Summary
Background
Many cells that migrate during normal embryonic development or in metastatic cancer first detach from an epithelium. However, this step is often difficult to observe directly in vivo and the mechanisms controlling the ability of cells to leave the epithelium are poorly understood. In addition, once cells detach, they must assume a migratory phenotype, involving changes in cytoskeletal and signaling dynamics. Drosophila border cells provide a model system in which forward genetics and live cell imaging can be combined to investigate the cellular and molecular mechanisms of epithelial cell detachment and migration in vivo.
Results
We identified the Drosophila homolog of the serine/threonine kinase PAR-1 (MARK/Kin1) in a screen for mutations that disrupt border cell migration. Previous studies identified two proteins, Apontic and Notch, which indirectly affect border cell detachment by regulating transcription of downstream targets. In contrast, PAR-1 directly modulates apical-basal polarity between border cells and epithelial cells to promote detachment. Furthermore, PAR-1, but not the apical polarity complex protein PAR-3, promotes the directionality of transient cell protrusions, which are essential for sensing the chemoattractant gradient followed by the border cells.
Conclusions
We conclude that PAR-1-dependent apical-basal polarity is required for proper detachment of migratory border cells from neighboring epithelial cells. Moreover, polarity controlled by PAR-1 influences the ability of migratory cells to sense direction, a critical feature of migration. Thus, this work reveals new insights into two distinct, but essential, steps of epithelial cell migration.
Introduction
During embryonic development many cells undergo dramatic movements necessary for normal organ formation. In the adult, cell migration is required for proper wound healing and immune system function. Misregulated cell migration can result in birth defects, failure of wounds to heal, and tumor invasion and metastasis. Despite advances in our understanding of the molecular regulation of cell migration, in particular mechanisms regulating actin polymerization and cell protrusion, many important questions remain unanswered. An especially critical but poorly understood step is the detachment of migratory cells from a polarized epithelium, yet it is often difficult to study this process in the natural tissue environment. Drosophila border cells represent a genetically tractable and elegant in vivo model system to dissect the mechanisms underlying detachment of a group of cells from an epithelium and their subsequent migration [reviewed in 1].
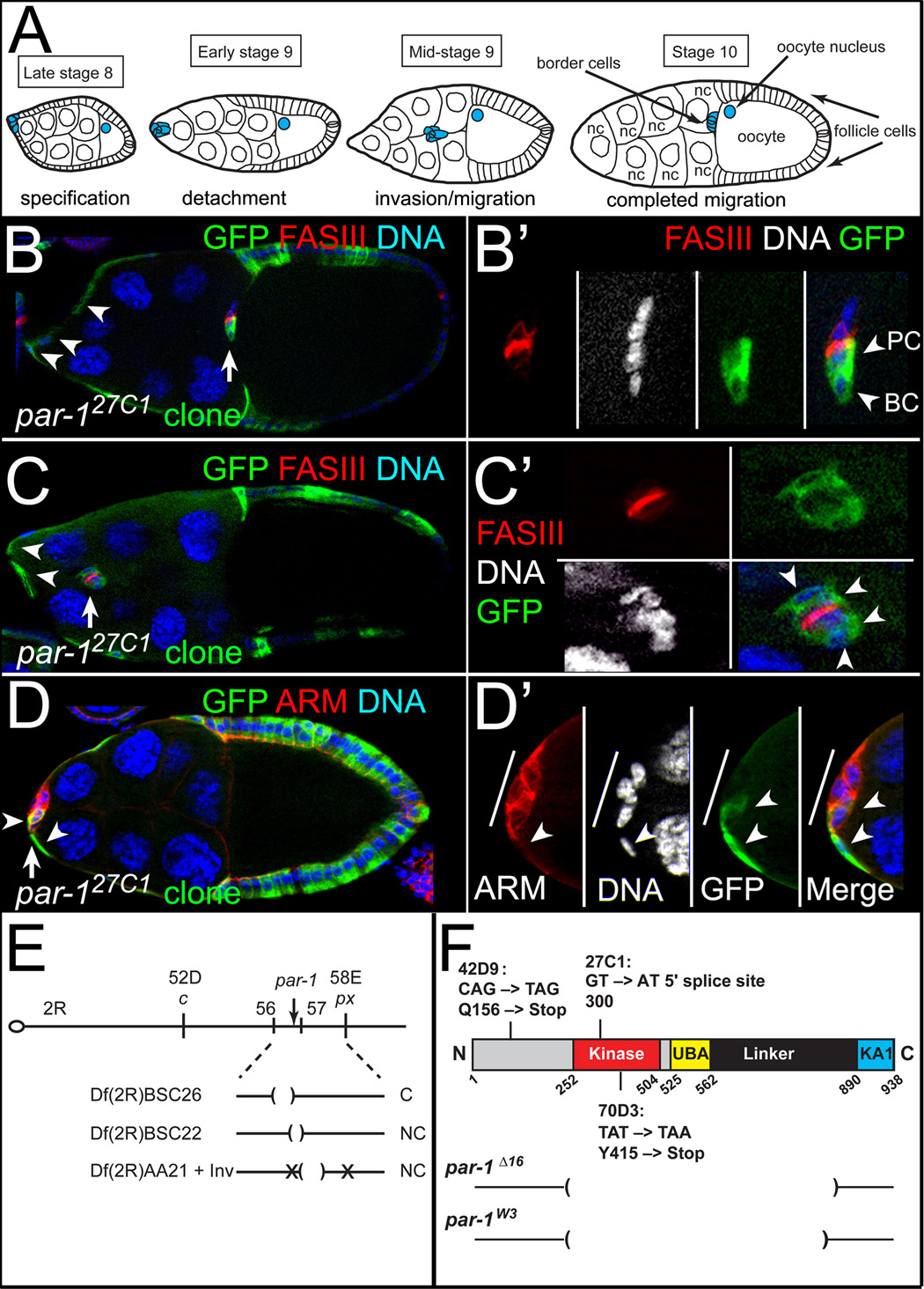
Border cells undergo a stereotypical and highly regulated cell migration during fly ovarian development (Figure 1A) [reviewed in 1]. The Drosophila ovary consists of strings of subunits called egg chambers, each of which will develop into a mature egg. In the center of each egg chamber are 15 germline-derived nurse cells and the oocyte, which are surrounded by ~650 somatically-derived follicle cells. The follicle cells form a monolayer epithelium, which late in oogenesis rearranges to cover the posterior oocyte and stretch around the anterior nurse cells. At the same time, the central pair of polar cells recruits 4 to 8 follicle cells at the anterior end to form the border cell cluster (Figure 1A). Border cells round up, break away from the follicular epithelium as a cohesive cluster, and migrate in-between the nurse cells until they reach the oocyte (Figure 1A).
Figure 1. par-1 is required for border cell migration.
