Abstract
β-Lactamases are one of the major causes of antibiotic resistance in Gram negative bacteria. The continuing evolution of β-lactamases that are capable of hydrolyzing our most potent β-lactams presents a vexing clinical problem, in particular since a number of them are resistant to inhibitors. The efficient inhibition of these enzymes is therefore of great clinical importance. Building upon our previous structural studies that examined tazobactam trapped as a trans-enamine intermediate in a deacylation deficient SHV variant, a novel penam sulfone derivative was designed that forms a more stable trans-enamine intermediate. We report here the 1.28 Å resolution crystal structure of wt SHV-1 in complex with a rationally designed penam sulfone, SA2-13. The compound is covalently bound to the active site of wt SHV-1 similar to tazobactam, yet forms an additional salt-bridge with K234 and hydrogen bonds with S130 and T235 to stabilize the trans-enamine intermediate. Kinetic measurements show that SA2-13, once reacted with SHV-1 β-lactamase, is about 10 fold slower at being released from the enzyme compared to tazobactam. Stabilizing the trans-enamine intermediate represents a novel strategy for the rational design of mechanism-based class A β-lactamase inhibitors.
Introduction
Resistance to β-lactam antibiotics mediated by bacterial β-lactamases (EC 3.5.2.6) is a global problem 1;2. β-Lactamases hydrolyze and inactivate penicillins and cephalosporins, thereby rendering these β-lactams inactive before they reach their intended bacterial targets, the penicillin binding proteins (PBPs). Hence, β-lactamases ensure bacterial survival against the therapeutic challenge of β-lactam antibiotics. To overcome this threat, β-lactam antibiotics are often administered with β-lactamase inhibitors (clavulanate, sulbactam, and tazobactam)3. This successful strategy has prolonged the clinical utility of penicillin antibiotics, especially against Class A β-lactamases. Unfortunately, not all β-lactamases are susceptible to inhibitors and of those that are, resistance against such inhibitors is occurring more frequently than before 4 thus stimulating the need to design novel β-lactamase inhibitors 5-7.
The major members of class A β-lactamases are SHV-1, TEM-1, and CTX-M enzymes. These β-lactamases hydrolyze the inhibitors through a complex, multistep reaction pathway. The catalytic mechanism by which β-lactamase inhibitors react with β-lactamases has been of great interest yet the identity of the species giving rise to inhibition remains controversial 8-18. Figure 1a shows a reaction scheme as it has been proposed for tazobactam 19;20: following the non-covalent association step, S70 is acylated opening the β-lactam ring of the inhibitor. This is followed by cleavage of the C5-S bond opening the thiazolidine ring with concomitant formation of a C5=N imine. This imine intermediate partitions between three pathways: tautomerization to form both cis- and trans-enamines, covalent modification resulting in irreversibly inactivated enzyme, and hydrolysis which liberates active enzyme. If the rate constant for conversion to the trans-enamine competes with the other two processes, and if the rate constant for the conversion of the trans-enamine to imine is slow in comparison to the assay time, it can be detected as “reactivation” of the “dead” enzyme.
Figure 1.
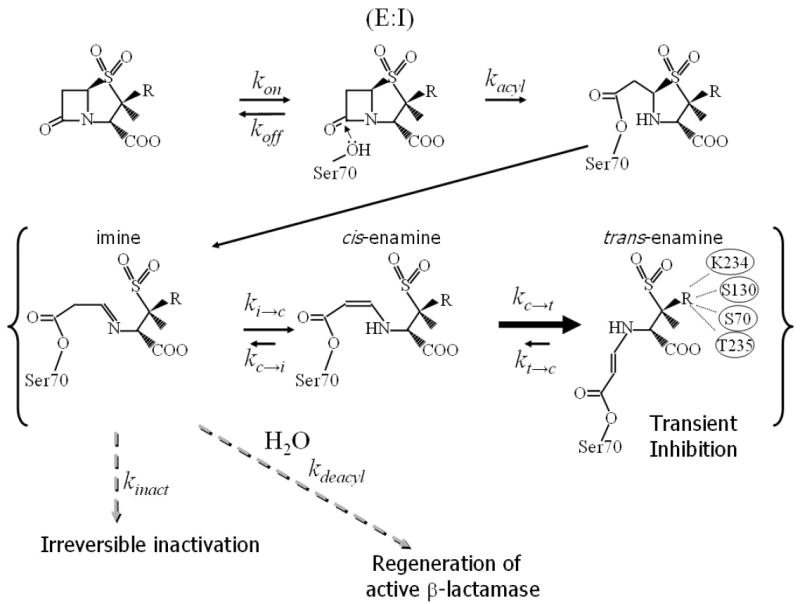

b. Schematic diagram of SA2-13 and tazobactam.
An approach to improving the efficiency of β-lactamase inhibitors is to slow down one or more of the rate constants by stabilizing a particular intermediate. Inhibitor resistant β-lactamase variants have been found to have decreased affinity for the inhibitors, increased turnover numbers and decreased kinact 21-24. Additionally these resistant enzymes display decreased complexity of inhibitory intermediates 25, and decreased trans-enamine formation 26. One approach to overcoming this problem is to decrease the rate constant for the deacylation step of β-lactamase inhibitors. For example, it was recently reported that a designed substrate penicillanic acid analog, with a 6α-hydroxylmethyl group in place of the ‘usual’ 6β-acylamino substituent, showed decreased deacylation 27. This modification was designed to displace a nucleophilic water (activated by the general base E166) needed for deacylation. Our groups recently interfered with the deacylation step of the reaction by mutating E166 to an alanine such that the nucleophilic deacylation water, normally positioned by this amino acid, is displaced. Through the use of Raman crystallography the resulting inhibition reaction 28 was tracked and showed the importance of the trans-enamine intermediate for sulbactam, clavulanic acid, and tazobactam, the latter being the most conformationally ordered 29;30.
