Summary
Cell growth (accumulation of mass) needs to be coordinated with metabolic processes that are required for the synthesis of macromolecules. The PI3-kinase/Akt signaling pathway induces cell growth via activation of complex 1 of the target of rapamycin (TORC1). Here we show that Akt-dependent lipogenesis requires mTORC1 activity. Furthermore, nuclear accumulation of the mature form of the sterol responsive element binding protein (SREBP1) and expression of SREBP target genes was blocked by the mTORC1 inhibitor rapamycin. We also show that silencing of SREBP blocks Akt-dependent lipogenesis and attenuates the increase in cell size in response to Akt activation in vitro. Silencing of dSREBP in flies caused a reduction in cell and organ size and blocked the induction of cell growth by dPI3K. Our results suggest that the PI3K/Akt/TOR pathway regulates protein and lipid biosynthesis in an orchestrated manner and that both processes are required for cell growth.
Keywords: HUMDISEASE, SIGNALING
Introduction
The serine/threonine kinase Akt (PKB/c-Akt) promotes cell growth (i.e., accumulation of cell mass) predominantly through activation of complex 1 of the mammalian target of rapamycin (mTORC1) (Sarbassov et al., 2005). mTORC1 is regulated by both nutrients and growth factors and plays a conserved role in cell growth control (Wullschleger et al., 2006). Glucose uptake and induction of glycolysis by Akt have been studied in the context of the metabolic effects of insulin signaling on cellular and organismal levels (Whiteman et al., 2002). However, the contribution of anabolic pathways other than protein biosynthesis to cell growth have only been considered recently (Plas and Thompson, 2005).
Akt regulates cell metabolism through several downstream targets (Manning and Cantley, 2007). Akt stimulates glucose uptake via induction of translocation of the glucose transporter 4 (GLUT4) to the plasma membrane and by increasing expression of GLUT1 (Kohn et al., 1996; Welsh et al., 2005). Akt promotes conversion of glucose into pyruvate through the glycolytic pathway by increasing the expression of glycolytic enzymes (Semenza et al., 1996) and by phosphorylating and activating hexokinase 2 (HK2) (Robey and Hay, 2006). Pyruvate can either be converted into lactate, or it can enter the tricarboxylic acid cycle (TCA cycle), where it is fully oxidized to generate ATP. In liver and adipose tissue, glucose is converted via pyruvate and citrate into lipids.
Akt activates mTORC1 through two distinct mechanisms. Phosphorylation of TSC2 by Akt (Inoki et al., 2002; Potter et al., 2002) results in activation of the Rheb GTPase while phosphorylation of the PRAS40 induces the dissociation of this inhibitory component from the mTORC1 complex (Oshiro et al., 2007; Sancak et al., 2007; Vander Haar et al., 2007). There are several lines of evidence that the Akt/mTORC1 axis is an essential regulator of cell growth in vivo. Akt1/Akt2 double-knockout mice exhibit severe growth deficiency (Peng et al., 2003) and mice deficient in S6-kinase 1 (S6K1) show a significant reduction in size accompanied by hypoinsulinaemia and glucose intolerance (Pende et al., 2004).
The family of sterol regulatory element binding proteins (SREBP) consists of three closely related members: SREBP1a, SREBP1c, and SREBP2 (Eberle et al., 2004). They have been identified as mediators of the effect of sterols on expression of enzymes involved in lipid and cholesterol homeostasis (Yokoyama et al., 1993). SREBPs belong to the family of basic helix-loop-helix-leucine zipper (bHLH-Zip) transcription factors. They are synthesized as inactive precursors and localize to the endoplasmic reticulum (ER), where they bind to the sterol cleavage activating protein (SCAP). The SREBP/SCAP complex binds to COPII proteins and translocates to the Golgi where a two-step proteolytic cleavage releases the N-terminal half of the SREBP protein and allows entry into the nucleus (Rawson, 2003). SREBPs bind to sterol regulatory element (SRE) and E box sequences found in the promoter regions of genes involved in cholesterol and fatty acid biosynthesis. Studies in knockout and transgenic mice have shown that SREBP1 preferentially regulates genes involved in fatty acid biosynthesis while SREBP2 mainly regulates genes of the cholesterol pathway (Horton et al., 2003).
Two key enzymes that are required to divert glycolytic carbon flux into lipid biosynthesis are ATP-citrate lyase (ACLY) and fatty acid synthase (FASN). ACLY converts cytosolic citrate into acetyl-CoA and oxaloacetate, thereby supplying the essential metabolite for lipid biosynthesis. Akt phosphorylates ACLY (Berwick et al., 2002), and ACLY activity is required for cell growth and tumorigenesis (Bauer et al., 2005; Hatzivassiliou et al., 2005). FASN catalyzes the condensation of acetyl-CoA and malonyl-CoA to generate long-chain fatty acids. We have previously shown that activation of Akt induces expression of a number of lipogenic genes, including ACLY and FASN (Porstmann et al., 2005). We show here that activation of SREBP and Akt-dependent induction of lipid biosynthesis requires the activity of mTORC1. Furthermore, activation of SREBP contributes to Akt-dependent cell growth in mammalian cells in vitro and in Drosophila melanogaster in vivo. Our data demonstrate the importance of lipogenesis for cell growth in two distinct model systems.
