Abstract
Background & Aims
The Raf kinase inhibitor protein (RKIP) has been identified as a suppressor of the mitogen-activated protein kinase (MAPK) pathway. Loss of RKIP function promotes tumor metastasis in prostate cancer and melanoma. The IGF-I mediated MAPK cascade is often activated in hepatocellular carcinoma (HCC), but the role of RKIP in the molecular pathogenesis of these tumors is unknown. This study was performed to evaluate the role of RKIP in HCC development.
Methods
The levels of RKIP expression in HCC tumor and corresponding peritumoral tissues were determined by immunohistochemistry and Western blot analysis. The underlying mechanisms of RKIP were assessed with immunoblot analysis, Raf kinase activity assay, cell proliferation and migration assays after either overexpression or knockdown of RKIP expression in HCC cell lines.
Results
RKIP expression is downregulated in human HCC compared to adjacent peritumoral tissues. Low RKIP levels were correlated with enhanced extracellular-signal-regulated-kinase (ERK)/MAPK pathway activation. Reconstitution experiments antagonized IGF-I mediated MAPK pathway activation resulting in reduced nuclear accumulation of phospho-ERK. In contrast, knockdown of RKIP expression using siRNA induced activation of the ERK/MAPK pathway. Ectopic expression of RKIP altered HCC cell proliferation and migration.
Conclusions
Our findings indicate that downregulation of RKIP expression is a major factor in activation of the IGF-I/ERK/MAPK pathway during human hepatocarcinogenesis.
Introduction
Hepatocellular carcinoma (HCC) accounts for 80–90% of primary liver tumors, and is one of the most common and devastating malignant diseases worldwide. The major risk factors for the development of HCC are chronic hepatitis B or C infection.1,2 Tumor development is associated with the failure of coordinated responses to growth factors and cytokines, which lead to an impaired balance of the proliferation-apoptosis process. Therefore, the deregulated expression of growth factors and cytokines may be important contributors to this mutistep process,3–6 of which insulin-like growth factors (IGF-I and II) appear to play a key role.7 One study reports altered IGF signaling in 90% of HCC, including the autocrine production of IGFs, IGF binding proteins (IGFBPs), IGFBP proteases, and IGF receptors expression.8 The binding of IGF-I to the extracellular domain of IGF-I receptor (IGF-IR) induces a conformational change that results in auto-phosphorylation of the receptor converting to the active form. This event triggers the initiation of multiple downstream signaling pathways including the MAPK and phosphatidylinositol 3’-kinase (PI3-K) signaling cascades, that result in cellular proliferation, transformation, and inhibition of apoptosis.9–11
The mitogen-activated protein kinase (MAPK) signaling pathways are highly conserved and involved in cell growth, differentiation, survival, and invasion.12,13 There are three major MAPK pathways: the extracellular-signal-regulated kinases (ERKs); the c-Jun N-terminal kinase (JNK or SAPK1); and p38 MAPK (SAPK2/RK). In general, ERK1/2 are the key transducers of proliferation signals and are often activated by mitogens. In contrast, SAPK/JNK and p38 are poorly stimulated by mitogens but strongly activated by cellular stress. Many different growth factor receptors, including insulin receptor and IGF-IR, activate the ERK/MAPK pathway through the small G protein Ras, which consequently binds Raf-1 kinase and thereby recruits Raf-1 to the inner surface of the cell membrane. After this event, Raf-1 phosphorylates MEK, which in turn phosphorylates and activates ERK. Phosphorylated ERK translocates into the nucleus and regulates gene expression via interaction with various transcription factors such as CREB, AP-1, Ets, and c-Myc.14 It has been shown that this pathway is activated in many malignant tumors including HCC.15–17 Moreover, activation of this pathway confers a chemoresistance phenotype and induces rapid tumor cell proliferation. Interruption of this cascade may increase drug sensitivity and promote apoptosis.14,18,19
The Raf kinase inhibitor protein (RKIP) was identified as an inhibitor of the MAPK signaling pathway.20–25 The RKIP is a conserved cytosolic protein with wide tissue expression and does not share significant homology with other kinase inhibitors.26,27 Yeung et al.24,25 showed that RKIP directly interacts with both Raf-1 and MEK, and disrupts the Raf-1/MEK interaction, thereby preventing the activation of MEK and downstream components of the signaling cascade. Overexpression of RKIP suppressed MAPK signaling and down-regulation of RKIP had the opposite effect.
Aberrant RKIP expression may play a critical role in the malignant process.28–30 Down-regulation of RKIP was observed in metastatic prostate as compared to non-metastatic cell lines. Immunohistochemical staining showed moderate to high RKIP expression in normal prostate and primary prostate tumors, whereas low or undetectable levels were observed in metastatic foci. Restoration of RKIP reduced spontaneous lung metastasis.31,32 These observations define RKIP as one of thirteen known metastasis suppressor genes.33 Schuierer et al.22 also reported that significant down-regulation of RKIP has been found in melanoma cell lines in comparison with normal melanocytes. Stable transfection of melanoma cells with RKIP revealed significant inhibition of invasiveness in vitro.
Although the molecular mechanism by which RKIP inhibits the ERK/MAPK signaling pathway has been partially delineated, little is known about the role of RKIP in human hepatocarcinogenesis and how RKIP may be regulated in HCC cells. In the present investigation, we demonstrate that the level of RKIP expression is a critical factor for ERK/MAPK signaling in HCC, and defines a molecular mechanism involved in HCC growth.
Materials and Methods
HCC Tissues and Cell Lines
Seventeen discarded HCC tumor tissues were obtained at the time of surgery and were frozen in liquid nitrogen within 30 minutes of excision Histologic confirmation of both tumor and corresponding peritumoral tissues was performed. Human HCC cell lines FOCUS, Huh7, Hep3B, and HepG2 were grown in Eagle’s Minimal Essential Medium (MEM) with 10% fetal bovine serum (FBS), L-glutamine (Life Technologies, Gaithersburg, MD), and MEM nonessential amino acids (Sigma, St. Louis, MO).
Immunohistochemistry
Formalin-fixed, paraffin-embedded tumor and adjacent uninvolved peritumoral tissue sections were screened for RKIP protein expression by immunohistochemistry. Sections were deparafinized, rehydrated, and subsequently incubated with anti-RKIP antibody (Upstate Biotechnology, Lake Placid, NY) overnight at 4°C. Biotin-labeled anti-rabbit IgG was incubated for 30 minutes at room temperature; detection of immunoreactivity was by the streptavidin–biotin method using horseradish peroxidase and diaminobenzidine as the chromogen (Histostain Kit; Zymed, San Francisco, CA). Sections were counterstained using hematoxylin (Zymed) and examined under a light microscopy. Immunostaining intensity was scored as negative (−), weak (+), moderate (++), or strong (+++) by 2 independent observers.
Quantitative Real-Time RT-PCR Assay
Total RNA was extracted from the 17 pairs of liver tissues or HCC cell lines using TRIzol® Reagent (Invitrogen™, Carlsbad, CA). First-strand complementary DNA (cDNA) was synthesized from 250 ng of total RNA using First Strand cDNA Synthesis Kit for reverse-transcription polymerase chain reaction (RT-PCR) (AMV) (Roche Diagnostics, Indianapolis, IN). To determine the levels of human RKIP mRNA expression, real-time RT-PCR reactions were performed using iCycler iQ Multi-Color Real Time PCR Detection System (Bio-Rad, Hercules, CA) with a mixture composed of SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA), 400 nM of each primer and 5 ng of cDNA (equivalent total RNA) from unknown samples. The primer sequences were as follows: RKIP, GACATCAGCAGTGGCACAGT and GTCACACTTTAGCGGCCTGT, 18S rRNA, GGACACGGACAGGATTGACA and ACCCACGGAATCGAGAAAGA. PCR products were excised and cloned into the pCR 2.1 Vector (Invitrogen™), and sequenced in both directions. After sequence confirmation, standards for real-time RT-PCR were prepared with 10-fold dilutions of each PCR product. The copy numbers of RKIP mRNA were quantified by determining the Ct values, followed by normalization to 18S rRNA and the standard curves for each gene. Experiments were performed in triplicate.