(A) Schematic of oogenesis showing stages of border cell migration. (B–D) Loss of par-1 disrupts border cell migration. Stage 10 par-127C1 MARCM mosaic mutant egg chambers stained for GFP (green) to mark homozygous mutant clones, FAS III to mark polar cells (red, B–C), Armadillo (ARM) to mark cell membranes (red, D), and DAPI to mark DNA/nuclei (blue/white). (B, B’) Border cells (arrow) completed their migration; anterior follicle cells are mutant (arrowheads). (B’) Magnified view of border cell cluster; a border cell (BC) and polar cell (PC) are mutant. (C, C’) Border cells (arrow) migrated partway to the oocyte; anterior follicle cells are mutant (arrowheads). (C’) Magnified view of mutant border cells (arrowheads); polar cells are wild-type. (D, D’) Undetached border cells (arrow); several border cells and an adjacent follicle cell are mutant (arrowheads). (D’) Magnified view of border cells (line). (E) Diagram of chromosome 2R and mapping results with indicated deficiencies (Df); C, complementing; NC, non-complementing. Location of inversion breakpoints, par-1 and meiotic mapping markers are indicated. (F) Schematic of PAR-1 protein with indicated point mutations of par-1 alleles. Brackets, deleted regions of par-1 Δ16 [11] and par-1W3 [19]. For this and subsequent figures: anterior towards the left; genotypes listed in the Supplement.
One key early step of border cell migration is their detachment from neighboring follicle cells. Initially the cluster rounds up, becoming distinct from the non-migratory epithelial follicle cells, and forms protrusions. Border cells subsequently separate from the epithelium and sever an attachment between the trailing border cell and an anterior follicle cell. Live-cell imaging reveals that detachment from the epithelium is slower and more variable than expected, with the time from initial rounding and protrusion to final severing taking up to two hours [2]. In some egg chambers, the cluster migrates almost halfway to the oocyte before the attachment is broken, whereas in other egg chambers complete detachment occurs as soon as the cluster migrates. Once border cells completely break away, the remaining follicle cells at the anterior end stretch toward each other to preserve the epithelial layer [3]. Apontic and Notch indirectly regulate border cell separation from epithelial cells by regulating transcription [2–4]. However, the identities of proteins that directly promote detachment remain elusive. Genetic screens have been carried out to isolate mutations that disrupt border cell migration [5–8], yet in most cases the affected genes are unknown. We now identify par-1, a member of the par polarity family, as a gene required for border cell detachment.
The serine/threonine kinase PAR-1 is best known for regulating the polarity of a wide variety of cells in different biological settings, such as apical-basal polarity in epithelial cells and anterior/posterior polarity in C. elegans embryos and Drosophila oocytes [reviewed in 9]. PAR-1 cellular localization is usually complementary to PAR-3 and its binding partners PAR-6 and atypical Protein Kinase C (aPKC) [reviewed in 9]. In epithelial cells, PAR-1 localizes to basolateral membranes, where it phosphorylates and inhibits apical Bazooka [10], the Drosophila homolog of PAR-3. PAR-1 also regulates microtubule stability [11–13] and WNT signaling [14, 15]. Among common polarity proteins, aPKC has been implicated in cell migration, specifically at the leading edge of migrating astrocytes and at the growing tips of axons in cell culture [reviewed in 9]. PAR-1 contributes to neuronal migration in mice, possibly by regulating microtubules [16]; however, its ability to regulate the migration of other cells, particularly epithelial cells, has not been reported. Our understanding of the roles that polarity proteins play in cell migration is incomplete. Nor is it known whether their modes of action will be similar in different cell types. In this study, we report that Drosophila PAR-1 is a key regulator of two critical aspects of epithelial cell migration, cell detachment and directional protrusion.
Results
Identification of PAR-1 as a Regulator of Border Cell Migration
In a large-scale screen for mutants on chromosome 2R that regulate border cell migration, we identified a mutant allele, l(2)27C1, that exhibited frequent migration defects in mosaic egg chambers containing homozygous mutant follicle cell clones (Figures 1C–D) [6]. Although most clusters containing one or two mutant border cells completed their migration to the oocyte (Figure 1B; 90%, n = 59), clusters containing two or more mutant cells either had incomplete migration or never migrated away from the anterior (Figures 1C–1D’; 58%, n = 24). These results suggested that the mutated gene played an early role in the development and/or migration of border cells.
Three additional alleles (36G4, 42D9, 70D3) were isolated in a mutagenesis screen (see Supplemental Experimental Procedures) and used to map the affected gene to chromosomal region 56D–F (Figure 1E). Complementation testing with P-elements from this region identified a hypomorphic par-1 allele, par-16323, that was semi-lethal with l(2)27C1. Strong par-1 alleles, par-1W3 and par-1Δ16 (Figure 1F), failed to complement the new alleles and exhibited migration defects in mosaic mutant clones (not shown; n = 62 for par-1Δ16; n = 26 for par-1W3). We identified alterations to the par-1 coding region in three of the four new alleles (Figure 1F and Figure S1A–B). par-127C1 is likely to be a strong loss-of-function allele because PAR-1 protein was undetectable in mutant cells (Figure S1C). Markers of border cell identity, Focal Adhesion Kinase (FAK), nuclear STAT, and Singed (SN) [5, 7, 17, 18], were expressed normally in par-1 mutant border cells and were not expressed ectopically in mutant follicle cells (Figure S2 and not shown).
PAR-1 is Required for Border Cell Detachment from the Epithelium
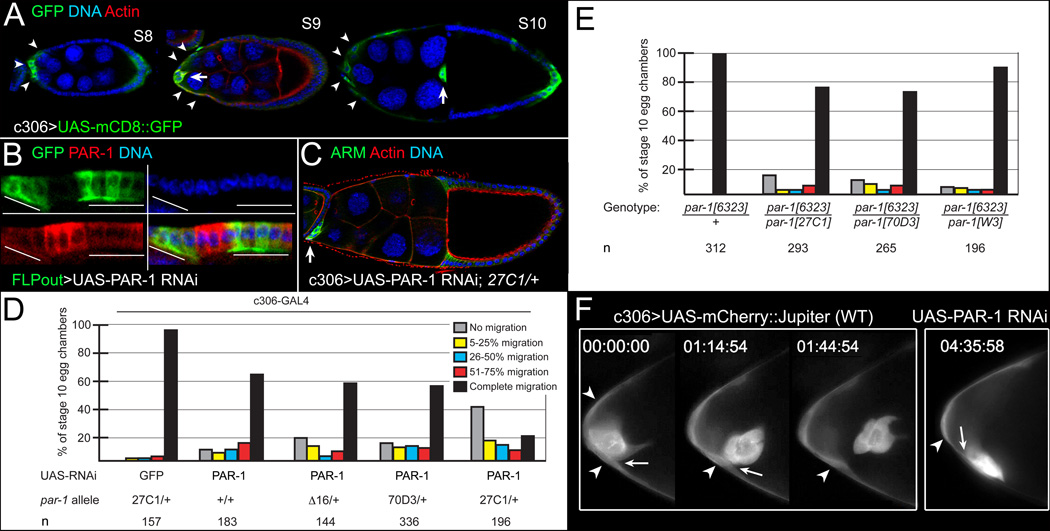
To assess PAR-1 function in migration in more detail, and to analyze clusters in which all border cells had reduced PAR-1, we constructed transgenic flies expressing GAL4-inducible PAR-1 dsRNA (see Supplemental Experimental Procedures). Expression of PAR-1 RNAi in the ovary reduced PAR-1 protein levels (Figure 2B). Border cell migration was disrupted when PAR-1 RNAi was expressed in all border cells and adjacent epithelial ‘stretched’ follicle cells using c306-GAL4 (Figures 2A, 2C, and 2D). Removing one copy of par-1 increased the severity of the migration defects (Figures 2C and 2D). Viable trans-heterozygous combinations of par-1 alleles, which disrupt oocyte polarity but not epithelial polarity [19], also disrupted border cell migration (Figure 2E). Although the overall migration defect was less frequent than that observed with PAR-1 RNAi, many phenotypic border cells remained at the anterior end of the egg chamber (Figure 2E).