Results
Role of mTORC1 in Akt-Dependent Cell Growth and Lipid Biosynthesis
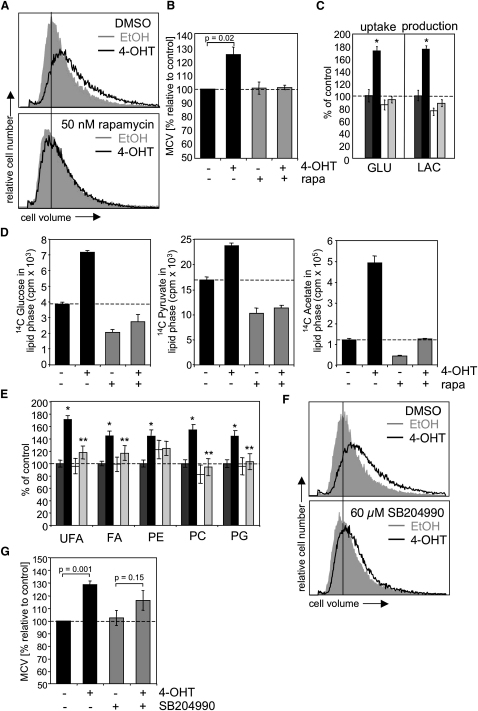
Akt is involved in the regulation of cell growth in both mammalian and invertebrate systems by activation of protein biosynthesis through the mTOR/S6K pathway (Sarbassov et al., 2005). To investigate the involvement of de novo lipid biosynthesis in the regulation of cell growth by Akt, we used immortalized human retinal pigment epithelial cells (RPE) expressing a conditional allele of the Akt1 kinase (myrAkt-ER) (Porstmann et al., 2005). Cells were cultured in medium supplemented with 1% lipoprotein deficient serum (LPDS) to reduce activation of endogenous Akt and limit the availability of exogenous lipids. Activation of myrAkt-ER by addition of 4-hydroxytamoxifen (4-OHT) caused a 20%–30% increase in median cell volume (MCV) (Figures 1A and 1B). Simultaneous treatment with the specific mTORC1 inhibitor rapamycin abolished the Akt-dependent increase in cell volume. Rapamycin did not affect cell size in the absence of Akt activation since the experiment was performed under serum starvation conditions. However, rapamycin markedly reduced the Akt-dependent increase in cellular protein content (Figure S1). Importantly, we did not observe significant changes in cell-cycle distribution upon Akt activation under the conditions used here (data not shown).
Figure 1.
mTORC1 Is Required for Akt-Dependent Cell Growth and Induction of Lipid Synthesis
(A) RPE myrAkt-ER cells were treated with 100 nM 4-hydroxytamoxifen (4-OHT) or solvent (ethanol) for 24 hr in medium with 1% lipoprotein deficient serum (LPDS) in the presence or absence of 50 nM rapamycin. In (A), cell size was determined by measuring electronic cell volume.
(B) Changes in median cell volume (MCV) induced by Akt activation relative to solvent control.
(C) RPE myrAkt-ER cells were treated with ethanol (dark gray bars), 4-OHT (black bars), rapamycin (open bars), or rapamycin and 4-OHT (light gray bars) for 48 hr in medium containing 1% LPDS. Glucose uptake and lactate production was determined by measuring metabolite concentrations in culture supernatant using NMR spectroscopy. Metabolite levels were normalized to cell number and compared to medium without cells. Values represent mean percent changes relative to ethanol-treated cells. (∗) p ≤ 0.01 ethanol versus 4-OHT.
(D) RPE myrAkt-ER cells were treated as in (A), and incorporation of D-[6-14C]glucose, [2-14C]-pyruvate, and [1-14C]-acetate into cellular lipids was measured.
(E) Cellular lipids were extracted from cells treated as in (C) and analyzed by NMR spectroscopy. Metabolite concentrations were normalized to cell number and values represent mean percent changes relative to ethanol treated cells. (∗) p < 0.01 ethanol versus 4-OHT; (∗∗) p < 0.05 4-OHT versus 4-OHT plus rapamycin. UFA, unsaturated fatty acids; FA,saturated fatty acids; PE = phosphatidylethanolamine; PC = phosphatidylcholine; PG = phosphatidylglycerol.
(F) RPE myrAkt-ER cells were stimulated with 4-OHT for 24 hr in the presence or absence of 60 μM SB20490, and cell volume was determined.
(G) Changes in MCV after activation of Akt in the presence or absence of 60 μM SB20490. In (B), (D), and (G), error bars represent standard deviation (SD). In (C) and (E), error bars represent standard error of the mean (SEM).
We next used nuclear magnetic resonance spectroscopy (NMR) to measure changes in medium metabolite concentration before and after myrAkt-ER activation. Akt activation induced an almost 2-fold increase in glucose uptake and lactate production in RPE cells (Figure 1C) consistent with a role for Akt in activation of glucose uptake and glycolysis. Akt-dependent glucose uptake and lactate production were completely blocked in the presence of rapamycin (Figure 1C). Likewise, we observed a reduction in Akt-dependent amino acid uptake and cellular amino acid content by rapamycin treatment (Figure S2). Analysis of incorporation of radioactive glucose, pyruvate, or acetate into cellular lipids showed that Akt activation leads to a substantial increase in glucose-dependent lipogenesis. Crucially, this was completely blocked by rapamycin treatment (Figure 1D), indicating that activation of de novo lipid synthesis by Akt requires mTORC1 function.
We have previously shown that Akt induces accumulation of intracellular lipids including phosphoglycerides (Porstmann et al., 2005). Figure 1E shows that inhibition of mTORC1 by rapamycin prevents Akt-dependent accumulation of unsaturated (UFA) and saturated fatty acids (FA) as well as phosphatidylcholine (PC) and phosphatidylglycerol (PG) levels (Figure 1E).
We next asked whether induction of lipid biosynthesis is required for Akt-dependent increase in cell size. Fatty acid biosynthesis requires cytoplasmic acetyl-CoA, which is generated by the enzyme ATP-citrate lyase (ACLY). ACLY function is required for tumor growth in vivo (Bauer et al., 2005; Hatzivassiliou et al., 2005). Inhibition of ACLY using the inhibitor SB204990 attenuated Akt-dependent increase in cell volume in RPE cells (Figures 1F and 1G).
