (SeMARNat) provided permission to perform scientific research and collections in Mexico (permit # FAUT-0007 to MBL). Nicole Fisher, William Godsoe, Shantel Tank and Jeremy Yoder assisted in data collection. Jack Sullivan and Chris Drummond advised us on phylogenetic analysis. Fieldwork in Mexico was funded by the National Geographic Society and field and laboratory work by grants DEB-051684 (with REU supplement), DEB 02-42783 and DEB 00-75871 from the National Science Foundation's Ecology and Systematic Biology programmes.
Footnotes
Electronic supplementary material is available at http://dx.doi.org/10.1098/rspb.2007.1405 or via http://journals.royalsociety.org.
Supplementary Material
(1) Detailed Materials and Methods; (2) Table S1: Primers used in this study and PCR conditions; (3) Table S2: Collections Data and GenBank Accessions. Locality information and botanical authorities are provided for data first published here; (4) Table S3: ParaFit cospeciation analysis of congruence between cpDNA and AFLP datasets; (5) Table S4: Node ages and support values; (6) Supplemental Figure S1: Complete chronogram of the Agavaceae based on Bayes consensus tree derived from 153 cpDNA sequences from the trnL gene and the the trnL – trnF intergenic spacer; (7) Supplemental Figure S2: Chronograms of the pruned topologies used to calculate speciation rates in Agave sensu latissimus and Yucca
References
- Althoff D.M, Segraves K.A, Leebens-Mack J, Pellmyret O. Patterns of speciation in the yucca moths: parallel species radiations within the Tegeticula yuccasella species complex. Syst. Biol. 2006;55:398–410. doi: 10.1080/10635150600697325. doi:10.1080/10635150600697325 [DOI] [PubMed] [Google Scholar]
- Baker R.H, DeSalle R. Multiple sources of character information and the phylogeny of Hawaiian drosophilids. Syst. Biol. 1997;46:654–673. doi: 10.1093/sysbio/46.4.654. doi:10.2307/2413499 [DOI] [PubMed] [Google Scholar]
- Baldwin B.G, Sanderson M.J. Age and rate of diversification of the Hawaiian silversword alliance (Compositae) Proc. Natl Acad. Sci. USA. 1998;95:9402–9405. doi: 10.1073/pnas.95.16.9402. doi:10.1073/pnas.95.16.9402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogler D.J, Simpson B.B. A chloroplast DNA study of the Agavaceae. Syst. Bot. 1995;20:191–205. doi:10.2307/2419449 [Google Scholar]
- Bogler D.J, Neff J.L, Simpson B.B. Multiple origins of the yucca–yucca moth association. Proc. Natl Acad. Sci. USA. 1995;92:6864–6867. doi: 10.1073/pnas.92.15.6864. doi:10.1073/pnas.92.15.6864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogler D.J, Pires C, Francisco-Ortega J. Phylogeny of the Agavaceae based on ndhF, rbcL, and ITS sequences: implications of molecular data for classification. In: Columbus J.T, Friar E.A, Porter J.M, Prince L.M, Simpson M.G, editors. Monocots: comparative biology and evolution. vol. 1. Missouri Botanical Garden Press; St Louis, MI: 2006. pp. 311–326. [Google Scholar]
- Clary K.H, Simpson B.B. Systematics and character evolution of the genus Yucca L. (Agavaceae): evidence from morphology and molecular analyses. Bol. Soc. Bot. Mex. 1995;56:77–88. [Google Scholar]
- Darwin, C. 1874 Letter to J D Hooker, April 7, 1874. In A calendar of the correspondence of Charles Darwin, 1821–1882 (eds F. Burkhardt & S. Smith). Cambridge, UK: The Press Syndicate of the University of Cambridge.
- Eguiarte L, Souza V, y Silva-Montellano A. Evolución de la familia Agavaceae: filogenia, biologia reproductiva, y genética de poblaciones. Bol. Soc. Bot. Mex. 2000;66:131–150. [Google Scholar]
- Gaunt M.W, Miles M.A. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol. Biol. Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- Good-Avila S.V, Souza V, Gaut B.S, Eguiarte L.E. Timing and rate of speciation in Agave (Agavaceae) Proc. Natl Acad. Sci. USA. 2006;103:9124–9129. doi: 10.1073/pnas.0603312103. doi:10.1073/pnas.0603312103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant V. Pollination systems as isolating mechanisms in angiosperms. Evolution. 1949;3:82–97. doi: 10.1111/j.1558-5646.1949.tb00007.x. doi:10.2307/2405454 [DOI] [PubMed] [Google Scholar]
- Hanson, M.A. 1993 Dispersed unidirectional introgression from Yucca schidigera into Yucca baccata (Agavaceae). PhD dissertation, The Claremont Graduate School, Claremont, California.
