Abstract
Purpose
To evaluate hypotheses which explain phenotypic variability in sex determining region Y positive 46,XX males. We investigate two 46,XX males without gonadal ambiguities.
Methods
Cytogenetic and molecular analyses were used to identify the presence of Y chromosome material and to map the translocation breakpoint. Finally, the pattern of X chromosome inactivation was studied using the methylation assay at the androgen receptor locus.
Results
The presence of Y chromosome material, including the sex determining region Y gene, was demonstrated in both men. However, the amount of translocated Y chromosome material differed between the patients. Different X chromosome inactivation patterns were found in the patients; random in one patient and non-random in the other.
Conclusions
We found a lack of association between phenotype and X chromosome inactivation pattern. Our cytogenetic and molecular analyses show support for the position effect hypothesis explaining the phenotypic variability in XX males.
Keywords: Azoospermia, Position effect, X chromosome inactivation, XX male, X/Y translocation
Introduction
Testicular differentiation in humans is determined by the sex determining region Y (SRY) gene located on the Y chromosome. The SRY gene is first expressed in the male genital ridge, with continued expression into adulthood in Sertoli cells and germ cells [1, 2]. The sex chromosomes pair during paternal meiosis, where meiotic recombination facilitates the exchange of genetic material between the pseudoautosomal regions [3]. However, the erroneous exchange between homologous but normally non-recombining regions can also occur resulting in the transfer of differing amounts of Y chromosome material, including the SRY gene, most often to the short arm of the X chromosome, leading to a 46,XX male phenotype [4, 5].
XX maleness is a rare genetic condition affecting one in 24,000 newborn males [6]. XX maleness is a genetically heterogeneous condition. The SRY gene is present in the majority of patients [7], although several studies have shown that approximately 10% of patients lack Y material, including the SRY gene [8, 9]. While the majority of XX males are carriers of an X/Y translocation, XX maleness may also arise as a consequence of a genetic mutation permitting testicular differentiation in the absence of SRY, or result from undetected mosaicism for a cell line carrying the Y chromosome [4, 10-12]. Previously, we found a 46,XX male infant without the presence of the SRY gene, derived from intracytoplasmic sperm injection (ICSI) [13]. Whether ICSI had enhanced the production of this 46,XX male is unclear.
The phenotype of the XX male varies greatly. Some XX males may have normal internal and external male gonads [14], whereas others may have small testes, abnormal secondary sexual characteristics [6], and hypospadias [15–17]. Most men are diagnosed in adulthood due to infertility caused by azoospermia [6]. A small proportion of SRY positive XX individuals are true hermaphrodites [18, 19]. It is not yet clear how one genotype can give rise to different phenotypes. It has been proposed that the variation in phenotype observed in SRY positive XX individuals is primarily dependent on X chromosome inactivation (XCI) pattern and the amount of Y material that has been translocated to the X chromosome, or a combination of both [18, 20, 21]. Therefore, a more masculine phenotype is expected when the Y bearing X chromosome preferentially escapes inactivation and when more Y chromosome material is present on the X, presumably protecting the SRY gene from the spread of inactivation [18, 20, 21]. However, more recently, another mechanism, known as the position effect, has been proposed to delineate the observed phenotypic differences [ 22]. Under this hypothesis, the phenotypic differences are dependent on the proximity of the breakpoint to the SRY gene as well as the presence or absence of cryptic rearrangements affecting the expression of the SRY gene [22].
Here we report on two 46,XX males with a different amount of Y chromosome material on the short arm of one of the X chromosomes. The correlation between phenotype, the location of the breakpoint on the short arm of the Y chromosome and the pattern of X chromosome inactivation is discussed. We evaluate the proposed hypotheses that explain the phenotype-genotype correlation in SRY positive XX patients.
Materials and methods
Patient information
Patient 1
A 24-year-old male with a karyotype of 46,X,add(X)(p22.3) was diagnosed by cytogenetic analysis of G-banded peripheral lymphocytes. The presence of SRY was confirmed by fluorescence in situ hybridization (FISH) analysis making the final karyotype 46,X,add (X)(p22.3).ish der (X)t(X;Y)(p22.3;p11.32)(SRY). The patient underwent cytogenetic analysis because of primary infertility and azoospermia. Physical analysis revealed gynecomastia and small testes, each less than 5cc. He had a seminal volume of 0.5 ml with elevated FSH (55.4 U/L) and LH (28.4 U/L), and normal testosterone level (39.7 pmol/L). The patient is a carrier of the I148T cystic fibrosis mutation.
