Summary
Efficient enzyme catalysis depends on exquisite details of structure beyond those resolvable in typical medium- and high-resolution crystallographic analyses. Here we report synchrotron-based cryocrystallographic studies of natural substrate complexes of the flavoenzyme human glutathione reductase (GR) at nominal resolutions between 1.1 and 0.95 Å that reveal new aspects of its mechanism. Compression in the active site causes overlapping van der Waals radii and distortion in the nicotinamide ring of the NADPH substrate, which enhances catalysis via stereoelectronic effects. The bound NADPH and redox-active disulfide are positioned optimally on opposite sides of the flavin for a 1,2-addition across a flavin double bond. The new structures extend earlier observations to reveal that the redox-active disulfide loop in GR is an extreme case of sequential peptide bonds systematically deviating from planarity, a net deviation of 53° across 5 residues. But this apparent strain is not a factor in catalysis as it is present in both oxidized and reduced structures. Intriguingly, the flavin bond lengths in oxidized GR are intermediate between those expected for oxidized and reduced flavin, but we present evidence that this may not be due to the protein environment but instead to partial synchrotron reduction of the flavin by the synchrotron beam. Finally, of more general relevance, we present evidence that the structures of synchrotron-reduced disulfide bonds cannot generally be used as reliable models for naturally reduced disulfide bonds.
Keywords: enzyme mechanism, radiation damage, steric compression, stereoelectronic control, peptide planarity
Introduction
The short lifetimes of reaction intermediates makes difficult any detailed structural study of steps involved in enzyme catalysis. The use of transition-state analogs and other inhibitors has allowed for much useful insight into enzyme mechanism, but because the inhibitors are not true substrates, the results may not always apply in detail. Only in a few cases has it been possible to examine authentic reaction intermediates in sufficient detail to visualize fine details of catalysis at atomic resolution, commonly defined as 1.2 Å resolution or better1. These cases include horseradish peroxidase2, which used time-resolved crystallography, and D-2-deoxyribose-5-phosphate aldolase3.
Understanding flavoenzymes—enzymes using FAD or FMN in catalysis—is particularly complex because of flavin’s involvement in a wide diversity of chemical reactions and its many possible redox and protonation states, each of which has unique properties and can be stabilized or destabilized by the protein environment4. Proteins apparently modulate flavin reactivity via a variety of mechanisms, including bending the flavin away from planarity and varying the degree of stabilization of negative charges at the flavin N1/O2α locus and at other loci5–10. Nevertheless, despite many investigations of flavoenzymes in their native state and after reconstitution with modified flavins4–11, much remains poorly understood about how proteins modulate flavin reactivity.
The flavoenzyme glutathione reductase (GR) is a dimeric disulfide oxidoreductase that converts oxidized glutathione (GSSG) to two molecules of reduced glutathione (GSH) using an NADPH cofactor and an FAD prosthetic group. Glutathione plays a critical role in maintaining the cell’s reducing environment and battling oxidative stress. Human erythrocyte GR is a homodimer of 52 kD monomers, each with three domains: an NADPH-binding domain, an FAD-binding domain, and a dimerization domain12. The NADPH- and FAD-binding domains meet at the active site, in which both monomers participate.
Much of our understanding of how GR works comes from combining steady-state and presteady-state analyses of catalysis by GR and related disulfide reductases13–18 with a series of 2 Å resolution structures of GR in various redox states and bound to combinations of natural substrates: GRNative, GRGSSG/NADP, GRGSH and GRNADPH19. Figure 1 illustrates the consensus mechanism derived from these studies. First, NADPH binds and transiently reduces the flavin. Tyr197, which swings out of the way so NADPH can bind, is proposed to act as a spring in forcing the nicotinamide into the flavin. The reduced flavin then reduces the Cys58-Cys63 disulfide bond by forming a short-lived covalent intermediate with Cys63, followed by formation of a stable charge-transfer complex between the flavin and the Cys63 thiolate. Calculations have shown that the pairwise overlap of molecular orbitals in the X-ray structure is optimal for hydride transfer between the nicotinamide and the flavin and also for covalent catalysis of electron transfer between the flavin and Cys6320. After formation of the charge-transfer complex, NADP+ dissociates and is replaced by another NADPH. This constitutes the reductive half-reaction leading to the enzyme form known as EH2. The oxidative half-reaction begins with the binding of GSSG. Cys58 in GR, which is activated similarly to serine or cysteine proteases by the His467′-Glu472′ pair (primes denote residues coming from the second subunit of the dimer), attacks CysI of GSSG to form a mixed disulfide between GSI and Cys58. After the freed GSHII leaves, Cys58 and Cys63 re-form a disulfide releasing the second molecule of GSH product.
Figure 1.
Catalytic cycle of glutathione reductase. The native state with no substrate bound is not part of the cycle but merely forms an entrance point. Dotted lines indicate charge-transfer complexes between NADPH, FAD, and the sulfur of Cys63. The substrate and product binding and dissociation may occur with different timing than that shown. The four crystal structures reported here provide information about the catalytic intermediates as follows: 1 is GRNative; 2 are derived from GRNative and GRNADPH; 3 is derived from GRNADPH and GRGSSG/NADP; 4 is GRNADPH; 5 is derived from GRGSSG/NADP and GRNADPH; 6 is derived from GRGSH and GRNADPH; 7 is derived from GRNative and GRNADPH, with GRGSSG/NADP and GRGSH providing an idea for the GSHI binding site. Created in Inkscape.
Despite the extensive enzymatic and structural studies of GR catalysis, uncertainties remain that more detailed pictures of catalysis might resolve. For example, in the native structure determined at 1.54 Å resolution12, marginally reliable deviations in peptide planarity of the active-site disulfide loop were suggested to indicate strain that would favor disulfide reduction. Similarly, very small deviations in covalent flavin geometry were suggested as possible evidence as to how the protein modulates flavin reactivity. But both of these observations were near the limits of coordinate accuracy and need confirmation. Also, the more accurate determination of the geometry of the nicotinamide-flavin approach provided by atomic-resolution analysis will help us to better understand the hydride-transfer step. Finally, atomic-resolution analysis could yield additional insight by providing a direct visualization of the protonation states of the active site Cys and His residues at various stages of catalysis.
The advent of synchrotron sources and cryocrystallography has induced an explosion in atomic-resolution structures: over 220 structures with >45 residues determined at resolutions of ≤1 Å now exist in the Protein Data Bank21 as compared to only five in 1996. Of these proteins, only two are flavoenzymes: cholesterol oxidase22 and pentaerythritol tetranitrate reductase23. Here, we use the same GR crystal form previously studied19 to extend the structural analysis of the GR catalytic cycle to near 1 Å resolution.
Results and Discussion
Structure determination
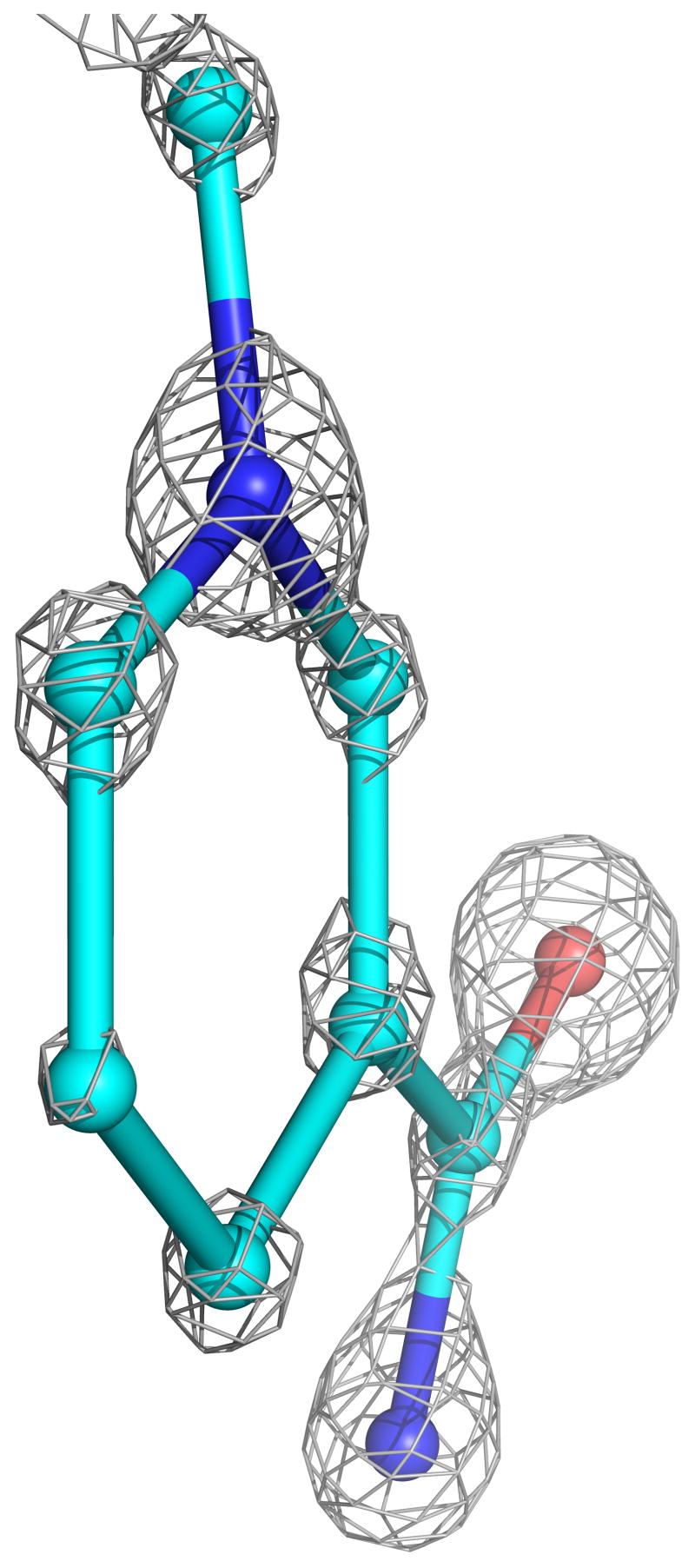
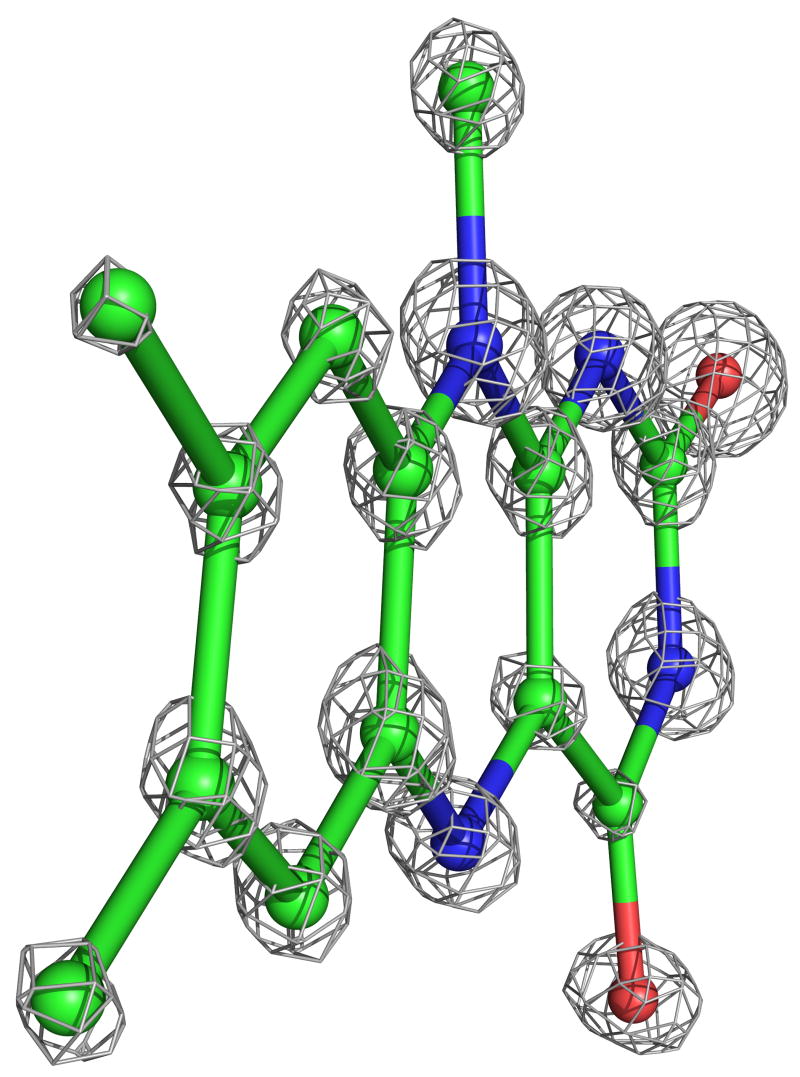
