Abstract
Very long chain fatty acids (VLCFA) and branched-chain fatty acids (BCFA) are potent inducers of the peroxisome proliferator-activated receptor PPARα, a nuclear receptor that enhances transcription of peroxisomal enzymes mediating β-oxidation of these potentially toxic fatty acids. However, it is not known whether the respective free fatty acids or their activated metabolites, i.e. CoA thioesters: (i) are the endogenous high-affinity PPARα ligands, (ii) alter PPARα conformation, and (iii) alter recruitment of coregulatory proteins to PPARα. As shown by quenching of PPARα intrinsic amino acid fluorescence, PPARα exhibited high affinity (3-29nM Kds) for the CoA thioesters of the common (C20-C24) VLCFA. In contrast, with the exception of arachidonic acid (Kd=20nM), PPARα only weakly bound the VLCFA. PPARα also exhibited higher affinity for the CoA thioesters of BCFA (phytanoyl-CoA, pristanoyl-CoA; Kds near 11nM) than for the respective free branched-chain fatty acids. As shown by circular dichroism, the high affinity VLCFA-CoA and BCFA-CoA strongly altered PPARα conformation. Likewise, the high affinity VLCFA-CoA and BCFA-CoA altered co-factor recruitment to PPARα as shown by co-immunoprecipitation from liver homogenates. In contrast, nearly all the respective free fatty acids elicited only weak conformational changes in PPARα and did not alter co-factor recruitment to PPARα. In summary, the CoA thioesters of very-long-chain and branched-chain fatty acids are much more potent PPARα ligands than the free acids, resulting in altered PPARα conformation and cofactor recruitment. Since these are hallmarks of ligands-activated nuclear receptors, this suggests that the CoA thioesters are the active forms of these PPARα ligands.
Keywords: PPARα, β-oxidation, fatty acyl-CoA, phytanic acid, fluorescent binding assays, circular dichroism, nuclear transcription factors
Peroxisome proliferator-activated receptors (PPAR) are ligand-activated members of the steroid/thyroid nuclear hormone receptor superfamily. PPAR have been identified in several species, with at least three isotypes recognized (α, β/δ, γ). Each isotype, encoded by a different gene, is differentially expressed in select tissues and expression levels depend on cellular processes (rev. in (1). Although there is some overlap in ligand specificity, unique isotype binding occurs by a variety of xenobiotics depends upon specific amino acid substitutions within the PPAR ligand-binding pocket (1,2). Independent of ligand binding, a basal level of transcription occurs by PPAR forming a heterodimer with the retinoid X receptor (RXR) which binds to a specific DNA response element (PPRE) within the target genes (3,4). Ligand binding to one and, even more so, both receptors greatly potentiates transcription (rev. in (4). PPRE exist within many of the genes involved in fatty acid oxidation and cell differentiation, suggesting that PPAR are responsible for regulating transcription of these genes (5).
An especially important role of PPARα is inducing transcription of multiple peroxisomal enzymes required for β-oxidation of very-long-chain fatty acids (VLCFA) and branched-chain fatty acids (BCFA). VLCFA and BCFA accumulation are associated with human neurological disorders including Zellweger syndrome, X-linked adrenoleukodystrophy, Refsum disease, and MFP-2 deficiency (6-8). These genetic disorders arise from defects in enzymes of the peroxisomal β-oxidation pathway which result in accumulation of VLCFA, BCFA, and their thioesters (6-8). Both VLCFA and isoprenoid-derived BCFA preferentially undergo peroxisomal β-oxidation following acylation of the respective fatty acids to CoA thioesters. The classical peroxisomal β-oxidation pathway catalyzed by palmitoyl-CoA oxidase (AOX), L-bifunctional protein (L-PBE), and 3-ketoacyl-CoA thiolase (PTL) is responsible for the oxidation of straight-chain fatty acids (9). All three enzymes of the classical peroxisomal β-oxidation system are transcriptionally activated by PPARα ligands (rev. in (10). In contrast, although the branched-chain phytanic acid is considered a β-oxidation pathway precursor, a different set of peroxisomal enzymes are utilized for oxidation of BCFA. For example, phytanic acid must first undergo α-oxidation to produce pristanoyl-CoA before entering the β-oxidation cycle (11). BCFA-CoAs are then utilized by the branched-chain acyl-CoA oxidase, D-bifunctional protein (D-PBE), and sterol carrier protein-x (SCP-x). Both D-PBE (9) and SCP-x (12,13) are inducible upon administration of branched-chain PPARα ligands.
While there is evidence indicating that VLCFA and BCFA are peroxisome proliferators and may be PPARα ligands, it is not completely clear whether the unesterified VLCFA and BCFA directly and/or metabolites thereof activate PPARα. For example, early transactivation assays show: (i) PPARα is activated by polyunsaturated (20:4, 20:5, 22:6), but not monounsaturated (24:1, 22:1) VLCFA (3), (ii) n-3 polyunsaturated VLCFA elicited similar PPARα activation levels as n-6 polyunsaturated VLCFA (3). These findings of VLCFA being potential PPARα ligands are supported by gel-shift assays (14) and co-activator-dependent receptor ligand assays (15). Likewise, transactivation studies with branched-chain fatty acids also show PPARα activation (11). It must be noted that most of these assays utilize very high concentrations of VLCFA (i.e. 30-100 μM) and BCFA (20 μM)--several orders of magnitude higher than concentrations of unesterified fatty acids detected in nuclei of living cells (16,17). These data suggested that affinities of PPARα for these ligands are very weak, i.e. Kds in the μM range, and the free fatty acids may not represent the endogenous ligands. The finding that, since unesterified fatty acids are potent detergents, they are rapidly activated/metabolized to their CoA thioesters in cells and tissues (rev. (18-20), suggests that metabolites such as the CoA thioesters may represent the endogenous PPARα ligands. This possibility is supported by recent findings in mice. For instance, peroxisome proliferators increase the level of CoA thioesters of the peroxisome proliferators in mice (21,22). Inhibitors of fatty acid thioesterification such as 2-bromopalmitic acid inhibit the effects of bezafibrate on peroxisomal proliferation (23). Mice with a disruption of the AOX gene exhibit peroxisomal proliferation, elevated serum levels of VLCFA, as well as elevated VLCFA-CoAs, hyperactivation of PPARα, and up-regulation of down-stream genes which contain PPRE (23). On the other hand, ablation of the adrenoleukodystrophy gene inhibits VLCFA transport into peroxisomes and results in elevated serum VLCFA, unaltered liver VLCFA-CoA, and normal PPARα activity (10). Taken together with other studies (24,25) these data in both normal and gene-targeted mice suggest that intermediates of peroxisomal β-oxidation are PPARα ligands.
In summary, while many of the above experiments are suggestive that the CoA thioesters of VLCFA and BCFA represent functional ligands for PPARα, it remains to be shown whether the VLCFA, BCFA, or their CoA thioesters exhibit characteristic hallmarks of endogenous ligands that bind to PPARα: (i) high (Kds in nM range) affinity, (ii) physically alter the structure/conformation of the PPARα protein, and (iii) affect co-factor recruitment PPARα (26). The objective of the present study was to begin to resolve these issues through use of direct ligand binding assays (i.e. non-fluorescent ligand binding assays based on quenching of tyrosine emission), circular dichroism to characterize potential ligand-induced changes in PPARα secondary structure, and co-immunoprecipitation to study co-factor recruitment.
EXPERIMENTAL PROCEDURES
Chemicals
Arachidic acid (C20:0), arachidonic acid (C20:4, n-6), eicosapentaenoic acid (C20:5, n-3), behenic acid (C22:0), docosapentaenoic acid (C22:5, n-3), docosahexaenoic acid (C22:6, n-3), lignoceric acid (C24:0), nervonic acid (C24:1, n-6), phytanic acid (3,7,11,15-tetramethyl-hexadecanoic acid), 2-fluoropalmitic acid, 1-Iodohexadecane, and coenzyme A were from Sigma (St. Louis, MO). Pristanic acid (3,7,11,15-tetramethyl-pentadecanoic acid) was from Larodan Fine Chemicals (Malmö, Sweden). A-bromopalmitate was from City Chemical LLD (West Haven, CT). Purity of fatty acids was examined by HPLC (27) and determined to be greater than 95%. All fatty acyl-CoAs were synthesized as previously described (28), purified by HPLC (27), and found to be >98% unhydrolyzed. Mouse anti-PPARα monoclonal antibodies, as well as rabbit anti-PPARα, anti-steroid receptor coactivator-1 (SRC-1), and anti-cyclic AMP-responsive enhancer binding protein binding protein (CBP) polyclonal antibodies, were from Affinity BioReagents (Golden, CO). Anti-rabbit IgG secondary antibodies-HRP conjugated were from Santa Cruz Biotechnology (Santa Cruz, CA). BCA protein assay, mammalian co-immunoprecipitation kit, chemiluminescent substrate, and film were from Pierce Chemical Co. (Rockford, IL).
Recombinant mouse PPARα protein
The cDNA encoding mouse PPARα with a deletion of the amino-terminal A/B domain (i.e. encoding PPARα amino acids 101-468) was cloned into a (His)6-tagged bacterial expression vector (pET-PPARαΔAB) and was a generous gift from Dr. Noa Noy (Cornell University, USA) (29). The recombinant protein was expressed, purified, and analyzed by SDS-PAGE and Western blotting as previously described (30). Protein concentration was determined by Bradford assay. Since Bradford assays underestimate the mass of proteins such as liver fatty acid binding protein, uncorrected Bradford assays can result in considerable disagreement in the number of binding sites obtained by solution binding assays (e.g. radioligand, fluorescence) as compared to crystallography (18,31,32). Since both X-ray crystallography (33,34) and fluorescence binding assays (29,30) are in complete agreement that PPARα has only a single binding site, this indicates that the Bradford assay is not underestimating PPARα protein mass.
Direct fluorescence binding assay: Quenching of PPARαΔAB aromatic amino acid residues by non-fluorescent ligands
The direct binding of non-fluorescent ligands to PPARαΔAB was carried out as described (30,35). Briefly, PPARαΔAB (0.1μM in 2ml of PBS, pH 7.4) was titrated with increasing ligand (0-2000nM). Both fatty acids and fatty acyl-CoAs are molecules with limited solubility in aqueous buffer as evidenced by critical micelle concentrations in the typical range of 1μM (36) and 60-200μM (20), respectively. For the majority of the binding assays, the ligand concentration examined was 0-300nM, well below the critical micelle concentration. Only in the rare exception where fatty acid binding was not seen at 300nM was the examined concentration increased above 500nM. PPARαΔAB aromatic amino acid excitation was carried out at 280nm and emission was measured from 300-400nm. Fluorescence emission spectra were obtained at 24°C with a PC1 photon counting spectrofluorometer (ISS Inc., Champaign, IL), corrected for background (buffer and solvent effects), and maximal intensities measured. The dissociation constant (Kd) and the number of binding sites (n) were calculated as previously described (30).