The patient’s family, his father, mother, brother and sister, also underwent cytogenetic analysis. The fertility status of the younger brother and sister are unknown.
Patient 2
A 24-year-old male underwent cytogenetic analysis for delayed puberty. Physical analysis of the patient revealed gynecomastia, small testes and a small penis. The patient had scanty auxiliary and pubic hair, which showed female distribution. He had no facial hair growth. Testicular ultrasound showed presence of both testes in the scrotal sac and an absence of female organs. The patient’s FSH and LH were elevated, while testosterone was normal. There was no sperm in the semen. The patient’s family history was unknown. Family members were unavailable for study.
This study was conducted with the approval of the Clinical Ethics Board at the University of British Columbia and included informed consent from all participants of the study.
Cytogenetic and molecular analysis
Cytogenetic analysis was carried out on stimulated peripheral lymphocytes by standard methods. Eleven G-banded metaphase spreads were analyzed for each individual. FISH analysis was performed on metaphase spreads obtained from patient 2. The following probes were used: CEPX (green), CEPY (orange), SRY gene (Yp11.1) (orange) (Vysis Inc., Downers Grove, IL, USA). DNA was extracted from peripheral lymphocytes from the patients and patient 1’s family members using standard salt extraction methods. As additional material was observed from cytogenetic analysis on one of the X chromosomes of patient 1, comparative genomic hybridization (CGH) was performed to characterize and confirm the origin of the rearrangement. CGH was performed as previously described [13].
Mapping and analysis of Y chromosome sequences
The Y chromosome breakpoint was mapped by the polymerase chain reaction (PCR) by amplifying sequence tagged sites (STS) spanning the Yp arm: sy14 (SRY), sy19 (ZFY), sy57, sy65, sy69 (PRKY), sy72 and sy78. Additional markers were tested in patient 2: sy24, sy35, sy43, sy48, sy52, sy173 and sy54. Amplification conditions and analysis were performed as previously described [23]. All primer sequences have been previously published and are available online from GenBank.
X chromosome inactivation
XCI pattern was measured for the patients using the methylation assay at the androgen receptor (AR) locus as previously described [13, 24]; however, the fluorescent primer was labeled with 6FAM (Sigma–Genosys, Oakville, ON). Genomic DNA was digested with methylation sensitive enzymes, HpaII (2.5U) (New England Biolabs, Ipswich, MA) with a secondary cutter RsaI (1U) (New England Biolabs, Ipswich, MA) in a total reaction volume of 10 µl. An undigested control was prepared using similar reagents except for the HpaII enzyme. The digests were incubated at 37°C overnight prior to PCR amplifications with a set of fluorescently labeled primers. The PCR products were analyzed on a 3130 Genetic Analyzer (Applied Biosystems Canada, Streetville, ON). Peaks were analyzed using the Peak Scanner Software v1.0 (Applied Biosystems Canada, Streetville, ON) and the degree of skewing was calculated according to the peak areas.
Results
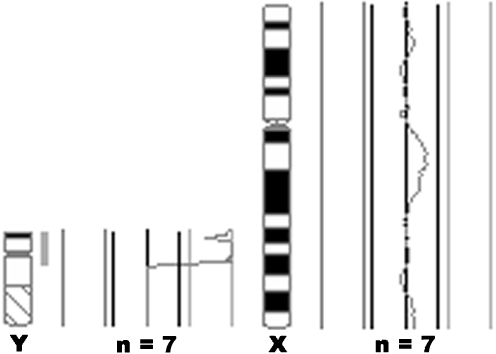
Additional material can be seen attached to the short arm of one of the X chromosomes at band p22.3 (Fig. 1) in patient 1. Patient 1’s family members all had a normal karyotype. CGH analysis of the patient’s DNA showed a gain of the Y chromosome p arm (Fig. 2), confirming the origin of the additional material identified on one of the X chromosomes by cytogenetic analysis.
