Abstract
The retinoblastoma (pRB) family proteins regulate the E2F transcription factors; their complexes regulate critical transitions through the cell cycle. The function of these pRBfam ily/E2F complexes, which includes p130/E2F4, in response to genotoxic agents, is not well understood. We investigated the role of E2F4 in the genotoxic stress response. Following radiation treatment, E2F4 colocalized with p130 in the nucleus during a radiation-induced stable G2-phase arrest. Arrested cells had significantly decreased expression of Cyclins A2 and B1 and decreased phosphorylation of mitotic protein monoclonal-2 (MPM-2) mitotic proteins. Small interference RNA (siRNA)-mediated knockdown of E2F4 sensitized cells to subsequent irradiation, resulting in enhanced cellular DNA damage and cell death, as determined by caspase activation and decreased clonogenic cell survival. Downstream E2F4 targets potentially involved in the progression from G2 into M phase were identified by oligonucleotide microarray expression profiling. Chromatin immunoprecipitation localized E2F4 at promoter regions of the Bub3 and Pttg1 mitotic genes following irradiation, which were among the downregulated genes identified by the microarray. These data suggest that in response to radiation, E2F4 becomes active in the nucleus, enforces a stable G2 arrest by target gene repression, and thus provides increased cell survival ability by minimizing propagation of cells that have irreparable DNA damage.
Keywords: E2F4 knockdown, G2 cell cycle arrest, apoptosis, ionizing radiation
Introduction
Progression through the cell cycle is dependent upon the integrated expression of factors that both positively and negatively regulate cell cycle transit. This complex regulatory network may either promote transitions or block progression through the initiation of checkpoints that occur during the G1 to S, or G2 to M-phase cell cycle transitions. In response to DNA damage, the G2 arrest is most prominent, which occurs in tumor cells that lack an effective G1 arrest.
The G2 arrest can occur via different pathways, including checkpoint activation. The checkpoint pathway implicates the DNA damage sensors, ataxia telangiectasia mutated/ataxia telangiectasia mutated and Rad3 related (ATM/ATR) and effectors, Cell Cycle Checkpoint Kinase 1/Cell Cycle Checkpoint Kinase 2 (CHK1/CHK2), which converge on the regulation of the CDC25C phosphatase and its downstream target, cyclin-dependent kinase 1 (CDK1) (Kastan and Bartek, 2004). In wild-type p53 cells, both a G1 and G2 arrest occur in response to DNA damage (Wahl and Carr, 2001). By inactivating p53, the G1 arrest is abolished and the G2 arrest is attenuated (Bunz et al., 1998; Chan et al., 2000; Taylor and Stark, 2001). A G2 arrest still occurs in ATM-, ATR-, and p53-deficient cells, suggesting that there are other pathways that mediate an effective and stable G2 arrest.
The E2F family of transcription factors controls the expression of genes implicated in the cell cycle (Dimova and Dyson, 2005). It is composed of nine proteins: E2F1, 2, 3(a, b), 4–8. E2Fs 1–3a are considered to be transcriptional activators and E2Fs 3b-8 are regarded as transcriptional repressors (Trimarchi and Lees, 2002). These proteins are regulated spatially and temporally throughout the cell cycle. Most E2Fs complex with their heterodimeric partners, DP1 (E2F Dimerization Partner 1; transcription factor DP1; TFD1) and DP-2 (TFD2), as well as with pRB and the related pocket proteins, p107 and p130, to control gene transcription (Takahashi et al., 2000). Cell synchronization experiments identified E2F cell cycle-phase specific complexes. Thus E2F4 and E2F5, in complex with p130, are present primarily in quiescent cells (Sardet et al., 1995). Decreasing levels of E2F4 are associated with pRB and p107-CDK2 during S-phase progression (Takahashi et al., 2000; Farkas et al., 2002).
Progression through the cell cycle is dependent on the degree of phosphorylation of pRB and its related pocket proteins. The phosphorylation status of pRB and the pocket proteins regulate their association with E2Fs, which can result in the deactivation of repressive complexes and cytoplasmic localization of E2F4 (Gaubatz et al., 2001; Farkas et al., 2002). Cell cycle-specific fluctuations in the levels of E2F4 bound to the promoters of its target genes actively repress transcription (Takahashi et al., 2000). Although less is known about the role of E2Fs during G2 transition, recent work indicates that cooperative promoter occupancy by multiple E2Fs, including E2F4, may contribute to the transcriptional control during this phase of the cell cycle (Zhu et al., 2004).
We and others have shown that pRB-deficient cells are characterized by increased mRNA and protein levels of genes that are under the transcriptional control of activating E2Fs (Almasan et al., 1995b; Herrera et al., 1996). By inactivating the pRB family members that complex with these activating E2Fs through introducing the E7 human papilloma virus (HPV) coupled with DNA damage, the capacity of colorectal cells to undergo G2 arrest is reduced (Polager and Ginsberg, 2003). Recent work has specifically implicated p130 in the mechanism of G2 arrest following treatment with either etoposide or adriamycin (Jackson et al., 2005). Cells that are p53-null are still capable of undergoing G2 arrest, but cells lacking p130 and p107 cannot (Jackson et al., 2005). In fact, as a consequence of the p130 and p107 deficiency, cells undergo cell death, which is similar to the response of pRB-deficient cells to antineoplastic drugs (Almasan et al., 1995b). Taken together, these findings provide support for an important role of the pRB family members and the E2F transcription factors in the G2 arrest. As the pRB family members interact with multiple binding partners and control a wide range of signaling pathways (Cobrinik, 2005), including cell cycle regulation, hypoxia signaling (Budde et al., 2005), and cancer development (Attwooll et al., 2004), the studies reported here were designed to determine whether E2F4 played a direct role in G2 arrest following ionizing radiation (IR).
Results
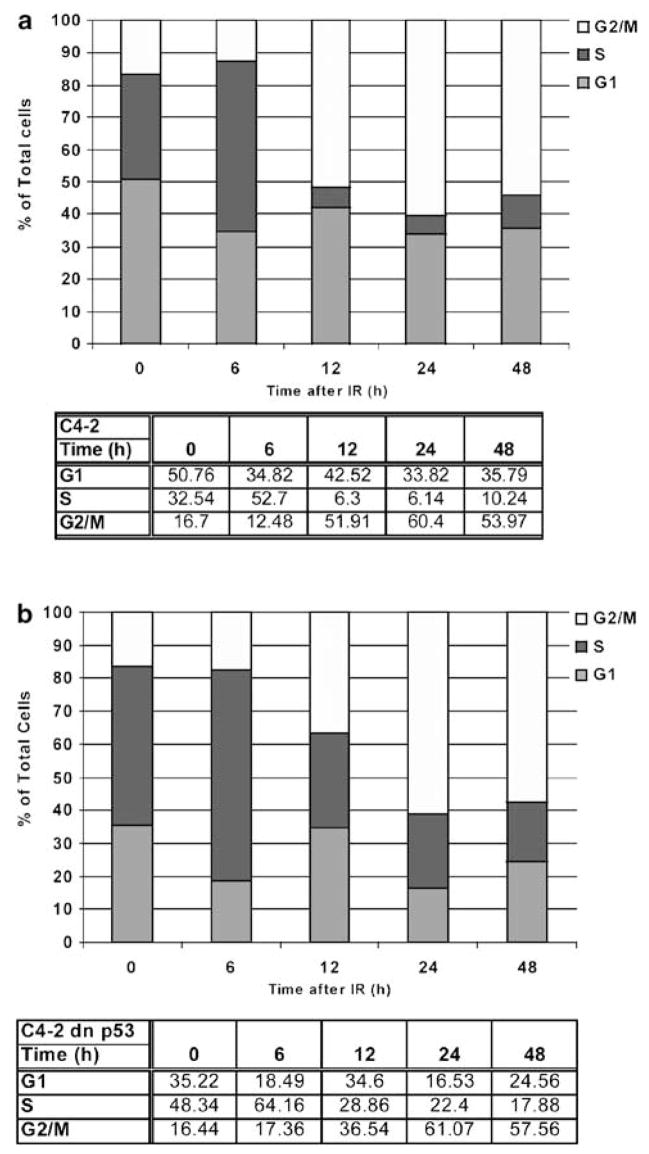
C4-2 prostate carcinoma cells undergo a stable G2/M-phase arrest following irradiation
Prostate carcinoma cells are quite resistant to IR treatment. To determine whether this resistance is dependent on cell cycle, the cell cycle distribution of LNCaP C4-2 was examined in response to irradiation. C4-2 cells started to accumulate with a 4C DNA content (G2/M) at 12 h post-IR with a maximal effect observed at 24 h (Figure 1a). Concomitantly, the proportion of cells in the G1 and S phases decreased; the effect was more pronounced in the S-phase cell population. At 24 h, the population of G2/M cells was still predominant, as compared to the proportion of S-phase cells. To determine if p53 activity was required for the radiation-induced cell cycle arrest, C4-2 cells were stably transfected with a C-terminal fragment dominant-negative (DN) p53 mutant (Ossovskaya et al., 1996), such that following irradiation, its transcriptional activity was abrogated, as shown by a dramatic decrease from abundant to undetectable levels of p21WAF1 (S. Ray et al., unpublished data). Similar to the parental C4-2, the derivative cells containing the stably expressed DN-p53 had a robust 4C arrest following irradiation (Figure 1b). The arrest response in the DN-p53 C4-2 cells was attenuated at 12 h, as compared to C4-2 cells. Moreover, the arrested cells did not return to a normal asynchronous cell cycle phase through 48 h post-IR. These data support a G1 arrest present in p53-proficient cells and reinforce the notion that human tumor cell lines arrest predominantly at 4C, regardless of their p53 status.