In an effort to slow the deacylation step by improving the stability of the trans-enamine intermediate in wt SHV-1, the tazobactam bound SHV-1 E166A structure 30 was used as the starting point for rational inhibitor design. Tazobactam makes numerous interactions within the active site. However, the triazolyl moiety of tazobactam is an obvious candidate for alteration since it does not make a direct hydrogen bond, only a water mediated hydrogen bond with S130 30. Based on a previous crystal structure, it is hypothesized that this triazolyl moiety might be beneficially replaced by a negatively charged carboxylate attached through an appropriately sized linker. This penam sulfone compound (SA2-13, Figure 1b) was synthesized and found to be a good inhibitor of SHV-1 β-lactamase. We report here the 1.28 Å resolution crystal structure of a designed tazobactam analog SA2-13 bound to wt SHV-1. The wt SHV-1 structure contains the inhibitor in a trans-enamine conformation stabilized via the carboxylate group positioned in close proximity to K234, S130, and the catalytic S70. This structure-based penam sulfone inhibitor captured in the active site offers new opportunities to exploit the trans-enamine intermediate for successful class A β-lactamase inhibition.
Materials and Methods
Design and synthesis of SA2-13
The starting point for the SA2-13 design are the coordinates of the trans-enamine intermediate of tazobactam trapped in the deacylation deficient E166A mutant of SHV-1 30. The position and interactions of the triazolyl moiety of tazobactam within the active site seemed sub-optimal since no direct hydrogen bonds were made. Tazobactam’s triazolyl moiety assumes a similar position to an ordered HEPES molecule observed in previous SHV-1 structures 29;31. The ability of SHV-1 to bind a negatively charged group within this active site region, such as the sulfonate group of HEPES, generated the basis for the modification of tazobactam. The triazolyl group was therefore replaced by a carboxylate moiety attached via a short linker (Figure 1). Such a linked-fragment approach 32 would allow the negatively charged group to interact with K234, S130, T235 in a similar fashion as the sulfonate moiety of HEPES. Experimental details of the synthesis and spectral data of SA2-13 are in the supporting information. Tazobactam was a kind gift of Wyeth Pharmaceuticals.
Kinetics of inactivation
Kinetic constants of the inactivation of the SHV-1 β-lactamases were determined at 25 °C (room temperature, RT), using an AgilentTM 8453 diode array spectrophotometer. Each experiment was performed in a 1 ml final volume using 20 mM phosphate-buffered saline at pH 7.4. Measurements were obtained using nitrocefin (BD Biosciences) (Δε 482 = 17400 M−1 cm−1). A direct competition assay was performed to determine the dissociation constant for the preacylation complex, KI or KD, of the inhibitors (SA2-13 and tazobactam). A final concentration of 100 μM nitrocefin was used as the indicator substrate and 7 nM SHV-1 β-lactamase in these determinations. The data were analyzed to account for the affinity of nitrocefin for the SHV-1 β-lactamase:
| (1) |
The first-order rate constant for enzyme inactivation, kinact, was determined by monitoring the reaction time courses in the presence of increasing concentrations of inactivators. A fixed concentration of enzyme, nitrocefin, and increasing nM concentrations of inactivator (tazobactam or SA2-13) were used in each assay. The kobs for inactivation was determined graphically as the reciprocal of the ordinate of the intersection of the straight lines obtained from the initial, v0, and final, vf, steady-state velocities. Each kobs was plotted versus inhibitor concentration, I, and fit to Equation 2 to determine kinact and kinact/KI (the second order rate constant for reaction of free enzyme with free inhibitor to give inactive enzyme):
| (2) |
The partitioning of the initial enzyme inhibitor complex between hydrolysis and enzyme inactivation, i.e. the turnover number (tn = kcat/kinact), was obtained in the following manner. First, increasing amounts of inhibitor (SA2-13 or tazobactam) were incubated with a fixed concentration of SHV-1 β-lactamase in a total volume of 600 μl of 20 mM phosphate-buffered saline, pH 7.4, at RT. After 24 h, an aliquot (40 μl) was removed from the mixture and the steady-state velocity was measured and compared with a control sample with no inactivator added. The proportion of SA2-13 or tazobactam relative to SHV-1 that resulted in 90% inactivation after 24 h was designated the turnover number, tn (as previously defined 19).
These values served as a guide to determine the ratio of inhibitor (I) to enzyme (E) in an experiment to determine kreact, the apparent first order rate constant for reactivation of the β-lactamase. To determine kreact, 13.9 μM SHV-1 was mixed with 1.25 mM SA2-13 or 1.25 mM tazobactam in a total volume of 80 μl for 30 minutes at RT (I/E ratio of SA2-13/SHV-1 = 90/1 and I/E ratio of tazobactam /SHV-1 = 90/1) forming a combination of inactive enzyme along with enzyme bound trans-enamine. This mixture was placed on a Macro Spin Column G10 (The Nest Group, Southborough, MA) and spun in an Eppendorf 5415D Table Top® centrifuge at 2000 rpm at RT for 30 s to remove unbound inactivator. Reactivation of the enzyme, corresponding to conversion of the enamine to free enzyme, was detected by incubating the 80 μl flow thru volume for 5, 30 and 1440 min at RT. At these designated time points, a 5 μl aliquot was removed and the transient recovery of activity monitored in the presence of 100 μM nitrocefin as an indicator substrate (final concentration in 1 ml cuvette is 7 nM SHV-1, 630 nM SA2-13 and 630 nM tazobactam). To serve as a control, 13.9 μM of SHV-1 without inhibitor was treated in an identical manner and measured at each time point. Enzfitter and Microsoft Excel were used as software programs for analysis and data is shown in Table 1.