Fig. 1.
G-banded sex chromosomes of patient 1. Additional material is visible attached to the short arm of one of the X chromosomes [add(X)(p22.3)], as indicated by the arrow. An X chromosome idiogram is shown on the left
Fig. 2.
CGH analysis of patient 1’s DNA. The average fluorescence profiles obtained from the analysis of seven cells are shown for the Y and X chromosomes where a female reference DNA was used. The CGH profile for the Y chromosome shows a gain of the Y chromosome short arm, as indicated by the shift to the right. The presence of two X chromosomes in the male patient is also shown, indicated by a 1:1 fluorescence profile
Cytogenetic analysis of patient 2 revealed a 46,XX karyotype. FISH analysis showed the presence of two copies of CEPX, absence of CEPY, and the presence of the SRY gene on one of the X chromosomes (result not shown). The karyotype was established to be 46,XX.ish der (X)t(X;Y)(p22.3;p11.2) (SRY). It is probable that both patients’ abnormal X chromosomes had originated from a de novo event, most likely occurring during meiosis I in spermatogenesis of their fathers.
PCR analysis of the Y chromosome confirmed the presence of the SRY gene (sy14) in both patients. The translocation breakpoint of the Y chromosome was mapped to the third interval of Yp in both patients. The breakpoint in patient 1 was more proximal to the centromere, located between markers sy57 and sy69; in patient 2 the breakpoint was located between markers sy52 and sy173 (Table 1). Due to a lack of Y-specific STSs between intervals 2B and 3B, more accurate breakpoint analysis in patient 2 could not be performed. In both cases a large portion of the Y chromosome p arm was translocated to the X chromosome, with the breakpoint localized away from the SRY gene. XCI analysis in the two patients showed two different patterns of inactivation. The XCI pattern in patient 1 was random as the skewing was estimated to be 56%, while the XCI pattern in patient 2 was non-random estimated to be 88%.
Table 1.
Yp breakpoint mapping through molecular analysis of STS sequences
| STS tested | Y interval | Patient 1a | Patient 2a |
|---|---|---|---|
| sy14 (SRY) | 1A1A | + | + |
| sy19 (ZFY) | 1A2 | + | + |
| sy24 | 1E | + | + |
| sy35 | 2A | + | + |
| sy43 | 2B | + | + |
| sy48 | 3A | + | + |
| sy52 | 3B | + | + |
| sy173 | 3C | + | - |
| sy54 | 3C | + | - |
| sy65 | 3C | + | - |
| sy57 | 3C | + | - |
| sy69 (PRKY) | 3G | - | - |
| sy72 | 4A | - | - |
| sy78 (centromere) | 4B | - | - |
Order of STS and the Y interval location are based on Vollrath et al. (1992) [39]
aPresence of STS (+); Absence of STS (-); Inferred presence of STS (+)
Discussion
The amount of the translocated Y chromosome likely determines whether the exchange is visible on cytogenetic analysis, such as in patient 1 reported here (Fig. 1). The additional material on the X chromosome in patient 1 was identified as Y chromosome in origin by CGH analysis (Fig. 2). Although CGH has been successfully used to identify Y chromosome material in XX cases [25], it is unable to identify the breakpoint region. More detailed molecular analysis allowed mapping of the Y chromosome breakpoint to interval 3 in both patients; however, a larger Yp fragment was translocated in patient 1 compared to patient 2 (Table 1). Based on the breakpoint location, the patients were classified as class 3 XX males [26]. Class 3 XX males are most common due to the presence of a recombination hotspot within this region [21, 27].