Figure 1.
Radiation induces a robust cell-cycle arrest in G2 and M independently of p53 function in prostate carcinoma C4-2 cells. Flow cytometry analyses, by staining for DNA content at different time points following irradiation, were performed as described in ‘Materials and methods.’ Percentages of C4-2 (a) and DN-p53-C4- 2 (b) cells in the different phases of the cell cycle analysed by flow cytometry are presented with respect to time. Data are representative of three separate and independent experiments.
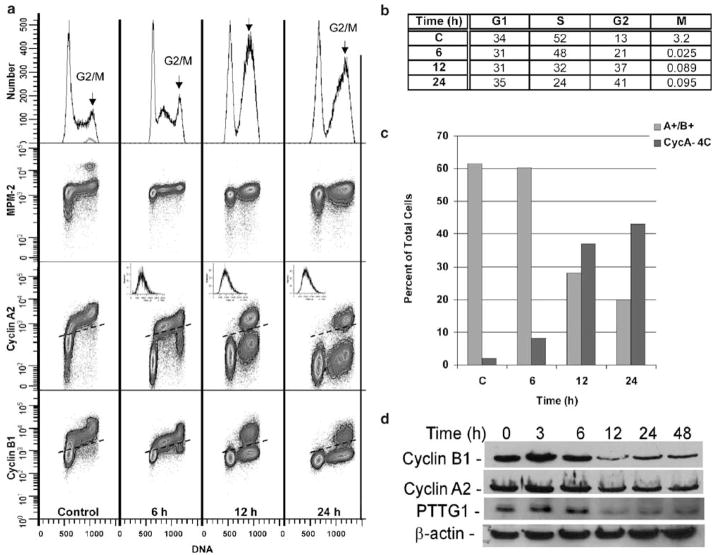
To distinguish between the G2 and M-phase cell populations having a similar 4C DNA content, a multi-parametric flow cytometric approach was used. The levels of Cyclins A2 and B1 were examined in combination with the phosphorylation of mitotic protein monoclonal-2 (MPM-2). MPM-2 phosphorylation occurs during mitosis and this assay is similar to the one that utilizes the phosphorylation status of histone H3 to detect M-phase cells (Yan et al., 2004). A fourth-dimensional analysis was provided by Hoechst staining for DNA content. The fraction of mitotic cells can be detected and counted by examining a plot of DNA content and MPM-2 immunoreactivity. In control cells, a 3.2% mitotic population (blue–green population) was present, which was depleted >99% following irradiation (Figure 2a, b). The decrease in the mitotic population confirms that cells were arrested in G2. The frequency patterns were consistent with an initial inhibition of G1, S, and G2 transit, which was followed by a stable arrest in G1 and G2 (Figure 2b). Beginning at 6 h, but clearly evident at 12 and 24 h, there are two clusters of 4C, non-mitotic cells. These represent 4C cells that either express both Cyclins A2 and B1 (bona fide G2 cells) or cells without significant levels of Cyclins A2 and B1 (Figure 2a, arrows). The Cyclin A2/B1-negative 4C clusters also show evidence of an additional population (recognizable by bimodality) that are shifted left with less DNA (arrows). These may represent apoptotic 4C cells. Examination of the frequencies of Cyclin A2/B1-positive and -negative 4C cells as a function of time indicates that cells progress from the cyclin-expressing state to the cyclin-negative state (Figure 2c). These results were confirmed by immunoblotting for Cyclins A2 and B1 (Figure 2d), which indicated that their levels were ~3-fold and 1.5-fold lower on average in irradiated cells at 12 h post-IR. Similarly, the levels of a critical mitotic protein PTTG1 (also called Securin) decreased approximately threefold compared to the untreated samples.
Figure 2.
Radiation-induced cell cycle arrest is specific to G2 phase. (a) Multiparametric staining was performed using the mitotic marker, MPM-2 (FSE, y-axis), Cyclin A2 (PE, y-axis), Cyclin B1 (Alexa 647, y-axis), and Hoechst 33342 (x-axis) for DNA content. The proportion of mitotic cells, highlighted with an arrow, was examined at the indicated times following irradiation. The light scatter profiles shown in the upper right-hand corners of the Cyclin A2 plot indicate that this population of A−/B− state is not apoptotic. (b) Proportion of cells in G1, S, and G2/M phase. (c) Bivariate analyses examined staining for Cyclin A2 and B1 in cells retaining 4C DNA content. For each staining combination and analysis, cells were collected before (0 h) and at 6, 12, and 24 h after irradiation. (d) Immunoblotting for Cyclin A, B1, and PTTG1 was performed using the respective primary antibodies. β-Actin was used as a loading control.
E2F4 nuclear colocalization with p130 following irradiation
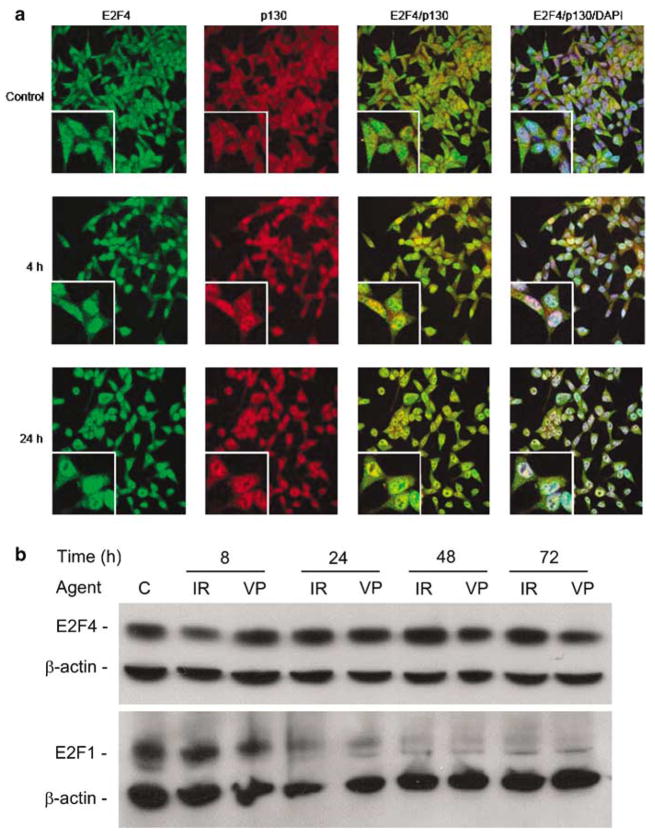
Irradiation induces E2F4, but not E2F5, to form a complex with the unphosphorylated form of p130 (DuPree et al., 2004). Examining the dynamics of E2F4 subcellular localization by confocal microscopy indicates that E2F4 and p130 were distributed throughout the cytoplasm and nucleus in untreated cells (Figure 3a). As early as 4 h post-IR (Figure 3a), E2F4 levels in the nucleus increased. p130 nuclear localization followed the same kinetics, the fluorescence signal overlay indicating its time-dependent nuclear colocalization with E2F4, that reached a peak at 24 h.
