Abstract
Through alternative processing of pre-mRNAs, individual mammalian genes often produce multiple mRNA and protein isoforms that may have related, distinct or even opposing functions. Here we report an in-depth analysis of 15 diverse human tissue and cell line transcriptomes based on deep sequencing of cDNA fragments, yielding a digital inventory of gene and mRNA isoform expression. Analysis of mappings of sequence reads to exon-exon junctions indicated that 92-94% of human genes undergo alternative splicing (AS), ∼86% with a minor isoform frequency of 15% or more. Differences in isoform-specific read densities indicated that a majority of AS and of alternative cleavage and polyadenylation (APA) events vary between tissues, while variation between individuals was ∼2- to 3-fold less common. Extreme or ‘switch-like’ regulation of splicing between tissues was associated with increased sequence conservation in regulatory regions and with generation of full-length open reading frames. Patterns of AS and APA were strongly correlated across tissues, suggesting coordinated regulation of these processes, and sequence conservation of a subset of known regulatory motifs in both alternative introns and 3′ UTRs suggested common involvement of specific factors in tissue-level regulation of both splicing and polyadenylation.
The mRNA and protein isoforms produced by alternative processing of primary RNA transcripts may differ in structure, function, localization or other properties1,2. AS in particular is known to affect more than half of all human genes, and has been proposed as a primary driver of the evolution of phenotypic complexity in mammals3,4. However, assessment of the extent of differences in mRNA isoform expression between tissues has presented substantial technical challenges5. Studies using expressed sequence tags (ESTs) have yielded relatively low estimates of tissue specificity, but have limited statistical power to detect differences in isoform levels6-8. Microarray analyses have achieved more consistent coverage of tissues9, but are constrained in their ability to distinguish closely related mRNA isoforms. High throughput sequencing technologies have the potential to circumvent these limitations by generating high average coverage of mRNAs across tissues while using direct sequencing rather than hybridization to distinguish and quantitate mRNA isoforms10,11.
Tissue-specific AS is usually regulated by a combination of tissue-specific and ubiquitously expressed RNA binding factors that interact with cis-acting RNA elements to influence spliceosome assembly at nearby splice sites1,2. Many factors can both activate or repress splicing, with activity often summarizable by an ‘RNA map’ describing dependence on the location of binding relative to that of core spliceosomal components12,13.
A digital inventory of gene and isoform expression
To assess gene and alternative mRNA isoform expression, the mRNA-SEQ protocol (Methods) was used to amplify and sequence between 12 and 29 million 32 bp cDNA fragments from 10 diverse human tissues and 5 mammary epithelial or breast cancer cell lines, generating over 400 million reads in total (Fig. S1a). Tissue samples were derived from single anonymous unrelated individuals of both sexes; for one tissue, cerebellar cortex, samples from six unrelated men were analyzed to assess variation between individuals (Table S1). In total, ∼60% of reads mapped uniquely to the genome allowing up to 2 mismatches, and an additional 4% mapped uniquely to splice junctions (SJs). Thus, about two-thirds of reads could be assigned unambiguously to individual genes; the frequency of mapping to incorrect genomic locations was estimated at ∼0.1% (Table S2).
Read density (coverage) was over 100-fold higher in exons than in introns or intergenic regions (Fig. S1c), and only ∼3% of reads mapped to ribosomal RNA genes, indicating that the great majority of reads derived from mature mRNA. Comparison of relative mRNA-SEQ read densities to published quantitative RT-PCR measurements for 787 genes in two reference RNA samples14 yielded a nearly linear relationship across ∼5 orders of magnitude (Fig. S1d), indicating that mRNA-SEQ read counts give accurate relative gene expression measurements across a very broad dynamic range10.
Alternative splicing is essentially universal in human multi-exon genes
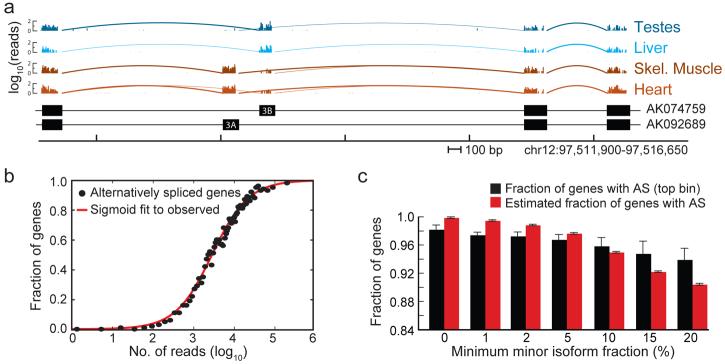
The mRNA-SEQ data were used to assess the expression of alternative transcript isoforms in human genes, as illustrated for the mitochondrial phosphate transporter gene SLC25A3 in Fig. 1a. Exons 3A and 3B of this gene are “mututally exclusive exons” (MXEs), meaning that transcripts from this gene contain one or the other of these exons, but not both. Far greater read coverage of exon 3A was seen in heart and skeletal muscle, with almost exclusive coverage of exon 3B in testes, liver (and other tissues studied), consistent with the predominant heart and muscle symptoms of exon 3A mutation15.