The phenotype of SRY positive XX males can vary greatly from a classical male phenotype, to a male with genital ambiguity, to a true hermaphrodite [18, 28]. However, factors responsible for the phenotypic variation have not yet been identified. The amount of translocated Y chromosome material and the pattern of X chromosome inactivation (XCI) have been proposed to play a role [21]. An association of skewed XCI against the Y-bearing X chromosome has often been observed in true hermaphrodites and in XX patients with gonadal ambiguity [7, 12, 14, 18, 29]. These patients normally have a small portion of the Y chromosome material translocated to the X, presumably allowing for XCI spreading and inactivating the SRY gene. A male phenotype without genital ambiguities is expected to result from a larger Yp SRY bearing fragment being translocated to the X chromosome, where the length of the Yp fragment may protect the SRY gene from silencing by the spread of XCI [18]. However, there is little evidence for XCI spreading into the Yp chromatin in humans [22], thus opposing the hypothesis of XCI affecting SRY expression. In the literature there is no consensus on the association of 46,XX males with a specific XCI pattern, as both random [12, 29, 30] and non-random [31] XCI patterns have been reported in 46,XX males with a normal male phenotype. A lack of association between XCI pattern and phenotype was also observed in the analysis of the two 46,XX males presented in this report. Both men did not have gonadal ambiguities, and had opposing XCI patterns, random in one (56%) and non-random (88%) in the other. However, we were unable to analyze the XCI pattern in the patients’ gonads as this would have required a testicular biopsy in patients with a compromised testicular environment, such as absence of germ cells and abnormal hormone production. It is possible that tissue specific XCI patterns exist and may differ between blood and testis. At this time we do not know whether the XCI patterns differ between blood and testis in the two patients. The implications of the possible difference on the phenotype are also unclear. In general, individuals with non-random XCI may be more prone to X-linked disorders than those with random XCI [32]. Even though the XCI pattern may not be associated with the XX male phenotype, there may be other implications for the pattern of skewing in XX individuals.
Phenotypic variability in XX males may be explained by the position effect. Normal expression of SRY may be disrupted by the close proximity of the Yp breakpoint and the presence of cryptic rearrangements affecting the expression of the SRY gene, which would result in genital ambiguities and true hermaphrodism [22]. Alternatively, a breakpoint situated either away from the SRY gene or in the absence of cryptic rearrangements would favor a male phenotype [22], as seen in our patients and other published data [20, 22]. The position effect could further be supported in our study by expression data, most importantly looking at the expression of the SRY gene. However, a fresh blood sample for gene expression analysis was not available from either one of the patients we studied.
XX males may come more often to the attention of fertility specialists because of the wider application of infertility treatments. These patients, however, are not only infertile but are sterile. The testicular biopsies performed on 46,XX males have revealed a complete lack of spermatogenic cells with only Leydig and Sertoli cells being present [7, 12, 14, 28]. Spermatogonia are present at birth but disappear in XX male patients before puberty [17, 18]. The lack of germ cells likely explains the presence of small testes often associated with XX maleness.
As part of infertility treatment, it has been recommended for infertile patients to receive genetic testing for cystic fibrosis. It is well accepted that mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene are associated with congenital bilateral aplasia of the vas deferens (CBAVD) and consequently azoospermia. However, the relationship between the occurrence of CFTR mutations in infertile men without CBAVD is not as clear. An increase in the incidence of CFTR mutations has been reported by some studies in patients affected by infertility [33, 34], but not by others [35, 36]. The I148T mutation, present in patient 1, is a rare CFTR mutation, and results in cystic fibrosis when present as a compound heterozygote with the 3199del6 mutation [37]. The association of a heterozygous I148T mutation with male infertility or a mild form of cystic fibrosis is possible but inconclusive and requires further study [37, 38]. Infertility in our patient carrier is due to the absence of the long arm of the Y chromosome and the AZF region genes, known to be crucial for spermatogenesis.
Conclusions
We demonstrated a lack of XCI pattern association with phenotype in two 46,XX males. This analysis shows support for the position effect hypothesis explaining the phenotypic variability in XX males based on the breakpoint proximity to the SRY gene and the absence of cryptic rearrangements. We recognize that the XCI pattern was only tested in the leukocytes of the patients and may be tissue specific. Molecular analysis of the Yp fragment in XX male patients will allow accurate comparison of the published cases to further support or refute the hypothesis.
Acknowledgements
We gratefully acknowledge the patients and the family members involved in this study. Financial support was provided by the Canadian Institute of Health Research (CIHR) (MOP53067 to SM) and The Hospital for Sick Children Foundation (XG 02-086 to SM). CH is a recipient of a graduate studentship award from the Interdisciplinary Women’s Reproductive Health Research (IWRH). AM is a recipient of a graduate studentship award from IWRH and of a postgraduate scholarship from the Natural Sciences and Engineering Research Council of Canada (NSERC).