Figure 3.
E2F4 has sustained levels after irradiation and colocalizes with p130 at the time of G2 arrest. (a) Immunofluoresscence confocal microscopy (×40 magnification) with antibodies specific for E2F4 and p130 indicated colocalization following irradiation. Merging the fluorescence signals provided by the two antibodies indicated their nuclear colocalization following irradiation. (b) irradiation and VP-16 treatments lead to sustained levels of E2F4 protein expression. C4-2 cells were left untreated (control), treated with 10 μM VP-16 or irradiated, and harvested at the time points indicated. Proteins in cell lysates were resolved by 10% SDS– PAGE, before immunoblotting with anti-E2F4 and -E2F-1 antibodies, with β-actin used as a loading control.
Examination of E2F4 expression revealed that its levels were slightly decreased at 8 h following irradiation, increased above control levels, and were then maintained through at least 72 h post-irradiation (Figure 3a). Similarly, treatment with another DNA-damaging agent, the topoisomerase II inhibitor VP-16, did not lead to significant changes in E2F4 levels. In great contrast, the protein levels of E2F1, which may counterbalance the activity of E2F4 (Crosby and Almasan, 2004), were dramatically downregulated by 24 h. Decreased E2F1 levels were sustained through at least 72 h post-irradiation or VP-16 treatment (Figure 3b). As Cyclin B1 and Pttg1 are potential E2F-regulated genes, these results indicate that reduced E2F1 and sustained levels of active (nuclear) E2F4/p130 complexes could both play an active role in G2-phase arrest in response to irradiation.
E2F4 is required for a stable G2 arrest in response to genotoxic stress
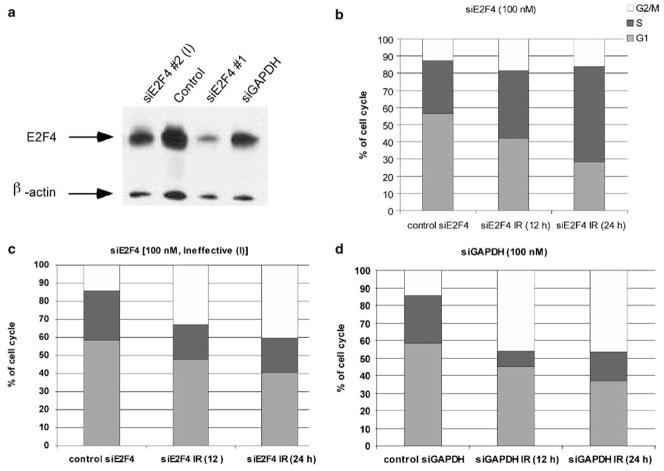
To investigate the functional role of E2F4 nuclear translocation, we examined the cell cycle arrest profile following irradiation in cells that had diminished E2F4 levels. Cells exposed to E2F4 small interference RNA (siRNA) optimized for effective E2F4 knockdown (DuPree et al., 2004), exhibited a substantial decrease in the levels of E2F4 (Figure 4a). When cells pretreated (24 h) with small interfering RNA (siRNA) against E2F4 (siE2F4) were irradiated, the cell cycle phase distribution was affected mostly in G1 and S; the proportion of the 4C cells at 12 and 24 h post-IR was increased only slightly (Figure 4b). This was in contrast to the cells treated with an ineffective E2F4 siRNA (siE2F4 #2 [I]) or glyceraldehyde 3-phosphate dehydrogenase (GAPDH)-specific siRNA (siGAPDH) before irradiation, which displayed the expected increase in the proportion of 4C cells at 12 and 24 h post-IR (Figure 4c) similar to earlier results (Figure 1a). Cell treatment with effective or ineffective (siE2F4 and siE2F4 #2 [I], respectively), vehicle control, or siGAPDH alone, did not result in any substantial changes in the cell cycle distribution (Figure 4c and d).
Figure 4.
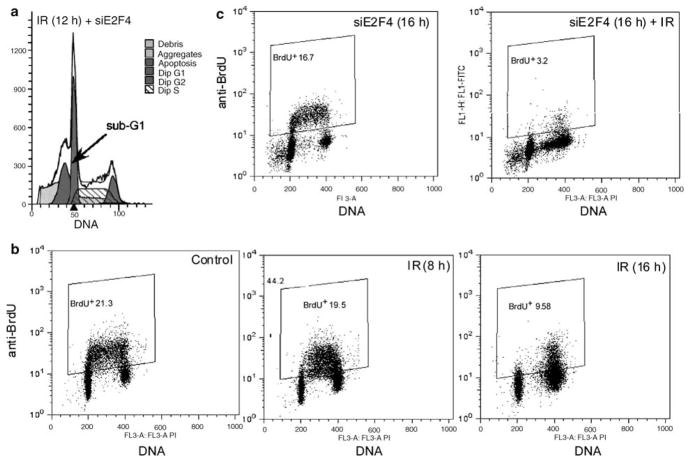
E2F4 downregulation causes abrogation of radiation-induced G2 cell cycle arrest and an apparent increase in S. (a) Cells were treated with vehicle control, siGAPDH, an ineffective or a selective siRNA against E2F4 at 24 h before collection and immunobloting, indicating that the specific siRNA against E2F4 (#1) was highly effective. (b) A non-specific siRNA to E2F4 [siE2f4 (I)] or specific to GAPDH (d) did not cause a marked decrease in the G2 arrest compared to data above (Figure 1a). (c) No changes in 4C and an apparent increase in the S-phase population in cells treated with siE2F4 before irradiation.
When E2F4 knockdown cells were irradiated, the DNA content distribution demonstrated a large accumulation of cells with sub-G1 content (Figure 5a, arrow, indicating apoptotic cells) and a pronounced accumulation of cells with late S-phase DNA content (Figure 5a). Cells also accumulated with <4C DNA content in the irradiated populations (Figure 2). In cells treated with siE2F4 that were not irradiated, this population was not apparent, although a slight perturbation of the frequency distribution in S is evident (Supplementary Figure). The <4C population is consistent with either a decrease in S-phase transit in late S or apoptosis originating from 4C G2 cells. In multiparametric analyses of irradiated cells, this population was shown to be both cyclin A and cyclin B1 negative at the 12 and 24 h time points (Figure 2a). Additionally, in irradiated cells at 6 h, a cluster of S-phase cells that accumulated in early-mid S were cyclin A and cyclin B1 positive, suggesting that cells responding to S-phase checkpoints retain expression of the mitotic cyclins. To further address the nature of these cells, bromodeoxyuridine (BrdU)-pulse labeling was used to distinguish S-phase cells from cells with a similar DNA content that were not synthesizing DNA. Following irradiation, BrdU incorporation decreased in a time-dependent manner, which is consistent with G1 and G2 arrest and a small sub-4C population of BrdU-negative cells (Figure 5b). This sub-G2 BrdU-negative population was present in irradiated cells; however, it was markedly enhanced in E2F4 knockdown irradiated cells (Figure 5c). The clustered early-mid S phase cells that accumulate at 8 h incorporated BrdU, suggesting that these ‘arrested’ cells are progressing slowly. Therefore, we tentatively conclude that in the absence of E2F4, cell death from G2 arrest is elevated and the likely trajectory is that cells arrest in G2, rapidly degrade the mitotic cyclins (few cells exist in intermediate states), then undergo apoptosis. This is the simplest model accounting for the combined results presented in Figures 2 and 5. We recognize that apoptosis has not been proven in this context.
Figure 5.