Figure 1. Frequency and relative abundance of AS isoforms in human genes.
a, mRNA-SEQ reads mapping to a portion of the SCL25A3 gene locus. The number of mapped reads starting at each nucleotide position is displayed (log10) for the tissues listed at right. Arcs represent junctions detected by SJ reads (bold arcs for junctions supported by >10 reads). Below – exon/intron structures of representative transcripts (GenBank accessions shown at right). b, Mean fraction of multi-exon genes with detected AS in bins of 500 genes, grouped by total read count per gene. A gene was considered as alternatively spliced if SJ reads joining the same 5′SS to different 3′SS, or joining the same 3′SS to different 5′SS were observed. The true extent of AS was estimated from the upper asymptote of the best-fit sigmoid curve (red). c, Frequency of AS in top bin and after estimation (as in b), considering only events with relative expression of less abundant (minor) splice variant exceeding given threshold. Bars indicate SEM.
The genome-wide extent of AS was assessed by searching against known and putative splicing junctions using stringent criteria that required each alternative isoform to be supported by multiple independent SJ reads with different alignment start positions. Binning the multi-exon genes in the Refseq database (94% of all Refseq genes) by read coverage and fitting to a sigmoid curve enabled estimation of the asymptotic fraction of AS genes in this set as ∼98% when excluding cell line data (Fig. S2), and ∼100% when using all samples (Fig. 1b). This analysis indicated that AS is essentially universal in human multi-exon genes, which comprise 94% of genes overall, with the important qualification that a portion of detected AS events may represent allele-specific splicing16,17.
Some of these events may involve exclusively low frequency AS isoforms. However, fully 92% of multi-exon genes were estimated to undergo AS when considering only events for which the relative frequency of the minor (less-abundant) isoform exceeded 15% in one or more samples (Fig. 1c). Thus, 0.92 × 0.94 or ∼86% of human genes were estimated to produce appreciable levels of two or more distinct populations of mRNA isoforms. Conversely, no evidence of AS was detected in the 6% of Refseq genes annotated as consisting of a single exon, even when searching against junctions between predicted exons in these genes.
Novel exons and splice junctions not previously seen in transcript databases were identified by mapping the reads against predicted exons and junctions. This approach yielded a set of 1413 high-confidence novel exons (Table S3), with an estimated false discovery rate (FDR) of <1.5% (Supp. Info.), and thousands of putative novel SJs (not shown). Thus, mRNA-SEQ has substantial potential for novel exon discovery, though very substantial read depth is required to efficiently detect low abundance isoforms (Fig. S3).
Extent of tissue-specific regulation of alternative RNA processing
To explore the extent of tissue-regulation of alternative transcripts, we examined eight common types of “alternative transcript events”1,2, each capable of producing multiple mRNA isoforms from human genes through AS, APA and/or alternative promoter usage (Fig. 2). Event types considered included skipped exons (SE) and retained introns (RI), in which a single exon or intron is alternatively included or spliced out of the mature message, and MXEs, described above. Also included were alternative 5′ splice site (A5SS) and alternative 3′ splice site (A3SS) events, which are particularly difficult to interrogate by microarray because the variably included region is often quite small. Tandem UTRs and mutually exclusive 3′ UTRs (3pMXE), in which alternative use of a pair of polyadenylation sites results in shorter or longer 3′ UTR isoforms, or in distinct terminal exons, respectively, were also considered. Finally, we considered mutually exclusive 5′ exons (5pMXE), in which alternative promoter use results in mRNA isoforms with distinct 5′ UTRs.
Figure 2. Pervasive tissue-specific regulation of alternative mRNA isoforms.