Table 1.
Kinetic measurements for SA2-13 and tazobactam
| KD (μM) | kinact (s-1) | kinact/KD | kreact(5’) (s-1) | kreact(30’) (s-1) | |
|---|---|---|---|---|---|
| tazobactam | 0.1 ± 0.025 | 0.10 ± 0.006 | 1.0 ± 0.3 | 0.034 ± 0.001 | 0.040 ± 0.016 |
| SA2-13 | 1.7 ± 0.1 | 0.05 ± 0.001 | 0.03 ± 0.002 | 0.004 ± 0.001 | 0.004 ± 0.010 |
| % residual v (5’) | % residual v (30’) | % residual v (24h) | tn | ||
| tazobactam | 4 | 4 | 0 | 5 | |
| SA2-13 | 4 | 4 | 47 | 120 |
Crystallization and Soaking
SHV-1 β-lactamase was expressed and purified according to previously described protocols 28. Crystals of SHV-1 were grown using the sitting drop method as described previously 33. The final 10 μL drop was prepared using 4 μl protein solution, 1 μl 5.6 mM Cymal-6 (Hampton Research), and 5 μl 30% w/v PEG 6000, 100 mM HEPES pH 7.0. The reservoir solution contained 30% w/v PEG-6000 in 100 mM HEPES pH 7.0 and crystals grew in 2-3 days at RT. To determine the best soaking time to trap the intermediate, Raman crystallography was used to identify and track the intermediate (which led us to increase the soaking concentration to 50 mM to obtain maximal occupancy). SHV-1 crystals were transferred to 10μl drops containing 30% PEG 6000 in 0.1 M HEPES pH 7.0 with 0.56 mM Cymal-6 and 50 mM SA2-13 compound and allowed to soak for 10 minutes. This was immediately followed by cryo-protection of the crystals via transfer to mother liquor containing 25% 2-methyl-2,4-pentanediol (MPD) for 1-2 minutes including the 50 mM SA2-13 prior to cryo-quenching by immersing the crystal in liquid nitrogen using a cryo-loop (Hampton Research, Inc.). Data were collected at the Advanced Photon Source APS (SBC beamline 19-ID) at 1.0332 Å wavelength to 1.28 Å resolution and were processed using SCALEPACK and DENZO 34 (Table 2).
Table 2.
Data collection and refinement statistics for wt SHV-1 complxed to SA2-13
| Data Collection | |
| Space group | P212121 |
| Unit cell dimensions (Å) | 49.8, 55.2, 84.4 |
| Wavelength (Å) | 1.0332 |
| Resolution (Å) | 50-1.28 (1.33-1.28) |
| Redundancy | 5.9 |
| Unique reflections | 59,323 |
| /<σ(I)> | 41.5 (10.4) |
| Rmerge(%) | 3.6 (16.6) |
| Completeness(%) | 97.7 (96.5) |
| Refinement | |
| Resolution range (Å) | 50-1.28 |
| R-factor | 15.4% |
| R-free | 16.7% |
| RMSD deviations from ideality | |
| Bond lengths (Å) | 0.0076 |
| Angles (°) | 1.50 |
| Average B-factor | |
| Protein | 8.3 Å2 |
| Inhibitor | 16.6 Å2 |
| Water molecules | 20.4 Å2 |
Crystallographic refinement
Structure determination of the SA2-13 soaked SHV-1 crystal was carried out using the isomorphous crystal structure of wt SHV-1 β-lactamase (PDB id 1G56)33. The program CNS 35 was used for crystallographic refinement. The starting model, consisting of only the protein coordinates, was subjected to initial rigid body refinement, followed by model minimization, temperature factor refinement, and simulated annealing. After several rounds of refinement, the Cymal-6 and water molecules were placed into difference density and included in additional rounds of refinement. Finally, the compound SA2-13 in the trans-enamine form was readily built into clear electron density (Figure 2) and added to the model for refinement. Throughout the refinement, an overall anisotropic B-factor was applied. Refinement topology and parameter files for SA2-13 and Cymal-6 were generated using the PRODRG2 webserver 36, and the program O was used for interactive graphical model rebuilding 37. Crystallographic refinement was monitored using the programs DDQ 38 and PROCHECK 39 and the final refinement statistics are shown in Table 2.
Figure 2.
Electron density of the SA2-13 compound bound in the active site of wt SHV-1 β-lactamase. Side-by-side stereo view of 1.28 Å resolution |Fo|-|Fc| omit electron density contoured at 2.5 σ is depicted in green. Atom types are color coded accordingly: oxygen (red), nitrogen (blue), sulfur (yellow), and carbon (light and dark grey for the protein and SA2-13, respectively). Waters labeled W1-4 are waters numbered 339A, 339B, 154, and 203, respectively, in the deposited coordinate set.