The four complexes of GRNative, GRGSSG/NADP, GRGSH, and GRNADPH previously studied at 2 Å resolution at room temperature were structurally analyzed at cryotemperatures and refined with SHELXL to R-factors near 12% and Rfree near 15% at nominal resolutions of 0.95, 1.1, 1.0 and 1.0 Å, respectively (Table 2). For each model, riding hydrogen atoms were included and individual anisotropic B-factors were refined. The inclusion of each of these led to drops in Rfree of >2%, indicating that their inclusions were justified (see Methods). In addition, multiple conformations were modeled for about 20% of the side chains in all of the atomic-resolution structures and for a few stretches of the backbone (see below). For the highest-resolution, GRNative structure, leaving out the hydrogens resulted in >2σ difference peaks for only about 30% of the peptide backbone NH atoms, so we conclude that this structure does not have sufficient information content to provide reliable evidence for the presence or absence of specific hydrogen atoms. Based on Cruickshank’s DPI error estimator24, the coordinate error for atoms with average B factors is ~0.02 Å. Consistent with this level of coordinate error and the nominal resolution of the analysis, atoms are clearly discernible as discrete peaks in the final electron density map (Figure 2).
Table 2.
Data collection and refinement statisticsa
| GRNative | GRGSSG/NADP | GRGSH | GRNADPH | |
|---|---|---|---|---|
| Cell dimensions (Å) | ||||
| a | 120.4 | 119.4 | 120.0 | 119.7 |
| b | 62.4 | 62.2 | 62.3 | 62.6 |
| c | 84.0 | 83.9 | 84.0 | 84.2 |
| β (°) | 122.0 | 121.9 | 121.9 | 122.3 |
| Resolution limit (Å) | 0.95 | 1.1 | 1.0 | 1.0 |
| Unique observations | 290866 | 192429 | 232314 | 249970 |
| Multiplicity | 4.1 | 3.5 | 2.8 | 3.2 |
| Average I/σ | 11.3 (2.1) | 7.6 (1.9) | 7.1 (3.0) | 7.2 (2.0) |
| Rmeas (%) | 7.6 (45.2) | 8.1 (42.4) | 9.4 (29.4) | 9.8 (30.7) |
| Completeness (%) | 88 (45.2)b | 91 (74.8) | 84.1 (55.7)b | 90.9 (64.5)b |
| Refinement | ||||
| Reflections with F > 0 σ | 276635 | 147475 | 184585 | 194474 |
| Protein atoms | 3835 | 3849 | 3842 | 3842 |
| Heteroatoms | 78 | 208 | 178 | 131 |
| Solvent atoms | 825 | 916 | 906 | 858 |
| Hydrogen atoms | 3317.8 | 3359 | 3351.5 | 3359 |
| rmsd bonds (Å) | 0.018 | 0.015 | 0.016 | 0.016 |
| rmsd angles (Å) | 0.038 | 0.034 | 0.033 | 0.033 |
| <Bprotein> (Å2) | 14.2 | 20.4 | 17.4 | 17.8 |
| <Bligand> (Å2) | 9.0 | 20.1 | 13.8 | 12.1 |
| Rcryst (Rfree) (%) | 12.2 (15.1) | 11.4 (16.4) | 11.3 (14.7) | 12.33 (15.7) |
| Coordinate error (Å)c | 0.018 | 0.028 | 0.022 | 0.022 |
Numbers in parentheses refer to the highest-resolution shell.
The resolution cutoffs for which local completeness exceeds 75% are 0.97 Å, 1.05 Å and 1.1 Å for GRNative, GRGSH and GRNADPH, respectively.
Coordinate estimated standard uncertainty calculated using the Rfree variant of Cruickshank’s DPI24, with the equation σ(x, Bavg) = 1.0(Ni/nobs)1/2C−1/3Rfreedmin, where Ni is the number of non-hydrogen atoms, nobs is the number of unique observations, and C is the completeness.
Figure 2.
Atomic-resolution electron density for the active-site cofactors. a) The nicotinamide ring of NADPH at 1.0 Å resolution (contour level 3.2*ρrms) from GRNADPH, with carbons (cyan), nitrogens (blue) and oxygens (red) having distinct electron density levels. The C4 atom, which transfers a hydride to FAD, is at the bottom. Pyramidalization of atom N1, at the top of the ring, is visible. A slight twist in the carboxamide relative to the ring is also visible. b) The flavin ring system of FAD at 0.95 Å resolution (contour level 3.8*ρrms) from GRNative, with coloring as in (a) except carbons are green. The N5 atom, where FAD receives electrons from NADPH, is at the bottom center. A small twist in the flavin is evident.
In addition, a 1.8 Å, room-temperature GRNADPH structure based on data collected using a laboratory X-ray source was solved and refined to R and Rfree near 14% and 19%, respectively (Table 2). This structure provides a direct image of NADPH binding at room temperature that replaces the previous best-resolved model of NADPH binding, which was derived by combining information from structures with a bound NADH or a bound NADP+19.
Because insights into catalysis are often based on small differences between the structures, we note that our refinement strategy involved fully refining the highest-resolution GRNative structure first and then using that as a starting model for generating the three other structures. This means that differences between each individual model and GRNative will tend to be underestimated, enhancing the confidence that can be placed in any significant structural differences observed.
The ultrahigh-resolution structures gave us no new insights into the binding of GSSG or GSH or the transfer of electrons from the redox-active disulfide to GSSG. In particular, we were unable to determine the protonation state of His467′ important for the oxidative half-reaction. Thus, we do not use space here to describe those aspects of the structures. Instead, the focus in this presentation is on novel structural results and catalytic insights related to the reductive half-reaction involving NADPH, FAD and the redox-active disulfide. In the following sections, we will describe the structural results, followed by insights relevant to catalysis.
Temperature-dependent changes in structure
Each of these new structures has been previously determined at lower resolution at room temperature, and comparisons were carried out to assess any consistent changes that appeared to be due to a change in temperature. As expected, the temperature factors in the cryo-structures were consistently lower, typically about 75% as large. In only a few segments does the cryo-structure have significantly higher temperature factors, and these are all surface loops involved in crystal contacts that shift somewhat and apparently become less ordered during the unit-cell changes that occur upon freezing. Interestingly, the most systematic exception to the general drop in B-factors is the set of residues that are the most ordered in the room temperature structures. For these residues with B~8 Å2, the B-factors stay largely the same, implying that these B-factors may be an indicator of lattice disorder in the crystal rather than intrinsic thermal motion of the atoms themselves.
Also as expected, the cryo-structures had many more ordered water molecules, with the GRNative structure going from 523 modeled water sites at room temperature to 832 at low temperature, including many partially occupied water sites involved in definable alternate hydrogen-bonding networks. With regard to alternate conformations of protein atoms, the 1.54 Å resolution room temperature structure of GRNative, showed evidence for alternate conformations of 12 side chains (see Table 8 of Karplus & Schulz 1987). In the 0.95 Å resolution cryo-stucture of the same crystal form, the discrete disorder of 8 of these residues is confirmed, but for 4 residues—Thr119, Ser190, Ser231 and Lys420—it is not. Discrete disorder is also modeled for an additional 69 residues. As these residues mostly had relatively high B-factors in the room-temperature structure, the observation of discrete disorder could be simply a resolution effect rather than a temperature effect. The one exception is the redox-active disulfide, which in the cryo-structure is modeled in both an open (reduced) conformation and a closed (oxidized) conformation. This is a result of radiation-induced opening of the disulfide rather than a temperature effect and is discussed further below. As no examples were found of atoms that were well-defined in both structures but with distinctly different conformations, we conclude that there are no notable conformational changes due to freezing itself.