RKIP Expression and Transfection Studies
For transient transfection experiments, HCC cells in 6 well plates at 60–70% confluence were transfected with 2 µg of CMV5-HA-RKIP plasmid25 or empty vector plasmid (control) using LipofectAMINE 2000 Reagent (Invitrogen). To obtain FOCUS cell lines transfected stably with either empty vector or RKIP plasmid, selection was initiated 48 hours after transfection using 800 µg/mL of G418 (Life Technologies, Gaithersburg, MD). The selection medium was changed every 4 days. Clones of G418-resistant cells were isolated and expanded for further characterization.
Western Blot Analysis
Eight of the 17 paired frozen tumor and peritumoral tissues or HCC cells were lysed and Western blot analysis was performed as previously described.34 Cells were starved with serum-free medium for 24 hours followed by stimulation with 100 nM IGF-I for 10–30 minutes. To examine nuclear translocation of phospho-ERK, cytosolic and nuclear extracts were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce, Rockford, IL). Following primary antibodies were used: RKIP (Upstate, Waltham, MA); c-Raf, phospho-c-Raf (Ser338), MEK1, phospho-MEK1, ERK and phospho-ERK (Cell Signaling, Beverly, MA); actin and Histone-H1 (Santa Cruz, Santa Cruz, CA). The primary antibody was followed by incubation with horseradish peroxidase-conjugated secondary antibody, and the Western Lighting System (PerkinElmer™ Life Sciences, Boston, MA). When necessary, the membranes were stripped with Restore™ Western Blot stripping buffer (Pierce) and reprobed. For protein quantification, bands were scanned, quantified using NIH Image 1.61 software, and normalized to an actin control. The results are reported as the mean of triplicate assays.
RKIP-siRNA
Control small interfering RNA (siRNA) (siCONTROL non-targeting siRNA) and siRNA specific for human RKIP (siGENOME SMARTpool reagent, human PBP) were purchased from Dharmacon (Lafayette, CO) and transfected into HepG2 HCC cells at a concentration of 100 nM using the DharmaFECT 4 transfection reagent (Dharmacon). Forty-eight hours after transfection, cells were starved with serum-free media for 24 hours and activated with or without IGF-I for 15 minutes.
Raf Kinase Activity Assay
The Raf kinase activity was analyzed with a non-radioactive Raf kinase assay kit (Upstate) with some modification. Briefly, 500 µg of total protein in each cell lysate was incubated with 2 µg of mouse monoclonal anti-Raf-1 antibody (Santa Cruz) overnight at 4°C, followed by incubation with UltraLink Immobilized Protein A/G gel (Pierce Biotec). After an extensive wash, the gels were incubated with inactive MEK-1 substrate at 30°C for 30 minutes. The phosphorylated MEK-1 in the reaction mixture was detected by Western blot with rabbit antiphospho-MEK-1 antibody (Upstate).
Immunofluorescent Staining
FOCUS cells were grown in a chamber slides (Nalge Nunc, Naperville, IL). Cells were fixed in cold methanol, and blocked with 5% goat and horse sera. For double-label immunofluorescent staining, cells were incubated with anti-RKIP and anti-phospho-ERK antibodies in blocking solution overnight at 4°C. Cells were rinsed and FITC-conjugated anti-mouse IgG and Texas red-conjugated anti-rabbit IgG (Vector Laboratories, Berlingame, CA) secondary antibodies were applied for 30 minutes at room temperature. Finally, coverslips were mounted with antifade Vectashield medium with DAPI (Vector Laboratories) to counterstain nuclei and examined under an Olympus IX70 fluorescence microscope (DSC Optical Services, Newton, MA).
Cell Proliferation Assay
We used the CellTiter 96 AQeous Non-Radioactive Cell Proliferation Assay (Promega, Madison, WI). Briefly, FOCUS cells transfected stably with either CMV5-HA-RKIP or empty plasmids were plated in 96-well plates at a density of 1000 cells/well in serum-free MEM with or without 100 nM IGF-I. The cells were incubated for 3 days at 37°C and the media were changed every 24 hours. Cell proliferation was measured after incubation for 2 hours with a combined MTS/PMS solution (Promega); a colorimetric method for determination of viable proliferating cells. Results are presented as the average absorbance of six wells in one experiment, and reported as the mean of triplicate assays.
Cell Migration Assay
A luminescence-based assay was used to evalulate cell migration, as previously described.35 FOCUS cells transfected stably with either CMX5-HA-RKIP or empty plasmids were used for this cell migration assay. Briefly, 5 × 104 cells in 0.5 mL of MEM containing 1% BSA were seeded into the upper chamber of 12-well plates separated by a polycarbonate membrane with 8-µm pores (Transparent PET Membrane; BD Biosciences, San Jose, CA). The lower chamber was filled with 0.75 mL of MEM containing 1% BSA and 100 nM IGF-I. After incubation for 3 hours at 37°C, the cells on the upper surface of the membrane (non-migrated) were harvested with a sterile cotton swab and placed into a well containing lysis buffer. The membrane (migrated adherent cells) was placed into another well and the suspension of migrated non-adherent cells in the lower chamber was placed into a third well containing lysis buffer. ATPLite substrate (PerkinElmer™) was added and luminescent counts per second were measured in a TopCount Microplate reader (Packard Instrument Company, Meriden, CT). The results were expressed as a percent of migrated cells to total cells and the experiments were performed in triplicate.
Wound Healing Assay
The empty vector transfected FOCUS cell line and stable RKIP transfected clones were plated into six-well plates. Confluent monolayers were scraped with a sterile plastic micropipette tip. The wound closure, as an index of migration, was observed over a 24 hours period and photographed microscopically at 0, 6, 12 and 18 hours.
Statistical analysis
Data were analyzed by the Student’s t test. A P value less than 0.05 was considered to be statistically significant.
Results
RKIP Protein Expression Is Downregulated in Human HCC Tumors
The expression level of RKIP protein was evaluated by immunohistochemistry in 17 paired human HCC tumors and adjacent uninvolved peritumoral tissues (Table 1). RKIP staining was detected in 83% (14/17) peritumoral tissues, but in only 12% (2/17) of HCC tumor tissues (P < 0.001). Figure 1 shows a representative immunohistochemical staining result. Moreover, immunoblot analysis of 8 of the 17 paired HCC and adjacent uninvolved tissue samples showed decreased RKIP protein levels in 7/8 HCC compared to adjacent peritumoral tissues (P < 0.0001) (Figure 2A, B). Consistent with these observations was the finding that phospho-ERK was also elevated in 7 of 8 HCC tissues with decreased expression of RKIP (Figure 2A). We next evaluated RKIP mRNA levels by real-time RT-PCR. Unexpectedly, RKIP mRNA levels were decreased in only 41%, increased in 47% and showed no change in 12% of the HCC. Overall, there was no significant difference in RKIP mRNA expression levels (P > 0.5) between HCC tumors and corresponding peritumoral tissues (Figure 2C). These results suggest that some of the differences observed in RKIP protein levels in HCC are probably due to post-transcriptional mechanisms.