Secondary Structure Determination: Effect of Ligand Binding on PPARαΔAB Circular Dichroism (CD)
Circular dichroic spectra of PPARαΔAB (0.8μM in 125μM HEPES, pH 8.0, 12.5μM DTT, 5mM KCl, 0.3% glycerol) were taken in the presence and absence of ligands (20μM) with a J-710 spectropolarimeter (JASCO Inc., Easton, MD) at 23°C in a 1-mm cuvette as described (30,35). Spectra were recorded from 280 to 185nm with a bandwidth of 2nm, sensitivity of 10 millidegrees, scan rate of 50nm/min, and a time constant of 1s. Ten scans were averaged for percent composition of α-helices, β-strands, turns, and unordered structures with the software package CDPro (downloaded from the website: http://lamar.colostate.edu/~sreeram/CDPro), which allows the percent calculation of various secondary structures by three different methods; SELCON3, CDSSTR, and CONTIN/LL (37).
Co-immunoprecipitation: Effects of ligand binding on co-factor recruitment
Livers from C57BL/6 male mice were homogenized in M-PER® buffer (Pierce Chemical Co., Rockford, IL) containing 150mM sodium chloride and protease inhibitors, and centrifuged to pellet insoluble debris. Protein concentration was determined by BCA protein assay. Ligands (100μM) were added to 2mg of liver homogenate, and co-immunoprecipitation procedures with a PPARα monoclonal antibody SDS-PAGE were conducted with the ProFound™ mammalian co-immunoprecipitation kit according to the manufacturer’s instructions (Pierce Chemical Co., Rockford, IL). Protein transfer and western blotting were performed using standard techniques (30). Proteins were quantified by densitometry, utilizing a single-chip charge-coupled device video camera FluorChemimager and accompanying FluorChem image analysis software from Alpha Innotech (San Leandro, CA). Co-immunoprecipitated protein values were standardized to PPARα values, and standardized values were normalized for each ligand sample to the level of interaction observed in the absence of ligands, which was set equal to 1. Statistical analysis was performed using the Student’s t test.
RESULTS
Direct Binding of Very-Long-Chain Fatty Acids to PPARα: Quenching of PPARα Intrinsic Aromatic Amino Acid Fluorescence
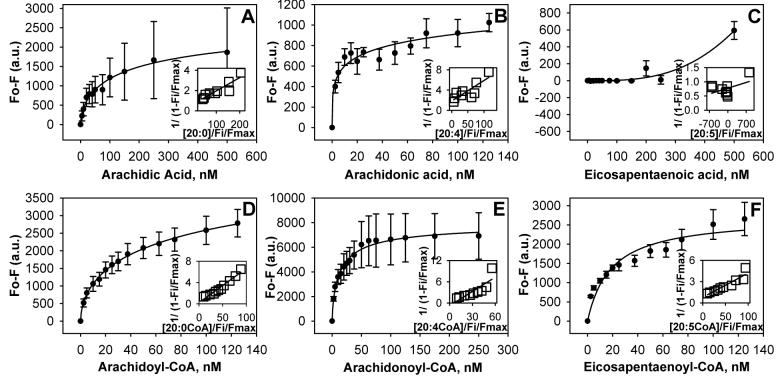
Since early radioligand fatty acid and fatty acyl-CoA binding assays are know to underestimate the affinities of binding proteins by several orders of magnitude (rev. in (18,19), an assay not dependent upon an exogenous radiolabeled or fluorescent ligand or upon physical separation of bound from free ligand was utilized. This assay, recently developed by our laboratory (30,35), takes advantage of PPARα’s intrinsic aromatic amino acid fluorescence which is quenched upon ligand binding. In the case of saturated VLCFA, saturation binding curves were obtained for the 20-carbon arachidic acid (Fig. 1A), but not the longer chain 22-carbon behenic acid (Fig. 2A) and 24-carbon lignoceric acid (Fig. 3A). Analysis of multiple replicates showed that PPARα bound arachidic acid with a Kd of 76±12nM at a single binding site (Table 1). This was confirmed by Hill plot (Fig. 1A) which also yielded one arachidic binding site (n = 1.0) for PPARα. Thus, saturated VLCFA were poorly bound by PPARα.
FIG. 1. Interaction of naturally-occurring C20 fatty acids and fatty acyl-CoAs with PPARαΔAB: Direct binding based on quenching of PPARαΔAB aromatic amino acid fluorescence emission.
The data are presented as the change in fluorescence intensity (Fo - F) plotted as a function of ligand concentration. PPARαΔAB (0.1μM) was titrated with 0-600nM of the following ligands: panel A, arachidic acid (C20:0); panel B, arachidonic acid (C20:4); panel C, eicosapentaenoic acid (C20:5); panel D, arachidoyl-CoA (C20:0-CoA); panel E, arachidonoyl-CoA (C20:4-CoA); panel F, eicosapentaenoyl-CoA (C20:5-CoA). Values represent the mean ± SE, n=4-5. Insets, linear plot of the binding curve from each panel.
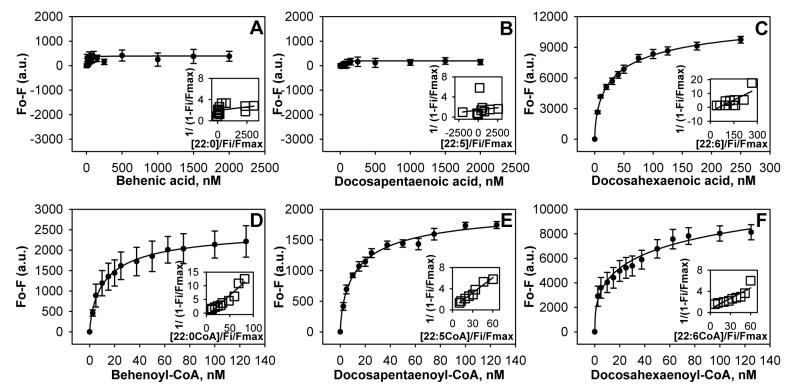
FIG. 2. Interaction of naturally-occurring C22 fatty acids and fatty acyl-CoAs with PPARαΔAB: Direct binding based on quenching of PPARαΔAB aromatic amino acid fluorescence emission.
The data are presented as the change in fluorescence intensity (Fo - F) plotted as a function of ligand concentration. PPARαΔAB (0.1μM) was titrated with 0-2500nM of the following ligands: panel A, behenic acid (C22:0); panel B, docosapentaenoic acid (C22:5); panel C, docosahexaenoic acid (C22:6); panel D, behenoyl-CoA (C22:0-CoA); panel E, docosapentaenoyl-CoA (C22:5-CoA); panel F, docosahexaenoyl-CoA (C22:6-CoA). Values represent the mean ± SE, n=4-5. Insets, linear plot of the binding curve from each panel.
FIG. 3. Interaction of naturally-occurring C24 fatty acids and fatty acyl-CoAs with PPARαΔAB: Direct binding based on quenching of PPARαΔAB aromatic amino acid fluorescence emission.
The data are presented as the change in fluorescence intensity (Fo - F) plotted as a function of ligand concentration. PPARαΔAB (0.1μM) was titrated with 0-2500nM of the following ligands: panel A, lignoceric acid (C24:0); panel B, nervonic acid (C24:1); panel C, lignoceroyl-CoA (C24:0-CoA); panel D, nervonoyl-CoA (C24:1-CoA. Values represent the mean ± SE, n=4-5. Insets, linear plot of the binding curve from each panel.
Table 1. Affinity of PPARαΔAB for very-long-chain, branched-chain, and non-metabolizable fatty acids and their CoA thioesters as determined by quenching of PPARαΔAB aromatic amino acid fluorescence.
Binding parameters were determined from fluorescent quenching of PPARαΔAB by naturally-occurring, non-fluorescent ligands as described in “Experimental Procedures”. The Kd (dissociation constant) was calculated from the reciprocal plots as described in “Experimental Procedure”. Values represent the mean ± SE (n = 4-5). Weak refers to detection of very low levels of quenching, but insufficient for determination of binding parameters; --- refers to no noticeable quenching
| Ligand | Chain length:double bonds (position) | Kd (nM) FA | Kd (nM) CoA |
|---|---|---|---|
| Arachidic acid/CoA Arachidonic acid/CoA Eicosapentaenoic acid/CoA |
20:0 20:4 (n-6) 20:5 (n-3) |
76 ± 12 20 ± 5 Weak |
16 ± 1 7 ± 2 29 ± 4 |
| Behenic acid/CoA Docosapentaenoic acid/CoA Docosahexaenoic acid/CoA |
22:0 22:5 (n-3) 22:6 (n-3) |
--- Weak 145 ± 48 |
6 ± 1 10 ± 1 11 ± 2 |
| Lignoceric acid/CoA Nervonic acid/CoA |
24:0 24:1 (n-9) |
Weak 769 ± 296 |
3 ± 1 9 ± 1 |
| Pristanic acid/CoA Phytanic acid/CoA |
15:tetrametyl 16:tetramethyl |
19 ± 2 34 ± 4 |
12 ± 1 11 ± 1 |
| 2-Bromopalmitic acid 2-Fluoropalmitic acid S-hexadecyl-CoA |
Br-16:0 F-16:0 S-16:0-CoA |
808 ± 195 219 ± 33 |
10 ± 1 |
In general, inclusion of unsaturation in the VLCFA enhanced the abilities of 20-carbon, and to a much lesser extent 22- and 24-carbon, VLCFA to be bound by PPARα. For example, nervonic acid (24-carbon fatty acid containing a single double bound) exhibited a modest saturation curve (Fig. 3B) and very weak Kd of 769nM (Table 1). VLCFA with larger numbers of double bonds were bound better, but in a pattern highly dependent on chain-length and position of the double bonds. For instance, the n-6 polyunsaturated 20-carbon fatty acid arachidonic acid (C20:4) (Fig. 1B) exhibited a steeper saturation curve than the n-3 polyunsaturated 22-carbon fatty acid docosahexaenoic acid (C22:6) (Fig. 2C), while the n-3 eicosapentaenoic acid (C20:5) did not exhibit significant binding except at very high concentrations (Fig. 1C) and the n-3 22-carbon docosapentaenoic acid (C22:5) was not bound (Fig. 2B). This was reflected in much higher affinity exhibited by PPARα for the n-6 arachidonic acid (Kd=20±5nM) than for the n-3 docosahexaenoic acid (Kd=145±48nM), eicosapentaenoic acid, and docosapentaenoic acid whose Kds were so weak as not to be measurable (Table 1). In summary, with the exception of the n-6 polyunsaturated arachidonic acid, affinities for the other saturated and n-3 VLCFA were relatively weak (arachidic acid, docosahexaenoic acid) or not detectable (eicosapentaenoic acid, docosapentaenoic acid, lignoceric acid), regardless of the presence of double bonds. These data were not consistent with most of the VLCFA being physiologically significant PPARα ligands.