Figure 2. par-1 is required for border cell detachment.
(A–D) Downregulation of PAR-1 by UAS-PAR-1 RNAi disrupts border cell detachment. (A) Stage 8–10 egg chambers showing c306-GAL4 driver pattern, visualized by UAS-mCD8::GFP (green), in anterior stretched follicle cells (arrowheads) and border cells (arrows). (B) Follicle cells expressing UAS-PAR-1 RNAi in FLPout GAL4 clones, marked by GFP (green, lines), have reduced PAR-1 (red). (C) Stage 10 par-127C1/+ egg chamber expressing PAR-1 RNAi driven by c306-GAL4, stained for ARM (green) and F-actin (red) to label cell membranes and DAPI (blue) to label nuclei. The border cells (arrow) did not migrate. (D, E) Quantification of border cell migration defects in wild-type or par-1/+ egg chambers expressing UAS-PAR-1 RNAi driven by c306-GAL4 (D), and in viable par-1 mutant egg chambers (E). Percentage of border cells that did not migrate/detach (gray) or migrated 5–25% (yellow), 26–50% (blue), 51–75% (red), or 76–100% (black) of normal distance to the oocyte; n, number of egg chambers examined. (F) Frames from time-lapse movies of stage 9 control (Movie S1) or PAR-1 RNAi (Movie S2) egg chambers at the indicated time (h:min:s). Control border cells detached at ~1.5 hours; PAR-1 RNAi border cells (n = 10) stayed attached (arrows) to follicle cells (arrowheads).
To determine why par-1 mutant border cells did not move, we performed time-lapse imaging of live egg chambers expressing c306-GAL4; UAS-mCherry::Jupiter (see Supplementary Experimental Procedures). Wild-type border cells visibly detached from the epithelium and migrated to the oocyte (Figure 2F and Movie S1). We imaged the behavior of phenotypic border cell clusters that expressed PAR-1 RNAi in a par-127C1 heterozygous mutant background or were viably mutant for par-1. Similar to what was observed in fixed egg chambers (Figures 2D and 2E), some clusters completed their migration whereas others did not, even by late oogenesis. Although border cell clusters formed normally and individual border cells moved around within the cluster, the most severely affected clusters did not move forward or away from the anterior end (Figure 2F and Movie S2 and Movie S3). Furthermore, when anterior stretched follicle cells were discernible within the focal plane of the movie, border cells remained visibly attached to these cells (Figure 2F). Although some clusters never detached during the movie, others detached several hours late (not shown). These results indicate that loss of par-1 disrupted the ability of many border cells to detach from the follicular epithelium.
Border cells did not detach when PAR-1 RNAi was expressed using c306-GAL4 (Figures 2A, 2C, and 2D), suggesting that PAR-1 functions in anterior follicle cells and border cells. Furthermore, we observed border cell migration defects when mutant clones were induced strictly in follicle cells using the GAL4/UAS system [6] (not shown). To investigate par-1 cell autonomy further, we used the MARCM mosaic clone method to label homozygous mutant cells with GFP (see Supplemental Experimental Procedures). We did not observe migration defects when only anterior stretched follicle cells (n = 47 egg chambers) or when polar cells were mutant (n = 28 egg chambers). However, in every case in which border cell migration was impaired, both border cells and anterior stretched follicle cells were mutant (20/20 examples examined; Figures 1C and 1D). In undetached clusters, we observed a mix of mutant and wild-type border cells (7/7 examples examined). In more than 150 egg chambers containing par-1 clones, none were observed in which only border cells were mutant, therefore we do not know if this would lead to migration or detachment defects. Taken together, these results are consistent with a requirement for par-1 in border cells and possibly adjacent anterior follicle cells.
Apical-Basal Polarity is Required for Detachment
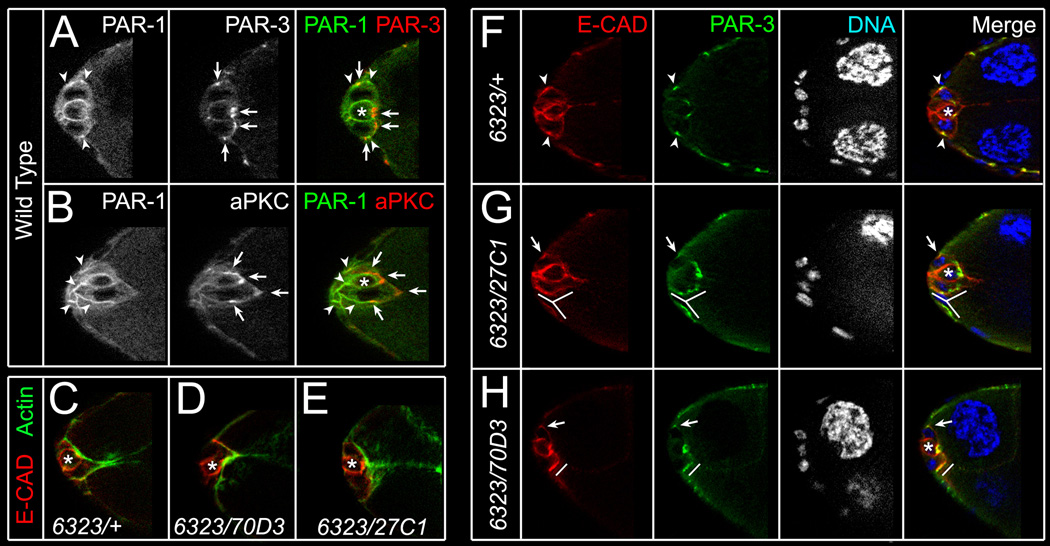
At all stages of border cell migration, PAR-1 protein primarily localizes to basolateral cell membranes [11, 19] (Figure S3). Before detaching, border cells exhibit apical-basal polarization similar to the epithelial follicle cells [20, 21], with apical facing inside the egg chamber. At this stage, PAR-3 and aPKC accumulated in apical junctions between follicle cells and border cells whereas Par-1 concentrated outside the junctions at lateral membranes (Figures 3A and 3B). Thus, PAR-1 and PAR-3/aPKC were largely non-overlapping, similar to their localization in follicle cells.
Figure 3. PAR-1 regulates localization of PAR-3 and E-CAD between border and follicle cells.
(A–B) Stage 9 wild-type border cells stained for PAR-1 (green, arrowheads) and PAR-3 (red, arrows; A) or aPKC (red, arrows; B). PAR-1, localized to basolateral cell membranes, is largely non-overlapping with apical PAR-3 and aPKC. (C–E) Stage 9 control (C) and viable par-16323/par-170D3 (D) or par-16323/par-127C1 (E) mutant border cell clusters stained for phalloidin (green) to visualize F-actin and E-cadherin (E-CAD; red) to label cell membranes. F-actin localizes normally. (F–H) Stage 9 control (F) and viable par-16323/par-127C1 (G) or par-16323/par-170D3 (H) mutant border cell clusters stained for E-CAD (red), PAR-3 (green) and DAPI to visualize nuclei (white/blue). (F) E-CAD and PAR-3 co-localize at foci between border cells and follicle cells (arrowheads; F). (G, H) In par-1 mutant egg chambers, E-CAD and PAR-3 are less enriched at foci (arrows; G, H), found in multiple foci (lines; G), or are broadly enriched (lines; H). Single optical sections shown in all panels; polar cells (*).