Results
Kinetic analysis
The kinetics of inactivation of SA2-13 and tazobactam are summarized in Table 1. The KD for SA2-13 compared to tazobactam is seventeen fold greater (0.100 ± 0.025 vs. 1.70 ± 0.1 μM, respectively). The first-order rate constants for enzyme and inhibitor complex inactivation (kinact) are two fold different. The slower kinact for SA2-13 compared to tazobactam is also apparent when performing Raman measurements on wt SHV-1 crystals (data not shown). This results in a 33 fold difference in the second order rate constant of inactivation. In contrast, the kreact following 5 min and 30 min incubations are markedly different. SA2-13 dissociates from the enzyme:inhibitor complex 8.5-fold slower at 5 min and 10 fold slower at 30 minutes. The consequence of this behavior is an equal residual velocity determination at 5 and 30 min (4%). When extended out to 1440 min, there was 47% recovery of activity in the SA2-13: SHV-1 inactivation mixture, while the tazobactam: SHV-1 mixture resulted in complete inactivation.
Structure refinement
The refined model of the wt SHV-1 β-lactamase soaked with SA2-13 bound to tazobactam contains residues 26-292, a covalently bound trans-enamine conformation of SA2-13 molecule, one partial and one entire Cymal-6 molecule, and 344 water molecules. Nine of the protein side chains had alternate conformations and one entire residue, N170, had an alternate conformation. The final model yielded an R-factor of 15.4% and R-free of 16.7% for data between 50-1.28Å (Table 2). The stereochemistry of the refined model was excellent with 93% of the residues falling within the most favored core of the Ramachandran plot and none within the disallowed region.
Like other inhibitor-SHV-1 complexes, the protein structure is very similar to that of previous SHV-1 structures. The root mean square difference for superposition of the Cα atoms of residues 26-292 is 0.33 Å between the uncomplexed wt SHV-1 β-lactamase and this SA2-13 complexed structure. The covalently bound SA2-13 has induced virtually no changes in the active site with the exception of residue N170 which has 2 alternate conformations: the ‘inward’ wt conformation pointing into the active site as observed in the uncomplexed SHV-1 structure where it binds the nucleophylic deacylation water 40, and an ‘outward’ conformation where it interacts with the carboxylate moiety attached to C3 of SA2-13 similarly to how tazobactam interacts in the E166A SHV-1 structure (Figure 2 & 3) 30. The alternate N170 conformations cause the nucleophilic deacylation water, that is normally held by E166 and N170, to have partial occupancy (waters w1 and w2 in Figure 3).
Figure 3.
Stereo diagram depictuing the interactions of SA2-13 within the active site of SHV-1 β-lactamase. Hydrogen bonds are depicted as black dashed lines. Water molecule are highlighted as red spheres. Residue N170 has two alternative conformations (shown as #1 and #2). This causes the catalytic deacylation water, in close proximity to N170 and E166, to also have two alternate positions that pair with each of the N170 conformations (indicated as W1 and W2, respectively).
SA2-13 conformation
The omit electron density is of excellent quality showing clear density for all SA2-13 moieties such as the sulfone, both carboxyl groups, and the carbonyl oxygen in the linker (Figure 2). This is remarkable since repeated attempts to soak in tazobactam, or other inhibitors, using identical soaking protocols have generated only empty active sites in wt SHV-1 (unpublished results). The linearized SA2-13 structure is observed in the trans-enamine conformation in an almost ring-like structure. The SA2-13 molecule is at one end covalently attached to S70 and at its other end, the carboxylate linker, anchored via a salt bridge and hydrogen bonds, bringing this end in close proximity to its covalent attachment point (Figures 2-4). The middle of the SA2-13 is observed to ‘loop’ out of the active site. The torsion angle of the enamine bond defined by atoms C7-C6=C5-N4 refined to a value of 166° which is similar to that observed for tazobactam (168°), which confirms its presence as a trans-enamine bond. The occupancy of the SA2-13 compound in the active site after the 10 minute soak was ≥90%, as estimated using occupancy/B-factor refinements carried out similarly to our previous tazobactam bound structure 30. The occupancy was therefore fixed at 1.0.
Figure 4.

Schematic diagram depicting the interactions of SA2-13 when bound to wt SHV-1 β-lactamase. Hydrogen bonds are depicted as dashed lines (distances are shown in Å). Van der Waals interactions are shown as a curved line with perpendicular stripes. The double bond between atoms C5 and C6 is in the trans configuration which, together with the intramolecular hydrogen bond involving the N4 atom, identify the trans-enamine intermediate for the covalently bound SA2-13. Interactions and active site changes that might contribute to enhanced stabilization of the trans-enamine intermediate of SA2-13 are indicated by i-iv and are discussed in the text.
Discussion
The trans-enamine intermediate of β-lactamase inhibitors, termed the “waiting-room” intermediate, has previously been observed in class A β-lactamases using spectroscopic measurements 8;19;26. Inhibitor resistant mutations can negatively affect this intermediate 26 making the stabilization of this intermediate a potential therapeutic strategy for inhibitors that are less receptive to β-lactamases with an inhibitor resistant phenotype.
The structure-based linked-fragment design of the SA2-13 β-lactamase inhibitor has resulted in stabilization of the trans-enamine intermediate of the transient inhibited state such that the enzyme’s recovery (kreact) via deacylation is decreased by about 10 fold within the first 30 minutes. This decreased deacylation rate constant for SA2-13 allows in vitro activity similar to that of tazobactam for time points up to 30 minutes despite a 17-fold drop in affinity.