Lack of BrdU incorporation reveals apoptotic cells with sub-G1 and sub-G2 DNA content. (a) A sub-G1 population of cells is generated at 12 h following radiation treatment in cells that have been transfected with siE2F4 24 h before irradiation. (b) Dual staining for incorporation of BrdU and DNA content following transfection and irradiation was performed to eliminate the possibility that the cells were re-entering S phase. Insets indicate the gating used for identifying BrdU+ cells (% in upper left-hand corner). (c) Cells were treated with vehicle alone or siE2F4, incubated for 24 h, irradiated, and labeled for 45 min before collection at either 8 or 16 h following irradiation. Although control cells show active incorporation of BrdU, those transfected and collected at 16 h following irradiation had a BrdU-negative S-phase population.
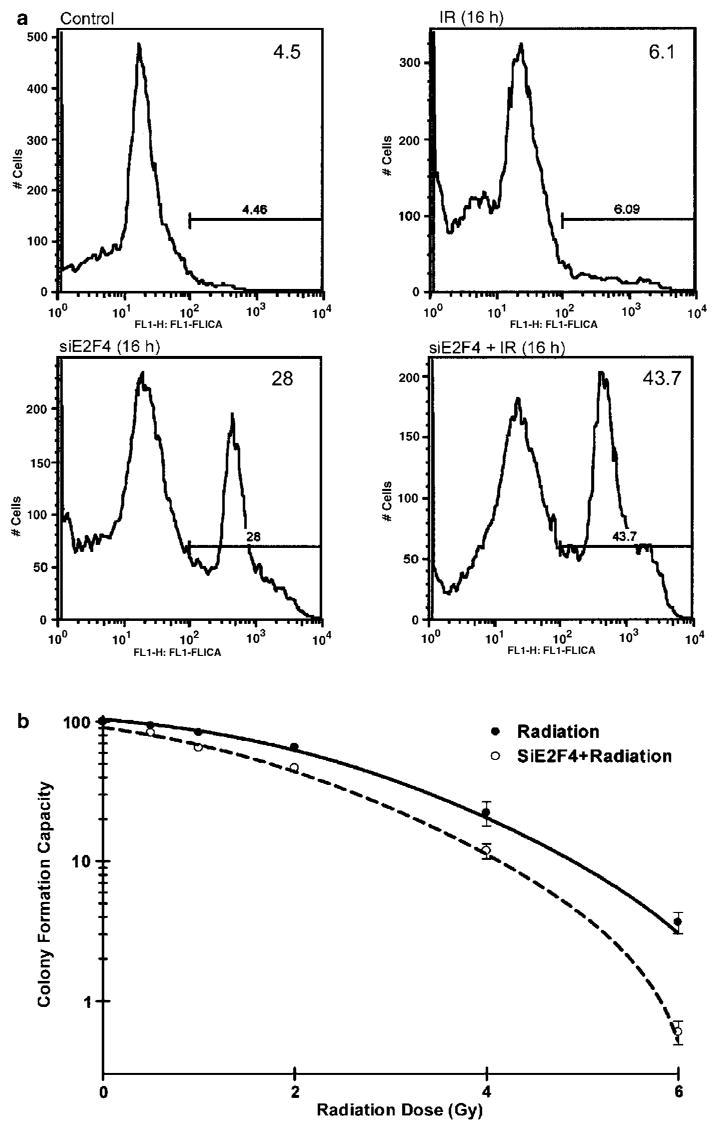
To confirm that apoptosis was elevated in siE2F4- treated cells that were irradiated, caspase activation was examined. Staining with CaspaTag™ indicated that 40% of the cells treated with siE2F4 had activated caspases at 16 h post-irradiation (Figure 6a). However, with siE2F4 transfection alone, ~15–20% of cells were apoptotic, as defined by this assay. This was also evident by sub-G1 measurements in non-irradiated cells (Figure 5c, left panel). These data suggest that E2F4 activity helps sustain G2 arrest by preventing cell death. To determine the long-term impact of the E2F4 knockdown, we examined clonogenic cell survival following treatment with different doses of radiation. siE2F4-treated cells were more sensitive to radiation, as indicated by the LD50 decrease from 2.1 to 1.1 Gy (Figure 6b). There was a significant (P⩾0.005) decrease in clonogenic survival at 4 Gy, which is a clinically therapeutic dose of radiation.
Figure 6.
E2F4 depletion leads to caspase activation and loss of clonogenic survival. (a) The CaspaTag™ assay was used to determine proteolytic caspase activation that results in free cysteine residues that covalently bind to the fluorescent substrate. (b) After treatment with siE2F4, cells were re-plated logarithmically (100–2500 cells/ml). After a 10-day incubation, colonies were fixed, stained and examined with respect to the untreated control. The significance was tested between irradiation and siE2F4+ irradiation by F-test using Prism software. Data represent the mean±s.d. of triplicate experiments.
Identification of putative E2F4 target genes involved in the radiation response
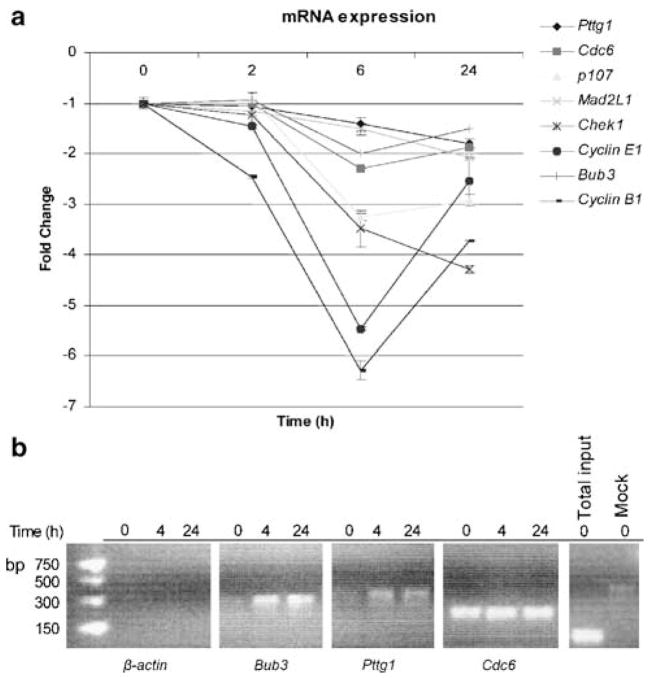
We next sought to identify genes whose expression was associated with the radiation-induced block in G2 to M transition by performing oligonucleotide microarray experiments. From this experiment (J Buchsbaum et al., unpublished data), we identified seven mitotic genes that were ⩾2-fold down-regulated in irradiated C4-2 cells at both 6 and 24 h following irradiation (Table 1). Expression levels of transcripts that were previously considered to be E2F targets having a role in the G2 to M checkpoint (Ren et al., 2002; Polager and Ginsberg, 2003) were validated by quantitative real-time polymerase chain reaction (RT-PCR) (Figure 7a). Both methods indicated a similar overall trend of down-regulation. At 6 h following irradiation, quantitative RT-PCR results indicate that all transcript levels were decreased ⩾2-fold, with most changes observed for Cyclin E1 (5.5-fold), Cyclin B1 (6.3-fold), and CHK1 (3.5-fold) expression. Interestingly, the decrease in levels of Pttg1 and Bub3 seemed to be delayed, with a continuing time-dependent decrease through the duration of the experiment. In comparison, other transcripts, particularly those for Cyclin B1 and E1, were downregulated rapidly by 6 h and then started to increase again by 24 h. Decreased RNA levels also correlated with decreased protein levels, as revealed by the effect of irradiation on two E2F target gene products. Cyclin B1 and Pttg1 showed a similar kinetics of downregulation following irradiation at 12 h post-IR (Figure 2b). In general, the changes in protein levels mimic those observed at the transcript level. However, while mRNA levels started to increase by 24 h, the protein levels did not recover through 48 h, suggesting a role for both transcriptional and post-transcriptional control mechanisms.
Table 1.