The enrichment of PAR-1 at border cell membranes suggested that it might regulate the localization of other membrane-associated proteins. F-actin accumulates at the apical side of wild-type border cells before they detach (Figure 3C) and this distribution was unchanged in viable par-1 mutant border cells (Figures 3D and 3E; n = 14 egg chambers). Both E-cadherin and PAR-3 are highly expressed in border cells and required for their migration [20, 21]. In wild-type egg chambers prior to detachment, E-cadherin and PAR-3 co-localized at foci found at the junctions between border cells and adjacent stretched follicle cells (Figure 3F; 8/9 egg chambers). In addition, E-cadherin localized to lateral membranes (Figure 3F) [5, 20]. However, in par-1 mutant border cells, individual foci enriched in E-cadherin were more difficult to discern at one or both junctions (Figures 3G and 3H; 23/27 egg chambers). PAR-3 was present in multiple foci (Figure 3G) or was uniformly localized between border cells and follicle cells (Figures 3G and 3H; 16/19 egg chambers). The apical protein Stardust (SDT) was also disrupted, although less frequently (not shown; 7/23 egg chambers).
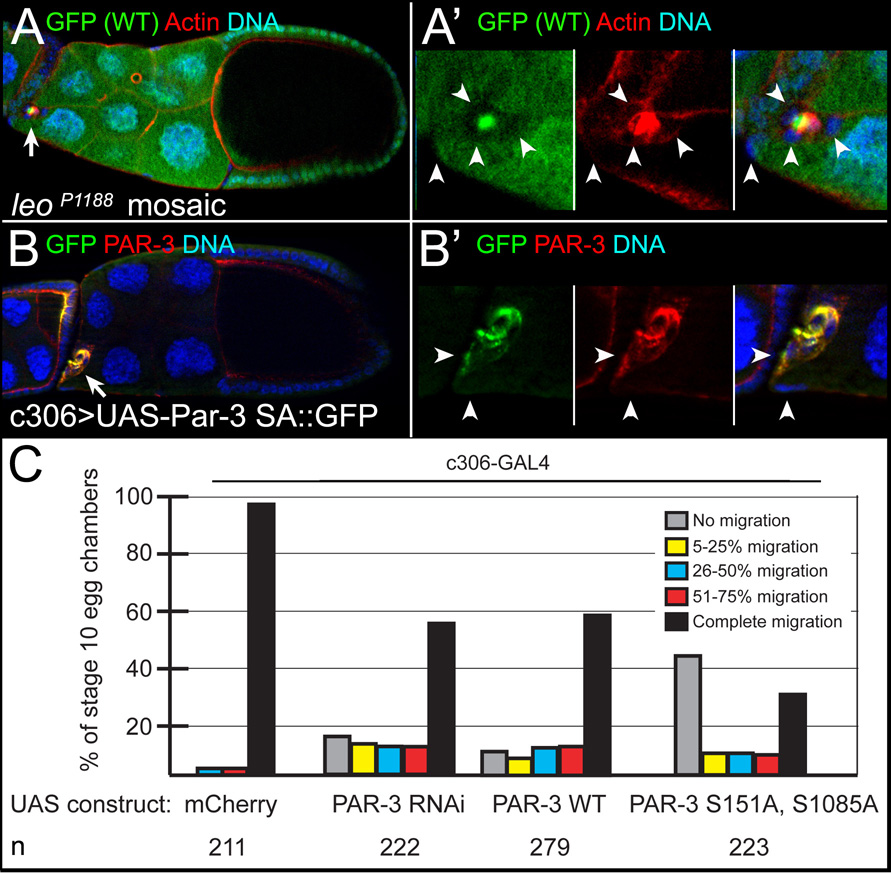
PAR-1 inhibits PAR-3 localization and activity during polarization of other cell types [10], suggesting that a similar mechanism functions in border cell detachment. At basolateral cell membranes, PAR-1 phosphorylates PAR-3 at two serines, S151 and S1085, creating binding sites for 14-3-3 protein [10, 22]. Binding of 14-3-3 prevents PAR-3 from forming a functional complex with aPKC and PAR-6 [10]. To test this model, we analyzed the function of Leonardo (LEO), the Drosophila homolog of 14-3-3ζ. A lacZ enhancer trap in leo (leoP1188/+) was highly expressed in border cells (not shown). Loss of leo caused reproducible migration defects in mosaic mutant clones (n = 42), some of which did not detach (Figures 4A and 4A’). We determined whether PAR-3 functioned in detachment, in addition to its known role in migration [21]. Border cells with reduced PAR-3 or overexpressing PAR-3 had mild detachment defects (Figure 4C). Overexpression of PAR-3 that cannot be phosphorylated by PAR-1 (PAR-3 S151A, S1085A; PAR-3SA mutant) [10] strongly enhanced the wild-type PAR-3 detachment phenotype (Figures 4B and 4C), even though both constructs were expressed at equivalent levels as assayed by anti-GFP and anti-Par-3 antibody staining (Figure 4B’ and not shown). Live time-lapse imaging of PAR-3 SA-expressing border cells revealed that many clusters did not move away from the epithelium (Movie S4), similar what was observed upon loss of par-1. These results suggest that having normal distributions and/or activities of the apical-basal polarity proteins PAR-3 and PAR-1 are necessary for efficient detachment.
Figure 4. PAR-1 inhibits PAR-3 in detachment.
(A, A’) leo is required for border cell migration. Stage 10 leoP1188 mosaic mutant egg chamber stained for GFP (green) to mark wild-type cells, F-Actin (red) to mark cell membranes, and DAPI to mark nuclei (blue). (A) leo mutant border cells (arrow, GFP-negative) did not migrate. (A’) Magnified view of mutant border cells (arrowheads); a follicle cell at the anterior is mutant, but on a slightly different focal plane (left arrowhead). (B, B’) PAR-3 SA overexpression in border cells (arrow in B) disrupts detachment. Stage 10 egg chamber expressing UAS-PAR-3 SA::GFP driven by c306-GAL4, stained for GFP (green), PAR-3 (red) and DAPI (blue) to mark nuclei. (B’) Magnified view of border cells and adjacent follicle cell (arrowheads); PAR-3 SA is uniformly localized. (C) Quantification, as in Figure 2D, of border cell migration in egg chambers expressing indicated UAS-PAR-3 transgenes driven by c306-GAL4.