Table 1.
Expression Level of RKIP Protein in Human HCC Tissues.*
| Sample No. | pT | T |
|---|---|---|
| 1 | ++ | − |
| 2 | + | − |
| 3 | + | − |
| 4 | + | − |
| 5 | − | − |
| 6 | ++ | − |
| 7 | ++ | + |
| 8 | + | − |
| 9 | + | − |
| 10 | + | − |
| 11 | +++ | − |
| 12 | ++ | − |
| 13 | − | − |
| 14 | + | − |
| 15 | − | − |
| 16 | +++ | − |
| 17 | ++ | + |
| 17 | 82.3% (14/17) | 11.8% (2/17) |
T: HCC tumor tissue, pT: adjacent uninvolved peritumoral tissues
Samples were immunostained with anti-RKIP antibody and detected by ABC method. Immunostaining intensity was scored as negative (−), weak (+), moderate (++), or strong (+++) on the basis of staining intensity.
Figure 1.
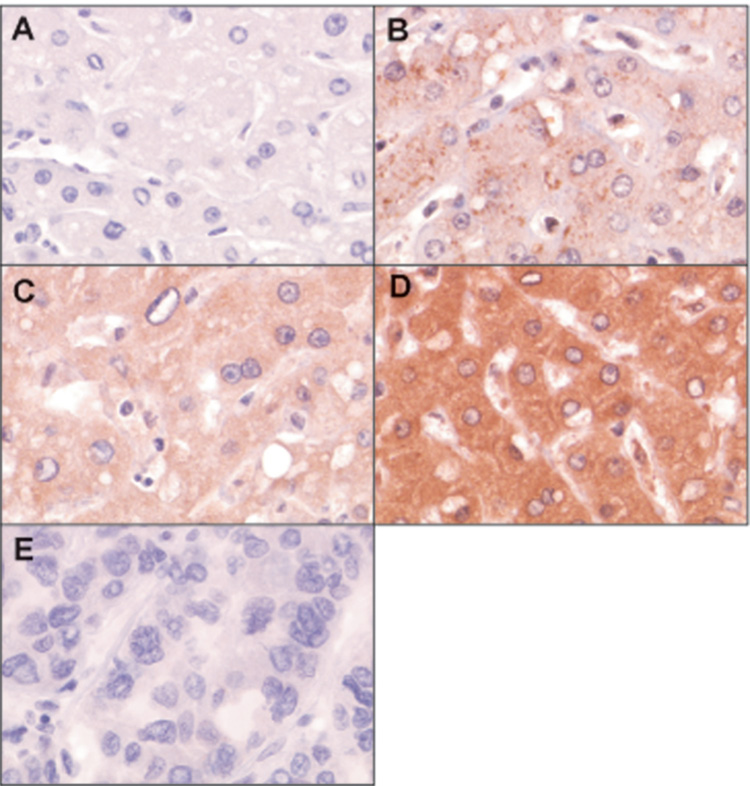
RKIP protein expression in human HCC tissue samples. Representative examples of HCC and peritumoral areas were immunostained with anti-RKIP antibody (brown color), and counterstained with hematoxylin (blue color). Magnification was 200 x. (A) Omission of the primary antibody served as the negative control. (B) Low (+) staining in peritumoral tissue. Less than 50% of hepatocytes showed immunoreactivity. (C) Moderate (++) staining in peritumoral tissue. Almost all of the hepatocytes showed homogenous moderate immunoreactivity. (D) Strong (+++) staining of peritumoral tissues. (E) Negative (−) staining of a HCC tumor.
Figure 2.
Expression of RKIP protein, mRNA, and phospho-ERK in HCC tissues. (A) Western blot analysis of the expression levels of RKIP and phospho-ERK. Cellular lysates derived from 8 pairs of HCC tumor (T) and peritumoral (pT) tissues were immunoblotted with anti-RKIP or anti-phospho-ERK antibodies. Actin served as a loading control. Note that phospho-ERK expression was increased in HCC tissues that had reduced expression of RKIP. (B) The bar graph represents the ratio of RKIP protein level (T/pT) obtained by densitometric scanning. Seven of the 8 HCC tumors had reduced RKIP expression (tumor versus peritumoral tissue: P < 0.0001). (C) Expression levels of RKIP mRNA in human HCC tissues, measured by quantitative real-time RT-PCR. The RKIP mRNA levels were normalized to 18S rRNA. The bar graph shows the ratio of RKIP mRNA level (T/pT). There was no difference in RKIP mRNA expression levels between HCC and peritumoral tissues (P > 0.5).
Expression of RKIP mRNA and Protein in HCC Cell Lines
We evaluated RKIP protein and mRNA expression levels in 4 HCC cell lines; FOCUS, Huh7, Hep3B, and HepG2 (Table 2). These 4 HCC cell lines have been previously characterized with respect to morphology, growth rate, production of albumin, anchorage independent growth in soft agar, and tumor formation in nude mice.36–38 Using these criteria, the differentiation status of these HCC cell lines was categorized as: FOCUS < Huh7 < Hep3B < HepG2 (undifferentiated < most differentiated). Interestingly, the least differentiated FOCUS cells showed the lowest level of RKIP protein, whereas the well-differentiated HepG2 cells expressed the highest level of RKIP. These results suggest a relationship between RKIP expression and HCC cellular differentiation (Table 2, Figure 3A). The RKIP mRNA expression levels were roughly correlated with its protein expression levels in HCC cells. However, the protein expression levels were different between HepG2 and Hep3B cells with similar mRNA levels, which suggest regulation by complex mechanisms (Table 2).
Table 2.
RKIP mRNA and Protein Expression in HCC Cell Lines
| HCC cell line | mRNA expression1 (copy number) | mRNA expression2 | Protein expression2 |
|---|---|---|---|
| FOCUS | 56.4 ± 24.7 | 25.0 | 3.6 |
| Huh7 | 119.0 ± 17.6 | 44.2 | 16.1 |
| Hep3B | 263.0 ± 3.2 | 92.9 | 44.6 |
| HepG2 | 283.0 ± 1.6 | 100 | 100 |
copy number per 107 18S rRNA accessed by qPCR
relative to the expression level of HepG2 (%)
Figure 3.
The role of RKIP in IGF-I-induced ERK/MAPK pathway activation in HCC. (A) Expression levels of RKIP protein and downstream components of the ERK/MAPK pathway in HCC cell lines. Cell lysates of FOCUS, Huh7, Hep3B and HepG2 cells containing 100 µg of total protein were immunoblotted for RKIP, MEK1, ERK and their phosphorylated forms; actin served as a loading control. (B) The bar charts represent the results of densitometric analysis and reveal the ratio of RKIP and actin, phospho-MEK1 and total MEK1, and phospho-ERK and total ERK. (C) Ectopic expression of RKIP blocked IGF-I-induced ERK/MAPK signaling pathway in FOCUS cells. Cells were transiently transfected with either RKIP or vector plasmid. After 24 hours serum starvation, cells were treated with 100 nM of IGF-I (+) for 15 minutes or untreated (−). Cell lysates were prepared and immunoblotted for RKIP and downstream signaling components of the pathway (c-Raf, MEK1, ERK and phosphorylated form); actin served as a loading control. (D) The bar charts depict the results of densitometric analysis from (C), and shown as the ratio of RKIP and actin, phospho-c-Raf and total c-Raf, phospho-MEK1 and total MEK1, and phospho-ERK and total ERK. The results are presented as the mean ± SE of triplicate experiments. (E) Knock-down of RKIP expression with siRNA induced the activation of IGF-I-induced ERK/MAPK signaling in HepG2 cells. Cells were transfected with either control si-RNA (C-siRNA) or RKIP-siRNA using the DharmaFECT 4 transfection reagent. Cells were treated with 100 nM of IGF-I (+) for 15 minutes or untreated (−) 24 hours after serum starvation followed by immunoblot for RKIP and downstream signaling components of the pathway (c-Raf, MEK1, ERK and phosphorylated form); actin served as a loading control. (F) The bar charts depict the results of densitometric analysis from (E), and are expressed as the ratio of RKIP and actin, phospho-c-Raf and total c-Raf, phospho-MEK1 and total MEK1, and phospho-ERK and total ERK.