Activation of SREBP1 by Akt
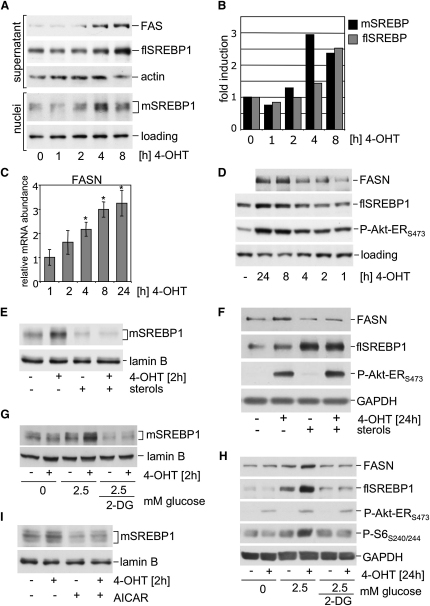
Studies in knockout mice have shown selective involvement of SREBP1a and 1c in the regulation of lipid biosynthesis (Liang et al., 2002) and promoters of lipogenic genes are preferentially bound by SREBP1a in vivo (Bennett et al., 2004). Mature SREBP1 can be detected in nuclear lysates as a diffuse band migrating between 60 and 65 kDa. Figures 2A and 2B show that accumulation of nuclear mSREBP1 can be detected within 2 hr of Akt activation. Induction of mSREBP1 precedes the increase in full-length SREBP1 (flSREBP1), which can be detected after 8 hr of Akt activation (Figures 2A, 2B, and S3). An increase in FASN mRNA and protein expression can be detected after 4 and 8 hr of Akt activation, respectively (Figures 2C and 2D). Induction of nuclear mSREBP1 as well as FASN expression in response to Akt activation is blocked by sterols (Figures 2E and 2F). Sterols induce a conformational change in SCAP that induces INSIG binding, thereby preventing ER/Golgi translocation of the SREBP/SCAP complex and SREBP processing (Sun et al., 2005). The observed induction of flSREBP in response to sterol treatment is most likely due to activation of the liver X receptor (LXR) (Repa et al., 2000) (Figure 2F).
Figure 2.
Akt Induces Rapid Accumulation of mSREBP1 in the Nucleus
(A) RPE myrAkt-ER cells were cultured in medium with 1% LPDS for 16 hr and treated with 100 nM 4-OHT for the indicated times. Nuclear extracts were prepared and analyzed for expression of mature SREBP1 (mSREBP1). Full-length SREBP1 (flSREBP) was detected in the supernatant.
(B) Quantitative representation of levels of mature and flSREBP1 in response to Akt activation in RPE cells.
(C and D) Cells were treated with 100 nM 4-OHT for the indicated times. Relative mRNA abundance of FASN was determined by qPCR. Values are normalized to GAPDH and are shown relative to solvent treated control. A representative of two independent experiments performed in duplicate is shown (D). Whole cell lysates prepared from cells treated in parallel to (C) were used to determine abundance of FASN, flSREBP1, and phospho-Akt-ER.
(E) Cells were cultured in medium with 1% LPDS and treated with 100 nM 4-OHT or solvent in the presence or absence of a mixture of 10 μg/ml cholesterol and 1 μg/ml 25-hydroxycholesterol (sterols) for 2 hr, and nuclear extracts were analyzed for expression of mSREBP1.
(F) Cells were treated with 100 nM 4-OHT in the presence or absence of sterols for 24 hr. Whole cell lysates were analyzed for expression of FASN, flSREBP1, and phospho-Akt-ER.
(G) Cells were cultured in medium with 1% LPDS for 16 hr and then placed in glucose-free medium or medium containing 2.5 mM glucose. Cells were treated with 2-deoxy-glucose (2-DG) and 4-OHT as indicated. Nuclear extracts were prepared and analyzed for the presence of mSREBP1.
(H) Cells were placed in glucose-free medium containing 1% LPDS or medium supplemented with 2.5 mM glucose or glucose and 2-DG for 30 min prior to stimulation with 100 nM 4-OHT for 24 hr. Whole cell lysates were analyzed for expression of FASN and full-length SREBP1.
(I) Cells were treated with 4-OHT for 2 hr with or without pretreatment with AICAR for 30 min. Nuclear extracts were analyzed for the presence of mSREBP1. Error bars represent SD.
Akt induces glucose uptake by inducing glucose transporter expression and membrane localization, and enhances glycolytic activity by activating hexokinase (Manning and Cantley, 2007). We next asked whether glucose is required for Akt-dependent activation of SREBP1. Absence of glucose or treatment with 2-deoxyglucose, an inhibitor of glycolysis, blocked Akt-dependent accumulation of mSREBP1 (Figure 2G) and prevented induction of SREBP target genes (Figure 2H). Energy depletion after glucose starvation or inhibition of glycolysis is likely to lead to activation of AMPK (Hardie and Carling, 1997). Activation of AMPK by AICAR also blocked accumulation of mSREBP1 in response to Akt activation (Figure 2I) and prevented induction of SREBP target genes (Figure S4). These results indicate that activation of SREBP1 by Akt is regulated by cellular energy status.
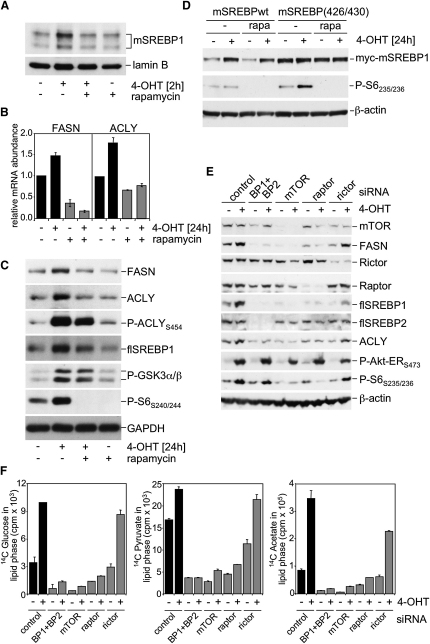
SREBP1 Activation and Lipogenesis Require mTORC1
The activity of mTORC1 is regulated by growth factor signaling as well as nutrient availability and energy homeostasis (Sarbassov et al., 2005). We therefore tested whether the regulation of SREBP1 by Akt involves mTORC1. Nuclear accumulation of mSREBP1 in response to Akt activation was prevented by rapamycin (Figure 3A). Likewise, induction of FASN and ACLY mRNA and protein levels was blocked by rapamycin treatment (Figures 3B and 3C). Importantly, treatment with rapamycin completely abolished induction of phosphorylation of the ribosomal protein S6, but only had a minor effect on phosphorylation of GSK3 or ACLY, indicating that the activity of the myrAkt-ER fusion protein is not affected by rapamycin (Figure 3C). Furthermore, rapamycin treatment also blocked insulin-dependent induction of FASN protein and mRNA (Figures S5A and S5B) and ectopic expression of mTOR was sufficient to induce mSREBP1 as well as FASN expression in the absence of myrAkt-ER activation in U2OS cells (Figure S5C).