Overall anisotropic motions
The large data-to-parameter ratio at atomic resolution allows for consideration of nonspherical anisotropic displacement parameters (ADPs) rather than a single isotropic B factor. Anisotropy can be quantified by a single number that is the ratio of the smallest to the largest elements of the 3 × 3 ADP matrix25. This measure ranges from 1 (perfectly isotropic) decreasing toward zero with increasing anisotropy. Atomic anisotropies in GRNative were roughly normally distributed with a mean of 0.32 and σ=0.1, and the three other GR structures showed higher means of ~0.43 and σ=0.1. In all distributions, very few atoms had anisotropies >0.8. This indicates that although high-resolution data are needed to reveal anisotropy, its presence is the rule and isotropically vibrating atoms are the rare exceptions. Separate distributions for just protein atoms, just heteroatoms or just solvent atoms are similar (means within 0.03 of the mean for all atoms). Except for GRNative, these results are consistent with Merritt’s analysis of all structures known at 1.4 Å or better (mean 0.45 and σ=0.15)26. The degree of anisotropy in GRNative is slightly higher than any protein in Merritt’s study; the closest was lysozyme (PDB ID 1lks) with a mean of 0.35. Described later are details of anisotropic motions in the active site and related to catalysis.
Synchrotron reduction fails to model natural reduction
Synchrotron radiation has been seen not only to cause a generic gradual decay in the diffraction strength of protein crystals but also to cause specific structural changes such as the cleavage of disulfide bridges27–30 and reduction of active site metallocenters31. These changes are thought to be caused by X-ray generated solvated electrons31. Many groups, including ours, have taken advantage of this “radiation-induced reduction,” assuming it provided a view of catalytically relevant reduced enzyme forms that allowed insights into enzyme mechanism2,31–34.
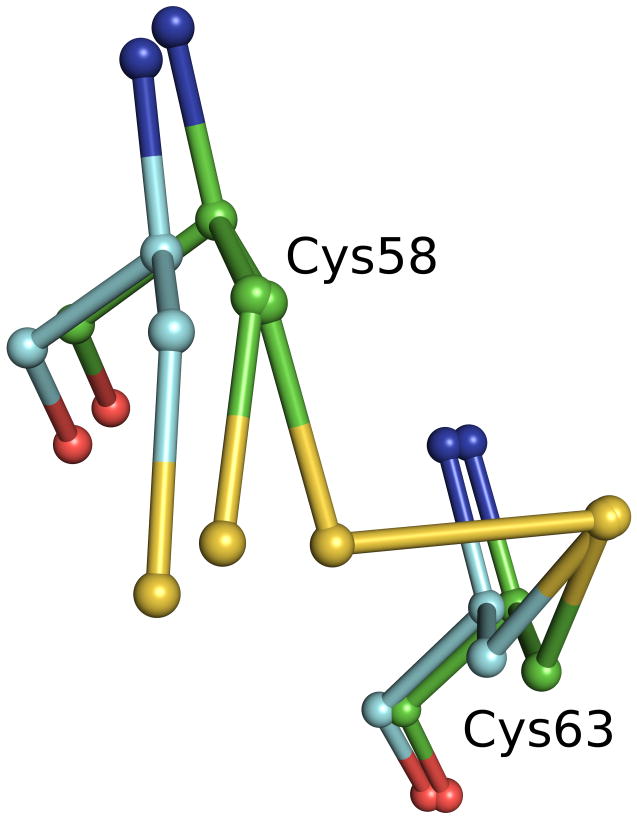
In both oxidized GR crystal forms analyzed here, GRNative and GRGSSG/NADP, synchrotron radiation partially cleaved the active-site (Cys58-Cys63) disulfide. Refinement gave occupancies of 0.53/0.47 (GRNative) and 0.38/0.62 (GRGSSG/NADP) for disulfide/open forms of the two structures. Interestingly, when these synchrotron-reduced structures are compared with the structures of GRGSH and GRNADPH that have been chemically reduced at room temperature, the positions of Cys58 differ significantly (Figure 3). The synchrotron-based reduction merely involved a χ1 sidechain rotation such that the Cys58 sulfur moved 1.0–1.3 Å away from Cys63; in contrast, the chemically reduced structures show an accompanying shift of up to 0.75 Å by the backbone atoms of Cys58 and nearby residues. Our explanation is that at the cryotemperatures of data collection, any larger-scale motions involving the protein backbone are blocked. We note that a recent published structure of the thioredoxin-like protein cDsbD35 showed a similar discrepancy between synchrotron reduction and chemical reduction, although the authors did not point out this difference. Also supporting the hypothesis that motions are limited at the cryotemperatures of data collection is that at temperatures near 200 K, the motions required for enzyme catalysis are hindered36. In any case, independent of the explanation, the discrepancy between between the synchrotron-cleaved and chemically reduced conformations seen for GR and cDsbD proves that one cannot assume a synchrotron-generated reduced form of an enzyme accurately reflects active-site changes that occur during normal catalysis. Consequently, any insights into reaction mechanisms based on radiation-reduced structures at cryotemperatures must be reexamined.
Figure 3.
Disulfide bonds reduced by radiation at cryotemperatures are different from those reduced chemically. The GRNative (green carbons) structure shows both the native disulfide and an alternate conformation for Cys58 due to radiation damage at cryotemperatures. GRNADPH (cyan carbons) shows the structure resulting from chemical reduction at room temperature. The overlay shows a clear difference in the backbone relaxation of Cys58 that depends upon the mode of reduction.
The redox-active disulfide loop has large ω-angle deviations
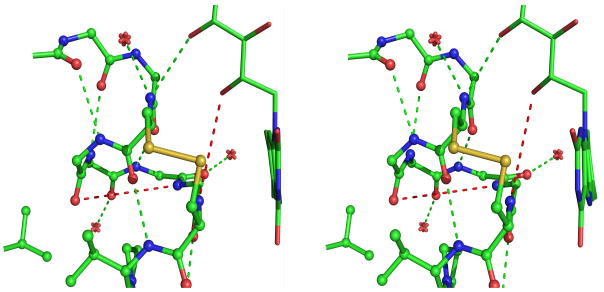
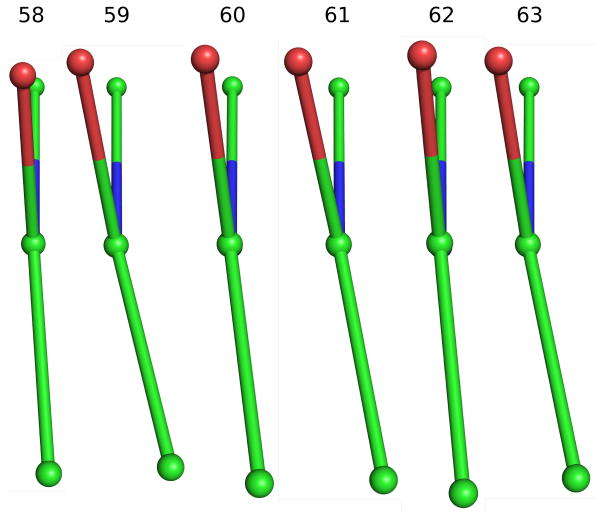
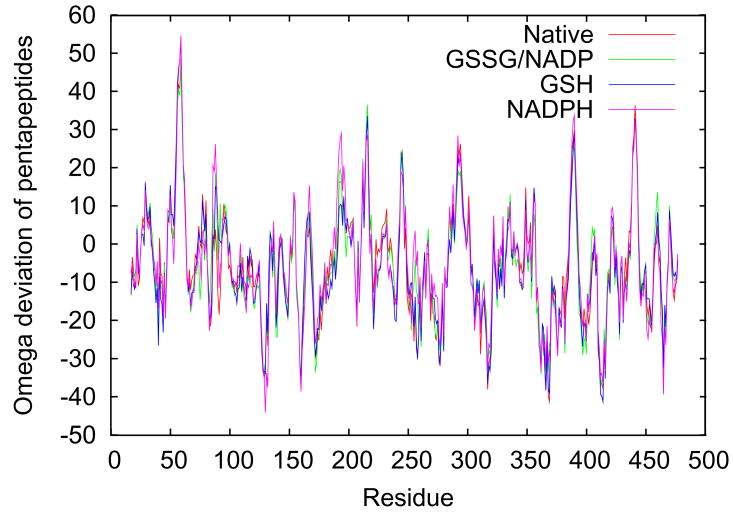
One of the goals of these atomic resolution refinements was to better assess the preliminary observation of conformational strain in the redox-active disulfide loop seen in native GR at 1.54 Å resolution, with the six peptide bonds from residue 58 to 64 all having negative ω-values with an average value of −175° 12 even though the maximal deviations from planarity were only about 10°. It is now well-documented that ω-deviations are strongly underestimated in lower-resolution structures but can be determined with about a 3° accuracy in atomic-resolution structures37. Consistent with this, all four of the atomic-resolution GR structures show ω-values that deviate from planarity by up to ~20°, and the rms deviations in the four structures are all less than 3°. Considering the redox-active disulfide loop (Figure 4a), the six peptides from Cys58 to Cys63 all deviate from planarity in the same direction with an average nonplanarity of ~10° (Figure 4b). The systematic non-planarity of near 50° for a 5-residue segment (Figure 4c) is nearly twice as large as seen for any other region in GR.
Figure 4.
Peptide non-planarity in the active-site disulfide loop. a) Stereoview of the disulfide loop with standard hydrogen bonds (green dotted lines) and unusually long “hydrogen bonds” (red dotted lines) shown. b) Views down each peptide bond in the loop in GRNative visually reveal the magnitude of omega deviations from planarity, which are 4°, 13°, 7°, 10°, 5°, and 11° for residues 58–63. c) A plot of smoothed (N=5) omega deviations from planarity shows the disulfide loop (residues 59–63 in particular) as the most consistently non-planar region in GR. Omega deviations in this loop are similar in all four structures. d) Histogram of pentapeptide stretches in atomic-resolution structures with deviations from planarity (see Methods). The level of nonplanarity of this GR loop (ranging from 46° to 53° in the four GR structures) is unusual, with only two other examples of similarly deviating loops (see Results & Discussion).