In order to explain the difference in kinetic behavior between SA2-13 and tazobactam, the following model illustrated in Figure 1a is proposed. Our data show that forming the Michaelis complex (E:I, Figure 1a) is favored for tazobactam over SA2-13 since tazobactam’s KD is 17 fold lower. The first-order rate constant for acylation of S70 is also twice as fast with tazobactam vs. SA2-13. Nevertheless, SA2-13 and tazobactam SHV-1 complexes have the same residual activity (4%) at 5 and 30 minute time points once the Michaelis complex is established. This is likely a result of SA2-13 being able to form more favorable trans-enamine interactions in the active site of SHV-1 than tazobactam. This is manifested by the longer time required to regenerate the free enzyme (kreact 8-10 fold longer). In a biologically relevant time scale (up to 30 minutes, the half life of bacterial cell division), these results are significant. But whether these inhibitory properties are yet of clinical utility, since antibiotic:inhibitor combinations are dosed every 6-8 hours, remains to be established since a larger proportion of free enzyme is generated after 24 h with SA2-13 compared to tazobactam (see Table 1). This indicates that the partitioning ratio for imine intermediate to form enamine or inactivate, the ki→t /kinact, is greater for SA2-13. The turnover number is also higher for SA2-13 indicating that the kdeacyl/kinact is greater as well for SA2-13. This suggests that that the pathway of irreversible inhibition is less ‘traveled’ by SA2-13 compared to tazobactam. A possible explanation is that the irreversible pathway might need the acylated inhibitor to be more dynamic in the active site to either fragment or react with nearby side chains such as S130. SA2-13 is more ordered in the trans-enamine intermediate state favoring the transient inhibitory species yet perhaps thereby limiting its irreversible inhibition pathway. Alternatively, the triazolyl moiety of tazobactam might make the inhibitor more prone to fragmentation and irreversible inhibition compared to the carboxyl linker of SA2-13. The addition of the carboxyl linker in SA2-13 has thus two effects, lengthening the trans-enamine state and decreasing the irreversible inhibition state but these effects seem to cancel out at time points of 30 minutes and less.
The atomic structure of the SHV-1 β-lactamase complexed with SA2-13 is surprisingly similar to those of the E166A:tazobactam structure and the wt SHV-1 structure except for a critical change involving residue N170 and a water molecule. The remarkable ease of obtaining a well-defined SA2-13 compound in the wt SHV-1 active site is in agreement with the decreased kreact since such relatively short soaks did not yield occupied active sites for tazobactam, or other inhibitors, unless much higher soaking concentrations in combination with a much longer time point was used 33 or a deacylation deficient E166A variant was used 29 30. The SA2-13 inhibitor structure bound to wt SHV-1 has possible implications for enzyme function and inhibitory mechanisms that will be discussed next.
Active site conformational changes
The only significant difference in the active site is that N170 adopts two alternate conformations upon SA2-13 binding (Figure 3). This N170 residue in wt SHV-1 normally adopts a single conformation involved in interacting with a nucleophilic water molecule held in place by both N170 and E166 33;40 which is also observed in numerous other class A β-lactamase structures. This water is thought to be involved in the deacylation step of the reaction 41 since it is in close proximity to the S70-acyl bond. One of the N170 conformations found in this SA2-13 complex (conf #1 in Figure 3) is in the ‘standard’ inward conformation priming the nucleophilic water which is also present, albeit at partial 50% occupancy water (w1 in Figure 3). The second conformation of N170 (#2 in Figure 3) is facing outward and interacts with the carboxylate moiety attached to C3 of SA2-13. This second conformation allows room for w1 to move 1.34 Å to the w2 position which is also partially occupied. Both the w2 position and the outward conformation of N170#2 have been previously observed in the E166A SHV-1 tazobactam bound structure 30. Since the N170 and E166 residues (indirectly) interact with each other to prime the nucleophilic water, the ability of N170 to adopt a different conformation in the E166A structure was not too surprising since E166 had been mutated. However, the ability of the novel SA2-13 compound to partially change the conformation of N170 in wt SHV-1 is unexpected and might have implications for inhibitor design as will be discussed later. N170 mutations have a drastic effect on activity and deacylation in class A β-lactamases 42 suggesting that, by extrapolation, inhibitor induced conformational changes in N170 could similarly have an effect on activity and deacylation.
SA2-13 active site interactions
The SA2-13 forms a linearized acyl-enzyme trans-enamine intermediate in which both the β-lactam ring and thiazolidine ring are opened (shown schematically in Figures 1 and 4). The compound makes a number of stabilizing interactions (Figures 3 and 4): its O8 atom, positioned in the oxyanion hole, forms hydrogen bonds with the backbone nitrogens of S70 and A237, the standard carboxylate group attached to the C3 atom makes a hydrogen bond with N132 as well as with N170 in one of the conformations observed (conf #2 in Figure 3), the sulfone moiety makes an intra-molecular hydrogen bond with N4 of SA2-13. The designed addition of SA2-13, the carboxyl-linker moiety, makes a number of intended interactions: a salt-bridge with K234, hydrogen bonds with S130, T235, and a water mediated interaction (Wat#3) with R244 (Figure 3 & 4). As intended, these latter interactions are similar to interactions that the sulfonate of HEPES makes in some of the SHV crystal structures (Figure 5). Although not predicted, one of the oxygens in the linker makes a water mediated interaction with S130 (via Wat#4, Figure 4).
Figure 5.

Overlay of SA2-13 and HEPES when bound to SHV-1. The active sites of the HEPES bound SHV-2 structure and that of our SA2-13 wt SHV-1 structure were superimposed. The figure shows the active site of wt SHV-1 depicting both SA2-13 (blue stick) and HEPES (yellow stick) illustrating the similar position of the sulfone+linker moiety of HEPES and that of the carboxyl+linker moiety of SA2-13.