The Level of RKIP Expression Influenced IGF-I-induced Activation of the ERK/MAPK Pathway
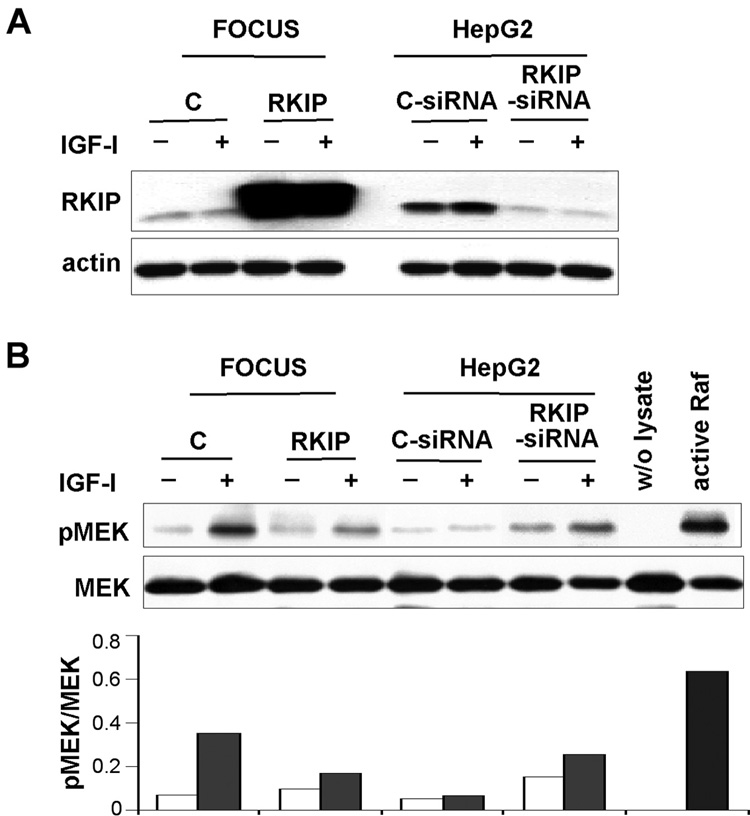
RKIP binds to both Raf-1 and MEK, and interferes with their interaction and inhibits downstream signaling.24,25 We explored signaling through this pathway in the four HCC cell lines. In FOCUS cells, which express low RKIP protein levels, we observed high MEK and ERK phosphorylation. In contrast, MEK and ERK phosphorylation was low in HepG2 cells, which express high levels of RKIP protein (Figure 3A and B). These results imply that RKIP may directly influence activation of the ERK/MAPK pathway. To further examine the effect of RKIP on the ERK/MAPK cascade, we first restored RKIP expression in FOCUS cells. As expected, IGF-I stimulation of FOCUS cells increased the phosphorylation of the ERK/MAPK pathway components Raf, MEK, and ERK. Ectopic expression of RKIP inhibited IGF-I-induced phosphorylation of MEK and ERK, but not Raf since it is upstream of RKIP (Figure 3C and D). Suppression of RKIP expression by siRNA increased phosphorylation of MEK and ERK in HepG2 cells, which imply activation of ERK/MAPK pathway (Figure 3E and F). To further confirm effects of RKIP on the ERK/MAPK pathway in HCC cells, we performed a Raf kinase activity assay using MEK-1 as the substrate. In FOCUS cells, IGF-I stimulation increased Raf kinase activity, however overexpression of RKIP reduced this activity. Moreover, we found that inhibition of RKIP expression using siRNA in HepG2 cells induced Raf kinase activity when stimulated with IGF-I as compared to basal Raf kinase activity (Figure 4B). These results indicate that the level of RKIP was important for modulation of IGF-I-induced ERK/MAPK signaling in HCC cells.
Figure 4.
The level of RKIP expression influences Raf kinase activity. (A) Expression of RKIP protein in HCC cell lines. In FOCUS cells, empty vector (C) or RKIP expression plasmid was transfected. Control siRNA (C-siRNA) or RKIP-siRNA was transfected into HepG2 cells. Cells were treated with (+) or without (−) IGF-I for 10 minutes after 24 hours of serum-starvation. Western blot of actin was performed as a control to normalize protein concentrations. (B) Raf kinase activity assay. Five hundred micrograms of protein of each sample were immunoprecipitated with mouse monoclonal anti-Raf antibody, followed by a Raf kinase activity assay as described in Materials and Methods. The blot was stripped and reprobed sequentially for MEK, demonstrating that equal amounts of recombinant MEK-1 substrate were added. The ‘w/o lysate’ lane represents the assay without adding cell lysate and served as the negative control. The ‘active Raf’ lane represents the assay using immunoprecipitate obtained from 0.1 µg active Raf protein serving as the positive control. The phosphorylated MEK (pMEK) exhibited by immunoprecipitated Raf protein. The bar graph represents the level of pMEK obtained by densitometric scanning (pMEK versus MEK). Note that overexpression of RKIP reduced Raf kinase activity in FOCUS cells, while knockdown of RKIP induced the activity in HepG2 cells.