To explore how unusual this was among all proteins, we surveyed atomic resolution structures in the Protein Data Bank21 (see Methods). 516 five-residue segments in 142 proteins were selected by this search. Among 48,428 residues in 249 proteins, the ~45–55° net deviation from planarity in GR is an extreme case matched by only two other structures (Figure 4d). One of those two is the flavoenzyme cholesterol oxidase (PDB ID 1n4w), where the region of deviation occurs in a poorly conserved loop distant from the active site. In the other structure, deoxyribose-phosphate aldolase (PDB ID 1p1x), the deviation occurs in a well-conserved loop (residues 169–174) implicated in binding the phosphate group of the substrate38.
Karplus & Schulz12,19 hypothesized that the systematic deviation in peptide planarity in GR would stabilize the reduced form of the enzyme if upon reduction, the loop opening allowed a more relaxed conformation to be adopted. Also, the presence of conformational strain could enhance the kinetics of reduction because the reduction rate of disulfide bonds is exponentially dependent upon the force applied to those bonds43. However, these hypotheses are not supported because the GR structures with an open disulfide loop (GRGSH and GRNADPH) harbor the same level of peptide nonplanarity as oxidized GR (Figure 4c). Since the nonplanarity remains in the open loop, we hypothesize it is related to the local conformation, as this has been shown to have a systematic influence on peptide planarity39–40. Another unfavorable aspect of the disulfide loop conformation is that the hydrogen-bonding potential of some of its backbone atoms is rather poorly satisfied (Figure 4a). In two cases, for a peptide NH group, the closest potential hydrogen-bonding partners are quite distant, 4.5–5.0 Å away (red dotted lines in Figure 4a). As was seen for the peptide nonplanarity, the quality of hydrogen bonding does not improve upon disulfide reduction.
One additional observation is that the Cys58-Cys63 disulfide bond is unusually long in both atomic-resolution oxidized structures: 2.22 Å in GRNative and 2.32 Å in GRGSSG/NADP, compared with the standard value of 2.04 Å. We suspect this is an artifact of the radiation-induced partial disulfide reduction. In GRGSH, Cys63-SG moves 0.15 Å compared with its position in GRNative, suggesting that the single position modeled for Cys63-SG in GRNative (GRGSSG/NADP) is actually an average of two conformations ~0.30 Å apart. In this case, the true oxidized position need not have an unusual bond length.
NADPH binding and catalysis of hydride transfer
Karplus & Schulz19 derived the NADPH binding mode at ca. 2 Å resolution by making a composite of the NADH and NADP+ bound forms. Here, the direct analysis of the NADPH complexes at room temperature and at cryotemperatures not only confirms the general features of the composite model, but the ultrahigh-resolution cryo-structure also gives novel information about the detailed interactions at the catalytic center that provide insight into the roles played in hydride-transfer catalysis by compression and stereoelectronic effects. It is well-known that steric compression can in principle contribute to enzyme catalysis41–44, but its involvement in particular enzymes is difficult to document without ultrahigh-resolution structural analyses of catalytically relevant complexes. At 1.8 Å resolution, the X-ray structure of the NADPH complex of GR shows the C4 atom to be only 3.3 Å from the flavin N5 atom, closer than normal van der Waals interactions predict. Now at atomic resolution, we confirm this close approach and see additional evidence of compression that would facilitate hydride transfer.
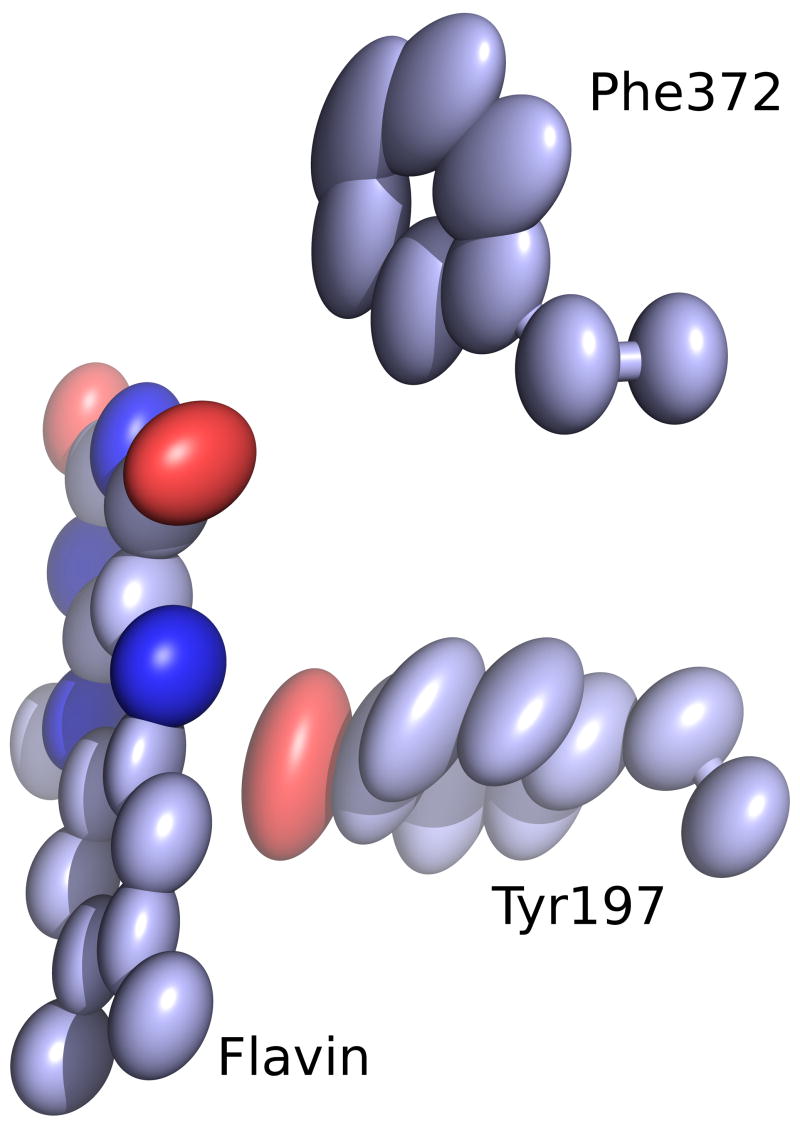
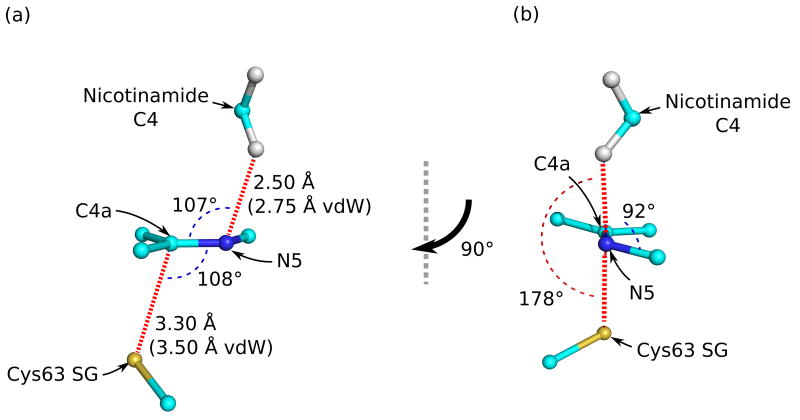
The first evidence of compression is a strong decrease in the level of motion of the active-site atoms in the NADPH complex. In the empty oxidized active site, the anisotropic thermal ellipsoids show that the flavin has significant freedom to shift perpendicular to the plane of the flavin, and Tyr197, the residue filling the nicotinamide pocket, also has freedom to anisotropically wag around its average position (Figure 5a). In contrast, when the nicotinamide displaces Tyr197 upon NADPH binding, both the absolute mobility and the anisotropy diminish markedly, indicating that the four groups—Tyr197, nicotinamide, flavin and the Cys63 thiolate—are tightly juxtaposed against one another (Figure 5b). The existence of compression is further supported by overlapping van der Waals radii in the active site (Figures 6 & 8). With a flavin N5 to nicotinamide C4 distance of 3.29 Å and a flavin C4a to Cys63-SG distance of 3.29 Å, the flavin is tightly fixed through compression from both sides. A third line of evidence for compression in the NADPH-bound complex is the shifting of the flavin ring system in the two reduced structures (Figure 6). In the GSH-reduced structure, which has the Cys-63 thiolate-flavin charge-transfer interaction but no bound nicotinamide, the flavin N5 is pushed toward the nicotinamide pocket by about 0.3 Å even though the Cys63 thiolate only moves half that distance. This specifies the relaxed distance of approach for the thiolate-flavin charge-transfer interaction. In contrast, in the NADPH-reduced structure, the flavin N5 is actually pushed by the presence of the nicotinamide 0.3 Å the other direction (toward the still-unmoved thiolate). These changes result in a 0.15 Å compression in the flavin-thiolate interaction (flavin C4a and Cys63S). This provides a structural explanation for how NADPH binding intensifies the thiolate-flavin charge-transfer intensity18. Assuming that the level of compression is evenly spread between the players, this implies that the nicotinamide/flavin interaction is similarly compressed. Finally, the fourth line of evidence for compression is a clearly visible distortion in the planarity of the nicotinamide group at atom N1 (Figures 2a & 7). Based on inspection of the structure, it appears that the pyramidalization of N1 occurs because if the nicotinamide ring were not puckered, it would extend in the direction of the ribose-N1 bond and the nicotinamide C4 atom would collide with the flavin N5 atom (Figures 6–8). This creates a loaded-spring effect, with the nicotinamide not only tightly packed but presumably strongly forced toward the flavin, aided by the backstop of Tyr197 (Figure 5b).
Figure 5.