- Hodges S.A. Rapid radiation due to a key innovation in columbines (Ranunculaceae: Aquilegia) In: Givnish T.J, Sytsma K.J, editors. Molecular evolution and adaptive radiation. Cambridge University Press; Cambridge, UK: 1997. pp. 391–405. [Google Scholar]
- Hodges S.A, Arnold M.L. Spurring plant diversification: are floral nectar spurs a key innovation? Proc. R. Soc. B. 1995;262:343–348. doi:10.1098/rspb.1995.0215 [Google Scholar]
- Holland J.N, Fleming T.H. Mutualistic interactions between Upiga virescens (Pyralidae), a pollinating seed-consumer, and Lophocereus schottii (Cactaceae) Ecology. 1999;80:2074–2084. [Google Scholar]
- Huelsenbeck J.P, Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. doi:10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Janzen D. How to be a fig. Annu. Rev. Ecol. Syst. 1979;10:13–51. doi:10.1146/annurev.es.10.110179.000305 [Google Scholar]
- Jiang Z.-F, Huang D.-W, Zhua C.-D, Zhen W.-Q. New insights into the phylogeny of fig pollinators using Bayesian analyses. Mol. Phylogenet. Evol. 2006;38:306–315. doi: 10.1016/j.ympev.2005.11.008. doi:10.1016/j.ympev.2005.11.008 [DOI] [PubMed] [Google Scholar]
- Kato M, Takimura A, Kawakita A. An obligate pollination mutualism and reciprocal diversification in the tree genus Glochidion (Euphorbiaceae) Proc. Natl Acad. Sci. USA. 2003;100:5264–5267. doi: 10.1073/pnas.0837153100. doi:10.1073/pnas.0837153100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakita A, Takimura A, Terachi T, Sota T, Kato M. Cospeciation analysis of an obligate pollination mutualism: have Glochidion trees (Euphorbiaceae) and pollinating Epicephala moths (Gracillariidae) diversified in parallel? Evolution. 2004;58:2201–2214. doi: 10.1111/j.0014-3820.2004.tb01598.x. [DOI] [PubMed] [Google Scholar]
- Kiester A.R, Lande R, Schemske D.W. Models of coevolution and speciation in plants and their pollinators. Am. Nat. 1984;124:220–243. doi:10.1086/284265 [Google Scholar]
- Legendre P, Desdevises Y, Bazin E. A statistical test for host–parasite coevolution. Syst. Biol. 2002;51:217–234. doi: 10.1080/10635150252899734. doi:10.1080/10635150252899734 [DOI] [PubMed] [Google Scholar]
- Machado C.A, Jouselin E, Kjellberg F, Compton S.G, Herre E.A. Phylogenetic relationships, historical biogeography and character evolution of fig-pollinating wasps. Proc. R. Soc. B. 2001;268:685–694. doi: 10.1098/rspb.2000.1418. doi:10.1098/rspb.2000.1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado C.A, Robbins N, Gilbert M.T, Herre E.A. Critical review of host specificity and its coevolutionary implications in the fig/fig–wasp mutualism. Proc. Natl Acad. Sci. USA. 2005;102:6558–6565. doi: 10.1073/pnas.0501840102. doi:10.1073/pnas.0501840102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marussich W.A, Machado C.A. Host-specificity and coevolution among pollinating and nonpollinating New World fig wasps. Mol. Ecol. 2007;16:1925–1946. doi: 10.1111/j.1365-294X.2007.03278.x. doi:10.1111/j.1365-294X.2007.03278.x [DOI] [PubMed] [Google Scholar]
- Page R.D.M. Clocks, clades, and cospeciation: comparing rates of evolution and timing of cospeciation events in host–parasite assemblages. Syst. Zool. 1991;40:188–198. doi:10.2307/2992256 [Google Scholar]
- Pellmyr O. Systematic revision of the yucca moths in the Tegeticula yuccasella complex (Lepidoptera: Prodoxidae) north of Mexico. Syst. Entomol. 1999;24:243–271. doi:10.1046/j.1365-3113.1999.00079.x [Google Scholar]
- Pellmyr O. Yuccas, yucca moths and coevolution: a review. Ann. Mo. Bot. Gard. 2003;90:35–55. doi:10.2307/3298524 [Google Scholar]
- Pellmyr O, Leebens-Mack J.H. Forty million years of mutualism: evidence for Eocene origin of yucca–yucca moth association. Proc. Natl Acad. Sci. USA. 1999;96:9178–9183. doi: 10.1073/pnas.96.16.9178. doi:10.1073/pnas.96.16.9178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellmyr O, Segraves K.A, Althoff D.M, Balcázar-Lara M, Leebens-Mack J. The phylogeny of yuccas. Mol. Phylogenet. Evol. 2007;43:493–501. doi: 10.1016/j.ympev.2006.12.015. doi:10.1016/j.ympev.2006.12.015 [DOI] [PubMed] [Google Scholar]