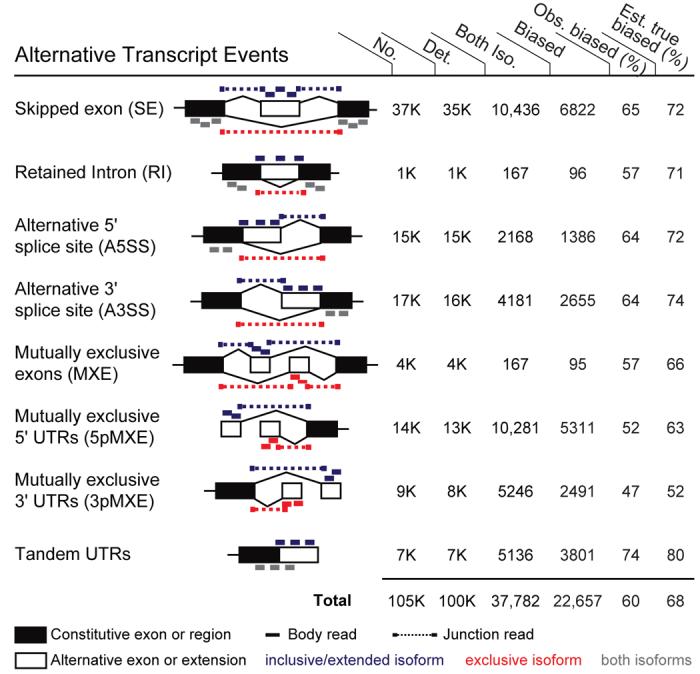
Rows represent the 8 different alternative transcript event types diagrammed. Mapped reads supporting expression of upper isoform, lower isoform or both isoforms are shown in blue, red, and gray, respectively. Columns 1-4 show numbers of events of each type: (1) supported by cDNA and/or EST data; (2) with ≥ 1 isoform supported by mRNA-SEQ reads; (3) with both isoforms supported by reads; and (4) events detected as tissue-regulated (Fisher's exact test) at an FDR of 5%. Columns 5-6 show: (5) the observed percentage of events with both isoforms detected that were observed to be tissue-regulated; and (6) the estimated true percentage of tissue-regulated isoforms after correction for power to detect tissue bias and for the FDR (Fig. S3). For some event types, ‘common reads’ (gray bars) were used in lieu of (for tandem 3′ UTR events) or in addition to ‘exclusion’ reads for detection of changes in isoform levels between tissues.
For each of these event types, reads deriving from specific regions can support the expression of one alternative isoform or the other (Fig. 2). The “inclusion ratio”, defined as the ratio of the number of “inclusion” (blue) reads to inclusion plus “exclusion” (red) reads, can be used to detect changes in the proportions of the corresponding mRNA isoforms. The fraction of mRNAs that contain an exon – the “Percent Spliced In” (PSI or Ψ) value – can be estimated as the ratio of the density of inclusion reads (i.e. reads per position in regions supporting the inclusion isoform) to the sum of the densities of inclusion and exclusion reads.
To assess tissue-regulated AS, a comprehensive set of ∼105K events of these 8 types was derived based on available human cDNA and EST data. Reads supporting both alternative isoforms were observed for more than a third of these events (Fig. 2), and the extent of tissue-specific regulation of these events was assessed by comparison of the inclusion ratio in each tissue relative to the other tissues, requiring a minimum 10% change in inclusion ratio (Fig. S4). Naturally, transcripts or isoforms identified as differentially expressed between tissues will reflect the combined effects of cell type specific differences in transcript levels, variation in the relative abundances of cell types between tissues, and variations between the individuals from whom the tissues derived.
Strikingly, a high frequency of tissue-specific regulation was observed for each of the 8 event types, including over 60% of the analyzed SE, A5SS, A3SS and tandem 3′ UTR events (Fig. 2, Table S4). In all, a set of over 22,000 tissue-specific alternative transcript events were identified, far exceeding previous sets of tissue-specific AS events that have typically numbered in the hundreds to low thousands6-9,18,19. Tissue-regulated SE and MXE events are listed in Tables S5 and S6, respectively. Binning events by expression level commonly yielded sigmoid curves for the fraction of tissue-regulated events of each type, enabling estimation of the true frequency of tissue-regulation for each event type (Fig. S5, S6). These estimates, ranging from 52% to 80% (Fig. 2), indicated that the majority of AS events are regulated between tissues, providing an important element of support for the hypothesis that AS is a major contributor to the evolution of phenotypic complexity in mammals.
Differences in alternative mRNA isoform expression between individuals
To assess the extent of AS isoform variation between individuals in comparison to tissue-regulated AS, the correlations between the vectors of inclusion ratios for all expressed SEs between pairs of samples were determined (Fig. 3), and similarly for other event types (not shown). In this analysis, strong clustering of the 6 cerebellar cortex samples was observed, with generally higher correlations among these samples than between pairs representing distinct tissues. Strong clustering of the 5 cell lines was also observed. This likely results from a combination of factors, including the common mammary epithelial origin of the cell lines studied, similar adaptations to culture conditions, and the high diversity of the tissues chosen.
Figure 3. The extent of individual-specific differences in alternative isoform expression.