Comparison of the trans-enamine SA2-13 and tazobactam structures
The interactions of SA2-13 in wt SHV-1 are, for the atoms in common, quite similar to those observed for tazobactam in E166A (Figure 6). These shared interactions include the intra-molecular hydrogen bond between the sulfone and N4 atom, the oxyanion hole interactions, and the carboxyl-C3 moiety:N170 hydrogen bond. In addition, the newly added carboxyl moiety of SA2-13 generates a salt-bridge with K234 and hydrogen bonds with S130 & T235 as was designed. A minor change involves the angle of the compound coming off S70 which is slightly different for SA2-13 compared to tazobactam leading to a slight shift of the sulfone/carboxyl part of about 1.0 Å (Figure 6). This angle has also been shown to vary more prominently in the sulbactam and clavulanic acid bound structures where there are fewer inhibitor:protein interactions 29.
Figure 6.
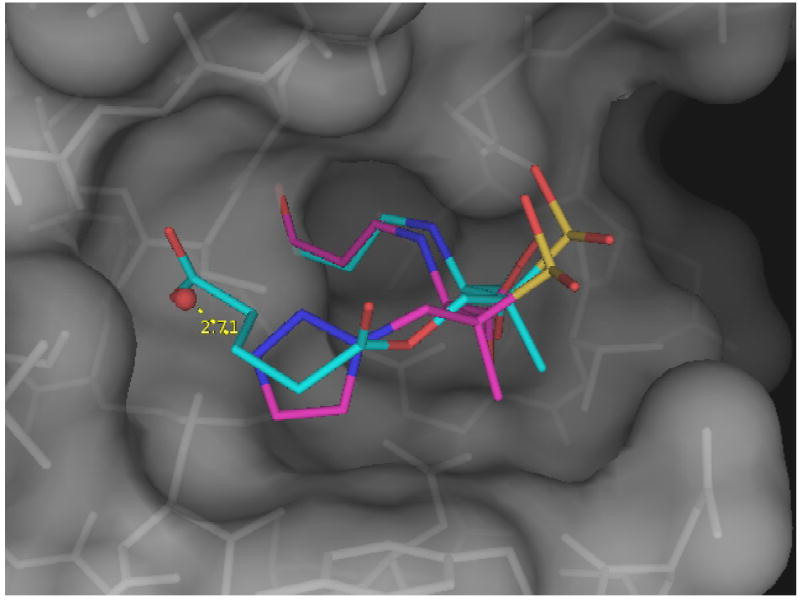
Superposition of the SA2-13 and tazobactam bound structures. Shown are SA2-13 (blue) when bound to wt SHV-1 and tazobactam (magenta) when bound to the deacylation deficient E166A variant of SHV-1. Only the protein atoms of the wt SHV-1:SA2-13 complex are shown. The water molecule involved in forming a hydrogen bond (yellow dashed line) with the triazolyl moiety of tazobactam when complexed to the E166A SHV-1 variant is depicted as a red sphere.
Implications for class A β-lactamase inhibitor design
The unprecedented ability of SA2-13 to form a stable, long-lived trans-enamine intermediate in wt SHV-1 is a novel and promising direction for future β-lactamase inhibitor design. There are 4 possible reasons for the lower deacylation efficiency of SA2-13 (shown in Figure 4 labeled i-iv): i) the newly added carboxyl-group provides a stabilizing force that keeps the original C3 attached carboxyl moiety in place such that N170 can swing out to interact so that the nucleophilic water is no longer catalytically primed 100% of the time by N170; ii) the newly added carboxyl moiety interacts with residues K234 and S130 whose side chains might play a role in catalysis 25;43;44 and whose properties might be altered by interacting with the charged carboxyl group; iii) the newly added carboxyl moiety is positioned such that it is within ~3.5 Å of the acyl-enzyme bond and might therefore electrostatically alter this bond’s susceptibility to the water’s nucleophilic attack needed for deacylation; and iv) the additional stabilizing interactions of the carboxylate force the intermediate in the trans-enamine conformation such that it has less opportunity to tautomerize back to the imine intermediate (Figure 1). The latter intermediate is thought to be the form that undergoes deacylation since the carbonyl atom is no longer (stabilized) conjugated with the enamine double bond since this double bond has moved in this imine intermediate (Figure 1a). All four possible reasons could significantly contribute to the decreased kreact and trans-enamine stabilization of SA2-13 although the first reason could maximally only contribute roughly a factor 2 since the N170 side chain is displaced only part of the time since it is in alternate conformations that are roughly equally occupied.
Our linked fragment approach in stabilizing the trans-enamine conformation has some intended features that suggest that this could be further exploited for general use in optimizing Class A β-lactamase inhibitors. First, the SA2-13 compound makes few, if any, interactions with residues that are not critical for catalysis since that might potentially limit its utility due to increased likelihood of resistance development. The residues that interact with SA2-13 are conserved (except for S130 which is a glycine in several inhibitor resistant SHV variants). Second, a bound negatively charged group in the vicinity of K234 and S130 is quite often observed in class A β-lactamase crystal structures, not only in SHV-1 (via a HEPES molecule, Figure 5) but also in PER-1 (via a bound sulfate ion) 45 and TEM-1(via a sulfate ion) 46 β-lactamase crystal structures. These structures suggests that this anion binding site is conserved among Class A β-lactamases and could potentially be further exploited via rational inhibitor design. Furthermore, β-lactamase inhibitor design studies focusing on stabilizing the transition state, via boronic acid analogs, have placed a carboxylate moiety in this region to improve binding 47. These studies suggest that this design feature of SA2-13 could be of general interest to other class A β-lactamases since it involves a very conserved active site region that can attract negatively charged groups.