Putative E2F4 target genes identified by HU-95 Affymetrix array
| Gene\levels | Absolute 0 h | Fold change 6 h after IR | Fold change 24 h after IR |
|---|---|---|---|
| Bub1b | 1.4 | −2.5 | −16.1 |
| CenpE | 0.9 | −2.0 | −24.6 |
| CHK1 | 1.0 | −8.8 | −3.8 |
| Hec | 7.2 | −8.7 | −23.9 |
| Mad2L | 3.5 | −2.4 | −3.8 |
| Pttg1 | 28.7 | −4.4 | −10.2 |
| Cdc6 | 1.3 | −2.6 | −2.0 |
Abbreviation: IR, ionizing radiation.
Figure 7.
Irradiation induces downregulation of E2F4 target genes. (a) Quantitative RT-PCR was performed to validate the levels of mRNA of genes identified by the microarray (Table 1) using the primers described in Table 2 (Materials and methods) at various time points following irradiation. The data are expressed with respect to the endogenous GAPDH control, values reported being relative to the 0 h time point. (b) ChIP analyses confirmed dynamic target binding to Bub3 and Pttg1 genes and constitutive binding to CDC6. β-Actin primers were used to confirm that there was no non-specific E2F4 binding and to serve as a positive control for the ‘Total input.’ In the ‘Mock’ control, ChIP was carried out without chromatin as a control for non-specific binding.
To determine if E2F4 played a direct role in gene regulation, quantitative RT-PCR experiments were performed for promoter-binding analyses using chromatin immunoprecipitation (ChIP), as described (Crosby et al., 2004). The promoter sites examined were chosen based on previous studies, which investigated the putative role of E2F4 in various cell cycle phases, including G2 and M, through combined crosslinking DNA immunoprecipitation and microarray experiments (Ren et al., 2002). To examine the effect of E2F4 regulation in the absence of any perturbing factors, we investigated the promoter occupancy at the time when the E2F4/p130 complexes entered into the nucleus and the cells exhibited the most prominent G2 arrest. ChIP analyses indicated that E2F4 is recruited to the promoter-binding sites of Pttg1 and Bub3 at 4 h following irradiation. E2F4 localization on these promoter sites persisted through at least 24 h (Figure 7b). In contrast, E2F4 was bound constitutively to the Cdc6 promoter region. As expected, there was no non-specific binding to the β-actin promoter region. These data indicate that E2F4 has a direct role in the radiation-induced repression of multiple genes that encode important mitotic regulators, such as those of Pttg1 and Bub3, through its recruitment to their promoters.
Discussion
The G2 arrest represents a mechanism by which many cell types, including those of epithelial origin, are able to halt DNA segregation and thereby prevent an accumulation of mutations, which would otherwise promote carcinogenesis (Kastan and Bartek, 2004). This arrest serves to allow time for repair of DNA damage, which may then result in resumption of proliferation or, if the damage is too severe, permanent arrest (Linke et al., 1996) or senescence (Narita et al., 2003) to prevent further genetic instability (Almasan et al., 1995a; Bindra and Glazer, 2005; Bindra et al., 2005). Although this mechanism is beneficial to non-neoplastic tissues, in tumor cells it can promote resistance to therapies that rely on eliminating proliferating cells. Clearly, identifying key mediators of G2 arrest is critical to sensitization of otherwise therapy-resistant cells. Two mechanisms have been described. An early and transient G2 arrest is considered to be ATM dependent (Xu et al., 2002). A second G2 arrest, which occurs later in response to irradiation caused in the earlier phases of the cell cycle, is independent of ATM (Iliakis et al., 2003). E2F4 is a good candidate for regulating this second, sustained G2 arrest, as depletion of ATM by siRNA-mediated knockdown had no effect on this radiation-induced G2 arrest (S Ray et al., unpublished data).
Earlier studies seeking to identify key proteins implicated in G2 arrest indicated that it is dependent on the pRB family members (Chan et al., 2000; Taylor et al., 2001). For example, p130 was shown to interact with E2F4 and repress the Cdk1 promoter when p53 was overexpressed in the absence of any DNA damage (Taylor et al., 2001). Overexpression of p53 regulates the extent of the arrest via Cyclin B1 repression (Innocente et al., 1999). However, the arrest in C4-2 cells can occur independently of Cyclin B1 and its associated kinase activities, as our investigations indicated that following irradiation, there are minimal changes in the p53-deficient DN-p53-C4-2 cells while there are dramatically downregulated Cyclin B1 levels in C4-2 cells (S Ray et al., unpublished data). Yet, both cell lines have a comparable G2 arrest.
Previously, we have shown that E2F4 undergoes a dramatic cellular redistribution, by translocating to the nucleus following irradiation (DuPree et al., 2004). This study extends these findings and defines a novel E2F4 function in the DNA damage response under physiological conditions, as irradiation resulted in nuclear colocalization of E2F4 with p130. Importantly, the knockdown of E2F4 prevented the radiation-induced G2 arrest and promoted apoptosis. The decreased levels of E2F1 may also contribute to a lack of apoptosis and radiation resistance, as increased levels of E2F1 are known to lead to apoptosis (Rogoff et al., 2002; Stevens et al., 2003). By downregulating E2F4 levels before irradiation, the extent of the stable G2 arrest was greatly diminished, with an increase in the proportion of cells with late S-phase DNA content. These cells were not synthesizing DNA and are better characterized as sub-G2 apoptotic cells. The increased DNA damage observed is reminiscent of a similar observation made for E2F1-mediated DNA double-strand break accumulation following RB inactivation (Pickering and Kowalik, 2006). To our knowledge, this is the first demonstration that E2F4 is critical for a stable G2 arrest.
The E2F family of transcription factors has now been implicated in the control of all phases of the cell cycle, including the regulation of genes during the G2 to M-phase transition (Ishida et al., 2001; Giangrande et al., 2004). Previous ChIP studies indicated that E2F members occupied gene promoters in a cell cycle-specific manner, suggesting their specific roles during the cell cycle through coordinated targeting and regulation of specific genes (Takahashi et al., 2000; Wells et al., 2000; Rayman et al., 2002). Together with other reports indicating multiple promoter occupancies that lead to both positive and negative transcriptional activation thresholds (Giangrande et al., 2004), we suggest a physiological role for E2F4 in the radiation-dependent stable G2 arrest, just as exogenous E2F 1–3a can promote cell cycle progression. For example, we found that E2F4 is responsible for binding to the promoters of and thus repressing two critical mitotic genes, Bub3 and Pttg1, following irradiation. It was shown that E2F4 is indeed capable of binding to promoters that were regulated during the G2 to M transition that could also be occupied by the activating E2Fs (1–3a) (Zhu et al., 2004). Although defining the role of individual target genes responsible for carrying out the function of E2F4 is of interest, multiple genes are likely to be important for sustaining cooperatively the G2 arrest in many experimental systems (Chan et al., 2000), including that of irradiated prostate tumor cells (J Buchsbaum et al., unpublished data).
C4-2 cells are arrested in G2 following irradiation for an extended period of time, but eventually resume cell proliferation and form colonies. A 4C DNA content and low levels of Cyclins A2 and B1 have been suggested also for cells undergoing a G1 arrest, as they undergo endoreduplication, or those that proceed through mitosis inappropriately and, as a result, undergo mitotic catastrophe. However, the DNA content profiles and BrdU-incorporation experiments excluded additional rounds of DNA replication taking place. Moreover, there was no indication that cells reached mitosis based on examining specific Cyclins (A2 and B1) and general (MPM-2) mitotic markers for at least a 24 h time period. Had the cells been recovering from arrest, abnormal mitoses should have occurred, such as those characterized in mitotic catastrophe (Chang et al., 2000). Interestingly, the prostate cancer epithelial cells used in our investigation are known to not undergo such events (Roninson I, personal communication). As C4-2 cells have functional pRB and p21, whose deficiency has been associated with endoreduplication (Niculescu et al., 1998; Chang et al., 2000), these cells may be undergoing a cell cycle exit that is initiated in G2 (Baus et al., 2003; Jackson et al., 2005), which does not involve endoreduplication or mitotic catastrophe.