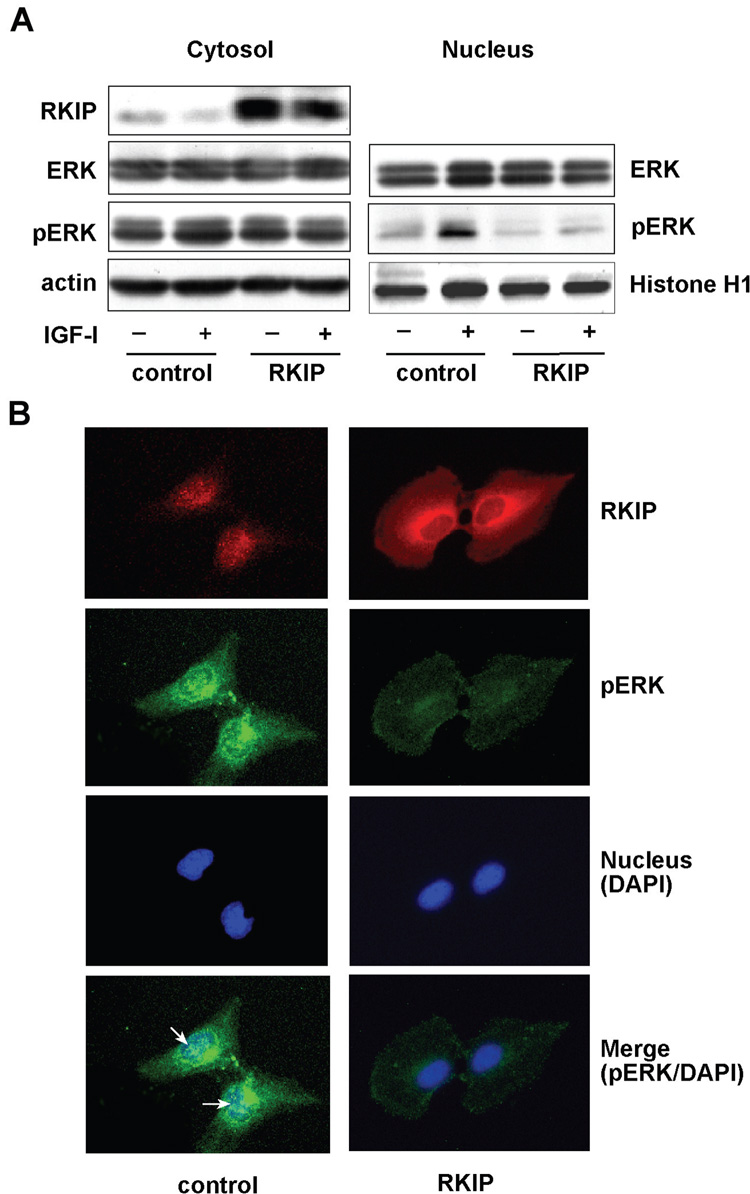
Activated-ERK translocates into the nucleus and regulates gene expression through the phosphorylation of transcriptional factors. We therefore investigated the effect of RKIP on nuclear phospho-ERK accumulation. IGF-I stimulation of FOCUS cells resulted in accumulation of phospho-ERK in the cytoplasm, which was abolished by restoration of RKIP (Figure 5A). In addition, IGF-I stimulation resulted in increased nuclear accumulation of phospho-ERK, which was inhibited by restoration of RKIP. The RKIP had no effect on the total amount of ERK present in the HCC cells (Figure 5A).
Figure 5.
Restoration of RKIP levels inhibited phospho-ERK nuclear accumulation in FOCUS cells. (A) RKIP or vector transfected FOCUS cells were stimulated with (+) or without (−) 100 nM of IGF-I for 15 minutes. Cytosolic and nuclear fractions were immunoblotted for RKIP, phospho-ERK (pERK) and total ERK expression; actin and Histone-H1 served as cytosolic and nuclear loading control, respectively. (B) Control and RKIP transfected FOCUS cells (grown on chamber slides) were immunostained with RKIP (red color) and phospho-ERK (green color), and the nucleus counterstained with the DAPI (blue color). The bottom panel reveals the merged images of phospho-ERK and DAPI staining indicating nuclear co-localization. The arrows indicate the nuclear pERK.
These observations were further confirmed by double-label immunofluorescent staining with anti-RKIP and anti-phospho-ERK antibodies of FOCUS cells transiently transfected with either RKIP or empty vector plasmid as a control. Strong cytoplasmic staining of RKIP was observed in RKIP-transfected compared to control cells (Figure 5B, top panel). Immunostaining of phospho-ERK revealed strong signals in the cytoplasm as well as nucleus in control cells and these levels were strikingly reduced by RKIP overexpression (Figure 5B, second panel). Nuclear staining of phospho-ERK was observed only in control cells but not in RKIP-transfected cells (Figure 5B, bottom panel). These observations demonstrate that restoration of RKIP blocks IGF-I induced activation of the ERK/MAPK pathway in FOCUS cells and leads to reduce nuclear phospho-ERK.
Restoration of RKIP Decreased HCC Cell Proliferation and Motility In Vitro
Activation of the ERK/MAPK pathway leads to cell proliferation, migration, and inhibition of apoptosis. Our results imply that ectopic expression of RKIP inhibits this pathway in FOCUS cells. We therefore evaluated the functional significance of RKIP expression in HCC cells. RKIP was overexpressed by stable transfection of FOCUS cells as revealed by Western blot analysis (Figure 6A). When control cells were stimulated with IGF-I, there was an increase in cell proliferation. However, RKIP overexpression suppressed this enhanced cell proliferation (Figure 6B) and no difference was found in apoptosis rates between control and RKIP overexpressing cells (data not shown). We then evaluated whether RKIP expression influenced HCC cell migration. Ectopic expression of RKIP significantly decreased FOCUS cell migration rates (28%) compared to control cells (56%)(Figure 6C). To further confirm the effect of RKIP on cell migration, control and RKIP overexpressing FOCUS cells were plated under confluence conditions and the monolayers were scrape-wounded. Control cells migrated rapidly and closed wound by 18 hours. In contrast, RKIP overexpression only closed the wound by 22% in the same time interval (Figure 7), further reinforcing the concept that RKIP is functionally linked to migration of HCC cells. Taken together, we conclude that RKIP overexpression antagonizes IGF-I activation of the ERK/MAPK cascade in HCC, and that the downstream biological consequences are reduced proliferation and migration.
Figure 6.
Restoration of RKIP reduced cell proliferation and migration in FOCUS cells. FOCUS cells were transfected stably with either empty vector (control) or RKIP expression plasmid. (A) Demonstration of RKIP protein expression by Western blot analysis. (B) RKIP expression inhibits FOCUS cell proliferation. The cells were grown in serum-free MEM with or without 100 nM IGF-I. Cell proliferation wasdetemined using CellTiter 96 AQeous Non-Radioactive Cell Proliferation Assay. The results are expressed as the mean ± SE of triplicate assays. IGF-I stimulation increased cell proliferation in control cells [C-IGF-I (+) versus C-IGF-I (−)], but there was no change of cell proliferation rates in RKIP-transfected FOCUS cells [RKIP-IGF-I (+) versus RKIP-IGF-I (−)]. P < 0.05 (day 2 and 3) (C) RKIP expression inhibits FOCUS cell migration. To assess the ability of the cells to cross a polycarbonate membrane (8-µm pore size), 5 × 104 cells in 0.5 mL of serum-free MEM containing 1% BSA was placed in the upper chamber and the lower chamber was filled with 0.75 mL of serum-free MEM containing 1% BSA and 100 nM IGF-I. After incubation for 3 hours at 37°C, the cells on the upper chamber and those attached to the membrane or migrated to the lower chamber were harvested and placed in lysis buffer, respectively. ATPLite substrate was added to each fraction and luminescent counts per second were measured in a TopCount Microplate reader. The results were expressed as a percentage of migrated cells to total cells and experiments were triplicated. P < 0.001 (control versus RKIP)
Figure 7.
RKIP inhibited FOCUS cell proliferation and motility. The control FOCUS cells and stable RKIP transfectants were placed into six-well plates. A wound was created in confluent monolayers with a sterile plastic micropipette tip and the percent of wound closure was revealed via the area of the wound that remained open at each time and plotted below. Note that ectopic RKIP expression in FOCUS cells strikingly reduced cell migration.