LKB1 Functions in PAR-1-mediated Border Cell Detachment
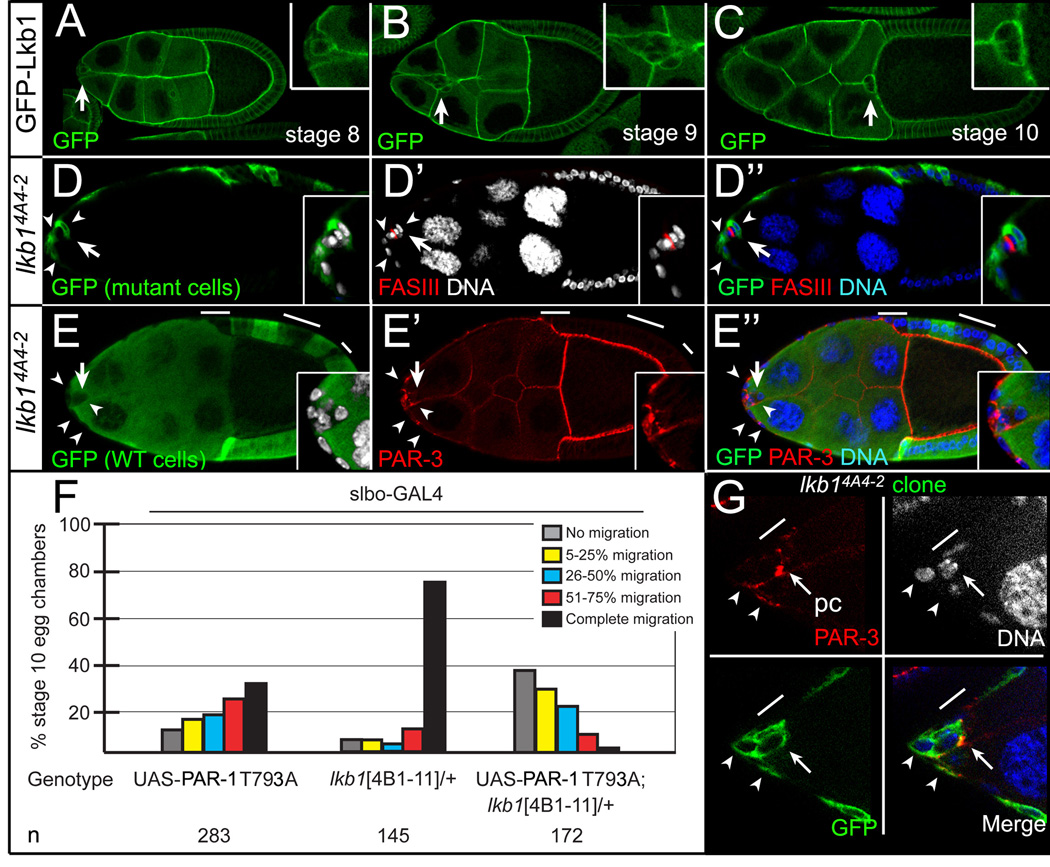
LKB1 (PAR-4) is a conserved serine/threonine kinase that phosphorylates and activates PAR-1 in many cellular contexts [23–25]. LKB1 is required for PAR-1-dependent events during Drosophila development [26, 27], suggesting that it could function similarly in border cells. GFP-tagged LKB1 was expressed uniformly at border cell membranes (Figures 5A–C) [26]. Mosaic mutant clones of lkb14A4-2, a strong loss-of-function allele, resulted in either no border cell migration (Figures 5D and 5E) or incomplete migration (not shown). When border cells did not complete their migration, border cells and anterior stretched follicle cells were mutant (45/45 egg chambers). lkb1 mutant border cells were closely associated with mutant anterior stretched follicle cells when clusters did not detach (12/12 examples; Figures 5D, 5E, and 5G). Similar to the phenotype observed upon loss of par-1, ectopic PAR-3 was evident between lkb1 mutant border cells and adjacent follicle cells (Figure 5G; n = 7 egg chambers).
Figure 5. lkb1 is required for border cell detachment.
(A–C) GFP-LKB1 expression in stage 8 (A), 9 (B), and 10 (C) egg chambers. GFP-LKB1 (green) localizes to cell membranes, including border cells (arrows; insets). (D, E) Defective migration (arrows) in stage 10 lkb14A4-2 mosaic mutant egg chambers stained for FAS III to mark polar cells (red, D) or PAR-3 to mark cell membranes (red, E) and DAPI (blue/white) to mark nuclei. (Insets) Magnified views of border cell clusters. (D-D”) Positively marked lkb1 mosaic mutant border cells (GFP-positive, green; arrowheads) did not detach; border cells and an adjacent follicle cell are mutant. (E-E”) Negatively marked lkb1 mosaic mutant border cells (GFP-negative; arrowheads) did not detach. Border cells and adjacent follicle cells are mutant; nurse cells are wild-type (green). (F) Loss of one copy of lkb14B1-11 significantly enhances migration defects caused by UAS-PAR-1::T793A driven by slbo-GAL4, which is expressed in border cells and adjacent follicle cells. Quantification as in Figure 2D. (G) Stage 9 positively marked lkb14A4-2 mosaic mutant border cells stained for PAR-3 (red) and DAPI (white/blue). Mutant border cells (GFP-positive, green) have broad distribution of PAR-3 at border cell/follicle cell junctions (arrowheads); pc, polar cells (arrow); line, non-mutant adjacent follicle cell.
We identified a genetic interaction between lkb1 and par-1 (Figure 5F). A conserved threonine in the C-terminus of PAR-1 (T793) is phosphorylated by aPKC in vertebrates; mutation of this residue to alanine causes the protein to mislocalize to apical cell membranes in addition to its normal basolateral localization [28–30]. Overexpression of PAR-1 T793A strongly inhibited border cell migration (Figure 5F). Loss of one wild-type copy of the lkb14B1-11 allele resulted in a mild, dominant migration defect, but significantly enhanced the PAR-1 T793A defects (Figure 5F). The observed genetic interaction and close resemblance of the phenotypes caused by loss of lkb1 and par-1 suggest that these two genes function in a conserved pathway to regulate border cell detachment.
PAR-1 Regulates Border Cell Protrusion Direction and Morphology
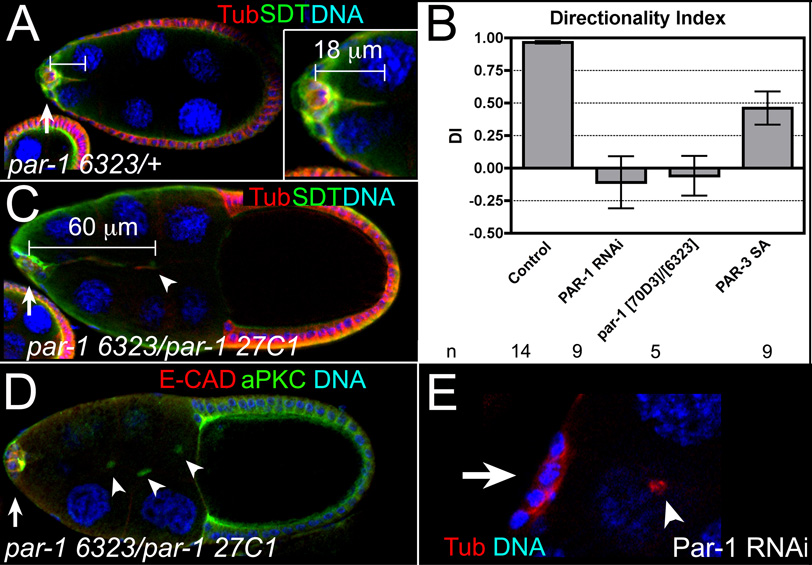
Border cells extend prominent cellular protrusions mainly in the direction of migration, which help them migrate and navigate to the oocyte [2, 31] (Figure 6A and Movie S1). Wild-type, par-1 mutant, and PAR-3 SA-expressing border cells formed protrusions measuring at least 18 µm from the center of the cluster (Figure 6A, Movie S1–Movie S4, Table S1). Whereas wild-type or PAR-3 SA-expressing border cells extended protrusions primarily in the direction of migration (Figures 6A and 6B; Movie S1 and Movie S4) [2], undetached par-1 mutant border cells extended more protrusions along the lateral sides of the egg chamber (Movie S2 and Movie S3). To quantify this effect, we calculated the directionality index (DI) of these protrusions (see Supplemental Experimental Procedures) [2]. The DI would be 1 if protrusions extended only in the forward direction, whereas it would be 0 if equal numbers of forward and backward/lateral protrusions formed. Wild-type border cell clusters had a DI of ~1, whereas PAR-1 RNAi and viable par-1 mutant border cells each had a DI close to 0 (Figure 6B). Thus, loss of par-1 decreased forward protrusions at the expense of backward/lateral protrusions. This phenotype was not observed following PAR-3 SA overexpression (Figure 6B).