Interestingly, a number of E2F4-bound genes originally identified through array analyses were also bound to E2F1 (Ren et al., 2002). This observation has been extended recently to the promoters of genes that are regulated during G2 and M, which contain distinct E2F4- and E2F1-binding sites (Zhu et al., 2004). Therefore, there is likely a balance between the E2Fs that occupy these promoters, indicating that having more E2F4 bound may result in cell cycle arrest (Crosby and Almasan, 2004). Conversely, having more E2F1 bound to the promoters is expected to support continued progression through the cell cycle. Following DNA damage, E2F1 levels may increase to promote cell death (Rogoff et al., 2002; Crosby and Almasan, 2004). In C4-2 cells subjected to genotoxic stress, the expression levels of E2F1 decrease and the levels E2F4 are sustained. Whereas increased levels of E2F1 are indicative of apoptosis, decreased levels may allow for increased cell survival as a result of failing to activate E2F1 apoptotic target genes. Thus, E2F4 inactivation and E2F1 activation may both promote apoptosis through increased gene expression. Apoptosis may be avoided through gene repression mediated by nuclear colocalization of E2F4 with p130.
In summary, these results reveal a novel role for E2F4 in regulating a stable G2 arrest that may play a critical role in the radiation resistance of prostate carcinoma, which is characterized by increased E2F4 expression (Waghray et al., 2001). E2F4, known earlier to regulate gene repression during G0, is now implicated in DNA damage-induced gene repression. Whereas E2F1 is known to have roles beyond its function to activate genes required for the G1/S transition, we now provide evidence that E2F4 acts in a similar manner, by having an important role beyond its established role in cellular quiescence. Therefore, the implications of E2F4 translocation to the nucleus and its association with p130 provide new insights into the G2 arrest response following genotoxic stress. The biological role for individual E2F4-repressed genes in the genotoxic stress response is of great interest for our future studies.
Materials and methods
Cell culture and treatment
Prostate carcinoma LNCaP C4-2 cells were maintained in a humidified incubator at 37°C, 5% CO2. The cells were grown in monolayer culture in RPMI-1640 with 10% heat-inactivated fetal bovine serum (Ray and Almasan, 2003). Exponentially growing cells were treated with IR(10 Gy unless indicated otherwise) from an X-ray irradiator (Pantak: 320 kVP, 20 A, half-value layer 2mm Cu; East Haven, CT, USA) emitting at a fixed dose rate of ~2 Gy/min.
Flow cytometry analyses
For propidium iodide (PI) staining, following trypsinization and washes in ice-cold phosphate-buffered saline (PBS), cells were fixed with 90% methanol (−20°C). Cells were then washed twice with PBS, treated with RNAse A (200 μg/ml; Gentra Systems, Minneapolis, MN, USA) for 20 min, and stained with 50 μg/ml PI (Sigma). Samples were analysed on a FACScan with a 488nm Argon laser (BD Biosciences, San Jose, CA, USA). Data were processed using FlowJo V6.1.1 software (Tree Star, Inc., San Carlos, CA, USA).
For multiparametric staining, 106 cells/time point, fixed as above, were incubated with antibodies that were labeled with fluorescent dyes before DNA staining, as previously described (Yan et al., 2004). The fixed cells were washed twice with ice-cold PBS and once with PBS/bovine serum albumin (BSA; 2% in PBS). Cells were stained for 1 h with Fluorescein-5-EX, succinimidyl ester-(FSE)-conjugated anti-MPM-2 (Upstate), Alexa 647-conjugated anti-Cyclin B1 (GNS1 clone; (Yan et al., 2004)), and Phycoerythrin (PE)-conjugated anti-Cyclin A2 (a gift from Vince Shankey, Beckman Coulter, Miami, FL, USA). Cells were washed three times with PBS/BSA and stained with Hoechst 33342 (Invitrogen, Carlsbad, CA, USA) for 30 min before analysis. The fluorescence measurements were done on an LSR II Flow Cytometer (BD Biosciences). As MPM-2 is specifically phosphorylated during the M phase of the cell cycle, DNA content and this mitotic marker allow for distinguishing between the G2 and M phases.
For BrdU/PI labeling, cells were pulse-labeled for 45 min with BrdU (10 μM; Sigma, St Louis, MO, USA). Cells (106) fixed in methanol overnight were washed twice with PBS and pellets treated with 4N HCl (in 0.1% Triton X-100) for 20 min at room temperature (RT). PBS/BSA in 0.1% Triton X-100 was added to the cells, which were then pelleted by centrifugation, before treatment with 0.1 sodium borate buffer (pH 8.5) for 2 min at RT and centrifugation at 700 g for 5 min. The pellet was washed once with PBS/BSA and twice with PBS/BSA in 0.1% Triton X-100. The cells were stained with fluorescein isothiocyanate (FITC)-conjugated anti-BrdU (BD Biosciences) for 1 h at 37°C in dark. Cells were washed twice with PBS/BSA, treated with RNAse A (200 μg/ml) (Gentra Systems) for 20 min, and stained with PI (50 μg/ml).
Caspase activation assay
The CaspaTag™ Pan-Caspase In situ Assay Kit, Fluorescein (Chemicon International, Temecula, CA, USA) was used to examine caspase activation in cells incubated with the Fluorochrome Inhibitors of Caspases (FLICA) reagent for 1 h before trypsinization, collection, and three rinses with the washing buffer. The inhibitor, a carboxyfluorescein-labeled fluoromethyl ketone peptide inhibitor of caspase (FAM-VAD-FMK), binds covalently to activated caspases’ reactive cysteine residue that is localized on the large subunit of the active caspase heterodimer. This reaction inhibits further enzymatic reactivity and the green fluorescence produced by the bound reagent is directly proportional to the amount of active caspases at the time of sample preparation. Samples were analysed for caspase positive (+) or negative (−) cells on a BD LSR II Flow Cytometer (BD Biosciences).
siRNA
Double-stranded siRNA against E2F4 (siE2F4) was prepared using sense and antisense RNA oligonucleotides as described (DuPree et al., 2004). Cells, grown to ~50% confluence, were transfected with siRNA-E2F4 (100 nM) using Oligofectamine (2.6 μl/ml) and Optimem (10 μl/ml) (Invitrogen) before irradiation. Ineffective [I] siE2F4 #2 (siE2F4 #2 [I]), a second siGAPDH control, and vehicle control (transfection reagents) were included as controls for non-specific siRNA transfection. Cells were harvested at 24 h post-IR, and mRNA levels analysed by quantitative RT-PCR. For cell cycle and BrdU labeling experiments, cells were transfected 24 h before irradiation.
Clonogenic assays
Exponentially growing cells (untreated or pretreated with siRNA against E2F4) were trypsinized. Cells (20 000–300 dilution) were inoculated (plating efficiency 80–90%) in Petri dishes and exposed to graded doses (0–10 Gy) of X-ray irradiation. Cells were then allowed to grow for 12 days, fixed and stained in methanol:acetic acid (75:25, v/v) containing 0.5% crystal violet (w/v) to visualize colonies of at least 50 cells.
Western blot analysis
Total protein, whose concentration was estimated by the Bio-Rad Protein Assay (Bio-Rad Laboratories Hercules, CA, USA) from whole-cell lysates, was electrophoresed (25 μg) on a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gel, as described (Crosby et al., 2004; DuPree et al., 2004). Proteins were then electrotransferred onto nitrocellulose membranes that were probed for E2F1 (C-20; 1:500), E2F4 (C-19; 1:200), Cyclin A2 (H-432; 1:1000), Cyclin B1 (GNS1; 1:1000), all from Santa Cruz Biotechnology, Santa Cruz, CA, USA, or PTTG1 (1:1000; MBL International Corporation, Woburn, MA, USA). β-Actin (Sigma) was used as a loading control. Blots were visualized with either sheep anti-rabbit or anti-mouse secondary antibodies, which were conjugated to horseradish peroxidase (HRP), and LumiGLO Chemiluminescent reagents (KPL, Gaithersburg, MD, USA).