- Pellmyr, O., et al In press. Phylogeny of the pollinating yucca moths, with revision of Mexican species (Tegeticula and Parategeticula; Lepidoptera, Prodoxidae). Zool. J. Linn. Soc
- Pybus O.G, Rambaut A, Holmes E.C, Harvey P.H. New inferences from tree shape: numbers of missing taxa and population growth rates. Syst. Biol. 2002;51:881–888. doi: 10.1080/10635150290102582. doi:10.1080/10635150290102582 [DOI] [PubMed] [Google Scholar]
- Ronsted N, Weiblen G.D, Cook J.M, Salamin N, Machado C.A, Savolainen V. 60 million years of co-divergence in the fig–wasp symbiosis. Proc. R. Soc. B. 2005;272:2593–2599. doi: 10.1098/rspb.2005.3249. doi:10.1098/rspb.2005.3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson M.J. A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol. Biol. Evol. 1997;14:1218–1231. [Google Scholar]
- Sanderson M.J. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Sanderson M.J, Donoghue M.J. Shifts in diversification rate with the origin of angiosperms. Science. 1994;264:1590–1593. doi: 10.1126/science.264.5165.1590. doi:10.1126/science.264.5165.1590 [DOI] [PubMed] [Google Scholar]
- Sanderson M.J, Donoghue M.J. Reconstructing shifts in diversification rates on phylogenetic trees. Trends Ecol. Evol. 1996;11:15–20. doi: 10.1016/0169-5347(96)81059-7. doi:10.1016/0169-5347(96)81059-7 [DOI] [PubMed] [Google Scholar]
- Sargent R.D. Floral symmetry affects speciation rates in angiosperms. Proc. R. Soc. B. 2004;271:603–608. doi: 10.1098/rspb.2003.2644. doi:10.1098/rspb.2003.2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D. Oxford series in ecology and evolution. Oxford University Press; New York, NY: 2000. The ecology of adaptive radiation. [Google Scholar]
- Shaw J, et al. The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005;92:142–166. doi: 10.3732/ajb.92.1.142. [DOI] [PubMed] [Google Scholar]
- Slowinski J.B, Guyer C. Testing the stochasticity of patterns of organismal diversity: an improved null mode. Am. Nat. 1989;134:907–921. doi:10.1086/285021 [Google Scholar]
- Swofford D. Sinauer Associates; Sunderland, MA: 2002. PAUP*. [Google Scholar]
- Tidwell W, Parker L. Protoyucca shadishii gen et sp nov, an arborescent monocotyledon with secondary growth from the middle Miocene of northwestern Nevada, USA. Rev. Palaeobot. Palynol. 1990;62:79–95. doi:10.1016/0034-6667(90)90018-E [Google Scholar]
- Verboom G.A, Linder H.P, Stock W.D. Phylogenetics of the grass genus Ehrharta: evidence for radiation in the summer-arid zone of the southern cape. Evolution. 2003;57:1008–1021. doi: 10.1111/j.0014-3820.2003.tb00312.x. [DOI] [PubMed] [Google Scholar]
- Weiblen G.D. How to be a fig wasp. Annu. Rev. Entomol. 2002;47:299–330. doi: 10.1146/annurev.ento.47.091201.145213. doi:10.1146/annurev.ento.47.091201.145213 [DOI] [PubMed] [Google Scholar]
- Weiblen G.D. Correlated evolution in fig pollination. Syst. Biol. 2004;53:128–139. doi: 10.1080/10635150490265012. doi:10.1080/10635150490265012 [DOI] [PubMed] [Google Scholar]
- Whittall J.B, Hodges S.A. Pollinator shifts drive increasingly long nectar spurs in columbine flowers. Nature. 2007;447:706–709. doi: 10.1038/nature05857. doi:10.1038/nature05857 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(1) Detailed Materials and Methods; (2) Table S1: Primers used in this study and PCR conditions; (3) Table S2: Collections Data and GenBank Accessions. Locality information and botanical authorities are provided for data first published here; (4) Table S3: ParaFit cospeciation analysis of congruence between cpDNA and AFLP datasets; (5) Table S4: Node ages and support values; (6) Supplemental Figure S1: Complete chronogram of the Agavaceae based on Bayes consensus tree derived from 153 cpDNA sequences from the trnL gene and the the trnL – trnF intergenic spacer; (7) Supplemental Figure S2: Chronograms of the pruned topologies used to calculate speciation rates in Agave sensu latissimus and Yucca