Figure 6. par-1 is required for normal border cell protrusions.
(A) Stage 9 control egg chamber stained for α-Tubulin (red) and SDT (green) to label border cells (arrow) and protrusions; DAPI marks nuclei (blue). (Inset) An 18 µm protrusion extends in direction of migration. (B) The directionality index was calculated for live control (0.96 ± 0.01), PAR-1 RNAi (−0.11 ± 0.20), viable par-1 (−0.06 ± 0.15), and PAR-3 SA (DI = 0.46 ± 0.13) mutant border cells. n, number of movies examined; error bars, SEM. (C) Stage 10 par-1 mutant egg chamber, stained as in (A). A long (60 µm), thin protrusion (arrowhead) extends from undetached border cell cluster (arrow). (D) Stage 10 par-1 mutant egg chamber stained for E-CAD (red), aPKC (green) and DAPI to mark nuclei (blue); three cytoplasts (arrowheads), of ~5.3 µm average diameter (n = 13), detached from the border cell cluster (arrow). (E) Stage 10 PAR-1 RNAi egg chamber stained for α-Tubulin (red) and DNA (blue); a cytoplast (arrowhead) detached from the border cells (arrow).
The overall length and lifetime of protrusions at stage 9 was similar between wild-type and PAR-1 RNAi border cells (Table S1). However, by stage 10, PAR-1 RNAi and viable par-1 mutant egg chambers exhibited defects in protrusion morphology. Wild-type protrusions were on average 22 µm in length (Figure 6A and Table S1; n = 49), whereas undetached par-1 mutant border cells had 40–67 µm long protrusions (Figure 6C; 9/25 examples). Live PAR-1 RNAi border cells extended misshapen protrusions (Movie S5; 4/9 movies). In addition to longer protrusions, we frequently observed 1–3 cell fragments along the migration pathway (Figure 6D, 6E, and Figure S4B; 47% of egg chambers, n = 113) that were never observed in front of wild-type clusters (n = 95). These fragments lacked nuclei and were not visibly attached to the cluster (Figure 6E and not shown), but were enriched in the border cell marker SN [17, 18] (not shown). Moreover, they strongly resembled ‘cytoplasts’ found in mal-d/mrtf mutants, which are proposed to develop from fragmented border cell protrusions [32]. Cytoplasts were occasionally observed in live PAR-1 RNAi-expressing egg chambers (not shown). Cytoplasts and long protrusions are not a general feature of border cells that do not migrate or detach, as they were not observed in egg chambers mutant for slow border cells (slbo) or expressing dominant-negative Kuzbanian [2, 33] (Figure S4A and not shown). Loss or overexpression of PAR-3 (WT or SA mutant) also did not affect protrusion morphology (Figure 4B and not shown). These data indicate that PAR-1, independent of apical PAR-3 polarity, specifically regulates the morphology and length of some border cell protrusions.
Discussion
Mechanisms of Epithelial Cell Detachment
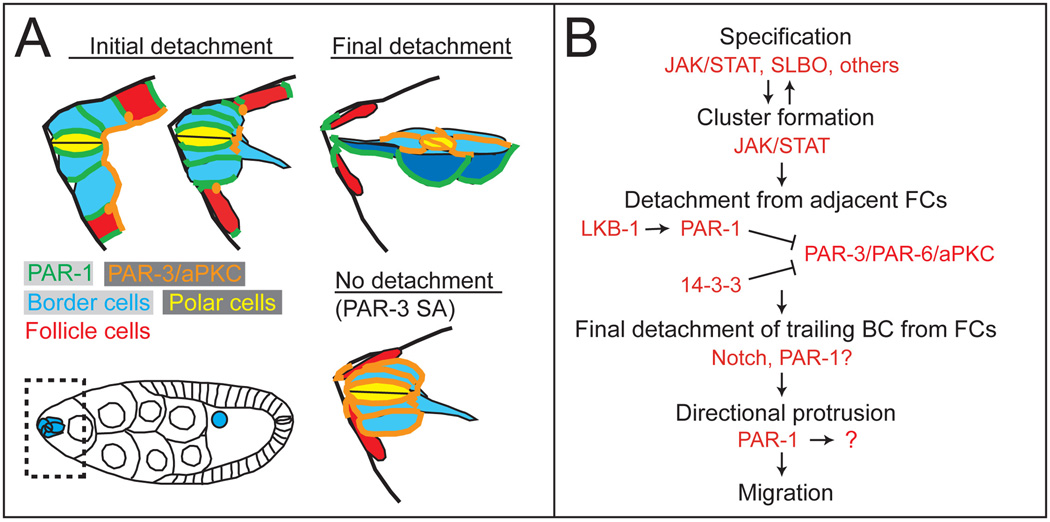
While detachment is critical for the separation of migratory cells from epithelia, the molecular and cellular mechanisms underlying this process are poorly understood. Here we demonstrate that disrupting apical-basal polarity inhibits the ability of border cells to detach from the follicle cell epithelium. Overexpression of a mutant form of PAR-3 that cannot be phosphorylated by PAR-1 phenocopied the loss of par-1. Although Drosophila LKB1 was initially reported to be downstream of PAR-1 [26], a recent study demonstrated that it phosphorylates PAR-1 in vitro and in vivo at a conserved threonine within the kinase domain [27]. Loss of lkb1 affected border cell detachment similar to par-1 mutants and exhibited a genetic interaction with par-1. We propose a model in which a conserved network of LKB1 and basolateral PAR-1 directly phosphorylates and inhibits apical PAR-3 between border cells and follicle cells, thereby promoting the separation of the two cell types (Figures 7A and 7B).
Figure 7. Model for PAR-1 function.
(A) Model for border cell detachment. Schematic of the anterior end of an egg chamber (boxed area in egg chamber). Basolateral PAR-1 restricts PAR-3 to the apical side, allowing border cells to separate from the epithelium. If PAR-3 expands to basolateral side, detachment does not occur. (B) Molecular pathway and steps required for early border cell cluster formation, detachment, protrusion, and migration. Note that specification occurs before the cluster forms and while it detaches [3].