Figure 3.
Akt-Dependent Activation of SREBP1 and Lipogenesis Requires mTORC1 Activity
(A) RPE-myrAkt-ER cells were cultured in medium with 1% LPDS for 16 hr and treated with 100 nM 4-OHT for 2 hr with or without pretreatment with 50 nM rapamycin for 30 min. Nuclear extracts were prepared and analyzed for expression of of mSREBP1.
(B) Cells were cultured in medium with 1% LPDS and treated with 100 nM 4-OHT or solvent for 24 hr in the presence or absence of 50 nM rapamycin. Total RNA was extracted and analyzed by qPCR for expression of ACLY and FASN. Values are normalized to GAPDH and are representative of one of three independent experiments.
(C) Whole cell lysates of cells treated as in (B) were used to determine the abundance of FASN, ACLY, and flSREBP1 as well as phosphorylation of ACLY and GSK3α/β.
(D) Cells were transfected with 1 μg of expression vector for myc-SREBP1 (aa 1-490) wild-type or the T426A/S430A mutant. 24 hr post-transfection, cells were placed in medium containing 1% LPDS and treated with 100 nM 4-OHT or solvent in the presence or absence of 50 nM rapamycin for 24 hr. Abundance of myc-mSREBP1 was determined using the 9E10 antibody.
(E) Cells were transfected with 100 nM siRNA oligonucleotides specific for SREBP1 and SREBP2 (BP1+BP2), mTOR, raptor, rictor or an unspecific control. 72 hr post-transfection, cells were placed in medium containing 1% LPDS and treated with 4-OHT for 24 hr. Lysates were used to confirm silencing efficiency and to determine FASN and ACLY expression.
(F) Cells treated in parallel to (E) were used to determine incorporation of D-[6-14C]glucose, [2-14C]-pyruvate or [1-14C]-acetate into lipid fractions. Error bars represent SD.
Mature SREBP proteins are highly unstable and are rapidly degraded through an ubiquitin-dependent pathway (Hirano et al., 2001). It has been shown that mSREBP can be phosphorylated by GSK3. This phosphorylation enables binding of the Fbw7 ubiquitin ligase and induces degradation of the SREBP protein. We next asked whether Akt affected the stability of mSREBP. Activation of Akt increased levels of transfected myc-tagged wild-type mSREBP1 (amino acids 1–490) (Figure 3D). This effect was abolished when two phosphorylation sites for GSK3 were mutated to alanine. Interestingly, Akt-dependent accumulation of exogenous wild-type mSREBP could still be observed in the presence of rapamycin, indicating that mTORC1 function is dispensable for stabilization of mSREBP1 in response to Akt activation. Furthermore, inhibition of GSK3 using the chemical inhibitor SB216763 did not block Akt-dependent accumulation of endogenous mSREBP1, induction of FASN protein, or FASN promoter activity in response to Akt activation (Figure S6). These results indicate that Akt-dependent activation of endogenous SREBP does not solely involve regulation of the stability of SREBP but suggest an additional, mTORC1-dependent pathway that contributes to SREBP activation.
It has previously been suggested that rapamycin treatment can also affect the activity of mTORC2 in certain cell types (Sarbassov et al., 2006). Our own results show that long-term treatment with rapamycin leads to activation of Akt phosphorylation by inhibition of a negative feedback loop in the cell line used here (Figure S7). To address any potential involvement of mTORC2 in the regulation of lipogenic gene expression, we used siRNA to selectively abolish expression of raptor or rictor, components of mTORC1 or mTORC2, respectively. Silencing of mTOR and raptor significantly reduces S6 phosphorylation (Figure 3E), while rictor silencing results in reduction of Akt phosphorylation on threonine 450 (Figure S8), which has been shown to be a substrate for mTORC2 (Facchinetti et al., 2008; Ikenoue et al., 2008). Silencing of mTOR or raptor, but not rictor, prevents Akt-dependent induction of FASN and SREBP1 expression (Figure 3E). Expression of SREBP2 was not affected by silencing of mTORC1 or mTORC2 components. Silencing of mTOR also blocked Akt- dependent FASN and ACLY induction in U2OS cells (Figure S9). We also addressed the role of SREBP in the induction of de novo lipid biosynthesis in response to Akt activation. Figure 3F shows that silencing of SREBP1 and 2 blocks incorporation of glucose, pyruvate, or acetate into cellular lipids. Furthermore, silencing of mTOR or raptor, but not rictor, abolished induction of lipogenesis in response to Akt activation (Figure 3F).
SREBP Is Required for Cell Growth in Mammalian Cells
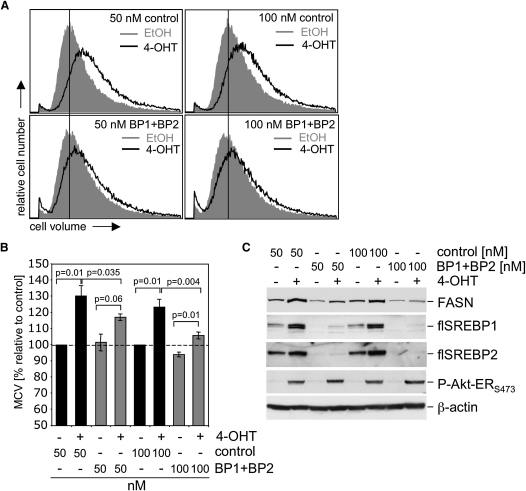
We next asked whether induction of lipogenic gene expression by SREBP is involved in the regulation of cell growth in mammalian cells. Transfection of siRNA oligonucleotides targeting both SREBP1 and 2 significantly reduced the increase in cell size observed in response to Akt activation in RPE cells in a dose-dependent manner (Figures 4A and 4B). Likewise, induction of FASN expression in response to Akt activation was reduced when SREBP1 and 2 were silenced (Figure 4C). Similar results were obtained when SREBP1 and 2 were silenced using different siRNA sequences (data not shown). These results indicate that activation of SREBP and expression of SREBP target genes is required for efficient cell growth in mammalian cells in vitro.