In summary, using the linked fragment design approach we have stabilized the trans-enamine intermediate in wt SHV-1 β-lactamase by replacing the triazolyl moiety of tazobactam with a carboxyl-linker moiety. Although one aspect of tazobactam was improved, enhancing its trans-enamine intermediate longevity, SA2-13 has lost some of tazobactam’s efficiency regarding KD and inhibition efficiency at the 24 hour time point. Both these latter inhibitor characteristics are important since tazobactam/piperacillin is dosed every 6-8 hours. SA2-13’s pharmacodynamic and pharmacokinetic properties are unknown and beyond the scope of this manuscript yet our mechanistic studies demonstrate a proof of concept of stabilizing this intermediate to obtain a good in vitro inhibitor. Whether this approach will lead to a potent in vivo inhibitor is unknown and it is likely that SA2-13 would need to be further improved. Future experiments are planned to test the inhibitory efficiency of SA2-13, and derivatives thereof, against other β-lactamases including inhibitor resistant and extended spectrum variants of class A β-lactamases.
Supplementary Material
Acknowledgments
The coordinates and structure factor files are deposited with the Protein Data Bank (PDB ID 2H5S). MSH is supported by a Department of Veterans Affairs Advanced Career Development Award. RAB is supported by the Merit Review Program and NIH grant R01AI063517-01. JDB acknowledges the support of the Robert A. Welch Foundation (grant N-0871) and the Centers for Disease Control (grant H75/CCH623342). PRC is supported by NIH GM54072. FvdA is supported by NIH grant AI062968. We thank Vivien Yee for critical reading of the manuscript and the personnel at the SBC beamline 19 at Advanced Photon Source for help with data collection.
Reference List
- 1.Helfand MS, Bonomo RA. Curr Drug Targets Infect Disord. 2003;3:9–23. doi: 10.2174/1568005033342181. [DOI] [PubMed] [Google Scholar]
- 2.Bush K. Curr Opin Investig Drugs. 2002;3:1284–1290. [PubMed] [Google Scholar]
- 3.Buynak JD. Biochem Pharmacol. 2006;71:930–940. doi: 10.1016/j.bcp.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Therrien C, Levesque RC. FEMS Microbiol Rev. 2000;24:251–262. doi: 10.1111/j.1574-6976.2000.tb00541.x. [DOI] [PubMed] [Google Scholar]
- 5.Georgopapadakou NH. Expert Opin Investig Drugs. 2004;13:1307–1318. doi: 10.1517/13543784.13.10.1307. [DOI] [PubMed] [Google Scholar]
- 6.Buynak JD. Curr Med Chem. 2004;11:1951–1964. doi: 10.2174/0929867043364847. [DOI] [PubMed] [Google Scholar]
- 7.Lee N, Yuen KY, Kumana CR. Drugs. 2003;63:1511–1524. doi: 10.2165/00003495-200363140-00006. [DOI] [PubMed] [Google Scholar]
- 8.Charnas RL, Fisher J, Knowles JR. Biochemistry. 1978;17:2185–2189. doi: 10.1021/bi00604a025. [DOI] [PubMed] [Google Scholar]
- 9.Brenner DG, Knowles JR. Biochemistry. 1981;20:3680–3687. doi: 10.1021/bi00516a003. [DOI] [PubMed] [Google Scholar]
- 10.Brenner DG, Knowles JR. Biochemistry. 1984;23:5839–5846. doi: 10.1021/bi00319a025. [DOI] [PubMed] [Google Scholar]
- 11.Brenner DG, Knowles JR. Biochemistry. 1984;23:5833–5839. doi: 10.1021/bi00319a024. [DOI] [PubMed] [Google Scholar]
- 12.Charnas RL, Knowles JR. Biochemistry. 1981;20:3214–3219. doi: 10.1021/bi00514a035. [DOI] [PubMed] [Google Scholar]
- 13.Fisher J, Charnas RL, Knowles JR. Biochemistry. 1978;17:2180–2184. doi: 10.1021/bi00604a024. [DOI] [PubMed] [Google Scholar]
- 14.Fisher J, Charnas RL, Bradley SM, Knowles JR. Biochemistry. 1981;20:2726–2731. doi: 10.1021/bi00513a004. [DOI] [PubMed] [Google Scholar]
- 15.Fisher J, Belasco JG, Khosla S, Knowles JR. Biochemistry. 1980;19:2895–2901. doi: 10.1021/bi00554a012. [DOI] [PubMed] [Google Scholar]
- 16.Fisher J, Belasco JG, Charnas RL, Khosla S, Knowles JR. Philos Trans R Soc Lond B Biol Sci. 1980;289:309–319. doi: 10.1098/rstb.1980.0048. [DOI] [PubMed] [Google Scholar]
- 17.Brown RP, Aplin RT, Schofield CJ. Biochemistry. 1996;35:12421–12432. doi: 10.1021/bi961044g. [DOI] [PubMed] [Google Scholar]
- 18.Bonomo RA, Liu J, Chen Y, Ng L, Hujer AM, Anderson VE. Biochim Biophys Acta. 2001;1547:196–205. doi: 10.1016/s0167-4838(01)00175-3. [DOI] [PubMed] [Google Scholar]