Confocal microscopy
Cells grown on coverslips were fixed in 3.7%. formaldehyde/PBS for 20 min at RT. Cells were then washed three times with PBS, blocked, and permeabilized for 5 min at RT with blocking buffer (2% goat serum in 0.3% Triton X-100 in PBS). Cells were incubated with anti-E2F4 and anti-p130 antibodies (1:100 in Triton X-100 blocking buffer) for 1 h at RT. Cells were again rinsed three times and incubated with Alexa 488- and 594-conjugated anti-rabbit/anti-mouse secondary antibodies (1:200, in Triton X-100 blocking buffer), respectively, for 45 min in dark. The coverslips were rinsed three times with blocking buffer, mounted with Vectashield containing 4, 6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA, USA), and the fluorescence signals were examined using a Leica TCS-SP2 (Leica Microsystems AG, Wetzlar, Germany) microscope at ×40 magnification.
Expression profiling with oligonucleotide arrays
Total RNA was extracted from cells by the Trizol® (Invitrogen) method and reverse-transcribed into double-stranded cDNA following isolation of Poly(A) RNA with RNAeasy (Qiagen, Valencia, CA, USA). The double-stranded cDNA was used to prepare biotinylated cRNA (Enzo Bioarray High Yield Kit, Affymetrix, Santa Clara, CA, USA). These cRNA probes were hybridized to the Affymetrix HU-95 oligonucleotide array. The GenePix 4000A laser scanner (Axon Instruments, Foster City, CA, USA) and GenePix v.3.0 scanning software were used to analyse the data. Quantitative RT-PCR experiments were performed to validate the levels of expression in the repressed gene targets. Total RNA was extracted from cells by the Trizol® method and reverse-transcribed into cDNA using Taqman Reverse Transcription Reagents (Roche for ABI, Branchburg, NJ, USA).
Quantitative RT-PCR
PCR amplification reactions were comprised of 1×iQ SYBR Green Supermix (Bio-Rad) and 4 μM each of sense and antisense primer. Reactions were carried out in a 96-well optical reaction plate with optical caps (Applied Biosystems, Foster City, CA, USA) in an iCycler iQ Real-Time PCR Detection System spectrofluorometric thermal cycler (Bio-Rad) with an initial 3 min incubation at 95°C followed by 40 cycles of amplification: 95°C for 15 s and 60°C for 1 min. Primers (Table 2) were synthesized by Integrated DNA Technologies, Inc. (IDT; Coralville, IA, USA).
Table 2.
PCR primer sequences for putative E2F4 target genes
| Gene | Sense primer (5′to 3′) | Antisense primer (5′to 3′) |
|---|---|---|
| Gapdh | CAC CAC TGA CAC GTT GGC AG | GAA ACT GTG GCG TGA TGG C |
| Bub3 | TTG GTG TAA GTC TGA ACC CAT CTT T | CAC AGT AAC TCT AAC ACA TCC CTT AGG G |
| Ccne1 | CCC ATC CTT CTC CAC CAA AG | CCC TGT TTG ATG CCA TCC AC |
| Cdc6 | GCC TCA GCC TCC CGA GTA G | TAG GGA GGC CAA GGT GGG |
| CHK1 | TGG TTG ACT TCC GGC TTT CT | TTC ACC AGG ATT CCC CAG AG |
| Ccnb1 | TGG TGA AGA GGA AGC CAT GG | AAG AGC TGT TCT TGG CCT CAGT |
| Hec | CAG AGG CAA AGA AGC GAT TGA | TTT GAC AAG GCA GTT GGC AC |
| Mad2L1 | TGA GGT CCT GGA AAG ATG GC | CAG TGG CAG AAA TGT CAC CG |
| p107 | ATC CAG GTA CCA CCG CCA T | TGG GTT GTG ACA TCA GGC TG |
| Pttg1 | TTT GAC CTG CCT GAA GAG CA | GAT TGG ATT CCC ATG GTG GA |
Abbreviation: PCR, polymerase chain reaction.
ChIP Assay
ChIP was performed as described (Crosby et al., 2004), with some modifications. Briefly, cells were incubated in culture media containing formaldehyde (1%) for 10 min at RT, with rotation, to crosslink the protein to the DNA. The reaction was stopped with glycine (0.125M, 5 min). Cells were trypsinized and collected by gentle centrifugation, washed once with ice-cold PBS and centrifuged at 4°C before lysis and chromatin sonication. Chromatin was pre-cleared with pre-blocked Pansorbin (Calbiochem, San Diego, CA, USA) (10–15 μl/107 cells). The Pansorbin was incubated with the chromatin for 15 min at RT and samples were centrifuged. Pre-cleared chromatin was incubated overnight (1 μg/sample) with anti-E2F4 antibody. As a mock control, the antibody was incubated with dialysis buffer (2mM EDTA, 50mM Tris-Cl. pH 8.0). Chromatin complexes were immunoprecipitated with Pansorbin. Two sequential elution steps with 50mM NaHCO3, 1% SDS were performed in which the complexes were spun on a rotating platform for 15 min and centrifuged at 500 g. The supernatants were collected and combined before adding 0.5M NaCl and heating for 4 h at 67°C to reverse crosslinking.
Following sequential RNAse A and proteinase K treatments and DNA precipitation, 40 cycles of quantitative RT-PCR were performed with 2–3 μl purified ChIP DNA/reaction. PCR primers used were as described (Ren et al., 2002): Bub3: (sense) 5′-GCC CAA AAT GGG ATT CTT GTG -3′, (antisense) 5′-TCA TTT TAC CAC CGC GCT GGG-3′; Pttg1: (sense) 5′-AAG ACC TGC GTG AGT GAA TGG-3′, (antisense) 5′-CCA GCT CTC AAA TCT TCC AGC-3′; Cdc6: (sense) 5′-AAA GGC TCT GTG ACT ACA GCC A-3′, (antisense) 5′-GAT CCT TCT CAC GTC TCT CAC A-3′; β-actin: (sense) 5′-AAC TCT CCC TCC TCC TCT TCC TC-3′, (antisense) 5′-GAG CCA TAA AAG GCA ACT TTC GG-3′. Amplified reactions were separated on a 2% agarose gel stained with ethidium bromide (Bio-Rad).
Acknowledgments
We thank Drs WHeston and A Gudkov (Cleveland Clinic) for valuable reagents and B Dynlacht (NYU) for advice. We also thank the Flow Cytometry and Imaging Core facilities staff at the Cleveland Clinic Foundation, and RM Sramkowski at the Case Comprehensive Cancer Center for their invaluable help. This work was supported by research grants from the US National Institute of Health (CA81504 and CA82858).