Figure 4.
Silencing of SREBP1 Restricts Akt-Induced Cell Growth in Mammalian Cells
(A) RPE-myrAkt-ER cells were transfected with either 50 or 100 nM siRNA oligonucleotides specific for SREBP1 and SREBP2 (BP1 + BP2) or an unspecific control. Seventy-two hours post transfection, cells were placed in medium containing 1% LPDS and treated with 100 nM 4-OHT or solvent for 24 hr, and cell size was determined.
(B) Changes in median cell volume (MCV) following Akt activation relative to solvent control.
(C) Whole cell lysates of cells treated in parallel to (A) were used to determine levels of FASN, SREBP1, SREBP2 and phospho-Akt-ER. Error bars represent SD.
Role of SREBP in Cell and Organ Size Control in Drosophila
HLH106 has been identified as the gene coding for a single SREBP isoform in flies, and dSREBP shows significant sequence homology to human SREBP1 within its DNA binding domain (Seegmiller et al., 2002).
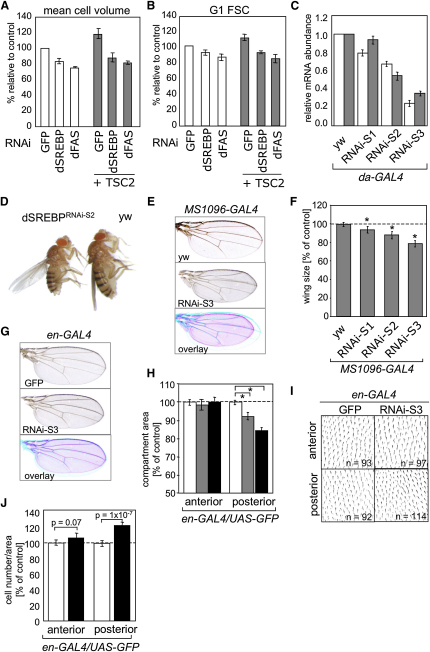
We first measured the effect of transient silencing of dSREBP or dFAS on cell volume in the Drosophila cell line Kc167. Figures 5A and 5B show that silencing of dSREBP or dFAS results in a significant reduction in mean cell volume and FSC of G1 cells. Simultaneous silencing of TSC2 did not rescue the observed reduction in cell size. Interestingly, insulin treatment of Kc167 cells resulted in increased levels of fatty acids and phosphoglycerides similar to the results obtained upon Akt activation in mammalian cells (Figure S10).
Figure 5.
Silencing of dSREBP Reduces Cell and Organ Size in Drosophila
(A) Kc167 cells were treated with dsRNAs corresponding to GFP, dSREBP, or dFAS in the absence or presence of dTSC2 dsRNA for 5 days, and electronic cell volume was determined.
(B) Cells treated as in (A) were used to determine FSC-A of the G1 population by FACS.
(C) dSREBP RNAi was expressed ubiquitously during embryogenesis using the daughterless-GAL4 (da-GAL4) driver. Expression of dSREBP (white bars) and dFAS (gray bars) in second instar larvae was determined by qPCR. Transcript levels are normalized to actin mRNA levels and represent three independent experiments. (RNAi-S1, single insertion, RNAi-S2 and RNAi-S3, double insertions).
(D) Representative phenotype of female flies expressing dSREBP RNAi using the da-GAL4 driver compared to control (yw).
(E) Wings of flies expressing dSREBPRNAi-S3 in the dorsal cell layer of the wing.
(F) Wing area analysis of control (yw), dSREBPRNAi-S1, dSREBPRNAi-S2, and dSREBPRNAi-S3. (∗) p ≤ 10−5.
(G) Wings of flies expressing dSREBPRNAi-S3 in the posterior compartment of the developing wing.
(H) Changes in wing area of control flies (white bars) or flies expressing SREBP RNAi-2 (gray bars) or SREBP RNAi-3 (black bars) in the posterior compartment of the wing. (∗) p ≤ 10−8.
(I) High-magnification images of the anterior and posterior compartments of wings shown in (G).
(J) Changes in wing epithelial cell number of flies expressing GFP (white bars) or dSREBPRNAi-S3 (black bars) in posterior compartment of the wing. Error bars represent SD.
To address the role of dSREBP in the regulation of cell growth in vivo, we generated a series of transgenic flies expressing an inverted repeat RNA under the control of the UAS promoter. Three different fly lines (RNAi-S1, RNAi-S2, and RNAi-S3) were used for the analysis. Induction of dSREBP RNAi expression using the daughterless (da)-GAL4 driver resulted in reduced expression of dSREBP and dFAS in larvae (Figure 5C). The three RNAi lines showed differences in silencing efficiency with RNAi-S3 having the strongest effect (Figure 5C). Silencing of dSREBP during embryogenesis caused a dose-dependent developmental delay and lethality during larval and pupal stages (Figure S11). Flies reaching adulthood showed a significant reduction in body size, reduced body weight, and wing area (Figures 5D and S12). The RNAi-S3 fly line did not produce any viable offspring when crossed to the da-GAL4 driver. This is consistent with an essential role of dSREBP during larval development (Kunte et al., 2006).