Direct Binding of Very-Long-Chain Fatty Acyl-CoAs to PPARα
To test PPARα’s specificity and affinity for the CoA thioesters of the above VLCFA, the effect of the respective VLCFA-CoAs on quenching of PPARα intrinsic aromatic amino acid fluorescence was examined. The results showed for the first time, contrary to the free VLCFA, the CoA thioesters of all the VLCFA bound with high affinity to PPARα and at a single site on PPARα. For example, each of the saturated VLCFA-CoAs exhibited saturation binding curves that were significantly steeper than the respective VLCFA: the 20-carbon arachidoyl-CoA (Fig. 1D) > arachidonic acid (Fig. 1A); the 22-carbon behenoyl-CoA (Fig. 2D) >>> behenic acid (Fig. 2A); the 24-carbon lignoceroyl-CoA (Fig. 3C) >>> lignoceric acid (Fig. 3A). Analysis of multiple binding curves yielded Kds in the following order: arachidonoyl-CoA (Kd=16±1nM) > behenoyl-CoA (Kd=6±1nM) > lignoceroyl-CoA (Kd=3±1nM) (Table 1). Likewise, the CoA thioester of nervonic acid was bound with high affinity (Kd=9±1nM)—85-fold better than the free nervonic acid (Table 1). In contrast to the free VLCFA, the CoA thioesters of both n-6 VLCFAs (arachidonoyl-CoA, Kd=16±1nM) and n-3 VLCFA (eicospentaenoyl-CoA, Kd=29±4nM; docosapentaenoyl-CoA, Kd=10±1nM; docosahexaenoyl-CoA, Kd=11±2nM) were bound with very high and essentially the same affinities by PPARα (Table 1). Thus, PPARα exhibited very high affinities for all the CoA thioesters of VLCFA. In contrast, PPARα exhibited very weak or no measurable affinity for the free VLCFA. Since polyunsaturated VLCFA (both n-6 and n-3) are known to activate PPARα in cells and animals (3), these data suggest that the respective VLCFA-CoAs, rather than the free acids, may be the active endogenous ligands for PPARα.
Direct Binding of Branched-Chain Fatty Acids and their CoA Thioesters to PPARα
Although phytanic acid is also a 20-carbon fatty acid, it differs markedly from the ≥20-carbon chain VLCFA. Unlike the straight-chain VLCFA, phytanic acid’s chain is only 16 carbons long and has 4 methyl side chains. Further, branched-chain fatty acids such as phytanic acid and its α-oxidation product pristanic acid are the most potent known naturally-occurring PPARα transcriptional activators—much more potent than even the unsaturated VLCFA (rev. in (38-40). However, it is not known whether: (i) phytanic acid or its α-oxidation product (i.e. pristanic acid) is the endogenous PPARα ligand, and (ii) the respective CoA thioesters of the branched-chain fatty acids are the endogenous PPARα ligands. These questions were resolved by the direct fluorescent binding assay described above. PPARα exhibited saturation binding curves for both phytanic acid (Fig. 4A) and pristanic acid (Fig. 4B). Double reciprocal plots were linear (inserts, Figs. 4A,B), indicating PPARα had a single binding site for each of these branched-chain fatty acids. Analysis of multiple binding curves indicated that PPARα bound branched-chain fatty acids with high affinities in the order: pristanic acid (Kd=19±2 nM) > phytanic acid (Kd=34±4nM) (Table 1). Although pristanic acid was a 2-fold higher affinity ligand, nevertheless the parent phytanic acid itself was a high affinity PPARα ligand. Thus, phytanic acid did not need to be metabolized to pristanic acid in order to be a high affinity ligand for PPARα. The CoA thioesters of branched-chain fatty acids were also bound with high affinity by PPARα in the order: pristanoyl-CoA (Kd=12±1nM) > phytanoyl-CoA (Kd=11±1nM) (Table 1). While the affinities of PPARα for the respective CoA thioesters of branched-chain fatty acids were higher than for the free acids, the difference was only 2-3 fold and PPARα exhibited high affinities (nM Kds) for both the free branched-chain fatty acids and their CoA thioesters. Thus, unlike the VLCFA, the branched-chain fatty acids as well as their CoA thioesters were high affinity ligands for PPARα.
FIG. 4. Interaction of branched-chain fatty acids and fatty acyl-CoAs with PPARαΔAB: Direct binding based on quenching of PPARαΔAB aromatic amino acid fluorescence emission.
The data are presented as the change in fluorescence intensity (Fo - F) plotted as a function of ligand concentration. PPARαΔAB (0.1μM) was titrated with 0-2500nM of the following ligands: panel A, phytanic acid (C16:0-branched); panel B, pristanic acid (C16:0-branched); panel C, bromopalmitate (C16:0-Br); panel D, phytanoyl-CoA (C16:0-CoA); panel E, pristanoyl-CoA (C16:0-CoA); panel F, S-hexadecyl-CoA (C16:0-S-CoA). Values represent the mean + SE, n=4-5. Insets, linear plot of the binding curve from each panel.
Direct Binding of Non-metabolizable Fatty Acids and Non-hydrolyzable Fatty Acyl-CoAs to PPARα
Since all the above fatty acids (saturated, unsaturated, and branched-chain) can be metabolized to their CoA thioesters and further transesterified to more complex lipids (41), the ability of PPARα to bind non-metabolizable saturated fatty acids, 2-bromopalmitic acid and 2-fluoropalmitic acid, was examined. PPARα exhibited a shallow saturation binding curve for 2-bromopalmitic acid (Fig. 4C) at a single site as indicated by the double reciprocal plot (Fig. 4C, insert). Similar results were obtained with 2-fluoropalmitic acid (binding curves not shown). Multiple replicates showed that PPARα’s affinity for both 2-bromopalmitic acid and 2-fluoropalmitic acid was weak, Kd=808±195nM and Kd=219±33nM, respectively (Table 1).
Conversely, PPARα is known to enhance the hydrolysis of fatty acyl-CoA thioesters (42) and therefore the possibility that the above findings of high affinity LCFA-CoA binding to PPARα might reflect in part the binding of the breakdown products of the fatty acyl-CoAs, i.e. the hydrolytically released free fatty acids, was considered. Although most of the straight-chain VLCFA do not bind to PPARα with high affinity, a few exceptions (arachidonic acid, phytanic acid, and pristanic acid) were bound by PPARα with high affinity. Therefore, the affinity of PPARα for a non-hydrolyzable fatty acyl-CoA analogue, S-hexadecyl-CoA (42), was examined. A sharp, saturable binding curve was observed upon titration of PPARα with increasing amounts of S-hexadecyl-CoA (Fig. 4F). Analysis of multiple binding curves showed that PPARα exhibited very high affinity for S-hexadecyl-CoA as indicated by a Kd=10±1nM (Table 1). Thus, the data obtained with the non-metabolizable fatty acids and non-hydrolyzable CoA analogue further substantiated the role of CoA thioesters as the active form of PPARα endogenous ligands.
Effect of Very-Long-Chain Fatty Acids on PPARα Secondary Structure: Circular Dichroism
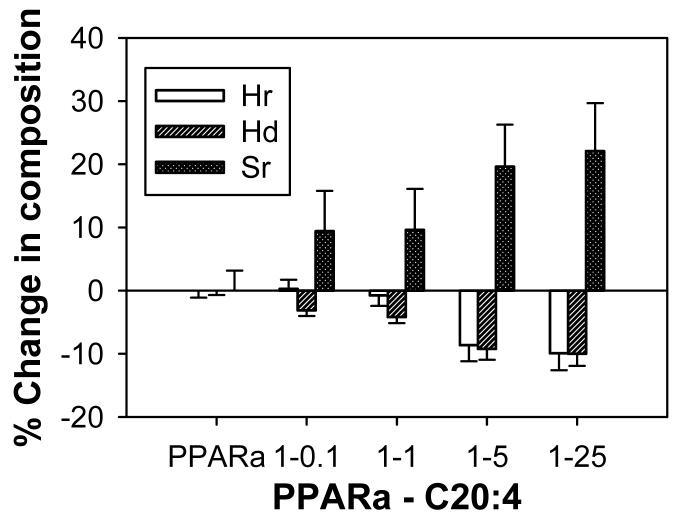
Ligand-induced conformational change is a hallmark of ligand-regulated nuclear transcription factors, including PPAR (34,35,41,43). The observation that binding of non-fluorescent ligands significantly quenched the emission of PPARαΔAB aromatic amino acid residues (Figs. 1-4) suggests that ligand binding altered the conformation of PPARα. To further examine whether the binding of naturally-occurring VLCFA alter PPARα structure, the effect of these ligands on the circular dichroic (CD) spectra of PPARα was examined. The far UV spectrum of PPARα exhibited two minima at 207 and 221nm and one maximum at 193nm (Fig. 5, filled circles; Fig. 6-7, open circles), suggesting the presence of substantial α-helical content in the PPARα polypeptide chain. This was confirmed by quantitative analysis of the CD spectra as described in “Experimental Procedures”, which indicated that PPARα was comprised of 40.2% α-helix, 14.5% β-sheets, 18.7% β-turns, and 26.6% unordered structures (Table 2). The addition of increasing concentrations of a high affinity ligand (arachidonic acid) caused conformational changes that increased in intensity until starting to plateau off at a ratio of PPARα:C20:4 of 1:5 (Fig. 5). The relative proportion of regular (Fig. 5, open bars) and distorted (Fig. 5, diagonal bars) α-helices decreased, concomitant with regular β-sheets increasing (Fig. 5, hatched bars) as the ratio of PPARα:C20:4 increased. Based upon these data and the fact that PPARα had varying affinities for the examined ligands, a PPARα/ligand ratio was chosen for the following CD experiments to allow comparison of maximal ligand-induced conformational changes, i.e. at saturated PPARα ligand binding site.
FIG. 5. Effect of increasing concentrations of ligand on PPARαΔAB conformation: Circular dichroic (CD) spectra.
Far ultraviolet (UV) circular dichroic (CD) spectra of PPARαΔAB in the presence of increasing arachidonic acid concentration were obtained and analyzed for percent composition as described in the “Experimental Procedures”. The effect of increasing PPARαΔAB:C20:4 ratios on regular α-helices (Hr, open bars), distorted α-helices (Hd, diagonal bars), and regular β-sheets (Sr, cross-hatched bars) are presented as percent change as compared to that obtained for PPARαΔAB in the absence of ligand.