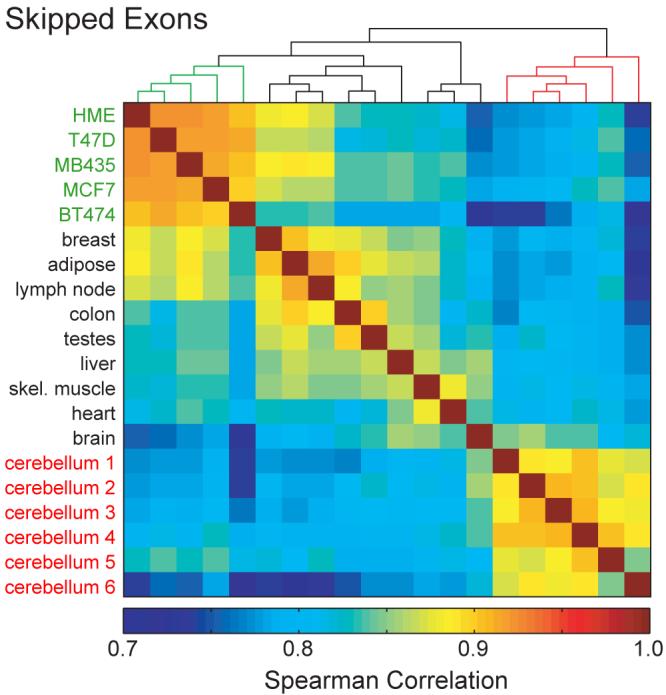
Spearman correlations of Ψ values for SEs in human tissues and cell lines. Correlations were computed separately for each pair of tissues and cell lines, and clustered according to similarity using average linkage hierarchical clustering.
The extent of variation in alternative isoform expression between individuals was also addressed by determining the number of differentially expressed exons among the 6 cerebellar cortex samples. Using the same approach as in Fig. 2, between ∼10% and 30% of alternative transcript events exhibited individual-specific variation, depending on the event type (Fig. S7), providing updated estimates of the scope of mRNA isoform variation between individuals16. These numbers are higher than estimates based on microarray analyses20, but are in general agreement with an integrated analysis of multiple data types which estimated that ∼21% of AS genes are affected by polymorphisms that alter the relative abundances of alternative isoforms17. However, these frequencies are still well below the 47-74% of events that exhibited variation between the 10 tissues (Fig. 2), and ∼2- to 3-fold less than the frequencies observed in comparisons between subsets of 6 tissues (Fig. S7), indicating that inter-individual variation is fairly common but still substantially less frequent than variation between tissues. Thus, most of the differences observed between tissue samples are likely to represent tissue-specific rather than individual-specific variation.
Distinctive properties of switch-like alternatively spliced exons
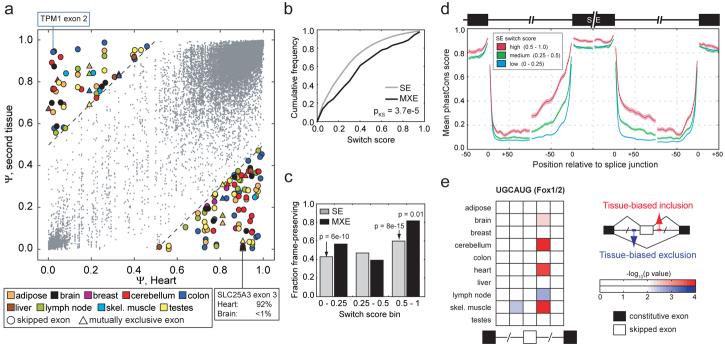
The quantitative nature of the mRNA-SEQ approach allowed assessment of both subtle and switch-like AS events. By comparing inclusion levels of SEs between tissues, a class of “switch-like” exons was observed that exhibited dramatically different inclusion levels between different tissues (shown for heart versus nine other tissues in Fig. 4a). The examples shown in color in Fig. 4a (e.g., TPM1 exon 2, with Ψ of 2% in heart and 95% in skeletal muscle, and the SLC25A3 MXE pair shown in Fig. 1a) underscore the flexibility of the splicing regulatory machinery, with a sizeable number of exons being recognized predominantly as exons in one tissue and predominantly as introns in another tissue, even for developmentally-related pairs of tissues such as heart and skeletal muscle.
Figure 4. Conservation and function of switch-like AS exons.
a, Scatter plot showing Ψ values of SEs and MXEs for which switch score was determined based on comparison of heart (x-axis) and a second tissue (y-axis). Exons with switch score > 0.5 are shown as filled symbols, others as small gray dots. b, Cumulative distribution functions of switch scores for SEs and MXE pairs (P-value based on Kolmogorov-Smirnov test). c, Reading frame preservation of SEs and MXEs grouped by switch score. SEs with lengths divisible by 3 and MXE pairs with lengths differing by 0 or a multiple of 3 were considered to preserve reading frame. P-values based on Fisher's exact test. d, Conservation in SEs and flanking intron regions grouped by SE switch score. The mean per-position phastCons score (from alignment of 4 mammalian genomes) and SEM are shown. e, Enrichment of UGCAUG motifs near tissue-regulated SEs. Colored squares represent −log10(P-value) for the enrichment of UGCAUG counts relative to cohorts of control 6mers in regions surrounding SEs with significantly increased (red) or decreased (blue) inclusion in each tissue with respect to other tissues.