How does the proper spatial restriction of apical polarity proteins lead to border cell detachment? The apical PAR-3/PAR-6/aPKC complex is required to form and stabilize epithelial adherens and tight/subapical junctions [34, 35]. Overexpression of PAR-3 in mammalian cells promotes tight junction assembly [35], suggesting a model in which PAR-1-dependent restriction of PAR-3 to the apical junction allows the disassembly and/or remodeling of adhesion between border cells and adjacent epithelial cells. Analysis of specific cell membrane markers revealed that loss of par-1 caused mislocalization of apical PAR-3 and E-cadherin at the interface between border cells and follicle cells. In addition, overexpression of PAR-3 and PAR-6 in border cells disrupts the polarized distribution of membrane proteins, including the adherens junction proteins E-cadherin and Armadillo/β-Catenin [21]. Expansion of the apical PAR-3 domain inhibits border cell detachment, possibly by increasing adhesion and preventing disassembly of junctions between border cells and follicle cells (Figure 7A).
Our results suggest that the ability to detach is a property of the basolateral domain and requires restriction of the apical domain, which may be a general mechanism for promoting detachment of epithelial cells. During epithelial-to-mesenchymal transitions induced by TGFβ signaling, TGFβ receptors bind to and phosphorylate the apical polarity protein PAR-6 to allow recruitment of the ubiquitin ligase Smurf1; this in turn degrades RhoA, resulting in loss of tight junctions [36]. Furthermore, the oncogene ErbB2 directly blocks the formation of the PAR-3/PAR-6/aPKC complex to inhibit breast epithelial cell polarity and tissue organization, apart from its function in cell proliferation [37]. These studies demonstrate that downregulation of apical-basal polarity, which in turn disrupts cell-cell junctions, remodels epithelia during tissue morphogenesis and tumor progression. We propose that PAR-1 is another protein that remodels specific epithelial junctions to promote cell migration. However, other basolateral proteins such as Scribble and Discs large seem to suppress rather than promote detachment of epithelial cells [reviewed in 38]. Therefore, the relationship between apical/basal polarity and epithelial cell invasion and detachment remains to be fully understood.
PAR-1 Control of Leading/Lagging Edge Border Cell Cluster Polarity
Border cells are guided to the oocyte by secreted ligands that bind to multiple receptor tyrosine kinases [reviewed in 1]. The secreted proteins PVF1, Spitz and Keren are synthesized in the oocyte. The receptor for PVF1 is PVR, and the receptor for both Spitz and Keren is the Drosophila EGF Receptor (EGFR); both receptors are expressed on border cells. Border cells, like all migrating cells that undergo chemotaxis, polarize in response to these guidance cues. One of the most obvious manifestations of polarization is the extension of prominent protrusions primarily from the front or leading edge of the cluster. Inhibiting the functions of PVR and EGFR together causes border cells to extend protrusions in all directions, indicating that guidance receptor activity limits protrusion to the leading edge [2]. However, guidance signaling not only induces protrusion extension from the front, but also suppresses them from the rear of the cluster [2]. Thus, guidance receptor activation at the front polarizes the cluster. Similarly, loss of par-1 causes more protrusions to extend to the side and rear of the cluster, suggesting that directional polarity is lost.
As border cells initiate their migration, the leading edge coincides with the apical domain, although this orientation changes after the cluster detaches [20, 21]. PAR-1 localizes to the lateral sides of border cells, and not to the leading or apical side of the cluster. One possibility is that PAR-1 restricts the localization of an unknown protein to the leading edge, which in turn promotes the formation of stable protrusions at the front. Although PAR-1 restricts PAR-3 to the apical side, PAR-3 SA overexpression did not disrupt directional protrusion extension. Alternatively, PAR-1 may activate a protein at the side and/or back of the cluster, which either suppresses or destabilizes protrusions at the lagging edge. The known role for PAR-1 in regulating microtubule stability [11–13] could contribute directly to protrusion extension and dynamics. Although we did not observe gross changes in the levels or distribution of α-Tubulin (not shown), PAR-1 could have a subtle role in regulating microtubule dynamics of border cell protrusions. Thus, PAR-1 participates in generating polarity with respect to the leading and lagging edges of the migrating border cells, independent of its effect on PAR-3.
Whereas border cells detach and migrate as part of normal development, similar mechanisms occur in pathological conditions such as tumor invasion and metastasis. The work presented here establishes PAR-1-dependent apical-basal polarity as an essential mechanism for cell detachment from a polarized epithelium. Furthermore, PAR-1 plays a second role during cell migration by polarizing the border cell cluster, possibly in response to guidance signaling, and is required for normal protrusion morphology. Given the diversity of mechanisms that contribute to polarizing epithelial cells and migrating cells, and the high degree of functional conservation of polarity proteins, it will be of great interest to determine whether and in what contexts PAR-1 homologs induce epithelial cell detachment and protrusions during development or in tumor metastasis in vertebrates.
Supplementary Material
Supplemental Data include Experimental Procedures, four figures, one table, and five movies.
Acknowledgments
We thank D. St Johnston, A. Ephrussi, C.Q. Doe, E. Knust, D. Cox, Y.-N. Jan, A. Wodarz, R. Carthew, A. Page-McCaw, R. Palmer, S. Hou, the Bloomington Stock Center, the Vienna Drosophila RNAi Center and the Developmental Studies Hybridoma Bank for reagents. Special thanks to J. Bo, T. Harris, P. Vanguri and K. Devlin for technical assistance and M. Prasad for essential advice on live imaging. This work was supported by startup funds provided by the Cleveland Clinic to J.A.M. and by NIH grants R01GM46425 and R01GM073164 to D.J.M. and R01GM078526 to J.A.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jang AC, Starz-Gaiano M, Montell DJ. Modeling migration and metastasis in Drosophila. J. Mammary Gland Biol. Neoplasia. 2007;12:103–114. doi: 10.1007/s10911-007-9042-8. [DOI] [PubMed] [Google Scholar]
- 2.Prasad M, Montell DJ. Cellular and molecular mechanisms of border cell migration analyzed using time-lapse live-cell imaging. Dev. Cell. 2007;12:997–1005. doi: 10.1016/j.devcel.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 3.Starz-Gaiano M, Melani M, Wang X, Meinhardt H, Montell DJ. Feedback inhibition of Jak/STAT signaling by apontic is required to limit an invasive cell population. Dev. Cell. 2008;14:726–738. doi: 10.1016/j.devcel.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Adam JC, Montell D. Spatially localized Kuzbanian required for specific activation of Notch during border cell migration. Dev. Biol. 2007;301:532–540. doi: 10.1016/j.ydbio.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 5.Bai J, Uehara Y, Montell DJ. Regulation of invasive cell behavior by Taiman, a Drosophila Protein Related to AIB1, a steroid receptor coactivator amplified in breast cancer. Cell. 2000;103:1047–1058. doi: 10.1016/s0092-8674(00)00208-7. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Montell DJ. Identification of mutations that cause cell migration defects in mosaic clones. Development. 1999;126:1869–1878. doi: 10.1242/dev.126.9.1869. [DOI] [PubMed] [Google Scholar]