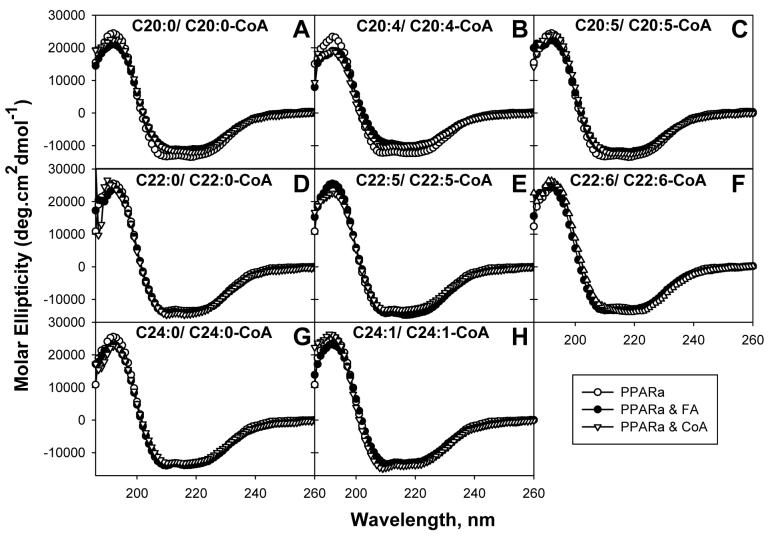
FIG. 6. Effect of VLCFA and VLCFA-CoA binding on PPARαΔAB conformation: Circular dichroic (CD) spectra.
Far ultraviolet (UV) circular dichroic (CD) spectra of PPARαΔAB in the absence or presence of added ligand were obtained as described in the “Experimental Procedures”. Panel A, CD spectrum of PPARαΔAB in the absence (empty circles) and presence of added ligand: arachidic acid (filled circles) or arachidoyl-CoA (empty triangles); panel B, in the presence of arachidonic acid (filled circles) or arachidonoyl-CoA (empty triangles); panel C, in the presence of eicosapentaenoic acid (filled circles) or eicosapentaenoyl-CoA (empty triangles); panel D, in the presence of behenic acid (filled circles) or behenoyl-CoA (empty triangles); panel E, in the presence of docosapentaenoic acid (filled circles) or docosapentaenoyl-CoA (empty triangles); panel F, in the presence of docosahexaenoic acid (filled circles) or docosahexaenoyl-CoA (empty triangles); panel G, in the presence of lignoceric acid (filled circles) or lignoceroyl-CoA (empty triangles); panel H, in the presence of nervonic acid (filled circles) or nervonoyl-CoA (empty triangles). Each spectrum represents an average of 10 scans for a given representative spectrum from three replicates.
FIG. 7. Effect of BCFA and BCFA-CoA binding on PPARαΔAB conformation: Circular dichroic (CD) spectra.
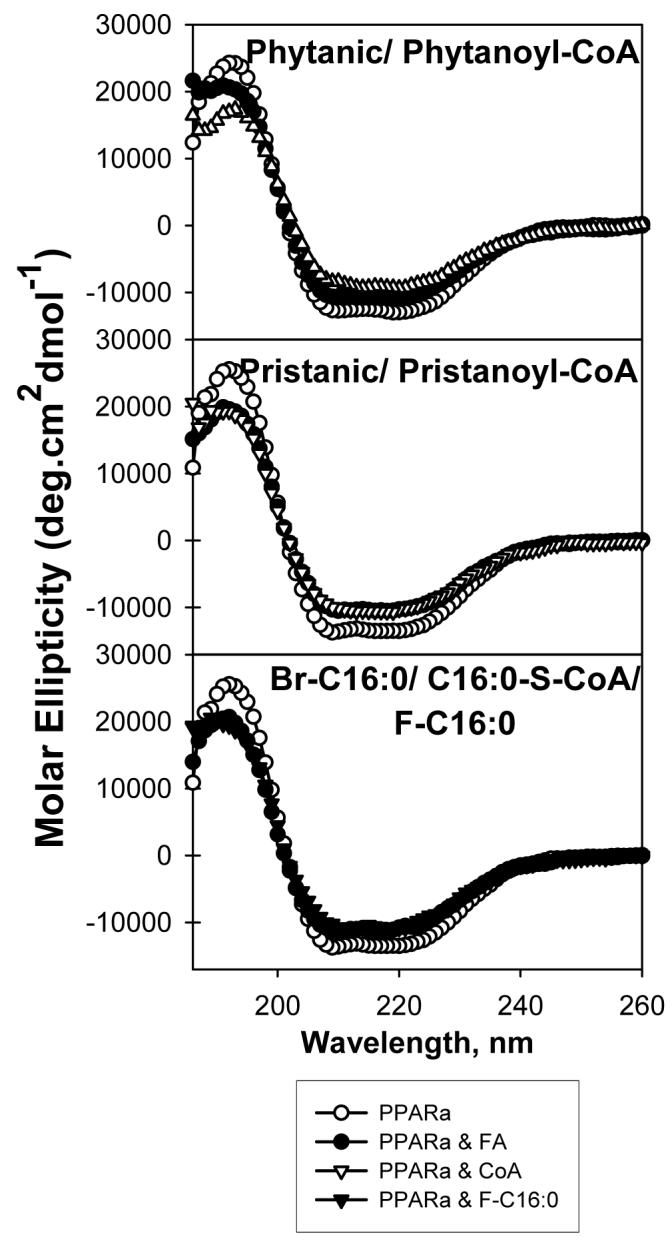
Far ultraviolet (UV) circular dichroic (CD) spectra of PPARαΔAB in the absence or presence of added ligand were obtained as described in the “Experimental Procedures”. Panel A, CD spectrum of PPARαΔAB in the absence (empty circles) and presence of added ligand: phytanic acid (filled circles) or phytanoyl-CoA (empty triangles); panel B, in the presence of pristanic acid (filled circles) or pristanoyl-CoA (empty triangles); panel C, in the presence of 2-bromopalmitic acid (filled circles), 2-fluoropalmitic acid (filled triangles) or S-hexadecyl-CoA (empty triangles). Each spectrum represents an average of 10 scans for a given representative spectrum from three replicates.
Table 2. Effect of very-long-chain, branched-chain, and non-metabolizable fatty acids and their CoA thioesters on the relative proportion of PPARαΔAB secondary structure determined by circular dichroism (CD).
The relative proportions of different types of secondary structure for PPARαΔAB in the absence and presence of added ligands was calculated as described in “Experimental Procedure”. These structures were as follows: H(r) represents regular α-helices, H(d) represents distorted α-helices, S(r) represents regular β-sheets, S(d) represents distorted β-sheets, Turns represent β-turns, and Unrd represents unordered structures. Asterisks represent significant differences between PPARαΔAB only and PPARαΔAB in the presence of added ligand
| Ligand | length:double bonds (position) | Hr (%) | Hd (%) | Sr (%) | Sd (%) | Turns (%) | Unordered (%) |
|---|---|---|---|---|---|---|---|
| PPARαΔAB | 24.2 ± 0.4 | 16.0 ± 0.1 | 8.0 ± 0.1 | 6.5 ± 0.1 | 18.7 ± 0.2 | 26. 6 ± 0.2 | |
| Arachidic acid Arachidoyl-CoA Arachidonic Acid Arachidonoyl-CoA Eicosapentaenoic Acid Eicosapentaenoyl-CoA |
20:0 20:0-CoA 20:4 (n-6) 20:4-CoA (n-6) 20:5 (n-3) 20:5-CoA (n-3) |
20.1 ± 0.4*** 21.5 ± 0.5*** 17.6 ± 0.6*** 17.4 ± 0.6*** 21.1 ± 0.4*** 22.1 ± 0.5** |
13.6 ± 0.1*** 14.3 ± 0.1*** 12.7 ± 0.3*** 13.0 ± 0.4*** 14.5 ± 0.1*** 14.6 ± 0.2*** |
10.4 ± 0.6*** 10.0 ± 0.5*** 12.0 ± 0.7*** 11.1 ± 0.6*** 10.0 ± 0.7*** 9.5 ± 0.3*** |
7.7 ± 0.1*** 7.2 ± 0.1*** 8.4 ± 0.3*** 8.5 ± 0.3*** 7.2 ± 0.1*** 7.3 ± 0.1*** |
20.1 ± 0.5** 19.3 ± 0.6 21.0 ± 0.7*** 21.5 ± 0.5*** 19.4 ± 0.6 19.2 ± 0.3 |
27.9 ± 0.5* 28.1 ± 0.8* 28.5 ± 0.8*** 28.8 ± 0.5*** 27.9 ± 0.7* 27.5 ± 0.5* |
| Behenic acid Behenoyl-CoA Docosapentaenoic acid Docosapentaenoyl-CoA Docosahexaenoic acid Docosahexaenoyl-CoA |
22:0 22:0 CoA 22:5 (n-3) 22:5-CoA (n-3) 22:6 (n-3) 22:6-CoA (n-3) |
24.6 ± 0.8 26.0 ± 1.1* 24.9 ± 0.7 22.6 ± 0.6* 24.3 ± 0.8 25.6 ± 0.8* |
16.1 ± 0.2 16.6 ± 0.3** 16.1 ± 0.2 15.3 ± 0.1*** 15.7 ± 0.2 16.6 ± 0.9 |
8.3 ± 0.2 8.5 ± 0.5 8.2 ± 0.1 8.9 ± 0.2** 9.0 ± 0.4* 8.7 ± 0.5* |
6.5 ± 0.1 5.7 ± 0.4** 6.7 ± 0.1 6.7 ± 0.1 6.6 ± 0.3 6.0 ± 0.6 |
19.0 ± 0.5 17.0 ± 0.9* 18.2 ± 0.5 18.9 ± 0.5 17.8 ± 0.7 16.9 ± 0.8** |
26.3 ± 0.3 26.0 ± 1.3 26.5 ± 0.3 27.3 ± 0.3 27.0 ± 0.5 26.3 ± 0.4 |
| Lignoceric acid Lignoceroyl-CoA Nervonic acid Nervonoyl-CoA |
24:0 24:0-CoA 24:1 (n-9) 24:1-CoA (n-9) |
23.4 ± 0.6 22.4 ± 0.6* 22.7 ± 0.6* 26.6 ± 0.6*** |
15.8 ± 0.2 15.2 ± 0.2*** 15.1 ± 0.1*** 17 ± 0.2*** |
8.5 ± 0.1* 9.1 ± 0.2*** 9.2 ± 0.2*** 6.5 ± 0.3*** |
6.8 ± 0.1 7.0 ± 0.1** 7.2 ± 0.1*** 5.8 ± 0.2** |
19.1 ± 0.5 19.3 ± 0.4 19.0 ± 0.5 17.7 ± 0.4* |
27.0 ± 0.3 27.2 ± 0.4 27 ± 0.3 26.2 ± 0.3 |
| Pristanic acid Pristanoyl-CoA Phytanic acid Phytanoyl-CoA |
15:tetramethyl 15:tetramethyl-CoA 16:tetramethyl 16:tetramethyl-CoA |
18.7 ± 0.4*** 18.2 ± 0.5*** 19.8 ± 0.5*** 16.1 ± 0.6*** |
13.5 ± 0.1*** 13.4 ± 0.1*** 13.9 ± 0.1*** 12.3 ± 0.3*** |
10.9 ± 0.6*** 11.9 ± 0.9*** 11.1 ± 0.6*** 13.0 ± 1.1*** |
7.7 ± 0.2*** 7.7 ± 0.3*** 7.7 ± 0.2*** 8.6 ± 0.3*** |
20.7 ± 0.5** 19.9 ± 0.8 19.8 ± 0.5* 21.5 ± 0.6*** |
28.8 ± 0.6*** 28.8 ± 0.9*** 27.6 ± 0.8 28.3 ± 0.7** |
| 2-Bromopalmitic acid 2-Fluoropalmitic acid S-hexadecyl-CoA |
Br-16:0 F-16:0 S-16:0-CoA |
20.0 ± 0.4*** 19.1 ± 0.5*** 16.9 ± 0.4*** |
13.9 ± 0.1*** 13.5 ± 0.1*** 12.6 ± 0.1*** |
10.3 ± 0.4*** 10.9 ± 0.5*** 12.1 ± 0.8*** |
7.7 ± 0.1*** 7.4 ± 0.2** 8.3 ± 0.2*** |
19.9 ± 0.4 19.9 ± 0.5 21.2 ± 0.6*** |
28.0 ± 0.8 29.2 ± 0.7*** 28.9 ± 0.8*** |
P<0.05
P<0.01
P<0.001
Addition of saturated VLCFA only minimally affected the structure of PPARα; as seen by the 20-carbon arachidic acid (Fig. 6A, filled circles), but not the 22-carbon behenic acid (Fig. 6D, filled circles) and 24-carbon lignoceric acid (Fig. 6G, filled circles). Only in the case of arachidic acid were PPARα’s molar ellipticity values at 207 and 221nm increased while concomitantly the values at 193nm decreased, indicating a reduction in PPARα α-helical content and an increase in β-sheets. Quantitative analysis of multiple CD spectra confirmed that the 20-carbon, but not the 22- or 24-carbon, saturated VLCFA significantly altered PPARα structure as shown by a 6.5% decrease in total α-helices (Hr and Hd) concomitant with 3.6% increases in total β-sheets (Sr and Sd), 1.4% increase in β-turns, and 1.3% increase in unordered structures (Table 2). Thus, with the exception of the 20-carbon arachidic acid the saturated VLCFA did not significantly alter PPARα secondary structure. Thus, these findings were entirely consistent with the relative affinities of PPARα for the respective saturated VLCFA: 20-carbon arachidic (modest binding/CD change) >>> 22-behenic acid (no binding/no CD change), 24-carbon lignoceric acid (no binding/no CD change).