To characterize functional features of such switch-like exons, SEs and MXEs were divided into groups depending on their ‘switch score’, defined as the maximum pairwise Ψ difference between tissues. Switch scores for pairs of MXEs were shifted toward higher values relative to SEs (P = 3.7×10−5, Kolmogorov-Smirnov test; Fig. 4b), suggesting that MXEs are more often involved in regulating highly tissue-specific functions. Preservation of reading frame in both isoforms was observed more commonly for exons with higher switch scores, both for SEs, consistent with ref.19, and to an even greater extent for MXEs (Fig. 4c). Thus, switch-like regulation appears to be used preferentially to express distinct ‘full-length’ protein isoforms in different tissues rather than as a means to switch off genes through production of truncated proteins or of messages subject to nonsense-mediated mRNA decay21. Indeed, genes containing SEs with high switch scores were enriched for Gene Ontology functional categories including ‘developmental processes’, ‘cell communication’, ‘transcriptional regulator activity’, and ‘regulation of metabolism’ that are likely to contribute to fundamental differences in the biology of different human tissues (Table S7).
Notably, those SEs with switch scores exceeding 0.5 showed higher sequence conservation in the regulated exon itself19 and in portions of the flanking introns than exons with lower switch scores (Fig. 4d). This observation suggested that such exons are of unusual biological importance and that switch-like regulation between tissues requires the presence of additional splicing regulatory sequence information.
Inference of a Fox-1/-2 tissue- and location-specific regulatory activity map
Among the best-characterized tissue-specific splicing factors are the Fox-1/Fox-2 proteins, which bind RNA cis-elements that contain UGCAUG hexanucleotides (6mers) or closely related sequences22-24. Analysis of UGCAUG frequencies revealed substantial enrichment in the intron immediately downstream of exons with increased inclusion in heart, skeletal muscle, brain, and cerebellar cortex (Fig. 4e), tissues where Fox proteins are highly expressed, suggesting common splicing activation activity in this location22-24. Enrichment of UGCAUG 6mers was also noted upstream of exons that had reduced inclusion in skeletal muscle, suggesting possible repressive activity in this context. This example illustrates the power of these expanded tissue-specific exon sets for inference of “tissue RNA maps”, summarizing both the location-dependent activity and tissue specificity of splicing regulatory elements.
Applying a similar approach to analyze enrichment of all 6mers in regions adjacent to tissue-specific exons identified 362 motif/tissue enrichment patterns (at an estimated 17% FDR), representing 6mers that exhibited significant enrichment adjacent to exons with increased or decreased inclusion in specific cell lines or tissues (Table S9). Enrichment of UGCAUG downstream of exons with high inclusion in skeletal muscle appears as the 3rd most significant motif/tissue pair, after enrichment of UCUCUC and CUCUCU (resembling the binding motifs of PTB and nPTB25) upstream of exons with increased inclusion in cerebellar cortex. The remaining motif/tissue pairs included a variety of known regulatory elements, including ACUAAC (see below), as well as putative novel regulatory motifs.
Coordination between splicing and cleavage/polyadenylation
Tandem 3′ UTR events exhibited an even higher frequency of tissue-regulated expression than SEs or other AS events studied (Fig. 2), yet little is known about how tissue regulation of tandem UTRs is accomplished, e.g., whether through APA or through differential stability of alternative UTR isoforms. Grouping tandem 3′ UTRs by switch score, the most switch-like events exhibited increased sequence conservation relative to those with lower switch scores in the vicinity of and upstream of the proximal (5′) polyadenylation signal (PAS), and also upstream of the distal (3′) PAS (Fig. 5a). While cis-regulatory elements contributing to differential stability should be located predominantly in the region unique to the long UTR isoform, APA could be regulated by elements located near either or both PAS. The observation of increased conservation around and upstream of the proximal PAS in switch-like tandem UTRs therefore supports a primary role for regulation at the level of APA.
Figure 5. Evidence for coordination between splicing and polyadenylation.