Deletion or overexpression of components of the insulin signaling pathway affects cell size in a cell-autonomous manner (Weinkove et al., 1999). We therefore asked whether silencing of dSREBP in specific compartments of the Drosophila wing could affect cell and organ size. Figures 5E and 5F show that expression of dSREBP RNAi cassette in the dorsal wing compartment (MS1096-GAL4) caused a dose-dependent reduction in adult wing area. Similarly, silencing of dSREBP using the engrailed (en)-GAL4 driver caused a specific reduction in the area of the posterior compartment of the wing (Figures 5G and 5H). Furthermore, reduced size of the posterior compartment was accompanied by an increase in density of wing epithelial cells (Figures 5I and 5J) indicating that silencing of dSREBP causes a reduction in cell size. We also investigated whether inhibition of dSREBP function by means other than gene silencing would cause similar growth inhibition. Deletion of the N-terminal transactivation domain in mammalian SREBP results in dominant inhibition of SREBP function (Sato et al., 1994). MS1096-GAL4-driven expression of N-terminally deleted dSREBP (DN75) or a mutant in which the transmembrane domain has been deleted in addition to the N terminus (DN75T) resulted in a substantial reduction in wing area (Figure S13).
SREBP Is a Target of the PI3K Pathway in Drosophila
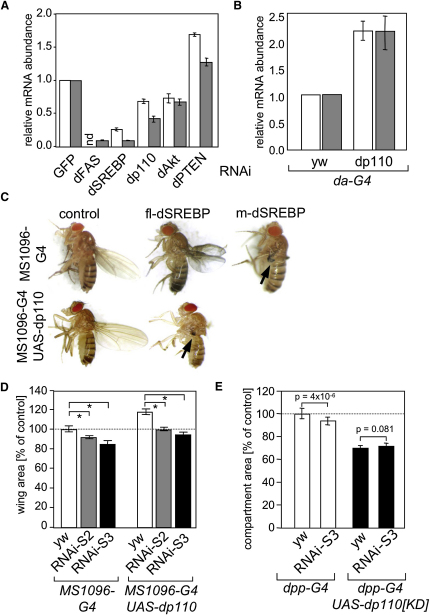
We next asked whether the PI3K/Akt pathway regulates the activity of SREBP in flies. Transient silencing of the catalytic subunit of PI3K, dp110, or dAkt reduced mRNA abundance of dFAS and dSREBP in Kc167 cells (Figure 6A). Conversely, silencing of dPTEN resulted in increased expression of both transcripts (Figure 6A). Ubiquitous expression of dp110 using the da-GAL4 driver resulted in enhanced dSREBP and dFAS expression in second instar larvae compared to controls (Figure 6B), indicating that the PI3K/Akt pathway activates dSREBP function.
Figure 6.
dSREBP Is Regulated by the PI3K Pathway and Required for dp110-Dependent Cell Growth
(A) Kc167 cells were treated with dsRNAs corresponding to GFP, dSREBP, dFAS, dp110, dAKT, and dPTEN for 5 days. Expression of dSREBP (white bars) and dFAS (gray bars) was determined by qPCR. Values are normalized to actin mRNA levels and show a representative of three experiments performed in duplicate.
(B) Second instar larvae expressing dp110 ubiquitously throughout development were analyzed for dSREBP (white bars) and dFAS (gray bars) by qPCR. Values are normalized to actin mRNA levels.
(C) Flies expressing fl-dSREBP or m-dSREBP under the control of the UAS promoter (UAS-fl-dSREBP1) were crossed with flies expressing GAL4 (MS1096-G4) or dp110 and GAL4 in the dorsal cell layer of the wing (MS1096-G4-UAS-dp110). Representative individual flies are shown. Arrows indicate severe malformation of the wing.
(D) Wing size analysis of flies expressing dSREBPRNAi-S2 or dSREBPRNAi-S3 with (MS1096-G4-UAS-dp110) or without (MS1096-G4) coexpression of dp110. n ≥ 7 wings; (∗) p ≤ 5 × 10−8.
(E) Quantification of wing area of flies expressing dSREBPRNAi-S3 with (dpp-G4-UAS-dp110[KD]) or without (dpp-G4) coexpression of kinase-dead dp110. Error bars represent SD.
Expression of mature dSREBP (m-dSREBP) using the MS1096-GAL4 driver resulted in significant lethality when flies were reared at 25°C (data not shown). When flies were reared at 18°C (resulting in a reduced activity of the GAL4/UAS system), severe deformation of the wing was observed (Figure 6C). Expression of the full-length form of dSREBP (fl-dSREBP) in the wing caused less severely misshapen wings even when flies were reared at 25°C. However, coexpression of dp110 and full-length dSREBP resulted in severely deformed wings, a phenotype that is similar to that caused by expression of m-dSREBP (Figure 6C). This result suggests that the activity of full-length dSREBP is enhanced by PI3K signaling.
Expression of dp110 causes an overgrowth phenotype in the wing, indicated by a 15%–20% increase in surface area of the wing (Leevers et al., 1996; Radimerski et al., 2002) (Figure 6D). Silencing of dSREBP attenuated the increase in wing size induced by expression of dp110 (Figure 6D). Similar results were obtained using a nonoverlapping RNAi sequence targeting dSREBP or by heterozygous deletion of the dSREBP gene (Figure S14).
Expression of a kinase domain mutant of PI3K (dp110[KD]) decreases cell and organ size in Drosophila (Leevers et al., 1996). Expression of dp110[KD] using the decapentaplegic (dpp)-GAL4 driver resulted in a 20%–30% reduction in the size of the dpp compartment. Expression of the dSREBP RNAi hairpin resulted in a small but significant decrease in wing area (Figure 6E). However, coexpression of dSREBP RNAi with dp110KD did not further decrease the size of this compartment. (Figure 6E). Taken together, these results suggest that dp110 and dSREBP are components of the same pathway in the regulation of cell growth in Drosophila.
Discussion
Cell growth is tightly controlled by mitogenic signaling pathways and requires the activation of biosynthetic pathways for the generation of macromolecules, including proteins and lipids. Activation of the mTOR complex 1 (mTORC1) increases translation and ribosome biogenesis (Wullschleger et al., 2006), but effects of mTORC1 on other anabolic processes are less well understood.