Unsaturated 20-carbon VLCFA altered the PPARα secondary structure much more than the longer chain unsaturated VLCFA. For instance, the n-6 polyunsaturated 20-carbon fatty acid arachidonic acid (C20:4) (Fig. 6B, filled circles) significantly altered the shape of the circular dichroic spectrum of PPARα, increasing the molar ellipticity values at 207 and 221nm while concomitantly decreasing the values at 193nm; indicating less α-helical content and more β-sheets. Quantitative analysis of multiple CD spectra confirmed that the 20-carbon n-6 arachidonic acid (20:4) significantly altered PPARα structure as shown by a 9.9% decrease in total α-helices (Hr and Hd) concomitant with a 5.9% increase in total β-sheets (Sr and Sd), 2.3% increase in β-turns, and 1.9% increase in unordered structures (Table 2). These were the largest CD changes elicited by any of the polyunsaturated VLCFA in the order: n-6 20:4 (arachidonic acid, Fig. 6B filled circles, Table 2) > n-3 20:5 (eicosapentaenoic acid, Fig. 6C filled circles, Table 2) >>> n-3 22:5 (docosapentaenoic acid, Fig. 6E filled circles, Table 2), n-3 22:6 (docosahexaenoic acid, Fig. 6F filled circles, Table 2). The presence of a single double bond in the 24-carbon nervonic acid altered the CD spectrum of PPARα (Fig. 6H) to alter the proportions of secondary structures (Table 2) very modestly. In summary, with the exception of the 20-carbon VLCFA (20:0 arachidic acid; n-6 20:4 arachidonic acid; n-3 20:5 eicosapentaenoic acid), the longer chain saturated and polyunsaturated VLCFA did not or only slightly altered the secondary structure of PPARα. These data were not consistent with many of the free VLCFA directly being physiologically significant PPARα ligands.
Effect of Very Long Chain Fatty Acyl-CoAs on PPARα Secondary Structure: Circular Dichroism
In contrast to the free acids, the CoA thioesters of all the VLCFA altered the secondary structure of PPARα as shown by significant changes in CD spectra (Figs. 6A-H, empty triangles). This qualitative observation was confirmed by quantitative analysis of multiple replicates (Table 2). However, the nature of the circular dichroic alterations induced by the VLCFA-CoAs was distinct for each type of VLCFA-CoA as follows: 20-carbon (20:4-CoA, 20:5-CoA, 20:0-CoA), some 22-carbon (22:5-CoA), and some 24-carbon (24:0-CoA) decreased the proportion of PPARα α-helices, while others among the 22-carbon (22:0-CoA; 22:6-CoA) and 24-carbon (24:1-CoA) VLCFA-CoAs increased the proportion of PPARα α-helices (Table 2). The results showed for the first time, contrary to the free VLCFA, the CoA thioesters of all the VLCFA altered the structure of PPARα. However, the type of structure change (i.e. increase or decrease in PPARα’s proportion of α-helices) was highly dependent on the nature of the VLCFA-CoA. Again, since the polyunsaturated VLCFA (both n-6 and n-3) are known to activate PPARα in cells and animals (3), these data suggest that the respective VLCFA-CoAs, rather than the free acids, may be the active endogenous ligands that alter PPARα conformation.
Effect of Branched-chain Fatty Acids and Acyl-CoAs on PPARα Secondary Structure: Circular Dichroism
Since phytanic acid and even more so pristanic acid are the most potent known naturally-occurring PPARα transcriptional activators—much more potent than even the unsaturated VLCFA (rev. in (38-40)—and both the free acids and their CoA thioesters were bound with high affinity by PPARα (Table 1), it was predicted that both these branched-chain fatty acids would also elicit the largest alterations in PPARα secondary structure. Both phytanic acid (Fig. 7A, filled circles) and pristanic acid (Fig. 7B, filled circles) elicited significant changes in molar ellipticity of PPARα. Although similar effects were noted for the free acid (filled circles) and the CoA thioester form (open triangles), the effects of the CoA were much more pronounced, especially for phytanoyl-CoA. These results were consistent with phytanic acid and phytanoyl-CoA significantly reducing the relative proportions of α-helical structures by 6.5% and 11.8%, respectively, and increasing the proportion of β-sheets by 4.3% and 7.1%, respectively, with smaller increases in β-turns and unordered structures (Table 2). Pristanic acid and pristanoyl-CoA also reduced the relative proportions of PPARα α-helices by 8% and 8.6%, respectively, and increased the proportion of β-sheets by 4.1% and 5.1%, respectively, with smaller increases in β-turns and unordered structures (Table 2). In summary, the data indicated that phytanic acid did not need to be metabolized to pristanic acid to significantly alter PPARα structure. In addition, unlike most of the VLCFA, both the free acids and CoA thioesters were very potent in inducing PPARα structural changes. The phytanoyl-CoA induced changes in PPARα structure were the strongest changes reported for any of the endogenous PPARα ligands examined.
Effect of Non-metabolizable Fatty Acids and Non-hydrolyzable Fatty Acyl-CoAs on PPARα Secondary Structure: Circular Dichroism
Despite their relatively weak binding to PPARα, the non-metabolizable 2-bromopalmitic acid and 2-fluoropalmitic acid significantly altered the CD spectra of PPARα (Fig. 7C, filled circles, filled inverted triangles), resulting in a 6-7% decrease in total α-helices (Hr and Hd) concomitant with a 3-4% increases in total β-sheets (Sr and Sd), with smaller increases in β-turns and unordered structures (Table 2). Consistent with the possibility of added substituents to the LCFA altering saturated LCFA binding to PPARα, adding an NBD- group to stearic acid increased its binding by PPARα more than 100-fold (30). The fact that 2-bromopalmitic acid and 2-fluoropalmitic acid both resulted in almost identical CD changes and were exceptions to the correlation of binding affinity with CD change may be due to the addition of the bromo and fluoro groups altering size, pKa values, stereo-structure, or a variety of other factors. Although addition of either a bromo or fluoro group to palmitic acid resulted in weak binding by PPARα, addition of the larger bromo group elicited weaker affinity than addition of the smaller fluoro group. However, despite its weaker affinity, the bigger size of the bromo- group added to palmitate resulted in similar overall CD changes as elicited by 2-fluoropalmitic acid. Interestingly, the non-metabolizable S-hexadecyl-CoA elicited the largest changes in PPARα CD spectra (Fig. 7C, empty triangles), indicating that the ability of the fatty acyl-CoA to alter PPARα’s structure did not require hydrolysis to the free acid.
Effect of Endogenous Very-Long-Chain Fatty Acids on co-factor Recruitment to PPARα: Co-Immunoprecipitation
While it is generally accepted that ligand activation of a nuclear receptor stimulates recruitment and interaction of co-activators while decreasing interactions with co-repressors (rev. in (44), there are exceptions and new co-activators and co-repressors are continually being identified (rev. in (45). To begin to examine the effect of VLCFA on PPARα co-factor recruitment, co-immunoprecipitation with the PPARα antibody was used to test the ability of native PPARα to bind co-activators (Fig. 8; SRC-1, black bars; CBP, white bars) by precipitation of both proteins from mouse liver homogenate in the presence or absence of select representatives of the three groups of PPARα ligands examined above: (i) saturated and unsaturated VLCFAs; (ii) VLCFA-CoAs; (iii) BCFAs and BCFA-CoAs.
FIG. 8. Effect of select ligands on PPARα co-factor recruitment: Co-Immunoprecipitation.