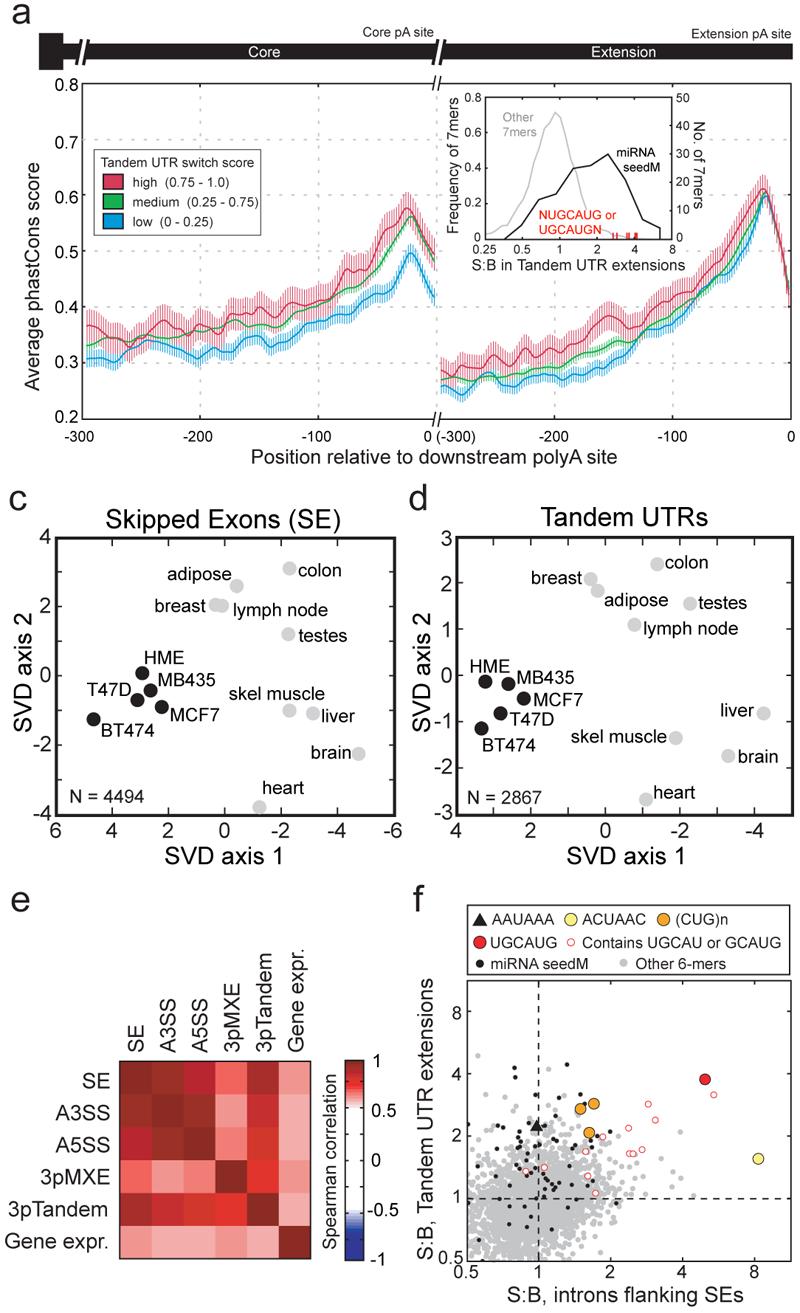
a, Mean and SEM of per-position phastCons score in the region 300 bp upstream of proximal and distal cleavage sites for tandem 3′ UTRs grouped by switch score. inset, Increased conservation of Fox-1/-2 motifs in tandem 3′ UTR extension regions. All non-CpG-containing 7mers (dotted line), miRNA seed matches (black), and 7mers containing UGCAUG (red) are shown. b, SVD analysis of SE inclusion levels across tissues and cell lines for SEs meeting minimum read coverage criteria in each of the 14 samples. Projections are shown in the dimensions corresponding to the two leading eigenvalues, which accounted for 25% of the variance. c, SVD analysis of tandem UTR inclusion levels (as in panel c). d, SVD analysis was conducted for the 14 samples based on Ψ values for the 5 indicated alternative transcript event types or based on gene expression values. Spearman correlations between corresponding pairwise distances in projections of the sort shown in panels b and c are shown. e, S:B ratios of non-CpG-containing 6mers in introns flanking SEs (x-axis) and in extended 3′ UTR regions (y-axis). The canonical PAS 6mer AAUAAA (black triangle), 6mers corresponding to seed matches to conserved mammalian miRNAs (black dots), and 6mers corresponding to binding motifs for the indicated splicing or 3′ UTR-binding factors (color) are shown.
In assessing the spectrum of cis-elements that may drive tissue regulation of tandem 3′ UTRs, a set of 7mers was identified that exhibited high conservation in the extension region of tandem 3′ UTRs (Fig. 5a, inset), with signal:background (S:B) ratios in 4 mammals26 exceeding 2:1. As expected, this set included the extended (7-base) seed matches to a number of conserved mammalian miRNAs26-28. Surprisingly, it also included all 8 of the 7mers that contain the Fox-1/-2 consensus binding motif, UGCAUG: all such 7mers had S:B ratios above 2.5:1, exceeding the S:B observed for seed matches to important miRNAs such as miR-7 and miR-181 (inset, Fig. 5a). Strong conservation of UGCAUG motifs in this location (>1 kbp on average from the nearest splice site) would not be expected on the basis of the canonical splicing regulatory activity of Fox-1/Fox-2 proteins. Instead, the high conservation observed in extended 3′ UTR regions suggests that these factors (or others with identical RNA binding specificity) play additional 3′ UTR-related roles, e.g., in APA, or in mRNA localization and/or translation.