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- Almasan A, Linke S, Paulson T, Huang L-c, Wahl GM. Genetic instability as a consequence of inappropriate entry and progression through S-phase. Cancer Metast Rev. 1995a;14:59–73. doi: 10.1007/BF00690212. [DOI] [PubMed] [Google Scholar]
- Almasan A, Yin Y, Kelly RE, Lee EY, Bradley A, Li W, et al. Deficiency of retinoblastoma protein leads to inappropriate S-phase entry, activation of E2F-responsive genes, and apoptosis. Proc Natl Acad Sci USA. 1995b;92:5436–5440. doi: 10.1073/pnas.92.12.5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwooll C, Denchi EL, Helin K. The E2F family: specific functions and overlapping interests. EMBO J. 2004;23:4709–4716. doi: 10.1038/sj.emboj.7600481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baus F, Gire V, Fisher D, Piette J, Dulic V. Permanent cell cycle exit in G2 phase after DNA damage in normal human fibroblasts. EMBO J. 2003;22:3992–4002. doi: 10.1093/emboj/cdg387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindra RS, Gibson SL, Meng A, Westermark U, Jasin M, Pierce AJ, et al. Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res. 2005;65:11597–11604. doi: 10.1158/0008-5472.CAN-05-2119. [DOI] [PubMed] [Google Scholar]
- Bindra RS, Glazer PM. Genetic instability and the tumor microenvironment: towards the concept of microenvironment-induced mutagenesis. Mutat Res. 2005;569:75–85. doi: 10.1016/j.mrfmmm.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Budde A, Schneiderhan-Marra N, Petersen G, Brune B. Retinoblastoma susceptibility gene product pRB activates hypoxia-inducible factor-1 (HIF-1) Oncogene. 2005;24:1802–1808. doi: 10.1038/sj.onc.1208369. [DOI] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- Chan TA, Hwang PM, Hermeking H, Kinzler KW, Vogelstein B. Cooperative effects of genes controlling the G(2)/M checkpoint. Genes Dev. 2000;14:1584–1588. [PMC free article] [PubMed] [Google Scholar]
- Chang BD, Broude EV, Fang J, Kalinichenko TV, Abdryashitov R, Poole JC, et al. p21Waf1/Cip1/Sdi1-induced growth arrest is associated with depletion of mitosis-control proteins and leads to abnormal mitosis and endoreduplication in recovering cells. Oncogene. 2000;19:2165–2170. doi: 10.1038/sj.onc.1203573. [DOI] [PubMed] [Google Scholar]
- Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24:2796–2809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- Crosby ME, Almasan A. Opposing roles of E2Fs in cell proliferation and death. Cancer Biol Ther. 2004;3:1208–1211. doi: 10.4161/cbt.3.12.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby ME, Oancea M, Almasan A. p53 binding to target sites is dynamically regulated before and after ionizing radiation-mediated DNA damage. J Environ Pathol Toxicol Oncol. 2004;23:67–79. doi: 10.1615/jenvpathtoxoncol.v23.i1.70. [DOI] [PubMed] [Google Scholar]
- Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- DuPree EL, Mazumder S, Almasan A. Genotoxic stress induces expression of E2F4, leading to its association with p130 in prostate carcinoma cells. Cancer Res. 2004;64:4390–4393. doi: 10.1158/0008-5472.CAN-03-3695. [DOI] [PubMed] [Google Scholar]
- Farkas T, Hansen K, Holm K, Lukas J, Bartek J. Distinct phosphorylation events regulate p130- and p107-mediated repression of E2F-4. J Biol Chem. 2002;277:26741–26752. doi: 10.1074/jbc.M200381200. [DOI] [PubMed] [Google Scholar]
- Gaubatz S, Lees JA, Lindeman GJ, Livingston DM. E2F4 is exported from the nucleus in a CRM1-dependent manner. Mol Cell Biol. 2001;21:1384–1392. doi: 10.1128/MCB.21.4.1384-1392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangrande PH, Zhu W, Schlisio S, Sun X, Mori S, Gaubatz S, et al. A role for E2F6 in distinguishing G1/S- and G2/M-specific transcription. Genes Dev. 2004;18:2941–2951. doi: 10.1101/gad.1239304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera RE, Sah VP, Williams BO, Makela TP, Weinberg RA, Jacks T. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol Cell Biol. 1996;16:2402–2407. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliakis G, Wang Y, Guan J, Wang H. DNA damage checkpoint control in cells exposed to ionizing radiation. Oncogene. 2003;22:5834–5847. doi: 10.1038/sj.onc.1206682. [DOI] [PubMed] [Google Scholar]
- Innocente SA, Abrahamson JL, Cogswell JP, Lee JM. p53 regulates a G2 checkpoint through cyclin B1. Proc Natl Acad Sci USA. 1999;96:2147–2152. doi: 10.1073/pnas.96.5.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, et al. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MW, Agarwal MK, Yang J, Bruss P, Uchiumi T, Agarwal ML, et al. p130/p107/p105Rb-dependent transcriptional repression during DNA-damage-induced cell-cycle exit at G2. J Cell Sci. 2005;118:1821–1832. doi: 10.1242/jcs.02307. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Linke SP, Clarkin KC, Di Leonardo A, Tsou A, Wahl GM. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- Narita M, Nunez S, Heard E, Lin AW, Hearn SA, Spector DL, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- Niculescu AB, III, Chen X, Smeets M, Hengst L, Prives C, Reed SI. Effects of p21(Cip1/Waf1) at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication. Mol Cell Biol. 1998;18:629–643. doi: 10.1128/mcb.18.1.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossovskaya VS, Mazo IA, Chernov MV, Chernova OB, Strezoska Z, Kondratov R, et al. Use of genetic suppressor elements to dissect distinct biological effects of separate p53 domains. Proc Natl Acad Sci USA. 1996;93:10309–10314. doi: 10.1073/pnas.93.19.10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering MT, Kowalik TF. Rb inactivation leads to E2F1-mediated DNA double-strand break accumulation. Oncogene. 2006;25:746–755. doi: 10.1038/sj.onc.1209103. [DOI] [PubMed] [Google Scholar]
- Polager S, Ginsberg D. E2F mediates sustained G2 arrest and down-regulation of Stathmin and AIM-1 expression in response to genotoxic stress. J Biol Chem. 2003;278:1443–1449. doi: 10.1074/jbc.M210327200. [DOI] [PubMed] [Google Scholar]
- Ray S, Almasan A. Apoptosis induction in prostate cancer cells and xenografts by combined treatment with Apo2 ligand/tumor necrosis factor-related apoptosis-inducing ligand and CPT-11. Cancer Res. 2003;63:4713–4723. [PubMed] [Google Scholar]
- Rayman JB, Takahashi Y, Indjeian VB, Dannenberg JH, Catchpole S, Watson RJ, et al. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 2002;16:933–947. doi: 10.1101/gad.969202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, et al. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogoff HA, Pickering MT, Debatis ME, Jones S, Kowalik TF. E2F1 induces phosphorylation of p53 that is coincident with p53 accumulation and apoptosis. Mol Cell Biol. 2002;22:5308–5318. doi: 10.1128/MCB.22.15.5308-5318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardet C, Vidal M, Cobrinik D, Geng Y, Onufryk C, Chen A, et al. E2F-4 and E2F-5, two members of the E2F family, are expressed in the early phases of the cell cycle. Proc Natl Acad Sci USA. 1995;92:2403–2407. doi: 10.1073/pnas.92.6.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens C, Smith L, La Thangue NB. Chk2 activates E2F-1 in response to DNA damage. Nat Cell Biol. 2003;5:401–409. doi: 10.1038/ncb974. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Rayman JB, Dynlacht BD. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- Taylor WR, Schonthal AH, Galante J, Stark GR. p130/E2F4 binds to and represses the cdc2 promoter in response to p53. J Biol Chem. 2001;276:1998–2006. doi: 10.1074/jbc.M005101200. [DOI] [PubMed] [Google Scholar]
- Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- Waghray A, Schober M, Feroze F, Yao F, Virgin J, Chen YQ. Identification of differentially expressed genes by serial analysis of gene expression in human prostate cancer. Cancer Res. 2001;61:4283–4286. [PubMed] [Google Scholar]
- Wahl GM, Carr AM. The evolution of diverse biological responses to DNA damage: insights from yeast and p53. Nat Cell Biol. 2001;3:E277–E286. doi: 10.1038/ncb1201-e277. [DOI] [PubMed] [Google Scholar]
- Wells J, Boyd KE, Fry CJ, Bartley SM, Farnham PJ. Target gene specificity of E2F and pocket protein family members in living cells. Mol Cell Biol. 2000;20:5797–5807. doi: 10.1128/mcb.20.16.5797-5807.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Kim ST, Lim DS, Kastan MB. Two molecularly distinct G(2)/M checkpoints are induced by ionizing irradiation. Mol Cell Biol. 2002;22:1049–1059. doi: 10.1128/MCB.22.4.1049-1059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T, Desai AB, Jacobberger JW, Sramkoski RM, Loh T, Kinsella TJ. CHK1 and CHK2 are differentially involved in mismatch repair-mediated 6-thioguanine-induced cell cycle checkpoint responses. Mol Cancer Ther. 2004;3:1147–1157. [PubMed] [Google Scholar]
- Zhu W, Giangrande PH, Nevins JR. E2Fs link the control of G1/S and G2/M transcription. Embo J. 2004;23:4615–4626. doi: 10.1038/sj.emboj.7600459. [DOI] [PMC free article] [PubMed] [Google Scholar]