Mouse liver homogenates were used to co-immunoprecipitate native PPARα protein with co-activators (SRC-1, black bars and CBP, white bars) in the presence of VLCFA (panel A), VLCFA-CoAs (panel B), and BCFA, BCFA-CoAs, 2-bromopalmitic acid, and S-hexadecyl-CoA (panel C). Protein values were standardized to PPARα protein levels and normalized for each ligand sample to the level of interaction observed in the absence of ligand (None), which was set equal to 1. Values represent mean protein quantities ± SE, n=4. Astericks (*) show significant differences between sample with ligand versus no ligand sample (P<0.05).
To test the effect of VLCFA which are poorly bound and only weakly alter structure of PPARα, the following were examined: saturated arachidic acid (C20:0), monounsaturated nervonic acid (C24:1), and polyunsaturated eicosapentaenoic acid (C20:5) and docosahexaenoic acid (C22:6). Among these only the polyunsaturated C22:6 caused any significant change to co-activator recruitment, and this resulted in a 2-fold increase in co-immunoprecipitation of SRC-1 (Fig. 8A). Since most of these ligands only weakly bound to PPARα, it was expected that there would be very little change to co-activator recruitment. However, the change in co-activator recruitment by the addition of C22:6 could not be correlated with high affinity binding of the free acid form of this ligand. Thus, the effect of the thioesters of these ligands was tested on co-activator recruitment.
Effect of Endogenous Very-Long-Chain Fatty Acyl-CoAs on Co-factor Recruitment to PPARα: Co-Immunoprecipitation
Although the preceding section showed the addition of C22:6 seemed to enhance activation, even though this ligand only weakly binds to PPARα, it must be considered that liver homogenates contain fatty acyl-CoA synthase activity (46,47). Therefore, the effect of the CoA thioesters of the above representative VLCFA on co-factor recruitment was examined. The saturated C20:0-CoA showed a 60% reduction in co-immunoprecipitation of both co-activators, while the monounsaturated C24:1-CoA resulted in a 50% decrease in CBP co-recruitment (Fig. 8B). Although the polyunsaturated C20:5-CoA did not result in any significant changes, the addition of C22:6-CoA increased co-immunoprecipitation of both co-activators by about 2.5-fold (Fig. 8B). The increase in co-activator recruitment of C22:6-CoA correlated with that of the free acid, although the effect was much stronger for the CoA thioester, suggesting that the effect of the free acid may be due to acylation of the free acid to its corresponding acyl-CoA. Further, the data suggests that saturated and monounsaturated VLCFA-CoAs may elicit repression of PPARα, while some polyunsaturated VLCFA-CoAs may induce strong activation, dependent upon the affinity of PPARα for each ligand. Since PPARα had a weaker affinity for C20:5-CoA (Table 1) than any of the other examined CoA thioesters, it was expected that this ligand would have a smaller effect on co-activator recruitment.
Effect of Branched-Chain Fatty Acids on Co-factor Recruitment to PPARα: Co-Immunoprecipitation
Since both the free acid form and the CoA thioesters of branched-chain fatty acids are high affinity PPARα ligands and alter PPARα conformation, the possibility that these ligands will also alter co-factor recruitment was examined. Phytanic acid significantly increased co-immunoprecipitation of SRC-1 from liver homogenates, approximately 2-fold, while no effect was seen on CBP (Fig. 8C). To determine if these effects might in part be due to the phytanoyl-CoA thioester, the studies were repeated with phytanoyl-CoA which decreased the co-immunoprecipitation of both co-activators more than 50% (Fig. 8C). Thus, as compared to the free phytanic acid, the phytanoyl-CoA exhibited opposite effects on co-activator recruitment. Thus, both the free acid and CoA thioester forms of the branched-chain phytanic acid alter co-factor recruitment, each in unique fashion.
Effect of Non-metabolizable Fatty Acids and Non-hydrolyzable Fatty Acyl-CoAs on Co-factor Recruitment to PPARα: Co-Immunoprecipitation
Since the effect of VLCFA in recruitment assays may be due to metabolism to their CoA thioesters (i.e. docosahexaenoic acid), the effect of the non-metabolizable fatty acids, 2-bromopalmitic acid and 2-fluoropalmitic acid, in co-activator recruitment was examined. No changes were noted to co-immunoprecipitation of either SRC-1 or CBP upon addition of either 2-bromopalmitic acid (Fig. 8C) or 2-fluoropalmitic acid (data not shown). However, addition of the non-hydrolyzable fatty acid analogue, S-hexadecyl-CoA, resulted in a 1.8-fold increase to SRC-1 and a 1.5-fold increase in CBP co-immunoprecipitation with PPARα. These data again substantiate the role of CoA thioesters as the active form of PPARα ligands.
DISCUSSION
Although many of the xenobiotics that undergo β-oxidation are PPARα activators, the nature of the endogenous ligands for PPARα is not completely resolved. While there is evidence indicating that VLCFA and BCFA are potent naturally-occurring peroxisome proliferators and may be PPARα ligands, it is not completely clear whether the unesterified VLCFA and BCFA directly, and/or metabolites thereof, represent the endogenous PPARα ligands/activators. For example, early transactivation assays show: (i) PPARα is activated by polyunsaturated VLCFA (C20:4, C20:5, C22:6), but not monounsaturated VLCFA (C24:1, C22:1) (3), (ii) n-3 polyunsaturated VLCFA elicited similar PPARα activation levels as n-6 polyunsaturated VLCFA (3). These findings of VLCFA being potential PPARα ligands were supported by gel-shift assays (14) and co-activator-dependent receptor ligand assays (15). Likewise, transactivation studies with branched-chain fatty acids also show PPARα activation by BCFA (11). It must be noted that most of these assays utilized very high concentrations of VLCFA (i.e. 30-100 μM) and BCFA (20 μM)--several orders of magnitude higher than concentrations of unesterified fatty acids detected in nuclei of living cells (16,17). These data suggested that affinities of PPARα for these ligands are very weak, i.e. Kds in the μM range, much lower than the concentrations of free fatty acids in nuclei of living cells (16,17), and thus the free acid forms of the VLCFA and BCFA may not represent the endogenous ligands. Since unesterified fatty acids are rapidly activated/metabolized to their CoA thioesters in cells and tissues (rev. (18-20), metabolites such as the CoA thioesters may represent endogenous PPARα ligands. Unfortunately, there is almost no data resolving whether VLCFA, BCFA, and/or their CoA thioesters exhibit the classic hallmarks of ligands that activate a nuclear receptor such as PPARα: (i) high affinity binding (i.e. nM Kds, in the range of nuclear concentrations of these ligands), (ii) alter conformation of the nuclear receptor, and (iii) alter co-factor recruitment to the nuclear receptor. The data presented herein help to clarify our understanding of these issues.
First, the current work presented evidence that the free acid forms of BCFA and only one free acid form of VLCFA bind to PPARα with high affinity. PPARα bound phytanic acid, pristanic acid, and some 20-carbon VLCFA with Kds of 19-76nM, well within the physiological range of free fatty acids detected in the nucleoplasm of living cells (16,17). Further, these data showed for the first time that phytanic acid did not need to be metabolized to pristanic acid to be a high affinity PPARα ligand. Both phytanic acid and pristanic acid, but not shorter chain metabolites, are the highly potent naturally-occurring PPARα activators in transactivation assays (rev. in (38-40) and in animals (12,13,48). The Kd values for both BCFA and 20-carbon VLCFA (arachidic acid, arachidonic acid) were in the same range as PPARα exhibited for unsaturated long chain fatty acids (LCFA) and peroxisome proliferators (bezafibrate) (29,30)—suggesting that BCFA, arachidic acid, and arachidonic acid are high affinity PPARα ligands that may be physiologically significant endogenous ligands.
Second, in contrast to the free acid forms, nearly all of the examined BCFA-CoAs and VLCFA-CoAs bind to PPARα with high affinities (i.e. 2-30nM Kds). This was not due to hydrolysis of bound CoA thioesters to the respective free acids since: (i) most of the free acid forms of VLCFA were not or only weakly bound by PPARα; (ii) PPARα bound the non-hydrolyzable S-hexadecyl-CoA with very high affinity (i.e. 10nM Kd) similar to that observed for the VLCFA-CoAs and BCFA-CoAs. Although nuclear fatty acyl-CoA concentration has been estimated to be less than 10nM, early methodology made it difficult to accurately measure acyl-CoA concentration in organelles (49). More recent data with fluorescent fatty acyl-CoAs in living cells has confirmed the earlier prediction that nuclearplasmic concentrations of fatty acyl-CoAs were indeed less than 10nM (17). However, this concentration can be increased several-fold by expression of cytoplasmic fatty acyl-CoA binding proteins (49,50). Thus, the CoA thioesters of VLCFA and BCFA are nearly all bound with affinities in the physiological range of fatty acyl-CoA concentrations in the nucleoplasm of living cells. The Kd values for all of the examined VLCFA-CoAs and BCFA-CoAs were in the same range as PPARα exhibited for long-chain fatty acyl-CoAs (LCFA-CoAs) and peroxisome proliferator drug-CoA thioesters (22,30). The finding that PPARα exhibits very high affinity (3-29nM Kds) for VLCFA-CoAs may help to explain the effects of n-3 and n-6 fatty acid supplementation on PPARα transactivation. In transactivation assays wherein cultured cells are supplemented with exogenous VLCFA, both n-3 and n-6 fatty acids enhance PPARα transactivation nearly equally well (3,14,15,40,51-53). Since PPARα exhibits high affinity (low nM Kds) for n-6 unsaturated fatty acids (29,30), as well as their CoA derivatives (30), either or both these ligands could contribute to PPARα activation. In contrast, since PPARα exhibits high affinity (low nM Kds) for n-3 fatty acyl-CoAs, but not for n-3 fatty acids, the CoA thioesters formed by intracellular fatty acyl-CoA synthases from dietary n-3 fatty acids may represent the high affinity endogenous ligands accounting for PPARα transactivation by these VLCFA. Thus, the ligand binding data obtained herein may reconcile the PPARα transactivation data obtaining similar transactivation effects with added n-3 and n-6 fatty acids. Taken together, these data suggest that VLCFA-CoAs and BCFA-CoAs are high affinity PPARα ligands and thus represent endogenous ligands. These data suggest that the VLCFA-CoAs and BCFA-CoAs, as well as some 20-carbon VLCFA and both BCFA, satisfy the high affinity criterion of physiologically significant ligands for PPARα.