Nicotinamide binding tightens the active site. a) Anisotropic mobility is shown as thermal ellipsoids for residues as seen in the active site of the GRGSSG/NADP complex (carbons violet). b) The same view for the GRNADPH complex (carbons cyan). In the NADPH complex, the motion is much lower and more isotropic.
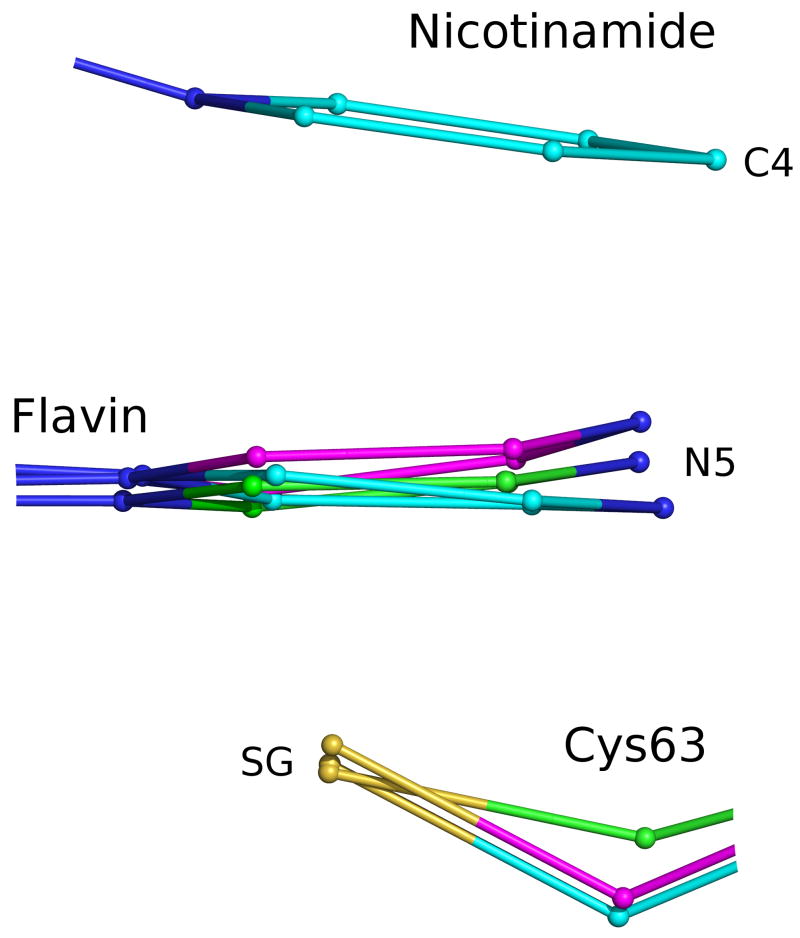
Figure 6.
Steric compression in nicotinamide-flavin interaction. Side view of overlaid active site centered on flavin, showing GRNative (green), GRGSH (magenta) and GRNADPH (cyan). In GRNADPH, NADPH binding above the flavin pushes it down into Cys63, and in GRGSH, GSH and the Cys63 thiolate below the flavin push it up. GRGSSG/NADP is not shown because its atoms are in the same positions as in GRNative. To conserve space the flavin-nicotinamide and the flavin-Cys63 separations are not to scale. Figure 8 shows these distances to scale.
Figure 8.
Stereoelectronic control in nicotinamide-flavin interaction. (a) A side view with the flavin N5-C4a bond in the plane of the paper and (b) a view down the flavin N5-C4a bond together show the optimal geometry for concerted 1–2 addition across the double bond. Compression in the form of shorter than van der Waals interactions is also shown in (a).
Figure 7.
The nicotinamide distortion and ribose conformation favor catalysis. a) The schematic shows the planes of the nicotinamide and flavin (solid black lines). The hypothesized partial boat is shown as a solid red line. Pyramidalization at the nicotinamide N1 places the lone pair on the flavin side where it (i) entropically favors the productive boat conformation to form and (ii) repels the hydride to be transferred (dashed red line). b) The ribose conformation relative to the nicotinamide stabilizes the electron-deficient NADP+ ring orbitals via hyperconjugative electron donation from the ribose. The glycosidic C-O bond position parallel to the nicotinamide ring also favors NADP+ over NADPH (see Results & Discussion).
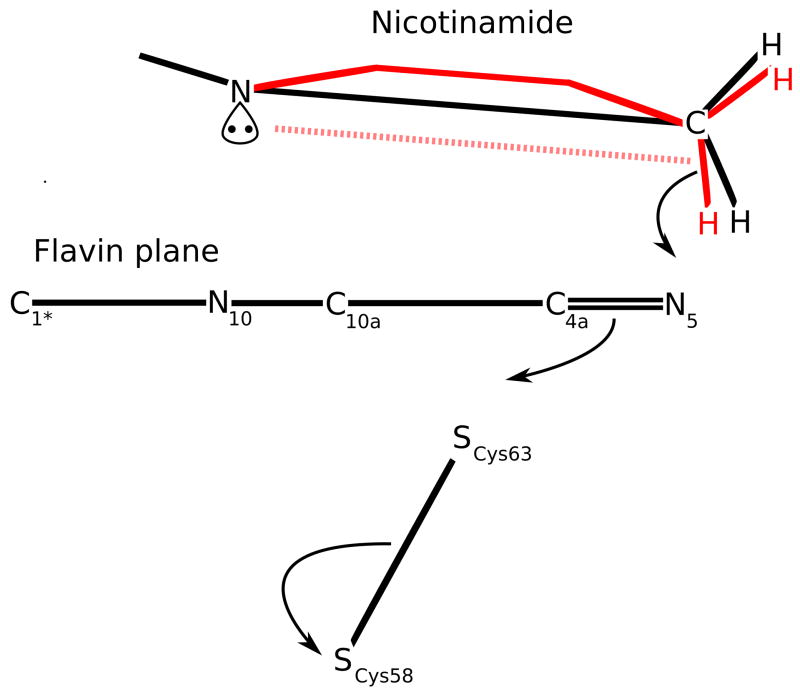
Interestingly, according to stereoelectronic theory, N1 pyramidalization can serve to optimize the nicotinamide for hydride transfer45, so that the observed distortion is likely to be a mechanism to chemically enhance the rate of hydride transfer. Normally, in the planar nicotinamide, the N1 lone-pair electrons form a conjugated system with the C=C bonds of the rings stabilizing NADPH46–49. In GR, however, the out-of-plane N1-C1′ bond (Figure 7a) pulls the N1 lone-pair electrons out of the resonance and moves them into a pseudoaxial orbital on the hindered flavin side. Importantly, this favors hydride transfer when the C4 atom fluctuates out of the plane, creating a boat-like conformation with the hydride to be transferred pseudoaxial and on the same side of the nicotinamide ring as the N1 lone pair43,46,49. This type of reaction, a 1,4-syn elimination, has been well-studied50. Furthermore, the N1 pyramidalization with the lone pair facing the flavin entropically restricts by half the conformations available to nicotinamide (N1 can only pucker in one direction). This makes the required boat-like conformation more likely and thus enhances the propensity for hydride transfer.
A second factor improving catalysis is the conformation of the ribose moiety relative to the nicotinamide, which has been shown by Wu & Houk47 to influence the NADPH redox potential. In GR, the ribose C-O bond is parallel to the nicotinamide ring, which stabilizes NADP+ by allowing hyperconjugative donation by the ribose σC-H and σC-C orbitals into the electron-deficient ring of NADP+47 (Figure 7b). Additionally, this parallel conformation minimizes NADPH stabilization via the anomeric effect—the ribose C-O is a better acceptor than C-H, so if the C-O were anti to the N1 lone pair, it would stabilize it with the σ*C-O orbital47. These two factors together serve to increase the NADPH reduction potential, favoring hydride transfer.
A third factor favoring catalysis proposed in the lower (1.54 Å) resolution analysis was that deviations in key flavin bond lengths indicated the protein environment predisposed the flavin toward reduction. The data in Table 1 indicate that changes in the redox states of small-molecule flavins are associated with changes in four bond lengths by >0.05 Å11. None of the four structures of the enzyme are expected to have formally reduced flavins. However, at atomic resolution, the two EH2 structures have all bond lengths close to those of reduced flavin, and the two oxidized state structures have some bonds that are reduced-like and others that are intermediate between oxidized and reduced states (Table 1). The strong tendency toward reduced-like bond lengths in all structures led us to ask whether they were influenced by synchrotron radiation. Synchrotron-induced reduction of FAD has been seen in DNA photolyase53, but in that case the flavin is naturally light-sensitive. An assessment of the other two flavoenzymes with structures known at ≤1 Å resolution, cholesterol oxidase and PETN reductase, showed that both appear reduced based on their bond lengths (Table 1). The PETN reductase structure was of a reduced form of the enzyme, but cholesterol oxidase was not, so the reduced bond lengths seen are unexpected22. Given these results, we are suspicious that X-ray reduction of these flavins has occurred to at least some extent and hesitate to draw conclusions about how the protein environment in GR influences flavin electronic structure. We further conclude that although ultrahigh-resolution structure determination has the potential to yield accurate bond-length information that can give insight into detailed influences of the protein environment on the flavin, because synchrotron radiation itself can influence the flavin structure, conclusions must be tempered unless the changes due to the protein environment and those due to perturbation by the experiment can be dissected.
Table 1.