Discussion
The insulin/IGF-I/IRS-1/MAPK cascade plays an important role in regulating liver regeneration following two/thirds hepatectomy, and during embryonic development.39,40 In situations of unrestrained growth, constitutive activation of the insulin/IGF-I/IRS-1/MAPK cascade due to enhanced IRS-1 expression has been identified in the majority of HCC.39 Indeed, IRS-1 overexpression is associated with activation of the ERK/MAPK cascade and results in increased HCC tumor size.41–43 The IRS-1 protein is the main substrate for the insulin/IGF-I receptor, and emits downstream signals through its interaction with SH-2 domain containing molecules. The role of insulin/IGF-I/IRS-1 signaling in human HCC is illustrated by studies demonstrating that overexpression of IRS-1 and/or activation of one or more of the components of this signaling pathway occurs in over 90 percent of tumors.44 Furthermore, inhibition of the insulin/IGF-I/IRS-1 mediated signaling by a dominant-negative IRS-1 mutant protein reversed the malignant phenotype of human HCC cells.45 Therefore, it is of great interest to determine if there are other molecular mechanisms that contribute to the activation of this pathway in addition to overexpression of IRS-1. In this regard, it appears that loss of RKIP function is a major event in the pathogenesis of HCC since approximately 80–90% of tumors evaluated here had reduced RKIP protein levels. More important, phospho-ERK expression was increased in 86% of HCC tissues that had reduced RKIP expression. In aggregate, these studies emphasize the importance of insulin/IGF-I signaling in the regulation of hepatic growth, and further indicate that aberrant activation plays an oncogenic role in the vast majority of HCC tumors.
The role of RKIP has been studied in several types of tumors including malignant melanoma and prostate tumor. However, there is no information on RKIP function in the liver and particularly in HCC. We hypothesize that RKIP may play a role in the molecular pathogenesis of HCC since loss of function would contribute to the activation of the ERK/MAPK pathway. Our experiments reveal that RKIP expression is downregulated in human HCC similar to what had been observed in melanoma and prostate tumor.22,29 Although it is believed that RKIP expression is important for suppression of metastasis, the cellular mechanisms of how RKIP protein levels are regulated have not yet been determined. On the basis of bioinformatic analysis of RKIP promoter sequences, expression may be regulated through several transcription factors such as AP-1, SP-1 and Ying and Yang-1 (YY1).21 In addition, Huerta-Yepez et al. found that treatment of prostate cancer cell lines with inhibitors for NF-κB or the transcription repressor YY1 led to upregulation of RKIP expression.46 In this study, although RKIP protein levels were clearly decreased in human HCC, RKIP mRNA levels were decreased in only 41% since the remaining tumor showed either an increased or no change. Therefore, RKIP protein expression is likely to be regulated at the post-transcriptional level at least in some human HCC. There are no prior reports that have characterized RKIP expression in different cell types as a function of differentiation. Our results demonstrate that RKIP protein level was directly correlated with the degree of HCC cell differentiation. Taken together, these observations suggest that RKIP expression may be modulated at different levels depending on the cell type and state of differentiation.
It has been clearly documented that the MAPK pathway was constitutively activated in several tumors types including HCC.47–53 However, the mechanism(s) contributing to this process are poorly understood. We demonstrated that enhanced activity of the ERK/MAPK pathway was correlated with lack of RKIP expression in FOCUS cells, the most malignant and poorly differentiated of the HCC cell lines evaluated. However, restoration of RKIP blocked activation of the IGF-I stimulated ERK/MAPK pathway through inactivation of MEK and ERK. This observation led us to address if restoration of RKIP protein could modulate HCC cell proliferation and migration. Indeed, the cell proliferation rate was reduced in FOCUS cells stably transfected with RKIP compared to the empty vector transfected control cells without any change in the rate of apoptosis. It has been previously reported that RKIP inhibited pancreatic β-cell growth by altering the cell cycle, rather than by promoting apoptosis.30 Although RKIP overexpression has been reported to sensitize RC1 cells (prostatic carcinoma cell line) and NHL cells to chemotherapeutic agents, it does not appear to increase the spontaneous apoptosis rate.21,28 In contrast, overexpression of RKIP triggers apoptosis in breast tumor cell lines.28 Therefore, the apoptotic response to RKIP overexpression may be context-dependent in the absence of chemotherapeutic agents. In addition to the effect of RKIP on cell proliferation, our studies revealed that restoration of RKIP significantly inhibited the migration and motility of FOCUS cells indicating that this pathway is important in hepatic oncogenesis. A recent study showed that pamidronate suppresses phosphorylation of ERK and reduced migration in HCC cell lines.54 Another investigation also demonstrated that activation of the ERK signaling pathway enhances cell migration in F9 parietal endodermal cells, which was suppressed by a MEK inhibitor.55 However, little is known about the mechanisms how ERK/MAPK pathway modulates cell migration and further studies will be required.
In summary, these observations provide evidence that down-regulation of RKIP expression in HCC contributes to constitutive activation of the ERK/MAPK pathway and promotes proliferation and migration of HCC cells. Furthermore, IGF-I stimulated activation of the ERK/MAPK pathway can be blocked by restoration of RKIP levels. In addition, RKIP expression probably contributes to HCC cell differentiation. The regulation of RKIP expression may involve both transcriptional and/or post-transcriptional mechanisms and further studies will be required. Finally, RKIP provides an attractive molecular target to regulate HCC proliferation and differentiation.
Acknowledgments
Supported in part by grants from National Institutes of Health CA-35711, AA-02666 (JRW), P20 RR015578 (MK), and by the Korean Association of the Study of Liver Disease Glaxo Wellcome Hepatologist Hepatitis Fellowship Fund (HCL).
Abbreviations used
- 18S rRNA
18S ribosomal RNA
- cDNA
complementary DNA
- HCC
hepatocellular carcinoma
- IGF
insulin-like growth factor
- MAPK
mitogen-activated protein kinase
- RKIP
Raf kinase inhibitor protein
- RT-PCR
reverse-transcription polymerase chain reaction
- siRNA
small interfering RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bosch FX, Ribes J, Diaz M, Cleries R. Primary Liver Cancer: Worldwide incidence and trends. Gastroenterology. 2004;127:S5–S16. doi: 10.1053/j.gastro.2004.09.011. 2004. [DOI] [PubMed] [Google Scholar]