Third, all the high affinity PPARα ligands (VLCFA-CoAs, BCFA-CoAs, BCFA, and some 20-carbon VLCFA) significantly altered the structure of PPARα. This was evidenced by two observations: (i) Binding of these ligands significantly quenched the PPARα aromatic amino acid fluorescence emission, consistent with a reorientation of one or more of these residues from the more hydrophobic interior of the protein to the more aqueous exposed protein surface. (ii) These ligand-induced structural changes in PPARα were confirmed by circular dichroism, which resulted in molar ellipticity changes in the presence of compounds demonstrated to bind with high affinity (low nM Kds) to PPARα. Circular dichroic spectra were unaltered in the presence of compounds that did not bind to PPARα. Interestingly, two types of ligand-induced PPARα conformational changes were observed; (i) The 20-carbon fatty acids (arachidic, arachidonic) and 20-carbon acyl-CoAs (arachidoyl-CoA, arachidonoyl-CoA), 22-carbon acyl-CoA (docosapentaenoyl-CoA), 24-carbon saturated acyl-CoA (lignoceroyl-CoA), 24-carbon monounsaturated fatty acid (nervonic acid), BCFA, and BCFA-CoAs produced an overall decrease in PPARα content of α-helices and a concomitant increase in β-sheets, similar to those obtained previously for LCFA-CoAs and unsaturated LCFA (30). In contrast, binding of a 22-carbon saturated acyl-CoA (behenoyl-CoA), 22-carbon n-3 polyunsaturated acyl-CoA (docosahexaenoyl-CoA), and a 24-carbon monounsaturated CoA (nervonoyl-CoA) increased the proportion of α-helices and concomitantly decreased β-sheets in PPARα. These findings with circular dichroism suggest that the high affinity VLCFA, VLCFA-CoAs, BCFA, BCFA-CoAs, and non-hydrolyzable S-hexadecyl-CoA all significantly altered the secondary structure of PPARα--thereby satisfying the second criterion that these are physiologically significant ligands for PPARα.
Fourth, the current work presented data demonstrating that high affinity ligands altered co-factor recruitment in a highly selective manner. The effect of ligands on co-activator (SRC-1, CBP) association with PPARα was examined by co-immunoprecipitation from liver homogenates using an anti-PPARα monoclonal antibody. The data demonstrated that most of the high affinity ligands significantly altered co-immunoprecipitation of at least one, if not both, of the co-activators, while most of the low affinity ligands did not. An interesting exception was docosahexaenoic acid which increased co-activator recruitment even though this ligand only weakly bound to PPARα. Furthermore, these data agree with the transactivation data indicating that addition of docosahexaenoic acid or phytanic acid to the medium(11) increases PPARα activation, while the addition of monounsaturated fatty acids does not (3). A potential explanation for the co-activator recruitment and activation activity of dietary docosahexaenoic acid may come from the observation that the CoA thioester of docosahexaenoic acid was both tightly bound by and increased co-activator recruitment to PPARα. This suggests that activation of PPARα in the presence of VLCFA such as docosahexaenoic acid may arise from endogenous fatty acyl-CoA synthase activity present in the homogenates and cells. Liver homogenates and hepatocytes represent a complex milieus wherein multiple endogenous fatty acyl-CoA synthases can convert the VLCFA and/or BCFA to their CoA thioesters (46,47). These data suggest that the VLCFA-CoAs and BCFA-CoAs, as well as some 20-carbon VLCFA and both BCFA, satisfy the co-factor recruitment criterion of physiologically significant ligands for PPARα.
With regards to ligand-induced conformational change in PPARα and co-factor recruitment, unique patterns emerged. While the VLCFA-CoAs were all high affinity PPARα ligands, the dissimilar circular dichroic changes obtained by the addition of behenoyl-CoA, docosahexaenoyl-CoA, and nervonoyl-CoA, as compared to the other high affinity PPARα ligands, suggested that the binding of these three ligands would elicit different co-factor recruitment than the other examined ligands. Indeed, nervonoyl-CoA binding seemed to decrease co-activator recruitment, similar to arachidoyl-CoA binding. However, docosahexaenoyl-CoA binding increased co-activator recruitment, similar to that seen for other PPARα ligands (29).
In conclusion, PPARα binds with highest affinities the VLCFA-CoAs and BCFA-CoAs, slightly weaker affinities the free BCFA and several 20-carbon free VLCFA (arachidic acid, arachidonic acid), but not longer chain free VLCFA. The high affinities (i.e. very low nM Kds) were in the range of fatty acyl-CoA concentrations observed in nucleoplasm of living cells (16,17). Furthermore, the high affinity binding of these ligands correlated with ligand-induced conformational changes in PPARα. Finally, high affinity ligand binding and ligand-induced conformational changes in general, elicited one or more alterations in co-activator recruitment. These data significantly added to our understanding of the action of VLCFA and BCFA on PPARα.
Abbreviations
- PPAR
peroxisome proliferator-activated receptor
- RXR
retinoid X receptor
- PPRE
peroxisome proliferator response element
- VLCFA
very-long-chain fatty acids
- BCFA
branched-chain fatty acids
- CoA
coenzyme A
- AOX
acyl-CoA oxidase
- PBE
bifunctional enzyme
- PTL
3-ketoacyl-CoA thiolase
- SCP-x
sterol carrier protein-x
- LCFA-CoA
long chain fatty acyl-CoA
- PPARαΔAB
peroxisome proliferator activated receptor α comprised of amino acids 101-468 (i.e. missing only the amino-terminal A/B domain)
- SRC-1
steroid receptor co-activator-1
- CBP
cyclic AMP-responsive enhancer binding protein (CREB) binding protein
- CD
circular dichroism
- HPLC
high performance liquid chromatography
- RAR
retinoic acid receptor
- LXR
liver X receptor
Footnotes
Acknowledgments: This work was supported in part by the USPHS National Institutes of Health DK41402 (FS and AK) and NIH National Research Service Award DK066732 (HAH).
REFERENCES
- 1.Hihi AK, Michalik L, Wahli W. PPARs: transcriptional effectors of fatty acids and their derivatives. Cell. Mol. Life Sci. 2002;59:645–650. doi: 10.1007/s00018-002-8467-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu HE, Lambert MH, Montana VG, Plunket KD, Moore LB, Collins JL, Oplinger JA, Kliewer SA, Gampe RT, McKee DD, Moore JT, Willson TM. Structural determinants of ligand binding slectivity between the peroxisome proliferator-activated receptors. Proc. Natl. Acad. Sci. 2001;98:13919–13924. doi: 10.1073/pnas.241410198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keller H, Dreyer C, Medin J, Mahfoudi A, Ozato K, Wahli W. Fatty acids and retinoids control lipid metabolism through activation of peroxisome proliferator activated receptor-retinoid X receptor heterodimers. Proc. Natl. Acad. Sci. 1993;90:2160–2164. doi: 10.1073/pnas.90.6.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desvergne B, Wahli W. Peroxisome proliferator activated receptors: nuclear control of metabolism. Endocrine Rev. 1999;20:649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 5.Francis GA, Fayard E, Picard F, Auwerx J. Nuclear receptors and the control of metabolism. Annu. Rev. Physiol. 2003;65:261–311. doi: 10.1146/annurev.physiol.65.092101.142528. [DOI] [PubMed] [Google Scholar]
- 6.Fan C-Y, Pan J, Chu R, Lee D, Kluckman KD, Usuda N, Singh I, Yeldandi AV, Rao MS, Maeda N, Reddy JK. Hepatocellular and hepatic peroxisomal alterations in mice with a disrupted peroxisomal fatty acyl-coenzyme A oxidase gene. J. Biol. Chem. 1996;271:24698–24710. doi: 10.1074/jbc.271.40.24698. [DOI] [PubMed] [Google Scholar]
- 7.Fourcade S, Savary S, Albet S, Gauthe D, Gondcaille C, Pineau T, Bellenger J, Bentejac M, Holzinger A, Berger J, Bugaut M. Fibrate induction of the adrenoleukodystrophy-related gene (ABCD2). Promoter analysis and role of the peroxisome proliferator activated receptor PPARalpha. Eur. J. Biochem. 2001;268:49319–49325. doi: 10.1046/j.1432-1327.2001.02249.x. [DOI] [PubMed] [Google Scholar]
- 8.Idel S, Ellinghaus P, Wolfrum C, Nofer J-R, Gloerich J, Assmann G, Spener F, Seedorf U. Branched chain fatty acids induce nitric oxide-dependent apoptosis in vascular smooth muscle cells. J. Biol. Chem. 2002;277:49319–49325. doi: 10.1074/jbc.M204639200. [DOI] [PubMed] [Google Scholar]
- 9.Hashimoto T. Peroxisomal beta-oxidation enzymes. Neurochem. Res. 1999;24:551–563. doi: 10.1023/a:1022540030918. [DOI] [PubMed] [Google Scholar]
- 10.Reddy JK, Hashimoto T. Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu. Rev. Nutr. 2001;21:193–230. doi: 10.1146/annurev.nutr.21.1.193. [DOI] [PubMed] [Google Scholar]
- 11.Zomer AWM, van der Burg B, Jansen GA, Wanders RJA, Poll-The BT, van der Saag PT. Pristanic acid and phytanic acid: naturally occurring ligands for the nuclear receptor peroxisome proliferator activated receptor alpha. J. Lipid Res. 2000;41:1801–1807. [PubMed] [Google Scholar]
- 12.Atshaves BP, Payne HR, McIntosh AL, Tichy SE, Russell D, Kier AB, Schroeder F. Sexually dimorphic metabolism of branched chain lipids in C57BL/6J mice. J. Lipid Res. 2004;45:812–830. doi: 10.1194/jlr.M300408-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Atshaves BP, McIntosh AL, Payne HR, Mackie J, Kier AB, Schroeder F. Effect of branched-chain fatty acid on lipid dynamics in mice lacking liver fatty acid binding protein gene. Am. J. Physiol. 2005;288:C543–C558. doi: 10.1152/ajpcell.00359.2004. [DOI] [PubMed] [Google Scholar]