To further investigate possible connections between tissue-specific regulation of AS and APA, global patterns of tissue-specific alternative isoform expression were compared. Applying singular value decomposition (SVD) (Supplemental Methods) to the vectors of inclusion ratios across samples for each AS and APA event type separately, a strong and consistent separation of the breast cell lines (4 cancer-derived and one immortalized cell line) from all tissue samples was observed (Fig. 5c,d). This separation implied the existence of a systematic difference in RNA processing regulation between cell lines and tissues that held for all types of AS events studied. For most AS and APA events, SVD analysis yielded similar groupings of tissues, e.g., with heart, skeletal muscle, brain and liver consistently clustered (Fig. S8). Consistent with this observation, pairwise distances between SVD projections for different types of AS events, e.g., SE, A5SS, and A3SS were all highly correlated (Fig. 5e), suggesting similarities in the regulatory control of these types of events1,2,13. More surprisingly, distances between SVD projections for tandem 3′ UTR events also correlated highly with those for events controlled purely at the level of splicing such as SEs (Fig. 5e). This observation raised the possibility that splicing and polyadenylation may be coordinately regulated across human tissues.
To explore possible regulatory connections between splicing and polyadenylation regulation (e.g., refs29-32), the conservation of 6mers adjacent to conserved AS and APA events was compared. While canonical 3′ UTR regulatory motifs such as the consensus PAS 6mer AAUAAA and various miRNA seed matches exhibited high S:B ratios, often 1.5:1 or higher, in extended 3′ UTR regions, these motifs generally had S:B ratios close to 1:1 in AS introns. However, a distinct subset of motifs with high S:B ratios in both UTRs and introns was also observed, several of which corresponded to well-known splicing-related motifs (Fig. 5h, Table S8). This set included not only the Fox-1/-2 motif UGCAUG and variations, consistent with the 7mer analysis of Fig. 5b, but also permutations of (CUG)n, which represent putative substrates of the CELF and MBNL families of muscle- and brain-specific splicing factors33. The highly significant S:B in both 3′ UTRs and introns suggested that these well known splicing-related motifs also commonly play 3′ UTR-related roles – e.g., control of APA or of mRNA stability, localization, or translation – as recently demonstrated for the Nova family of splicing factors34.
The 6mer ACUAAC, an excellent match to the consensus binding motifs of STAR family RNA binding factors, in particular Quaking/QKI35, was also notable. Not only did ACUAAC have significant S:B in 3′ UTRs, as expected from QKI's known role in control of mRNA stability36, but it also showed extremely high S:B in introns, exceeding 7:1. This extreme conservation suggested a common and important function in splicing regulation, a role which has been suggested but not yet directly demonstrated9,37. Motif enrichment analyses also suggested a possible role in brain-specific APA regulation (Fig. S9).
Discussion
We conclude that the coordination between tissue-specific AS and APA events implied by the correlated patterns of tissue bias observed in Fig. 5 may be mediated at least in part by tissue-specific RNA binding factors that play roles in regulation of both of these RNA processing steps. Such factors may include both canonical tissue-specific splicing factors, e.g., of the Fox-1/-2 and CELF families, moonlighting in 3′ UTR-related roles, and also canonical UTR-binding factors such as Quaking/QKI. Such functional duality has the potential to enable tightly coordinated regulation of polyadenylation and splicing, ensuring that the appropriate UTR regulatory sequences are expressed in conjunction with the coding regions for the relevant tissue-specific protein isoforms.
Methods Summary
Tissues and Cell Lines
Tissue samples from individual unrelated anonymous donors (Table S1) were obtained from Ambion for the following tissue types: adipose, whole brain, breast, colon, heart, liver, lymph node, skeletal muscle, testes. Cerebellar cortex samples were obtained from 6 anonymous unrelated donors were obtained according to NIH guidelines for confidentiality and privacy using protocols described previously38. HME is a human mammary epithelial cell line immortalized with hTERT39. The other cell lines are all breast cancer cell lines derived from invasive ductal carcinomas (ATCC). MCF-7, BT474 and T47D are estrogen receptor and progesterone receptor positive; MDA-MD435 is negative for both.
Library Preparation for Illumina Sequencing
Poly-T capture beads were used to isolate mRNA from 10 ug of total RNA. First strand cDNA was generated using random hexamer-primed reverse transcription, and subsequently used to generate second strand cDNA using RNase H and DNA polymerase. Sequencing adapters were ligated using the Illumina Genomic DNA sample prep kit. Fragments ∼200 bp long were isolated by gel electrophoresis, amplified by 16 cycles of PCR, and sequenced on the Illumina Genome Analyzer, as described40.
Computational Analyses of mRNA-SEQ Read Data
Computational and statistical methods used in analysis of the read data are described in Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Erik Anderson, Doug Black, Brad Friedman, and members of the Burge lab for helpful comments on the manuscript, Noah Spies for analyses, Joann Mudge, Gregory D. May, Neil A. Miller, Eric Vermaas, Tzvetana Kerelska, Juying Yan, and Victor Quijano for assistance in generating the mRNA-SEQ data, and Rosalinda C. Roberts and Nora Perrone-Bizzozero for supplying cerebellar cortex RNA samples. This research was supported by an NIH training grant (E.W.), and by grants from the Knut & Alice Wallenberg Foundation and the Swedish Foundation for Strategic Research (R.S.) and from the NIH (C.B.B.).