Footnotes
Capsule
A lack of association between phenotype and X chromosome inactivation pattern was found in two sterile SRY positive XX men without gonadal ambiguities.
References
- 1.Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–4. doi:10.1038/346240a0. [DOI] [PubMed]
- 2.Salas-Cortes L, Jaubert F, Barbaux S, Nessmann C, Bono MR, Fellous M, McElreavey K, Rosemblatt M. The human SRY protein is present in fetal and adult Sertoli cells and germ cells. Int J Dev Biol. 1999;43:135–40. [PubMed]
- 3.Rappold GA. The pseudoautosomal regions of the human sex. Hum Genet. 1993;92:315–24. doi:10.1007/BF01247327. [DOI] [PubMed]
- 4.Ferguson-Smith MA. X-Y chromosomal interchange in the aetiology of true hermaphrodism and of XX Klinefelter’s syndrome. Lancet. 1966;2:475–6. doi:10.1016/S0140-6736(66)92778-4. [DOI] [PubMed]
- 5.Weil D, Wang I, Dietrich A, Poustka A, Weissenbach J, Petit C. Highly homologous loci on the X and Y chromosomes are hot-spots for ectopic recombinations leading to XX maleness. Nat Genet. 1994;7:414–9. doi:10.1038/ng0794-414. [DOI] [PubMed]
- 6.de la Chapelle A. The etiology of maleness in XX men. Hum Genet. 1981;58:105–16. doi:10.1007/BF00284157. [DOI] [PubMed]
- 7.Boucekkine C, Toublanc JE, Abbas N, Chaabouni S, Ouahid S, Semrouni M, Jaubert F, Toublanc M, McElreavey K, Vilain E, Fellous M. Clinical and anatomical spectrum in XX sex reversed patients: relationship to the presence of Y specific DNA sequences. Clin Endocrinol (Oxf). 1994;40:733–42. doi:10.1111/j.1365-2265.1994.tb02506.x. [DOI] [PubMed]
- 8.Palmer MS, Sinclair AH, Berta P, Ellis NA, Goodfellow PN, Abbas NA, Fellous M. Genetic evidence that ZFY is not the testis-determining factor. Nature. 1989;342:937–9. doi:10.1038/342937a0. [DOI] [PubMed]
- 9.Abbas NE, Toublanc JE, Boucekkine C, Toublanc M, Affara NA, Job JC, Fellous M. A possible common mechanism of “Y-negative” human XX males and XX true hermaphrodites. Hum Genet. 1990;84:356–60. doi:10.1007/BF00196234. [DOI] [PubMed]
- 10.Page DC, Brown LG, de la Chapelle A. Exchange of terminal portions of X- and Y-chromosomal short arms in human XX males. Nature. 1987;328:437–40. doi:10.1038/328437a0. [DOI] [PubMed]
- 11.Van der Auwera B, Van Roy N, De Paepe A, Hawkins JR, Liebaers I, Castedo S, Dumon J, Speleman F. Molecular cytogenetic analysis of XX males using Y-specific DNA sequences, including SRY. Hum Genet. 1992;89:23–8. doi:10.1007/BF00207036. [DOI] [PubMed]
- 12.Fechner PY, Marcantonio SM, Jaswaney V, Stetten G, Goodfellow PN, Migeon CJ, Smith KD, Berkovitz GD, Amrhein JA, Bard PA, Lee PA, Reid CR, Tsalikian E, Urban MD. The role of the sex-determining region Y gene in the etiology of 46,XX maleness. J Clin Endocrinol Metab. 1993;76:690–5. doi:10.1210/jc.76.3.690. [DOI] [PubMed]
- 13.Ma S, Tang SS, Yuen BH, Bruyere H, Penaherrera M, Robinson WP. Cytogenetic and molecular study of a premature male infant with 46,XX derived from ICSI: case report. Hum Reprod. 2003;18:2298–301. doi:10.1093/humrep/deg462. [DOI] [PubMed]
- 14.Abdelmoula NB, Portnoi MF, Keskes L, Recan D, Bahloul A, Boudawara T, Saad A, Rebai T. Skewed X-chromosome inactivation pattern in SRY positive XX maleness: a case report and review of literature. Ann Genet. 2003;46:11–8. doi:10.1016/S0003-3995(03)00011-X. [DOI] [PubMed]