- 7.Silver DL, Montell DJ. Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell. 2001;107:831–841. doi: 10.1016/s0092-8674(01)00607-9. [DOI] [PubMed] [Google Scholar]
- 8.Mathieu J, Sung HH, Pugieux C, Soetaert J, Rorth P. A sensitized PiggyBac-based screen for regulators of border cell migration in Drosophila. Genetics. 2007;176:1579–1590. doi: 10.1534/genetics.107.071282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev. Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benton R, St Johnston D. Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003;115:691–704. doi: 10.1016/s0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- 11.Cox DN, Lu B, Sun TQ, Williams LT, Jan YN. Drosophila par-1 is required for oocyte differentiation and microtubule organization. Curr. Biol. 2001;11:75–87. doi: 10.1016/s0960-9822(01)00027-6. [DOI] [PubMed] [Google Scholar]
- 12.Doerflinger H, Benton R, Shulman JM, St Johnston D. The role of PAR-1 in regulating the polarised microtubule cytoskeleton in the Drosophila follicular epithelium. Development. 2003;130:3965–3975. doi: 10.1242/dev.00616. [DOI] [PubMed] [Google Scholar]
- 13.Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- 14.Ossipova O, Dhawan S, Sokol S, Green JB. Distinct PAR-1 proteins function in different branches of Wnt signaling during vertebrate development. Dev. Cell. 2005;8:829–841. doi: 10.1016/j.devcel.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 15.Sun TQ, Lu B, Feng JJ, Reinhard C, Jan YN, Fantl WJ, Williams LT. PAR-1 is a Dishevelled-associated kinase and a positive regulator of Wnt signalling. Nat. Cell Biol. 2001;3:628–636. doi: 10.1038/35083016. [DOI] [PubMed] [Google Scholar]
- 16.Sapir T, Sapoznik S, Levy T, Finkelshtein D, Shmueli A, Timm T, Mandelkow EM, Reiner O. Accurate balance of the polarity kinase MARK2/Par-1 is required for proper cortical neuronal migration. J. Neurosci. 2008;28:5710–5720. doi: 10.1523/JNEUROSCI.0911-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borghese L, Fletcher G, Mathieu J, Atzberger A, Eades WC, Cagan RL, Rorth P. Systematic analysis of the transcriptional switch inducing migration of border cells. Dev. Cell. 2006;10:497–508. doi: 10.1016/j.devcel.2006.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Bo J, Bridges T, Dugan KD, Pan TC, Chodosh LA, Montell DJ. Analysis of cell migration using whole-genome expression profiling of migratory cells in the Drosophila ovary. Dev. Cell. 2006;10:483–495. doi: 10.1016/j.devcel.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Shulman JM, Benton R, St Johnston D. The Drosophila homolog of C. elegans PAR-1 organizes the oocyte cytoskeleton and directs oskar mRNA localization to the posterior pole. Cell. 2000;101:377–388. doi: 10.1016/s0092-8674(00)80848-x. [DOI] [PubMed] [Google Scholar]
- 20.Niewiadomska P, Godt D, Tepass U. DE-Cadherin Is required for intercellular motility during Drosophila Oogenesis. J. Cell Biol. 1999;144:533–547. doi: 10.1083/jcb.144.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinheiro EM, Montell DJ. Requirement for Par-6 and Bazooka in Drosophila border cell migration. Development. 2004;131:5243–5251. doi: 10.1242/dev.01412. [DOI] [PubMed] [Google Scholar]
- 22.Hurd TW, Fan S, Liu CJ, Kweon HK, Hakansson K, Margolis B. Phosphorylation-dependent binding of 14-3-3 to the polarity protein Par3 regulates cell polarity in mammalian epithelia. Curr. Biol. 2003;13:2082–2090. doi: 10.1016/j.cub.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 23.Brajenovic M, Joberty G, Kuster B, Bouwmeester T, Drewes G. Comprehensive proteomic analysis of human Par protein complexes reveals an interconnected protein network. J. Biol. Chem. 2004;279:12804–12811. doi: 10.1074/jbc.M312171200. [DOI] [PubMed] [Google Scholar]
- 24.Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, Hawley SA, Udd L, Makela TP, Hardie DG, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spicer J, Rayter S, Young N, Elliott R, Ashworth A, Smith D. Regulation of the Wnt signalling component PAR1A by the Peutz-Jeghers syndrome kinase LKB1. Oncogene. 2003;22:4752–4756. doi: 10.1038/sj.onc.1206669. [DOI] [PubMed] [Google Scholar]
- 26.Martin SG, St Johnston D. A role for Drosophila LKB1 in anterior-posterior axis formation and epithelial polarity. Nature. 2003;421:379–384. doi: 10.1038/nature01296. [DOI] [PubMed] [Google Scholar]
- 27.Wang JW, Imai Y, Lu B. Activation of PAR-1 kinase and stimulation of tau phosphorylation by diverse signals require the tumor suppressor protein LKB1. J. Neurosci. 2007;27:574–581. doi: 10.1523/JNEUROSCI.5094-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurov JB, Watkins JL, Piwnica-Worms H. Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr. Biol. 2004;14:736–741. doi: 10.1016/j.cub.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki A, Hirata M, Kamimura K, Maniwa R, Yamanaka T, Mizuno K, Kishikawa M, Hirose H, Amano Y, Izumi N, et al. aPKC acts upstream of PAR-1b in both the establishment and maintenance of mammalian epithelial polarity. Curr. Biol. 2004;14:1425–1435. doi: 10.1016/j.cub.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 30.Vaccari T, Rabouille C, Ephrussi A. The Drosophila PAR-1 spacer domain is required for lateral membrane association and for polarization of follicular epithelial cells. Curr. Biol. 2005;15:255–261. doi: 10.1016/j.cub.2005.01.033. [DOI] [PubMed] [Google Scholar]
- 31.Fulga TA, Rorth P. Invasive cell migration is initiated by guided growth of long cellular extensions. Nat. Cell Biol. 2002;4:715–719. doi: 10.1038/ncb848. [DOI] [PubMed] [Google Scholar]
- 32.Somogyi K, Rorth P. Evidence for tension-based regulation of Drosophila MAL and SRF during invasive cell migration. Dev. Cell. 2004;7:85–93. doi: 10.1016/j.devcel.2004.05.020. [DOI] [PubMed] [Google Scholar]
- 33.Montell DJ, Rørth P, Spradling AC. slow border cells, a locus required for a developmentally regulated cell migration during oogenesis, encodes Drosophila C/EBP. Cell. 1992;71:51–62. doi: 10.1016/0092-8674(92)90265-e. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, Macara IG. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat. Cell Biol. 2005;7:262–269. doi: 10.1038/ncb1226. [DOI] [PubMed] [Google Scholar]
- 35.Hirose T, Izumi Y, Nagashima Y, Tamai-Nagai Y, Kurihara H, Sakai T, Suzuki Y, Yamanaka T, Suzuki A, Mizuno K, et al. Involvement of ASIP/PAR-3 in the promotion of epithelial tight junction formation. J. Cell Sci. 2002;115:2485–2495. doi: 10.1242/jcs.115.12.2485. [DOI] [PubMed] [Google Scholar]
- 36.Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL. Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science. 2005;307:1603–1609. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]
- 37.Aranda V, Haire T, Nolan ME, Calarco JP, Rosenberg AZ, Fawcett JP, Pawson T, Muthuswamy SK. Par6-aPKC uncouples ErbB2 induced disruption of polarized epithelial organization from proliferation control. Nat. Cell Biol. 2006;8:1235–1245. doi: 10.1038/ncb1485. [DOI] [PubMed] [Google Scholar]
- 38.Lee M, Vasioukhin V. Cell polarity and cancer-cell and tissue polarity as a non-canonical tumor suppressor. J. Cell Sci. 2008;121:1141–1150. doi: 10.1242/jcs.016634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data include Experimental Procedures, four figures, one table, and five movies.