Footnotes
Supplementary Information includes: Supplementary Methods, 9 Supplementary Figures, and 6 Supplementary Tables.
The reported sequence read data have been deposited to the Short Read Archive section of GEO at NCBI under accession numbers GSE12946 and SRA002355.1.
The authors have no competing financial interests except that S.L., I.K., L.Z. and G.P.S. are employees and shareholders of Illumina, Inc.
References
- 1.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–98. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 3.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Johnson JM, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–4. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 5.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 6.Xu Q, Modrek B, Lee C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 2002;30:3754–66. doi: 10.1093/nar/gkf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta S, Zink D, Korn B, Vingron M, Haas SA. Strengths and weaknesses of EST-based prediction of tissue-specific alternative splicing. BMC Genomics. 2004;5:72. doi: 10.1186/1471-2164-5-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugnet CW, et al. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput Biol. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008 doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 11.Sultan M, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–60. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 12.Ule J, et al. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–6. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Xiao X, Van Nostrand E, Burge CB. General and specific functions of exonic splicing silencers in splicing control. Mol Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi L, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–61. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayr JA, et al. Mitochondrial phosphate-carrier deficiency: a novel disorder of oxidative phosphorylation. Am J Hum Genet. 2007;80:478–84. doi: 10.1086/511788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graveley BR. The haplo-spliceo-transcriptome: common variations in alternative splicing in the human population. Trends Genet. 2008;24:5–7. doi: 10.1016/j.tig.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nembaware V, Wolfe KH, Bettoni F, Kelso J, Seoighe C. Allele-specific transcript isoforms in human. FEBS Lett. 2004;577:233–8. doi: 10.1016/j.febslet.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 18.Pan Q, et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol Cell. 2004;16:929–41. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Xing Y, Lee CJ. Protein modularity of alternatively spliced exons is associated with tissue-specific regulation of alternative splicing. PLoS Genet. 2005;1:e34. doi: 10.1371/journal.pgen.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwan T, et al. Genome-wide analysis of transcript isoform variation in humans. Nat Genet. 2008;40:225–31. doi: 10.1038/ng.2007.57. [DOI] [PubMed] [Google Scholar]
- 21.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A. 2003;100:189–92. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–16. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Auweter SD, et al. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. Embo J. 2006;25:163–73. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakahata S, Kawamoto S. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 2005;33:2078–89. doi: 10.1093/nar/gki338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oberstrass FC, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–7. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 26.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 27.Xie X, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–45. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majoros WH, Ohler U. Spatial preferences of microRNA targets in 3′ untranslated regions. BMC Genomics. 2007;8:152. doi: 10.1186/1471-2164-8-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 30.McCracken S, Lambermon M, Blencowe BJ. SRm160 splicing coactivator promotes transcript 3′-end cleavage. Mol Cell Biol. 2002;22:148–60. doi: 10.1128/MCB.22.1.148-160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castelo-Branco P, et al. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol Cell Biol. 2004;24:4174–83. doi: 10.1128/MCB.24.10.4174-4183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L, Lee JE, Wilusz J, Wilusz CJ. The RNA-binding protein CUGBP1 regulates stability of tumor necrosis factor mRNA in muscle cells: implications for myotonic dystrophy. J Biol Chem. 2008;283:22457–63. doi: 10.1074/jbc.M802803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ladd AN, Cooper TA. Finding signals that regulate alternative splicing in the post-genomic era. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-11-reviews0008. reviews0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Licatalosi D, et al. Mechanisms of alternative mRNA processing in the brain revealed by HITS-CLIP. Nature. 2008 in press. [Google Scholar]
- 35.Galarneau A, Richard S. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat Struct Mol Biol. 2005;12:691–8. doi: 10.1038/nsmb963. [DOI] [PubMed] [Google Scholar]
- 36.Kim HH, Gorospe M. GU-rich RNA: expanding CUGBP1 function, broadening mRNA turnover. Mol Cell. 2008;29:151–2. doi: 10.1016/j.molcel.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu JI, Reed RB, Grabowski PJ, Artzt K. Function of quaking in myelination: regulation of alternative splicing. Proc Natl Acad Sci U S A. 2002;99:4233–8. doi: 10.1073/pnas.072090399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paz RD, et al. Increased expression of activity-dependent genes in cerebellar glutamatergic neurons of patients with schizophrenia. Am J Psychiatry. 2006;163:1829–31. doi: 10.1176/ajp.2006.163.10.1829. [DOI] [PubMed] [Google Scholar]
- 39.Elenbaas B, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schroth GP, Luo S, Khrebtukova I. Methods Mol. Biol. 2008 in press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.