- 15.Lopez M, Torres LM, Mendez JP, Cervantes A, Perez-Palacios G, Erickson RP, Alfaro G, Kofman-Alfaro S. Clinical traits and molecular findings in 46,XX males. Clin Genet. 1995;48:29–34. [DOI] [PubMed]
- 16.Castineyra G, Copelli S, Levalle O. 46,XX male: clinical, hormonal/genetic findings. Arch Androl. 2002;48:251–7. [DOI] [PubMed]
- 17.Grigorescu-Sido A, Heinrich U, Grigorescu-Sido P, Jauch A, Hager HD, Vogt PH, Duncea I, Bettendorf M. Three new 46,XX male patients: a clinical, cytogenetic and molecular analysis. J Pediatr Endocrinol Metab. 2005;18:197–203. [DOI] [PubMed]
- 18.Kusz K, Kotecki M, Wojda A, Szarras-Czapnik M, Latos-Bielenska A, Warenik Szymankiewicz A, Ruszczynska-Wolska A, Jaruzelska J. Incomplete mascularization of XX subjects carrying the SRY gene on an inactive X chromosome. J Med Genet. 1999;36:452–6. [PMC free article] [PubMed]
- 19.Quiepo G, Zenteno JC, Pena R, Nieto K, Radillo A, Dorantes LM, Erana L, Lieberman E, Soderlund D, Jimenez AL, Ramon G, Kofman-Alfaro S. Molecular analysis in true hermaphroditism: demonstration of low-level mosaicism for Y-derived sequences in 46,XX cases. Hum Genet. 2002;111:278–83. doi:10.1007/s00439-002-0772-9. [DOI] [PubMed]
- 20.Numabe H, Nagafuchi S, Nakahori Y, Tamura T, Kiuchi H, Namiki M, Kohda N, Yoshimitsu Fukushima Y, Fuse H, Kusano M, Arai T, Matsuzaki Y, Fukutani K, Isurugi K, Kuroki Y, Ikeuchi T, Yoshida M, Minowada S, Nakagome Y. DNA analysis of XX and XY hypospadic males. Hum Genet. 1990;90:211–4. [DOI] [PubMed]
- 21.McElreavey K, Salas-Cortes L. X-Y tranlocations and sex differentiation. Semin Reprod Med. 2001;19:133–9. doi:10.1055/s-2001-15393. [DOI] [PubMed]
- 22.Sharp A, Kusz K, Jaruzelska J, Tapper W, Szarras-Czapnik M, Wolski J, Jacobs P. Variability of sexual phenotype in 46,XX(SRY) patients: the influence of spreading X inactivation versus position effects. J Med Genet. 2005;42:420–7. doi:10.1136/jmg.2004.022053. [DOI] [PMC free article] [PubMed]
- 23.Minor A, Wong EC, Harmer K, Ma S. Molecular and cytogenetic investigation of Y chromosome deletions over three generations facilitated by intracytoplasmic sperm injection. Prenat Diagn. 2007;27:743–7. doi:10.1002/pd.1772. [DOI] [PubMed]
- 24.Beever CL, Penaherrera MS, Langlois S, Robinson WR. X chromosome inactivation patterns in Russell-Silver syndrome patients and their mothers. Am J Med Genet Part A. 2003;123:231–5. doi:10.1002/ajmg.a.20317. [DOI] [PubMed]
- 25.Rigola MA, Carrera M, Ribas I, Egozcue J, Miro R, Fuster C. A comparative genomic hybridization study in a 46,XX male. Fertil Steril. 2002;78:186–8. doi:10.1016/S0015-0282(02)03165-5. [DOI] [PubMed]
- 26.Vergnaud G, Page DC, Simmler MC, Brown L, Rouyer F, Noel B, Botstein D, de la Chapelle A. A deletion map of the human Y chromosome based on DNA hybridization. Am J Hum Genet. 1986;38:109–24. [PMC free article] [PubMed]
- 27.Schiebel K, Winkelmann M, Mertz A, Xu X, Page DC, Weil D, Petit C, Rappold GA. Abnormal XY interchange between a novel isolated protein kinase gene, PRKY, and its homologue, PRKX, accounts for one third of all (Y)XX males and (Y-)XY females. Hum Mol Genet. 1997;6:1985–9. doi:10.1093/hmg/6.11.1985. [DOI] [PubMed]