- 14.Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc. Natl. Acad. Sci. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krey G, Braissant O, L’Horset F, Kalkhoven E, Perroud M, Parker MG, Wahli W. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol. Endocrinology. 1997;11:779–791. doi: 10.1210/mend.11.6.0007. [DOI] [PubMed] [Google Scholar]
- 16.Huang H, Starodub O, McIntosh A, Kier AB, Schroeder F. Liver fatty acid binding protein targets fatty acids to the nucleus: real-time confocal and multiphoton fluorescence imaging in living cells. J. Biol. Chem. 2002;277:29139–29151. doi: 10.1074/jbc.M202923200. [DOI] [PubMed] [Google Scholar]
- 17.Huang H, Starodub O, McIntosh A, Atshaves BP, Woldegiorgis G, Kier AB, Schroeder F. Liver fatty acid binding protein colocalizes with peroxisome proliferator receptor alpha and enhances ligand distribution to nuclei of living cells. Biochemistry. 2004;43:2484–2500. doi: 10.1021/bi0352318. [DOI] [PubMed] [Google Scholar]
- 18.McArthur MJ, Atshaves BP, Frolov A, Foxworth WD, Kier AB, Schroeder F. Cellular uptake and intracellular trafficking of long chain fatty acids. J. Lipid Res. 1999;40:1371–1383. [PubMed] [Google Scholar]
- 19.Gossett RE, Frolov AA, Roths JB, Behnke WD, Kier AB, Schroeder F. Acyl Co A binding proteins: multiplicity and function. Lipids. 1996;31:895–918. doi: 10.1007/BF02522684. [DOI] [PubMed] [Google Scholar]
- 20.Powell GL, Tippett PS, Kiorpes TC, McMillin-Wood J, Coll KE, Schultz H, Tanaka K, Kang ES, Shrago E. Fatty acyl CoA as an effector molecule in metabolism. Fed. Proc. 1985;44:81–84. [Google Scholar]
- 21.Berge R, Stensland E, Aarsland A, Tsegai G, Osmundsen H, Aarsaether N, Gjellesvik DR. Induction of cytosolic clofibroyl-CoA hydrolase activity in liver of rats treated with clofibrate. Biochim. Biophys. Acta. 1987;918:60–66. doi: 10.1016/0005-2760(87)90009-9. [DOI] [PubMed] [Google Scholar]
- 22.Aarsland A, Berge RK. Peroxisome proliferating sulphur- and oxy-substituted fatty acid analogues are activated to acyl coenzyme A thioesters. Biochem. Pharmacol. 1991;41:53–61. doi: 10.1016/0006-2952(91)90010-3. [DOI] [PubMed] [Google Scholar]
- 23.Fan C-Y, Pan J, Usuda N, Yeldandi AV, Rao MS, Reddy JK. Steatohepatitis, spontaneous peroxisome proliferation and liver tumors in mice lacking peroxisomal fatty acyl-CoA oxidase. J. Biol. Chem. 1998;273:15639–15645. doi: 10.1074/jbc.273.25.15639. [DOI] [PubMed] [Google Scholar]
- 24.Qi C, Zhu YJ, Pan J, Usuda N, Maeda N, Yeldandi AV, Rao MS, Hashimoto T, Reddy JK. Absence of spontaneous peroxisome proliferation in enoyl-CoA hydratase/L-3-hydroxylacyl-CoA dehydrogenase-deficient mouse liver-further support for the role of fatty acyl CoA oxidase in PPAR alpha ligand metabolism. J. Biol. Chem. 1999;274:15775–15780. doi: 10.1074/jbc.274.22.15775. [DOI] [PubMed] [Google Scholar]
- 25.Chevillard G, Clemencet M-C, Latruffe N, Nicolas-Frences V. Targeted disruption of the peroxisomal thiolase B gene in mouse: a new model to study disorders related to peroxisomal lipid metabolism. Biochemie. 2004;86:849–856. doi: 10.1016/j.biochi.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 26.Escher P, Wahli W. Peroxisome proliferator activated receptors: insights into multiple cellular functions. Mutat. Res. 2000;448:121–138. doi: 10.1016/s0027-5107(99)00231-6. [DOI] [PubMed] [Google Scholar]
- 27.Hubbell T, Behnke WD, Woodford JK, Schroeder F. Recombinant liver fatty acid binding protein interactions with fatty Acyl-Coenzyme A. Biochemistry. 1994;33:3327–3334. doi: 10.1021/bi00177a025. [DOI] [PubMed] [Google Scholar]
- 28.Kawaguchi A, Yohmura T, Okuda S. A new method for the preparation of acyl-CoA thioesters. J. Biochem. 1981;89:337–339. doi: 10.1093/oxfordjournals.jbchem.a133207. [DOI] [PubMed] [Google Scholar]
- 29.Lin Q, Ruuska SE, Shaw NS, Dong D, Noy N. Ligand selectivity of the peroxisome proliferator-activated receptor α. Biochem. 1999;38:185–190. doi: 10.1021/bi9816094. [DOI] [PubMed] [Google Scholar]
- 30.Hostetler HA, Petrescu AD, Kier AB, Schroeder F. Peroxisome proliferator activated receptor alpha (PPARα) interacts with high affinity and is conformationally responsive to endogenous ligands. J. Biol. Chem. 2005;280:18667–18682. doi: 10.1074/jbc.M412062200. [DOI] [PubMed] [Google Scholar]
- 31.Frolov A, Cho TH, Murphy EJ, Schroeder F. Isoforms of rat liver fatty acid binding protein differ in structure and affinity for fatty acids and fatty acyl CoAs. Biochemistry. 1997;36:6545–6555. doi: 10.1021/bi970205t. [DOI] [PubMed] [Google Scholar]
- 32.Thompson J, Winter N, Terwey D, Bratt J, Banaszak L. The crystal structure of the liver fatty acid-binding protein. J. Biol. Chem. 1997;272:7140–7150. doi: 10.1074/jbc.272.11.7140. [DOI] [PubMed] [Google Scholar]
- 33.Xu HE, Lambert MH, Parks VG, Blanchard SG, Lehmann JM, Wisely GB, Wilson TM, Kliewer SA, Milburn MV. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol. Cell. 1999;3:397–406. doi: 10.1016/s1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
- 34.Cronet P, Peterson JFW, Folmer R, Blomberg N, Sjoblom K, Karlsson U, Lindstedt E-L, Bamberg K. Structure of the PPARalpha and gamma ligand binding domain in complex with AZ 242; ligand selectivity and agonist activation in the PPAR family. Structure. 2001;9:699–706. doi: 10.1016/s0969-2126(01)00634-7. [DOI] [PubMed] [Google Scholar]
- 35.Petrescu AD, Hertz R, Bar-Tana J, Schroeder F, Kier AB. Ligand specificity and conformational dependence of the hepatic nuclear factor-4alpha (HNF-4α) J. Biol. Chem. 2002;277:23988–23999. doi: 10.1074/jbc.M201241200. [DOI] [PubMed] [Google Scholar]
- 36.Vorum H, Brodersen R, Kragh-Hansen A, Pedersen AO. Solubility of long chain fatty acid in phosphate buffer at pH 7.4. Biochem. Biophys. 1992;1126:135–142. doi: 10.1016/0005-2760(92)90283-2. [DOI] [PubMed] [Google Scholar]
- 37.Sreerama N, Woody R. Estimation of protein secondary structure from circular dichroism spectra; Comparison of CONTIN, SELCON, and DCSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 38.Adida A, Spener F. Intracellular lipid binding proteins and nuclear receptors inolved in branched-chain fatty acid signaling. Prost. Leukot. Essen. Fatty Acids. 2002;67:91–98. doi: 10.1054/plef.2002.0404. [DOI] [PubMed] [Google Scholar]
- 39.Hanhoff T, Wolfrum C, Ellinghaus P, Seedorf U, Spener F. Pristanic acid is activator of PPARalpha. Eur. J. Lipid Sci. 2001;103:75–80. [Google Scholar]
- 40.Wolfrum C, Ellinghaus P, Fobker M, Seedorf U, Assmann G, Borchers T, Spener F. Phytanic acid is ligand and transcriptional activator of murine liver fatty acid binding protein. J. Lipid Res. 1999;40:708–714. [PubMed] [Google Scholar]
- 41.Petrescu A, Huang H, Hertz R, Bar-Tana J, Schroeder F, Kier AB. Role of regulatory F-domain in hepatocyte nuclear factor-4alpha ligand specificity. J. Biol. Chem. 2005;280:16714–16727. doi: 10.1074/jbc.M405906200. [DOI] [PubMed] [Google Scholar]
- 42.Schroeder F, Huang H, Hostetler HA, Petrescu AD, Hertz R, Bar-Tana J, Kier AB. Stability of fatty acyl CoA thioester ligands of hepatocyte nuclear factor -4alpha and peroxisome proliferator-activated receptor alpha. Lipids. 2005;40:559–568. doi: 10.1007/s11745-005-1416-y. [DOI] [PubMed] [Google Scholar]
- 43.Dowell P, Peterson VJ, Zabriskie TM, Leid M. Ligand-induced peroxisome proliferator-activated receptor α conformational change. J. Biol. Chem. 1997;272:2013–2020. doi: 10.1074/jbc.272.3.2013. [DOI] [PubMed] [Google Scholar]
- 44.Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol. Rev. 2001;81:1269–1304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- 45.Perissi V, Rosenfeld MG. Controlling nuclear receptors: the circular logic of cofactor cycles. Mol. Cell. Biol. 2005;6:542–554. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- 46.Lewin TM, Kim J-H, Granger DA, Vance JE, Coleman RA. Acyl CoA synthetase isoforms 1, 4, and 5 are present in different subcellular membranes in rat liver and can be inhibited independently. J. Biol. Chem. 2001;276:24674–24679. doi: 10.1074/jbc.M102036200. [DOI] [PubMed] [Google Scholar]
- 47.Bronfman M, Inestrosa NC, Nervi F, Leighton F. Acyl CoA synthetase and the peroxisomal enzymes of beta-oxidation in human liver: Quantitative analysis of their subcellular localization. Biochem. J. 1984;224:709–720. doi: 10.1042/bj2240709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ellinghaus P, Wolfrum C, Assmann G, Spener F, Seedorf U. Phytanic acid activates the peroxisome proliferator-activated receptor alpha (PPARalpha) in sterol carrier protein-2-/sterol carrier protein x-deficient mice. J. Biol. Chem. 1999;274:2766–2772. doi: 10.1074/jbc.274.5.2766. [DOI] [PubMed] [Google Scholar]
- 49.Faergeman NJ, Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem. J. 1997;323:1–12. doi: 10.1042/bj3230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knudsen J, Jensen MV, Hansen JK, Faergeman NJ, Neergard T, Gaigg B. Role of acyl CoA binding protein in acyl CoA transport, metabolism, and cell signaling. Mol. Cell. Biochem. 1999;192:95–103. doi: 10.1007/978-1-4615-4929-1_11. [DOI] [PubMed] [Google Scholar]
- 51.Kliewer SA, Sundseth SS, Jones SA, Brown PJ, Wisely GB, Koble CS, Devchand P, Wahli W, Willson TM, Lenhard JM, Lehmann JM. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and y. Proc. Natl. Acad. Sci. 1997;94:4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc. Natl. Acad. Sci. 1999;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Banner CD, Gottlicher M, Widmark E, Sjovall J, Rafter JJ, Gustafsson J. A systematic analytical chemistry/cell assay approach to isolate activators of orphan nuclear receptors from biological extracts: characterization of peroxisome proliferator-activated receptor activators in plasma. J. Lipid Res. 1993;34:1583–1591. [PubMed] [Google Scholar]