Flavin covalent bond geometry
| N5-C4a (Å) | C4a-C4 (Å) | C4a-C10a (Å) | C10a-N1 (Å) | |
|---|---|---|---|---|
| Small-molecule flavinsa | ||||
| Oxidized | 1.30 | 1.49 | 1.47 | 1.32 |
| Oxidized-H+ | 1.30 | 1.49 | 1.42 | 1.36 |
| Oxidized(−) | 1.30 | 1.49 | 1.45 | 1.31 |
| Reduced | 1.42 | 1.40 | 1.37 | 1.38 |
| GR 2.0 Å structuresb | ||||
| GRNativec | 1.33 | 1.44 | 1.45 | 1.35 |
| GRGSSG/NADP | 1.30 | 1.49 | 1.44 | 1.32 |
| GRGSH | 1.32 | 1.48 | 1.47 | 1.33 |
| GRNADPH | 1.30 | 1.49 | 1.47 | 1.33 |
| GR 1.0 Å structuresd | ||||
| GRNative | 1.36 | 1.47 | 1.38 | 1.33 |
| GRGSSG/NADP | 1.39 | 1.47 | 1.36 | 1.38 |
| GRGSH | 1.38 | 1.39 | 1.40 | 1.36 |
| GRNADPH | 1.37 | 1.43 | 1.40 | 1.37 |
| Atomic-resolution flavoenzymes | ||||
| Cholesterol oxidasee | 1.40 | 1.41 | 1.41 | 1.36 |
| PETN reductasef | 1.39 | 1.40 | 1.40 | 1.37 |
Finally, a fourth factor favoring catalysis is that the nicotinamide C4 hydride and Cys63-SG are on opposite sides of the flavin plane and roughly in line with the N5-C4a bond (Figure 8), in position for good HOMO-LUMO overlap20,54–55 and also consistent with optimal geometry for 1,2-addition of the hydride and the sulfur across a double bond47,56–57. This optimal geometry for a 1,2-addition raises the interesting possibility that the reduction of GR is not a distinct two-step process with hydride transfer to the flavin occurring first, followed by disulfide reduction, but instead is concerted, with hydride transfer linked to and enhanced by disulfide reduction. If this is the case, stopped-flow studies would not show presence of a reduced flavin. Indeed, Huber and Brandt wrote of yeast GR13: “It is possible that electron transfer is relatively concerted so that discrete intermediates are not detected, as suggested by Matthews et al. (1979)57a for lipoamide dehydrogenase.” Furthermore, in wild-type E. coli GR, reoxidation of FAD obscured direct observation of FAD reduction18, with both steps at rates greater than 100 s−1. However, mutants can disrupt this tight linkage. Reduced flavin intermediates were observed in active-site mutants of E. coli GR by Rietveld et al.18 and mercuric reductase by Miller et al57b. In another E. coli GR mutant of the Cys63 equivalent to alanine to block the reaction at flavin reduction, only 10% of the flavin was reduced, indicating a possible influence of the disulfide acceptor on the thermodynamics of hydride transfer18. These points suggest a reductive half-reaction in disulfide reductases that, if not concerted, is at least tightly linked to hydride transfer both kinetically and thermodynamically.
Materials and Methods
Protein expression, purification, and crystallization
Recombinant human GR was provided by R.H. Schirmer, prepared in Escherichia coli as previously described14. Crystals were grown as previously described58. Briefly, they were grown reproducibly at room temperature using 10 μL hanging drops containing 20 mg/mL GR, 3% ammonium sulfate, 0.1 M potassium phosphate and 0.1% β-octyl glucoside at pH 7.0. The reservoir was 700 μL containing 21–23% ammonium sulfate and 0.1 M potassium phosphate at pH 8. Crystals used at cryotemperatures were frozen by soaking them for 1 minute in each of 5%, 10%, 15%, 25% glycerol solutions of the mother liquor before cryocooling in liquid nitrogen.
Crystals of GR grew to 1.0 × 0.7 × 0.4 mm3, but the larger crystals tended to lose diffraction quality under cryogenic conditions. The best data were measured from crystals measuring 0.5 × 0.3 × 0.2 mm3. To prepare complexes of GR with natural substrates, crystals of oxidized GR were soaked in mother liquor supplemented with 10 mM GSSG/5 mM NADP+ (24 hours for GRGSSG/NADP), 1–5 mM GSH (24 hours for GRGSH), or 1–5 mM NADPH (1–2 hours for GRNADPH). Reduction of crystalline GR was monitored by the ensuing color change from golden yellow to orange-red. Reduced crystals of GR were immediately cryocooled in liquid nitrogen and stored for data collection. Cryocooling was performed according to the method developed for native crystals and with the cryosolutions supplemented with the appropriate substrates to maximize ligand occupancies.
X-ray data collection and refinement for atomic-resolution structures
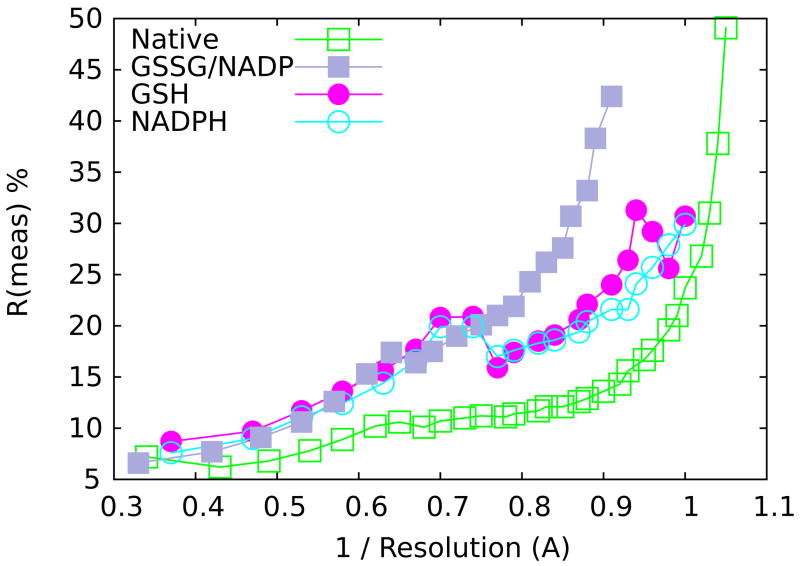
Crystals of GR belong to the space group C2. Diffraction data were collected at the Advanced Photon Source on BioCARS beamline 14-BM-C using an ADSC Quantum 4 detector at 100 K with λ=1.0 Å radiation. For each structure, data sets were collected from a single crystal. Using a detector offset in 2θ, data from several oscillation runs were collected to cover reciprocal space at high resolution, then data from a low-resolution pass was also collected with no detector offset and with a shortened exposure time to minimize the number of overloaded reflections. Data were processed and scaled using Denzo and Scalepack59. Rmeas and Rmerge were calculated from unmerged data and Rmeas >50% and I/σ <2.0 were used in defining resolution limit cutoffs (Figure 9). In some cases the highest resolution bin completeness is low (near 50%), but in all cases the completeness climbs to be above 75% within 0.1 Å of the reported resolution (Table 2). Finally, 5% of the native data were selected to set aside as a test set for Rfree calculations. Rfree test sets for all of the complexes were based on the native test set.
Figure 9.
Data quality as a function of resolution. Observed data quality is indicated by Rmeas values and plotted as a function of resolution−1. The four structures are colored as indicated in the key. These data were used to determine where to set the high-resolution cutoff (see Methods).
Refinement of GRNative began with PDB model 3grs transformed into the C2 unit cell and all water molecules removed. This 1.54 Å room temperature model was used a starting point (R=0.347, Rfree=0.352) for 40 cycles of conjugant gradient refinement at 1.5 Å resolution with the TNT package60. A quick visual inspection/correction of obvious changes and addition of 192 waters followed by an additional 40 cycles of refinement yielded a model with excellent geometry and R=0.253 Rfree=0.305. All subsequent refinement was performed with SHELX61.
SHELX refinement for GRNative followed the scheme described in the SHELX manual for high-resolution structures61. The resolution limit was extended to 1.0 Å in SHELX with a diffuse solvent correction (SWAT). Following this, individual atoms were refined anisotropically using the suggested SIMU/DELU restraints. The default value for DELU standard deviation was used, and the standard deviation of SIMU was moved up to 0.1 from 0.02. The ISOR restraint was only applied to solvent atoms and was increased to a standard deviation of 0.1 from 0.02. As refinement progressed, alternate conformations of sidechains and loops were modeled as indicated by the electron density maps. Water molecules were added as manually identified throughout refinement; in later rounds, this was supplemented with automated methods of SHELX. These suggested water molecules were manually checked for reasonable geometry before they were added to the model. In the final steps of refinement, some water molecules were reduced to half occupancy, riding hydrogens were added, and the resolution was extended to 0.95 Å. All of these steps consisted of multiple cycles of refinement and manual model building. 2Fo-Fc and Fo-Fc maps were monitored along with the peak list from SHELX, and geometry outliers were identified by ProCheck62. Iterative rounds of model building/refinement continued until convergence.
This produced the final GRNative model (R=0.122, Rfree=0.151) at 0.95 Å. This model was used as a starting point for refinement of the GRNADPH, GRGSH and GRGSSG/NADP complex structures. Further refinement and manual model building were performed as needed for each of these structures independently from this common starting point. Refinement results are shown in Table 2.
X-ray data collection and refinement for the room-temperature 1.8 Å GRNADPH,1.8 structure
This data set used another NADPH-soaked crystal, prepared identically to the above-described crystals but not frozen. The crystal was mounted in a glass capillary filled with the soak solution and G25 Sephadex to immobilize the crystal (as in Karplus & Schulz 1989). Data for the 1.8 Å, room-temperature GRNADPH was collected at our in-house X-ray source with Cu-Kα radiation (Rigaku RU-H3R rotating anode operating at 50 kV and 100 mA and an R-Axis IV image plate detector; λ=1.54 Å, Δϕ=0.3°, 400 10-min images) (Table 2). The structure was solved using molecular replacement of the 1.54 Å GRNative structure and refined with first CNS63, then Refmac64. Between refinement cycles, Fo-Fc and 2Fo-Fc electron-density maps were used for manual rebuilding with O65, and later with Coot66. To account for large-scale anisotropy shared among residue groups, TLS (translation, libration and screw) domains were added in Refmac based on a TLS refinement of malarial GR67 and checked using TLSMD68. The addition of TLS domains to the refinement resulted in decreases in R and Rfree of 0.7% and 1.3%, respectively.
Structural comparisons and analyses
Noncovalent interactions were analyzed using Coot’s66 interface to Reduce 3.0 and Probe 2.1169, which displays favorable and unfavorable van der Waals interactions as well as hydrogen bonds visually within Coot.
Global anisotropy was analyzed using PARVATI26, which produced a distribution of atomic displacement parameters (ADPs) across each structure and highlighted the most anisotropic residues. Local ADPs were analyzed using Coot or PyMol to display anisotropy with ellipsoids at 50% or 67% probability. Figure 5 shows anisotropy at 67% probability. Structural figures were also generated with PyMol70.