We show here that ectopic activation of Akt induces a significant increase in cell volume in the absence of mitogens and that Akt-dependent size increase is blocked by the mTORC1 inhibitor rapamycin. Analysis of medium metabolites by NMR revealed that Akt activation results in increased uptake of glucose and amino acids as well as secretion of lactate. The involvement of mTORC1 in Akt-dependent translocation of glucose and amino acid transporters to the plasma membrane has been demonstrated before (Edinger and Thompson, 2002). We observed significant accumulation of fatty acids and phosphoglycerides in response to Akt activation and enhanced incorporation of labeled glucose, pyruvate, or acetate into the lipid fraction in cells with activated Akt, all of which was blocked by rapamycin. These results show that Akt activates de novo lipogenesis and that mTORC1 activity is required for this effect.
Hepatocytes show increased SREBP1c mRNA levels in response to insulin treatment or Akt expression (Azzout-Marniche et al., 2000; Fleischmann and Iynedjian, 2000). We have previously shown that Akt activation results in nuclear accumulation of mSREBP1 and enhances expression of a number of SREBP target genes (Porstmann et al., 2005). Here, we show that nuclear accumulation of mSREBP1 is rapid and precedes the induction of transcription of SREBP target genes as well as the increase in flSREBP1. It should be noted that the srebf1 gene is a target of positive feed-forward regulation, since a functional SRE has been identified in its promoter region (Amemiya-Kudo et al., 2000). Thus, induction of flSREBP after Akt activation could be due to enhanced SRE-dependent transcription of the gene. Activation of SREBP1 by Akt was completely blocked by glucose starvation, inhibition of glycolysis, or AMPK activation by AICAR. AMPK is activated by an increase in AMP/ATP ratio following energy deprivation. AMPK regulates lipid metabolism by phosphorylating ACC and induces a switch from fatty acid biosynthesis to fatty acid oxidation (Hardie and Carling, 1997). Thus, regulation of SREBP activity by AMPK could reflect an additional adaptation to energy restriction.
Activity of mTORC1 is regulated by mitogens but also responds to cellular energy status (Sarbassov et al., 2005). AMPK activates TSC2 resulting in inhibition of mTORC1 (Inoki et al., 2003). It seems likely that inhibition of SREBP1 activity following glucose deprivation or AMPK activation is mediated by mTORC1. Our results show that nuclear accumulation of SREBP1 and induction of SREBP target genes requires mTORC1 activity. It has been suggested that induction of FASN and ACC expression by Her2 in breast cancer cells involves increased protein translation (Yoon et al., 2007). However, we observed an increase in FASN and ACLY mRNA abundance in response to Akt activation. Induction of FASN and ACLY mRNA was blocked in the presence of rapamycin indicating that mTORC1 is involved in the regulation of SREBP-dependent transcription. Furthermore, silencing of mTOR or raptor, but not rictor, abolished activation of expression of SREBP1, FASN, and ACLY, thus confirming that regulation of SREBP-dependent gene expression is exclusive to mTORC1.
It has been shown that GSK3 regulates the stability of mSREBP1 by inducing the association of the ubiquitin ligase Fbw7 resulting in rapid degradation of SREBP1 (Sundqvist et al., 2005). We observed an increase in the stability of exogenous mSREBP in response to Akt activation. This was dependent on the presence of two GSK3 phosphorylation sites but was still observed in the presence of rapamycin. It therefore seems likely that Akt regulates SREBP activity in at least two ways. Akt increases the stability of mSREBP1 by inhibiting GSK3-dependent phosphorylation. However, our results indicate that Akt also regulates SREBP1 activity in a GSK3-independent, mTORC1-dependent manner. It has been shown that dominant negative Akt prevents ER/Golgi translocation of SCAP (Du et al., 2006). TOR has been implicated in vesicle transport in yeast (Neufeld, 2007) and localization of mTOR to the ER and the Golgi plays an essential role in its regulation and downstream signaling (Drenan et al., 2004). Thus, it seems possible that mTORC1, or proteins downstream of mTORC1 signaling such as S6K or 4-EBP, could be involved in the regulation of SREBP processing within these compartments.
Our results clearly implicate mTORC1 in the regulation of lipogenesis in epithelial cells. Silencing mTOR or raptor, but not rictor, is sufficient to abolish Akt-dependent incorporation of glucose, pyruvate, or acetate into cellular lipids. This effect seems to be most pronounced for the incorporation of acetate, which only requires the activity of ACC and FASN. An involvement of raptor in the regulation of lipogenesis is also consistent with its essential function during mouse development (Guertin et al., 2006). Interestingly, silencing of SREBP1 and 2 was sufficient to prevent Akt-dependent lipogenesis and significantly reduced cell growth in response to Akt activation. This result confirms that SREBP-dependent gene expression represents an important mechanism in the regulation of lipogenesis during cell growth.
Consistent with a role for SREBP in cell size regulation in mammalian cells, we observed that silencing of dSREBP or its target gene dFAS resulted in a significant decrease in cell size in Drosophila Kc167 cells. Interestingly, dSREBP or dFAS silencing completely blocked the increase in cell volume caused by silencing of TSC2. Silencing of dSREBP in different compartments of the developing wing imaginal disc caused a reduction of the respective compartment in adult flies and was caused by a reduction in cell size rather than cell number.
It is somewhat surprising that expression of m-dSREBP in the wing causes a severe growth defect. It has been shown before that overexpression of mSREBP1a causes cell-cycle arrest through induction of the cdk inhibitors p21, p27, and p16 (Inoue et al., 2005; Nakakuki et al., 2007). It is also possible that overactivation of SREBP causes an imbalance between protein and lipid biosynthesis and suggests that SREBP activity needs to be tightly controlled to allow normal tissue development. Kunte and coworkers have detected dSREBP activation in the developing fat body, the midgut as well as in oenocytes, a specialized cell type that have hepatocyte-like functions in lipid metabolism (Gutierrez et al., 2007; Kunte et al., 2006). This expression pattern suggests a role in lipid metabolism similar to the function of SREBP1c in mammalian liver. Our results are consistent with a cell-autonomous role for dSREBP and suggest that dSREBP is required for normal growth of peripheral larval tissue.
dSREBP silencing restricted tissue growth induced by overexpression of dp110 in the wing and did not enhance the reduction in size of a wing compartment expressing kinase-dead dp110. These results suggest that both factors are components of the same pathway and indicate that activation of dSREBP is required for the induction of cell growth by the PI3K pathway in flies. It will be interesting to investigate whether regulation of dSREBP downstream of PI3K requires TOR. Further investigations are required to elucidate the exact mechanism of regulation of dSREBP activity by signaling pathways involved in growth regulation in flies.