- 2.Moradpour D, Blum HE. Pathogenesis of hepatocellular carcinoma. Eur. J. Gastro & Hepatol. 2005;17:477–483. doi: 10.1097/00042737-200505000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Fausto N, Laird A, Webber E. Role of growth factors and cytokines in hepatic regeneration. FASEB J. 1995;9:1527–1536. doi: 10.1096/fasebj.9.15.8529831. [DOI] [PubMed] [Google Scholar]
- 4.Park YN, Chae KJ, Kim YB, Park P, Theise N. Apoptosis and proliferation in hepatocarcinogenesis related to cirrhosis. Cancer. 2001;92:2733–2738. doi: 10.1002/1097-0142(20011201)92:11<2733::aid-cncr10126>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 5.Tannapfel A, Wittekind C. Genes involved in hepatocellular carcinoma: deregulation in cell cycling and apoptosis. Virch Arch. 2002;440:345–352. doi: 10.1007/s00428-002-0617-x. [DOI] [PubMed] [Google Scholar]
- 6.Thorgeirsson SS, Teramoto T, Factor VM. Dysregulation of apoptosis in hepatocelular carcinoma. Sem Liver Dis. 1998;18:115–122. doi: 10.1055/s-2007-1007148. [DOI] [PubMed] [Google Scholar]
- 7.Humbel RE. Insulin-like growth factors I and II. Eur J Biochem. 1990;190:445–462. doi: 10.1111/j.1432-1033.1990.tb15595.x. [DOI] [PubMed] [Google Scholar]
- 8.Scharf J, Braulke T. The role of the IGF axis in hepatocarcinogenesis. Horm Metab Res. 2003;35:685–693. doi: 10.1055/s-2004-814151. [DOI] [PubMed] [Google Scholar]
- 9.Alexia C, Fallot G, Lasfer M, Schweizer-Groyer Gc, Groyer A. An evaluation of the role of insulin-like growth factors (IGF) and of type-I IGF receptor signalling in hepatocarcinogenesis and in the resistance of hepatocarcinoma cells againist drug-induced apoptosis. Biochem Pharm. 2004;68:1003–1015. doi: 10.1016/j.bcp.2004.05.029. [DOI] [PubMed] [Google Scholar]
- 10.Radcliff K, Tang TB, Lim J, Zhang Z, Abedin M, Demer LL, Tintut Y. Insulin-like growth factor-I regulates proliferation and osteoblastic differentiation of calcifying vascular cells via extracellular signal-regulated protein kinase and phosphatidylinositol 3-kinase pathways. Cir Res. 2004;96:398–400. doi: 10.1161/01.RES.0000157671.47477.71. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Sun Y. Insluin-like growth factor receptor-1 as an anti-cancer target: Blocking transformation and inducing apoptosis. Curr Cancer Drug Targets. 2002;2:191–207. doi: 10.2174/1568009023333863. [DOI] [PubMed] [Google Scholar]
- 12.Nottage M, Siu LL. Rationale for Ras and Raf-kinase as a target for cancer theapeutics. Curr Pharm Design. 2003;8:2231–2242. doi: 10.2174/1381612023393107. [DOI] [PubMed] [Google Scholar]
- 13.O'Neill E, Kolch W. Conferring specificity on the ubiquitous Raf/MEK signalling pathway. Br J Cancer. 2004;90:283–288. doi: 10.1038/sj.bjc.6601488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang F, Steelman L, Lee J, Shelton J, Navolanic P, Blalock W, Franklin R, McCubrey J. Signal transduction mediated by the Ras/Raf/MEK/ERK pathway from cytokine receptors to transcription factors: potential targeting for therapeutic intervention. Leukemia. 2003;17:1263–1293. doi: 10.1038/sj.leu.2402945. [DOI] [PubMed] [Google Scholar]
- 15.Mitsui H, Takuwa N, Maruyama T, Maekawa H, Hirayama M, Sawatari T, Hashimoto N, Takuwa Y, Kimura S. The MEK1-ERK MAP kinase pathway and the PI 3-kinase-Akt pathway independently mediate anti-apoptotic signals in HepG2 liver cancer cells. Int J Cancer. 2001;92:55–62. [PubMed] [Google Scholar]
- 16.Schmidt CM, McKillop IH, Cahill PA, Sitzmann JV. Increased MAPK expression and activity in primary human hepatocellualr carcinoma. Biochem Biophys Res Comm. 1997;236:54–58. doi: 10.1006/bbrc.1997.6840. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt CM, McKillop IM, Cahill PA, Sitzmann JV. The role of cAMP-MAPK signalling in the regulation of human hepatocellular carcinoma growth in vitro. Eu J Gastro & Hepatol. 1999;1:1393–1399. doi: 10.1097/00042737-199912000-00009. [DOI] [PubMed] [Google Scholar]
- 18.Dent P, Grant S. Pharmacologic interruption of the Mitogen-activated Extracellular-regulated kinase/Mitogen-activated protein kinase siganl transduction pathway: potential role in promoting cytotoxic drug action. Clin. Cancer Res. 2001;7:775–783. [PubMed] [Google Scholar]
- 19.Weinstein-Oppenheimer CR, Henriquez-Roldan CF, Davis JM, Navolanic P, Saleh OA, Steelman L, Franklin R, Robinson PJ, McMahon M, McCubrey J. Role of the Raf signal transduction cascade in the in vitro resistance to the anticancer drug doxorubicin. Clin Cancer Res. 2001;7:2898–2907. [PubMed] [Google Scholar]
- 20.Keller ET, Fu Z, Brennan M. The role of Raf kinase inhibitor protein (RKIP) in health and disease. Biochem Pharm. 2004;68:1049–1053. doi: 10.1016/j.bcp.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 21.Odabaei G, Chatterjee D, Jazirehi AR, Goodglick L, Yeung KC, Bonavida B. Raf-1 kinase inhibitor protein: Stucture, function, regulation of cell signaling, and pivotal role in apoptosis. Adv Cancer Res. 2004;91:169–200. doi: 10.1016/S0065-230X(04)91005-6. [DOI] [PubMed] [Google Scholar]
- 22.Schuierer MM, Bataille F, Hagan S, Kolch W, Bosserhoff AK. Reduction in Raf kinase inhibitor protein expression is associated with increased Ras-extracellular signal-regulated kinase signaling in melanoma cell lines. Cancer Res. 2004;64:5186–5192. doi: 10.1158/0008-5472.CAN-03-3861. [DOI] [PubMed] [Google Scholar]
- 23.Trakul N, Rosner MR. Modulation of the MAP kinase signaling cascade by Raf kinase inhibitory protein. Cell Res. 2005;15:19–23. doi: 10.1038/sj.cr.7290258. [DOI] [PubMed] [Google Scholar]
- 24.Yeung KC, Janosch P, McFerran B, Rose DW, Mischak H, Sedivy JM, Kolch W. Mechanism of suppression of the Raf/MEK/Extracellular singal-regulated kinase pathway by the Raf kinase inhibitor protein. Mol Cell Biol. 2000;20:3079–3085. doi: 10.1128/mcb.20.9.3079-3085.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeung KC, Seitz T, Li S, Janosch P, McFerran B, Kaiser C, Fe F, Katsanakis KD, Rose DW, Mischak H, Sedivy JM, Kolch W. Suppression of Raf-1 kinase activity and MAP kinase signaling by RKIP. Nature. 1999;401:173–177. doi: 10.1038/43686. [DOI] [PubMed] [Google Scholar]
- 26.Banfield MJ, Barker JJ, Perry AC, Brady RL. Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure. 1998;6:1245–1254. doi: 10.1016/s0969-2126(98)00125-7. [DOI] [PubMed] [Google Scholar]
- 27.Serre L, de Jesus KP, Zelwer C, Bureaud N, Schoentgen F, Benedetti H. Crystal structures of YBHB and YBCL form Escherichia coli, two bacterial homologues to a Raf kinase inhibitor protein. J Mol Biol. 2001;310:617–634. doi: 10.1006/jmbi.2001.4784. [DOI] [PubMed] [Google Scholar]
- 28.Chatterjee D, Bai Y, Wang Z, Beach S, Mott S, Roy R, Braastad C, Sun Y, Mukhopadhyay A, Aggarwai BB, Darnowski J, Pantazis P, Wyche J, Fu Z, Kitagwa Y, Keller ET, Sedivy JM, Yeung KC. RKIP sensitizes prostate and breast cancer cells to drug-induced apoptosis. J Biol Chem. 2004;279:17515–17523. doi: 10.1074/jbc.M313816200. [DOI] [PubMed] [Google Scholar]