Structural overlays were created using the McLachlan algorithm71 as implemented in the program ProFit by iteratively overlaying structures using a subset of Cα atoms with a maximum per-atom RMSD of 0.1 Å until convergence was reached. When overlaying more than 2 structures, ProFit’s multiple overlay function was used, and structures were overlaid in order of decreasing resolution to minimize bias to the average structure. For overlays containing multiple conformations and substrates, the original ProFit-overlaid structure (lacking multiple conformations) had a complete structure (including multiple conformations) overlaid upon it in Coot using least-squares fit, resulting in an identically overlaid set of structures.
Protein Geometry Database
The Protein Geometry Database (PGD; Berkholz et al., unpublished), an in-house database derived from the PDBselect list of protein structures with nonredundant sequences72, was used to compare the GR structure with the rest of the Protein Data Bank. The PGD contains primarily covalent backbone geometry and is flexibly searchable, with results available via data dumps or a graphing module. Searches were performed on structures determined at 1.2 Å resolution or better, using a subset of the PGD containing no proteins with >90% sequence identity. Figures of the results were generated using gnuplot.
Protein Data Bank entry codes
Models were deposited into the RCSB Protein Data Bank with accession codes 3DK9, 3DK4, 3DK8, 3DJJ and 3DJG for GRNative, GRGSSG/NADP, GRGSH, GRNADPH, and GRNADPH,1.8, respectively.
Acknowledgments
This work was sponsored by National Science Foundation grant MCB-9982727 to P.A.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dauter Z, Lamzin VS, Wilson KS. The benefits of atomic resolution. Curr Opin Struct Bio. 1997;7(5):681–688. doi: 10.1016/s0959-440x(97)80078-4. [DOI] [PubMed] [Google Scholar]
- 2.Berglund GI, Carlsson GH, Smith AT, Szöke H, Henriksen A, Hajdu J. The catalytic pathway of horseradish peroxidase at high resolution. Nature. 2002;417(6887):463–8. doi: 10.1038/417463a. [DOI] [PubMed] [Google Scholar]
- 3.Heine A, DeSantis G, Luz JG, Mitchell M, Wong C, Wilson IA. Observation of covalent intermediates in an enzyme mechanism at atomic resolution. Science. 2001;294(5541):369–374. doi: 10.1126/science.1063601. [DOI] [PubMed] [Google Scholar]
- 4.De Colibus L, Mattevi A. New frontiers in structural flavoenzymology. Curr Opin Struct Bio. 2006;16(6):722–728. doi: 10.1016/j.sbi.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Miura R. Versatility and specificity in flavoenzymes: Control mechanisms of flavin reactivity. Chem Rec. 2001;1(3):183–194. doi: 10.1002/tcr.1007. [DOI] [PubMed] [Google Scholar]
- 5a.Lennon BW, Williams CH, Ludwig ML. Crystal structure of reduced thioredoxin reductase from Escherichia coli: structural flexibility in the isoalloxazine ring of the flavin adenine dinucleotide cofactor. Protein Sci. 1999;8(11):2366–2379. doi: 10.1110/ps.8.11.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraaije MW, Mattevi A. Flavoenzymes: diverse catalysts with recurrent features. Trends in Bioch Sci. 2000;25(3):126–132. doi: 10.1016/s0968-0004(99)01533-9. [DOI] [PubMed] [Google Scholar]
- 7.Massey V. Introduction: flavoprotein structure and mechanism. FASEB J. 1995;9(7):473–5. doi: 10.1096/fasebj.9.7.7737454. [DOI] [PubMed] [Google Scholar]
- 8.Massey V. The chemical and biological versatility of riboflavin. Biochem Soc Trans. 2000;28:283–296. [PubMed] [Google Scholar]
- 9.Fox KM, Karplus PA. The flavin environment in old yellow enzyme: An evaluation of insights from spectroscopic and artificial flavin studies. J Biol Chem. 1999;274(14):9357–9362. doi: 10.1074/jbc.274.14.9357. [DOI] [PubMed] [Google Scholar]
- 10.Ghisla S, Massey V. Mechanisms of flavoprotein-catalyzed reactions. FEBS J. 1989;181(1):1–17. doi: 10.1111/j.1432-1033.1989.tb14688.x. [DOI] [PubMed] [Google Scholar]
- 11.Karplus PA. Structural flavinology on the brink. In: Ghisla S, Kroneck P, Macheroux P, Sund H, editors. Flavins and flavoproteins 1999. Agency for Scientific Publ; 1999. pp. 233–238. [Google Scholar]
- 12.Karplus PA, Schulz GE. Refined structure of glutathione reductase at 1.54 Å resolution. J Mol Biol. 1987;195(3):701–29. doi: 10.1016/0022-2836(87)90191-4. [DOI] [PubMed] [Google Scholar]
- 13.Huber PW, Brandt KG. Kinetic studies of the mechanism of pyridine nucleotide dependent reduction of yeast glutathione reductase. Biochemistry. 1980;19(20):4568–4575. doi: 10.1021/bi00561a005. [DOI] [PubMed] [Google Scholar]
- 13a.Thorpe C, Williams CH. Spectral evidence for a flavin adduct in a monoalkylated derivative of pig heart lipoamide dehydrogenase. J Biol Chem. 1976;251(23):7726–7728. [PubMed] [Google Scholar]
- 14.Krauth-Siegel RL, Arscott LD, Schoenleben-Janas A, Schirmer RH, Williams CH. Role of active site tyrosine residues in catalysis by human glutathione reductase. Biochemistry. 1998;37(40):13968–13977. doi: 10.1021/bi980637j. [DOI] [PubMed] [Google Scholar]
- 15.Argyrou A, Blanchard JS, Palfey BA. The lipoamide dehydrogenase from Mycobacterium tuberculosis permits the direct observation of flavin intermediates in catalysis. Biochemistry. 2002;41(49):14580–14590. doi: 10.1021/bi020376k. [DOI] [PubMed] [Google Scholar]
- 16.Böhme CC, Arscott LD, Becker K, Schirmer RH, Williams CH. Kinetic characterization of glutathione reductase from the malarial parasite Plasmodium falciparum. Comparison with the human enzyme. J Biol Chem. 2000;275(48):37317–23. doi: 10.1074/jbc.M007695200. [DOI] [PubMed] [Google Scholar]
- 17.Vanoni MA, Wong KK, Ballou DP, Blanchard JS. Glutathione reductase: comparison of steady-state and rapid reaction primary kinetic isotope effects exhibited by the yeast, spinach, and Escherichia coli enzymes. Biochemistry. 1990;29(24):5790–5796. doi: 10.1021/bi00476a021. [DOI] [PubMed] [Google Scholar]
- 18.Rietveld P, Arscott LD, Berry A, Scrutton NS, Deonarain MP, Perham RN, et al. Reductive and oxidative half-reactions of glutathione reductase from Escherichia coli. Biochemistry. 1994;33(46):13888–13895. doi: 10.1021/bi00250a043. [DOI] [PubMed] [Google Scholar]
- 19.Karplus PA, Schulz GE. Substrate binding and catalysis by glutathione reductase as derived from refined enzyme: substrate crystal structures at 2 Å resolution. J Mol Biol. 1989;210(1):163–180. doi: 10.1016/0022-2836(89)90298-2. [DOI] [PubMed] [Google Scholar]
- 20.Sustmann R, Sicking W, Schulz GE. The Active Site of Glutathione Reductase: An Example of Near Transition-State Structures. Angew Chem Int Ed Eng. 1989;28(8):1023–1025. [Google Scholar]
- 21.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nuc Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lario PI, Sampson N, Vrielink A. Sub-atomic resolution crystal structure of cholesterol oxidase: What atomic resolution crystallography reveals about enzyme mechanism and the role of the FAD cofactor in redox activity. Journal of Molecular Biology. 2003;326(5):1635–1650. doi: 10.1016/s0022-2836(03)00054-8. [DOI] [PubMed] [Google Scholar]
- 23.Khan H, Barna T, Harris RJ, Bruce NC, Barsukov I, Munro AW, et al. Atomic resolution structures and solution behavior of enzyme-substrate complexes of Enterobacter cloacae PB2 pentaerythritol tetranitrate reductase: Multiple conformational states and implications for the mechanism of nitroaromatic explosive degradation. J Biol Chem. 2004;279(29):30563–30572. doi: 10.1074/jbc.M403541200. [DOI] [PubMed] [Google Scholar]
- 24.Cruickshank DWJ. Coordinate uncertainty. In: Rossmann MG, Arnold E, editors. International Tables for Crystallography. Dordrecht: Kluwer Academic Publishers; 2001. pp. 403–418. [Google Scholar]
- 25.Trueblood KN, Burgi HB, Burzlaff H, Dunitz JD, Gramaccioli CM, Schulz HH, et al. Atomic dispacement parameter nomenclature. Report of a subcommittee on atomic displacement parameter nomenclature. Acta Cryst A. 1996;52(5):770–781. [Google Scholar]
- 26.Merritt EA. Expanding the model: anisotropic displacement parameters in protein structure refinement. Acta Cryst D. 1999;55(6):1109–1117. doi: 10.1107/s0907444999003789. [DOI] [PubMed] [Google Scholar]
- 27.Burmeister WP. Structural changes in a cryo-cooled protein crystal owing to radiation damage. Acta Cryst D. 2000;56(3):328–341. doi: 10.1107/s0907444999016261. [DOI] [PubMed] [Google Scholar]
- 28.Weik M, Sussman J. Synchotron X-ray radiation produces specific chemical and structural damage to protein structures. Proc Nat Acad Scip. 2000;97:623–628. doi: 10.1073/pnas.97.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ravelli RBG, Garman EF. Radiation damage in macromolecular cryocrystallography. Curr Opin Struct Bio. 2006;16(5):624–629. doi: 10.1016/j.sbi.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Ravelli RBG, McSweeney SM. The ‘fingerprint’ that X-rays can leave on structures. Structure. 2000;8(3):315–328. doi: 10.1016/s0969-2126(00)00109-x. [DOI] [PubMed] [Google Scholar]