- 28.Margarit E, Soler A, Carrio A, Oliva R, Costa D. Vendrell, Rosell J, Ballesta F: Molecular, cytogenetic, and clinical characterization of six XX males including one prenatal diagnosis. J Med Genet. 1998;35:727–30. [DOI] [PMC free article] [PubMed]
- 29.Schempp W, Muller G, Scherer G, Bohlander SK, Rommerskirch W, Fraccaro M, Wolf U. Localization of Y chromosome sequences and X chromosomal replication studies in XX males. Hum Genet. 1989;81:144–8. doi:10.1007/BF00293890. [DOI] [PubMed]
- 30.Jakubowski L, Jeziorowska A, Constantinou M, Helszer Z, Baumstark A, Vogel W, Mikiewicz-Sygula D, Kaulzewski B. Xp;Yp translocation inherited from the father in an SRY, RBM, and TSPY positive true hermaphrodite with oligozoospermia. J Med Genet. 2000;37:E28. doi:10.1136/jmg.37.10.e28. [DOI] [PMC free article] [PubMed]
- 31.Vorona E, Zitzmann M, Gromoll J, Schüring AN, Nieschlag E. Clinical, endocrinological, and epigenetic features of the 46,XX male syndrome, compared with 47,XXY Klinefelter patients. J Clin Endocrinol Metab. 2007;92:3458–65. doi:10.1210/jc.2007-0447. [DOI] [PubMed]
- 32.Lyon MF. X-chromosome inactivation and human genetic disease. Acta Paediatr Suppl. 2002;439:107–12. doi:10.1080/080352502762458030. [DOI] [PubMed]
- 33.Schulz S, Jakubiczka S, Kropf S, Nickel I, Muschke P, Kleinstein J. Increased frequency of cystic fibrosis transmembrane conductance regulator gene mutations in infertile males. Fertil Steril. 2006;85:135–8. doi:10.1016/j.fertnstert.2005.07.1282. [DOI] [PubMed]
- 34.van der Ven K, Messer L, van der Ven H, Jeyendran RS, Ober C. Cystic fibrosis screening in healthy men with reduced sperm quality. Hum Reprod. 1996;11:513–7. [DOI] [PubMed]
- 35.Ravnik-Glavac M, Svetina N, Zorn B, Peterlin B, Glavac D. Involvement of CFTR gene alterations in obstructive and nonobstructive infertility in men. Genet Test. 2001;5:243–7. doi:10.1089/10906570152742308. [DOI] [PubMed]
- 36.Pallares-Ruiz N, Carles S, Des Georges M, Guittard C, Arnal F, Humeau C, Claustres M. Complete mutational screening of the cystic fibrosis transmembrane conductance regulator gene: cystic fibrosis mutations are not involved in healthy men with reduced sperm quality. Hum Reprod. 1999;14:3035–40. doi:10.1093/humrep/14.12.3035. [DOI] [PubMed]
- 37.Monaghan KG, Highsmith WE, Amos J, Pratt VM, Roa B, Friez M, Pike-Buchanan LL, Buyse IM, Redman JB, Strom CM, Young AL, Sun W. Genotype-Phenotype correlation and frequency of the 3199del6 cystic fibrosis mutation among I148T carriers: Results from a collaborative study. Genet Med. 2004;6:421–5. doi:10.1097/01.GIM.0000139507.20179.3A. [DOI] [PubMed]
- 38.Karpman E, Williams DH 4th, Wilberforce S, Lipshultz LI. Compound genetic abnormalities in patients with cystic fibrosis transmembrane regulator gene mutation. Fertil Steril 2007;87:1468.e5–e8. [DOI] [PubMed]
- 39.Vollrath D, Foote S, Hilton A, Brown LG, Beer-Romero P, Bogan JS, Page DC. The human Y chromosome: a 43-interval map based on naturally occurring deletions. Science. 1992;258:52–9. doi:10.1126/science.1439769. [DOI] [PubMed]