- 29.Fu Z, Smith PC, Zhang L, Rubin MA, Dunn RL, Tao Z, Keller ET. Effects of Raf kinase inhibitor protein expression on suppression of prostate cancer metastasis. J Nat Cancer Inst. 2003;95:878–889. doi: 10.1093/jnci/95.12.878. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L, Fu Z, Binkley C, Giordano T, Burant CF, Logsdon CD, Simeone DM. Raf kinase inhibitory protein inhibits β-cell proliferation. Surgery. 2004;136:708–715. doi: 10.1016/j.surg.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 31.Keller ET. Metastasis suppressor genes: a role for raf kinase inhibitor protein (RKIP) Anti-Cancer Drugs. 2004;15:663–669. doi: 10.1097/01.cad.0000136877.89057.b9. [DOI] [PubMed] [Google Scholar]
- 32.Keller ET, Fu Z, Brennan M. The biology of a prostate cancer metastasis suppressor protein: Raf kinase inhibitor protein. J Cell Biochem. 2005;94:273–278. doi: 10.1002/jcb.20169. [DOI] [PubMed] [Google Scholar]
- 33.Kauffman EC, Robinson VL, Stadler WM, Sokoloff MH, Rinker-Schaeffer CW. Metastasis suppression: The evolving role of metastasis suppressor genes for regulating cancer cell growth at the secondary site. J Uro. 2003;169:1122–1133. doi: 10.1097/01.ju.0000051580.89109.4b. [DOI] [PubMed] [Google Scholar]
- 34.Merle P, de la Monte S, Kim M, Herrmann M, Tanaka S, von dem Bussche A, Kew MC, Trepo C, Wands JW. Functional consequences of Frizzled-7 receptor overexpression in human hepatocellular carcinoma. Gastroenterology. 2004;127:1110–1122. doi: 10.1053/j.gastro.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 35.de la Monte SM, Lahousse SA, Carter J, Wands JR. ATP luminescence-based motility-invasion assay. Biotechniques. 2002;33:98–100. doi: 10.2144/02331rr01. [DOI] [PubMed] [Google Scholar]
- 36.Darlington G, Kelly J, Buffone G. Growth and hepatospecific gene expression of human hepatoma cells in a defined medium. In Vitro Cell Dev Biol. 1987;23:349–354. doi: 10.1007/BF02620991. [DOI] [PubMed] [Google Scholar]
- 37.He L, Isselbacher KJ, Wands JR, Goodman HM, Shih C, Quaroni A. Establishment and characterization of a new human hepatocellular carcinoma cell line. In Vitro. 1984;20:493–504. doi: 10.1007/BF02619623. [DOI] [PubMed] [Google Scholar]
- 38.Knowles B, Howe C, Aden D. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980;209:497–499. doi: 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- 39.Khamzina L, Gruppuso P, Wands JR. Insulin signaling through IRS-1 and IRS-2 in normal liver development. Gastroenterology. 2003;125:572–585. doi: 10.1016/s0016-5085(03)00893-x. [DOI] [PubMed] [Google Scholar]
- 40.Sasaki Y, Nishiyama ZX, Avruch M, Wands JR. Expression and phosphorylation of insulin receptor substrate 1 during rat liver regeneration. J Biol Chem. 1993;268:3805–3808. [PubMed] [Google Scholar]
- 41.Ito T, Sasaki Y, Wands JR. Overexpression of human insulin receptor substrate 1 induces cellular transformation with activation of mitogen-activated protein kinase. Mol Cell Biol. 1996;16:943–951. doi: 10.1128/mcb.16.3.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka S, Ito T, Wands JR. Neoplastic transformation induced by insulin receptor substrate-1 overexpression requires an interaction with both Grb2 and Syp signaling molecules. J Biol Chem. 1996;271:14610–14616. doi: 10.1074/jbc.271.24.14610. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka S, Mohr L, Schmidt EV, Sugimachi K, Wands JW. Biologic effects of human insulin receptor substrate-overexpression in hepatocytes. Hepatology. 1997;27:598–604. doi: 10.1002/hep.510260310. [DOI] [PubMed] [Google Scholar]
- 44.Wands JR, Maradpour DW. Molecular pathogenesis of hepatocellular carcinoma. In: a B.TD ZD, editor. Hepatology: A Textbook of Liver Disease. 5th Edition. WB Saunders; 2006. in press. [Google Scholar]
- 45.Tanaka S, Wands JR. A carboxy-terminal truncated insulin receptor substrate-1 dominant negative protein teverses the human hepatocellular carcinoma malignant phenotype. J Clin Invest. 1997;98:2100–2108. doi: 10.1172/JCI119016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huerta-Yepez S, Vega M, Jazirehi AR, Garban H, Hongo F, Cheng G, Bonavida B. Nitric oxide sensitizes prostate carcinoma cell lines to TRAIL-mediated apoptosis via inactivation of NF-kB and inhibition of Bcl-xL expression. Oncogene. 2004;23:4993–5003. doi: 10.1038/sj.onc.1207655. [DOI] [PubMed] [Google Scholar]
- 47.Alexia C, Lasfer M, Groyer A. Role of constitutively activated and insulin-like growth factor-stimulated ERK1/2 signaling in human hepatoma cell proliferation and apoptosis. Evidence for heterogeneity of tumor cell lines. Ann NY Acad Sci. 2004;1030:219–229. doi: 10.1196/annals.1329.028. [DOI] [PubMed] [Google Scholar]
- 48.Ito Y, Sasaki Y, Horimoto M, Wada S, Tanaka Y, Kasahara A, Ueki T, Hirano T, Yamamoto H, Fujimoto J, Okamoto E, Hayashi N, Hori M. Activation of mitogen-activated protein kinases/extracellular signal-regulated in human hepatocellular carcinoma. Hepatology. 1998;27:951–958. doi: 10.1002/hep.510270409. [DOI] [PubMed] [Google Scholar]
- 49.Mishima K, Yamada E, Masui K, Shimokawara T, Takayama K, Sugimura M, Ichijima K. Overexpression of the ERK/MAP kinases in oral squamous cell carcinoma. Mod Pathol. 1998;11:886–891. [PubMed] [Google Scholar]
- 50.Price DT, Rocca GD, Guo C, Ballo MS, Schwinn DA, Luttrell LM. Activation of extracellular signal-regulated kinase in human prostate cancer. J Uro. 1999;162:1537–1542. [PubMed] [Google Scholar]
- 51.Sivaraman VS, Wang H, Nuovo GJ, Malbon CC. Hyperexpression of mitogen-activated protein kinase in human breast cancer. J Clin Invest. 1997;99:1478–1483. doi: 10.1172/JCI119309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsuboi Y, Ichida T, Sugitani S, Genda T, Inayoshi J, Takamura M, Matsuda Y, Nomoto M, Aoyagi Y. Overexpression of extracellular signal-regulated protein kinase and its correlation with proliferation in human hepatocellular carcinoma. Liver Int. 2004;24:432–436. doi: 10.1111/j.1478-3231.2004.0940.x. [DOI] [PubMed] [Google Scholar]
- 53.Vicent S, Garayoa M, Lopez-Picazo JM, Lozano MD, Toled G, Thunnissen FB, Manzano RG, Montuenga LM. Mitogen-activated protein kinase phosphatase-1 is overexpressed in non-small cell lung cancer and is an independent predictor of outcome in patients. Clin Cancer Res. 2004;10:3639–3649. doi: 10.1158/1078-0432.CCR-03-0771. [DOI] [PubMed] [Google Scholar]
- 54.Wada A, Fukui K, Sawai Y, Imanaka K, Kiso S, Tamura S, Shimomura I, Hayashi N. Pamidronate induced anti-proliferative, apoptotic, and anti-migratory effects in hepatocellular carcinoma. J Hepatol. 2006;44:142–150. doi: 10.1016/j.jhep.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 55.Hong T, Grabel LB. Migration of F9 parietal endoderm cells is regulated by the ERK pathway. J Cell Biochem. 2005;97(6):1339–1349. doi: 10.1002/jcb.20728. [DOI] [PubMed] [Google Scholar]