- 31.Carugo O, Carugo KD. When X-rays modify the protein structure: radiation damage at work. Trends in Bioch Sci. 2005;30(4):213–219. doi: 10.1016/j.tibs.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Alphey MS, Attrill H, Crocker PR, van Aalten DMF. High resolution crystal structures of Siglec-7: Insights Into ligand specificity in the Siglec family. J Biol Chem. 2003;278(5):3372–3377. doi: 10.1074/jbc.M210602200. [DOI] [PubMed] [Google Scholar]
- 33.Roberts BR, Wood ZA, Jonsson TJ, Poole LB, Karplus PA. Oxidized and synchrotron cleaved structures of the disulfide redox center in the N-terminal domain of Salmonella typhimurium AhpF. Prot Sci. 2005;4(9):2414. doi: 10.1110/ps.051459705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kort R, Hellingwerf KJ, Ravelli RBG. Initial events in the photocycle of photoactive yellow protein. J Biol Chem. 2004;279(25):26417–26424. doi: 10.1074/jbc.M311961200. [DOI] [PubMed] [Google Scholar]
- 35.Stirnimann CU, Rozhkova A, Grauschopf U, Böckmann RA, Glockshuber R, Capitani G, et al. High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study. J Mol Biol. 2006;358(3):829–845. doi: 10.1016/j.jmb.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 36.Rasmussen BF, Stock AM, Ringe D, Petsko GA. Crystalline ribonuclease A loses function below the dynamical transition at 220 K. Nature. 1992;357(6377):423–424. doi: 10.1038/357423a0. [DOI] [PubMed] [Google Scholar]
- 37.Sevcik J, Dauter Z, Lamzin VS, Wilson KS. Ribonuclease from Streptomyces aureofaciens at Atomic Resolution. Acta Cryst D. 1996;52(2):327–344. doi: 10.1107/S0907444995007669. [DOI] [PubMed] [Google Scholar]
- 38.Heine A, Luz JG, Wong C, Wilson IA. Analysis of the class I aldolase binding site architecture based on the crystal structure of 2-deoxyribose-5-phosphate aldolase at 0.99 Å resolution. J Mol Biol. 2004;343(4):1019–1034. doi: 10.1016/j.jmb.2004.08.066. [DOI] [PubMed] [Google Scholar]
- 39.Karplus PA, Shapovalov MV, Dunbrack A, Jr, Dunbrack R, Jr, Berkholz DS. A forward-looking suggestion for resolving the stereochemical restraints debate: ideal geometry functions. Acta Cryst D. 2008;64(3) doi: 10.1107/S0907444908002333. (in the press) [DOI] [PubMed] [Google Scholar]
- 40.Karplus PA. Experimentally observed conformation-dependent geometry and hidden strain in proteins. Prot Sci. 1996;5(7):1406–1420. doi: 10.1002/pro.5560050719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rajagopalan PTR, Benkovic SJ. Preorganization and protein dynamics in enzyme catalysis. Chem Rec. 2002;2(1):24–36. doi: 10.1002/tcr.10009. [DOI] [PubMed] [Google Scholar]
- 42.Bruice T, Lightstone F. Ground state and transition state contributions to the rates of intramolecular and enzymatic reactions. Acc Chem Res. 1999;32(2):127–136. [Google Scholar]
- 43.Almarsson O, Bruice TC. Evaluation of the factors influencing reactivity and stereospecificity in NAD(P)H dependent dehydrogenase enzymes. J Am Chem Soc. 1993;115(6):2125–2138. [Google Scholar]
- 44.Bruice TC, Pandit UK. Intramolecular models depicting the kinetic importance of “fit” in enzymatic catalysis. Proc Nat Acad Sci. 1960;46(4):402–404. doi: 10.1073/pnas.46.4.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nambiar KP, Stauffer DM, Kolodziej PA, Benner SA. A mechanistic basis for the stereoselectivity of enzymic transfer of hydrogen from nicotinamide cofactors. J Am Chem Soc. 1983;105(18):5886–5890. [Google Scholar]
- 46.Young L, Post CB. Catalysis by entropic guidance from enzymes. Biochemistry. 1996;35(48):15129–15133. doi: 10.1021/bi961875m. [DOI] [PubMed] [Google Scholar]
- 47.Wu YD, Houk KN. Theoretical evaluation of conformational preferences of NAD+ and NADH: an approach to understanding the stereospecificity of NAD+/NADH-dependent dehydrogenases. J Am Chem Soc. 1991;113(7):2353–2358. [Google Scholar]
- 48.Wu YD, Houk KN. Theoretical study of conformational features of NAD+ and NADH analogs: protonated nicotinamide and 1, 4-dihydronicotinamide. J Org Chem. 1993;58(8):2043–2045. [Google Scholar]
- 49.Benner SA. The stereoselectivity of alcohol dehydrogenases: A stereochemical imperative? Cell Molec Life Sci. 1982;38(5):633–637. doi: 10.1007/BF02327092. [DOI] [PubMed] [Google Scholar]
- 50.Yates RL, Epiotis ND, Bernardi F. Importance of nonbonded attraction in the stereochemistry of the SN2′ reaction. J Am Chem Soc. 1975;97(23):6615–6621. [Google Scholar]
- 51.Stehle T, Claiborne A, Schulz GE. NADH binding site and catalysis of NADH peroxidase. FEBS J. 1993;211(1):221–226. doi: 10.1111/j.1432-1033.1993.tb19889.x. [DOI] [PubMed] [Google Scholar]
- 52.Sem DS, Kasper CB. Geometric relationship between the nicotinamide and isoalloxazine rings in NADPH-cytochrome P-450 oxidoreductase: implications for the classification of evolutionarily and functionally related flavoproteins. Biochemistry. 1992;31(13):3391–3398. doi: 10.1021/bi00128a013. [DOI] [PubMed] [Google Scholar]
- 53.Kort R, Komori H, Adachi S, Miki K, Eker A. DNA apophotolyase from Anacystis nidulans: 1.8 Å structure, 8-HDF reconstitution and X-ray-induced FAD reduction. Acta Cryst D. 2004;60(7):1205–1213. doi: 10.1107/S0907444904009321. [DOI] [PubMed] [Google Scholar]
- 54.Cavelier G, Amzel LM. Mechanism of NAD (P) H: Quinone reductase: Ab initio studies of reduced flavin. Prot Struct Func Gen. 2001;43(4):420–432. [PubMed] [Google Scholar]
- 55.Rivas P, Zapata-Torres G, Melin J, Contreras R. Probing the hydride transfer process in the lumiflavine–1-methylnicotinamide model system using group softness. Tetrahedron. 2004;60(19):4189–4196. [Google Scholar]
- 56.Liotta CL, Burgess EM, Eberhardt WH. Trajectory analysis. 1. Theoretical model for nucleophilic attack at pi-systems. The stabilizing and destabilizing orbital terms. J Am Chem Soc. 1984;106(17):4849–4852. [Google Scholar]
- 57.Miessler GL, Tarr DA. Inorganic Chemistry. Pearson Prentice Hall; 2004. Molecular orbitals; pp. 116–164. [Google Scholar]
- 57a.Matthews RG, Ballou DP, Williams CH. Reactions of pig heart lipoamide dehydrogenase with pyridine nucleotides. Evidence for an effector role for bound oxidized pyridine nucleotide. J Biol Chem. 1979;254(12):4974–4981. [PubMed] [Google Scholar]
- 57b.Miller SM, Massey V, Ballou D, Williams CH, Distefano MD, Moore MJ, Walsh CT. Use of a site-directed triple mutant to trap intermediates: demonstration that the flavin C(4a)-thiol adduct and reduced flavin are kinetically competent intermediates in mercuric ion reductase. Biochemistry. 1990;29(11):2831–2841. doi: 10.1021/bi00463a028. [DOI] [PubMed] [Google Scholar]
- 58.Savvides SN, Karplus PA. Kinetics and crystallographic analysis of human glutathione reductase in complex with a xanthene inhibitor. J Biol Chem. 1996;271(14):8101–8107. doi: 10.1074/jbc.271.14.8101. [DOI] [PubMed] [Google Scholar]
- 59.Otwinowski Z, Minor W. Processing of X-Ray Diffraction Data Collected in Oscillation Mode. Academic Press, New York. 1997:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 60.Tronrud DE. TNT refinement package. Meth Enzp. 1997;277:306–319. doi: 10.1016/s0076-6879(97)77017-4. [DOI] [PubMed] [Google Scholar]
- 61.Sheldrick G, Schneider T. SHELXL: High-resolution refinement. Meth Enzp. 1997;277:319–343. [PubMed] [Google Scholar]
- 62.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26(2):283–291. [Google Scholar]
- 63.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Cryst D. 1998;54(5):905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 64.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Meth Enzp. 2003;374:300–21. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 65.Jones TA. A graphics model building and refinement system for macromolecules. J Appl Cryst. 1978;11(4):268–272. [Google Scholar]
- 66.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst A. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 67.Sarma GN, Savvides SN, Becker K, Schirmer M, Schirmer RH, Karplus PA. Glutathione reductase of the malarial parasite Plasmodium falciparum: crystal structure and inhibitor development. J Mol Biol. 2003;328(4):893–907. doi: 10.1016/s0022-2836(03)00347-4. [DOI] [PubMed] [Google Scholar]
- 68.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J Appl Crystp. 2006;39:109–111. [Google Scholar]
- 69.Word JM, Lovell SC, LaBean TH, Taylor HC, Zalis ME, Presley BK, et al. Visualizing and quantifying molecular goodness-of-fit: small-probe contact dots with explicit hydrogen atoms. J Mol Biol. 1999;285(4):1711–1733. doi: 10.1006/jmbi.1998.2400. [DOI] [PubMed] [Google Scholar]
- 70.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 71.McLachlan AD. Rapid comparison of protein structures. Acta Cryst A. 1982;38(6):871–873. [Google Scholar]
- 72.Hobohm U, Sander C. Enlarged representative set of protein structures. Prot Sci. 1994;3(3):522. doi: 10.1002/pro.5560030317. [DOI] [PMC free article] [PubMed] [Google Scholar]