- 19.Bush K, Macalintal C, Rasmussen BA, Lee VJ, Yang Y. Antimicrob Agents Chemother. 1993;37:851–858. doi: 10.1128/aac.37.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y, Janota K, Tabei K, Huang N, Siegel MM, Lin YI, Rasmussen BA, Shlaes DM. J Biol Chem. 2000;275:26674–26682. doi: 10.1074/jbc.M002369200. [DOI] [PubMed] [Google Scholar]
- 21.Chaibi EB, Peduzzi J, Farzaneh S, Barthelemy M, Sirot D, Labia R. Biochim Biophys Acta. 1998;1382:38–46. doi: 10.1016/s0167-4838(97)00127-1. [DOI] [PubMed] [Google Scholar]
- 22.Lin S, Thomas M, Shlaes DM, Rudin SD, Knox JR, Anderson V, Bonomo RA. Biochem J. 1998;333(Pt 2):395–400. doi: 10.1042/bj3330395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delaire M, Labia R, Samama JP, Masson JM. J Biol Chem. 1992;267:20600–20606. [PubMed] [Google Scholar]
- 24.Meroueh SO, Roblin P, Golemi D, Maveyraud L, Vakulenko SB, Zhang Y, Samama JP, Mobashery S. J Am Chem Soc. 2002;124:9422–9430. doi: 10.1021/ja026547q. [DOI] [PubMed] [Google Scholar]
- 25.Pagan-Rodriguez D, Zhou X, Simmons R, Bethel CR, Hujer AM, Helfand MS, Jin Z, Guo B, Anderson VE, Ng LM, Bonomo RA. J Biol Chem. 2004;279:19494–19501. doi: 10.1074/jbc.M311669200. [DOI] [PubMed] [Google Scholar]
- 26.Sulton D, Pagan-Roderiguez D, Zhou X, Liu Y, Hujer AM, Bethel CR, Helfand MS, Thomson JM, Anderson VE, Buynak JD, Ng LM, Bonomo RA. J Biol Chem. 2005;280:35528–35536. doi: 10.1074/jbc.M501251200. [DOI] [PubMed] [Google Scholar]
- 27.Maveyraud L, Massova I, Birck I, Miyashita K, Samama JP, Mobashery S. J Am Chem Soc. 1996;118:7435–7440. [Google Scholar]
- 28.Helfand MS, Totir MA, Carey MP, Hujer AM, Bonomo RA, Carey PR. Biochemistry. 2003;42:13386–13392. doi: 10.1021/bi035716w. [DOI] [PubMed] [Google Scholar]
- 29.Padayatti PS, Helfand MS, Totir MA, Carey MP, Carey PR, Bonomo RA, van den AF. J Biol Chem. 2005;280:34900–34907. doi: 10.1074/jbc.M505333200. [DOI] [PubMed] [Google Scholar]
- 30.Padayatti PS, Helfand MS, Totir MA, Carey MP, Hujer AM, Carey PR, Bonomo RA, van den AF. Biochemistry. 2004;43:843–848. doi: 10.1021/bi035985m. [DOI] [PubMed] [Google Scholar]
- 31.Nukaga M, Mayama K, Hujer AM, Bonomo RA, Knox JR. J Mol Biol. 2003;328:289–301. doi: 10.1016/s0022-2836(03)00210-9. [DOI] [PubMed] [Google Scholar]
- 32.Verlinde CL, Rudenko G, Hol WG. J Comput Aided Mol Des. 1992;6:131–147. doi: 10.1007/BF00129424. [DOI] [PubMed] [Google Scholar]
- 33.Kuzin AP, Nukaga M, Nukaga Y, Hujer A, Bonomo RA, Knox JR. Biochemistry. 2001;40:1861–1866. doi: 10.1021/bi0022745. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 36.van Aalten DM, Bywater R, Findlay JB, Hendlich M, Hooft RW, Vriend G. J Comput Aided Mol Des. 1996;10:255–262. doi: 10.1007/BF00355047. [DOI] [PubMed] [Google Scholar]
- 37.Jones TA, Zou JY, Cowan SW. Kjeldgaard Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 38.van den Akker F, Hol WG. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 1):206–218. doi: 10.1107/S0907444998007161. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J Appl Cryst. 2001;26:283–291. [Google Scholar]
- 40.Kuzin AP, Nukaga M, Nukaga Y, Hujer AM, Bonomo RA, Knox JR. Biochemistry. 1999;38:5720–5727. doi: 10.1021/bi990136d. [DOI] [PubMed] [Google Scholar]
- 41.Hermann JC, Ridder L, Holtje HD, Mulholland A. J Org Biomol Chem. 2006;4:206–210. doi: 10.1039/b512969a. [DOI] [PubMed] [Google Scholar]
- 42.Zawadzke LE, Chen CC, Banerjee S, Li Z, Wasch S, Kapadia G, Moult J, Herzberg O. Biochemistry. 1996;35:16475–16482. doi: 10.1021/bi962242a. [DOI] [PubMed] [Google Scholar]
- 43.Ellerby LM, Escobar WA, Fink AL, Mitchinson C, Wells JA. Biochemistry. 1990;29:5797–5806. doi: 10.1021/bi00476a022. [DOI] [PubMed] [Google Scholar]
- 44.Thomas VL, Golemi-Kotra D, Kim C, Vakulenko SB, Mobashery S, Shoichet BK. Biochemistry. 2005;44:9330–9338. doi: 10.1021/bi0502700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tranier S, Bouthors AT, Maveyraud L, Guillet V, Sougakoff W, Samama JP. J Biol Chem. 2000;275:28075–28082. doi: 10.1074/jbc.M003802200. [DOI] [PubMed] [Google Scholar]
- 46.Jelsch C, Lenfant F, Masson JM, Samama JP. FEBS Lett. 1992;299:135–142. doi: 10.1016/0014-5793(92)80232-6. [DOI] [PubMed] [Google Scholar]
- 47.Ness S, Martin R, Kindler AM, Paetzel M, Gold M, Jensen SE, Jones JB, Strynadka NC. Biochemistry. 2000;39:5312–5321. doi: 10.1021/bi992505b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