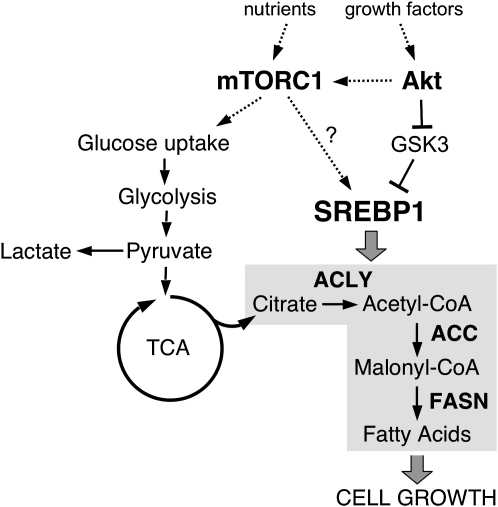
Taken together, our data are consistent with a model that places SREBP activation downstream of the intracellular signaling pathway that integrates mitogenic signaling with energy status and nutrient availability (Figure 7). It also suggests that protein biosynthesis and lipogenesis are regulated in a concerted manner during cell growth.
Figure 7.
Model of Regulation of Lipid Biosynthesis by Akt and mTORC1
Akt and mTORC1 regulate lipid biosynthesis on several levels. Activation of glucose uptake and induction of glycolysis is required for the generation of mitochondrial citrate. Activation of SREBP1 by Akt induces the expression of enzymes involved in lipid biosynthesis, including ACLY, FASN, and ACC. Akt stabilizes mSREBP1 by inhibition of GSK3. Inhibition of mTORC1 blocks accumulation of mSREBP1 in response to Akt activation through an unknown mechanism.
Experimental Procedures
Cell Culture and Reagents
RPE myrAkt-ER and U2OS myrAkt-ER cells have been described before (Porstmann et al., 2005). RPE myrAkt-ER cells were maintained in DMEM/HAMS F12 (1:1) medium with 10% FCS, glutamine, and sodium bicarbonate. U2OS myrAkt-ER cells were grown in DMEM with 10% FCS and glutamine. For culture at low sterol concentration, cells were grown in medium with 1% lipoprotein deficient serum (LPDS, Intracel). Kc167 cells were cultured in Schneider's medium (GIBCO) with 10% FCS, penicillin/streptomycin, and glutamine. A list of antibodies, chemicals, and plasmids is provided in the Supplemental Experimental Procedures.
Cell Size Determination and Flow Cytometry
RPE myrAkt-ER cells were detached using trypsin, washed, and resuspended in PBS. Median electronic cell volumes were determined in triplicate using a Z2 Coulter Counter (Multisizer II, Beckman-Coulter). Kc167 cells were collected by centrifugation and analyzed as above. For G1 specific cell size, KC167 cells were fixed and stained with propidium iodide and G1-FSC was determined. Experiments were repeated at least three times.
Metabolite Analysis
Two militers of culture medium samples were collected for the analysis of metabolite concentrations. Four hundred and fifty microliters of medium samples were mixed with 50 μl D2O, and 25 μl of 10 mM sodium TSP were added for chemical shift calibration and quantification. Intracellular metabolites were extracted using a dual-phase-extraction method as previously described (Tyagi et al., 1996). Lipid extracts were reconstituted in 600 μl deuterated chloroform, and 0.008% v/v tetramethylsilane was used as a chemical shift reference (0 ppm). Water-soluble metabolites were reconstituted in 600 μl D2O, and 25 μl of 1 mM TSP was added for quantitation and chemical shift calibration. Proton NMR spectra were acquired using a Bruker 600 MHz (Bruker Avance) NMR system. Spectral peaks for water-soluble and lipid metabolites were assigned according to Sitter et al. (2002) and Sze and Jardetzky (1990), respectively. Metabolite levels were normalized to cell number, and each experiment was repeated at least six times.
Lipid Synthesis
Cells were grown in medium with 1% LPDS in the presence of 100 nM 4-OHT or ethanol for 24 hr. 2.5 μCi/ml D-[6-14C]glucose (45 μM final concentration, Amersham), 2.5 μCi/ml [2-14C]-pyruvate (166 μM final concentration) or 10 μCi/ml [1-14C]-acetate (85 μM final concentration, both Perkin Elmer) was added, and cells were incubated for 4 hr at 37°C. Cell were washed three times in PBS and lysed in 0.5% Triton X-100. Lipids were extracted by successive addition of 2 ml methanol, 2 ml chloroform, and 1 ml dH2O. Phase separation was achieved by centrifugation at 1000 rpm for 15 min. The organic (lower) phase was recovered and dried. Lipids were dissolved in Ultima Gold LSC Cocktail (Perkin Elmer) and counted on a Beckman LS 6500 scintillation counter. Each measurement was performed in duplicate, results were normalized to total protein content, and experiments were repeated at least twice.
Cell Lysis and Immunoblotting
For total cell lysis, cells were washed with ice-cold PBS and dissolved in lysis buffer (1% Triton X-100, 50 mM Tris pH 7.5, 300 mM NaCl, 1 mM EGTA, 1 mM DTT, 1 mM NaVO4, and Protease-Inhibitor [Roche]). Nuclear lysates were prepared as described (Porstmann et al., 2005). For detection of mSREBP, cells were treated with 25 μg/ml ALLN for 1 hr prior to lysis. Equal amounts of cell lysates were separated on SDS-PAGE and blotted onto PVDF membrane (Immobilon). Proteins were detected by immunoblotting with ECL detection.
RNA Preparation and Quantitative PCR
Total RNA was isolated from RPE myrAkt-ER or Kc267 cells using the RNeasy kit (QIAGEN) and treated with DNase I (QIAGEN). For preparation of RNA from Drosophila larvae, 10–20 s instar larvae were homogenized in liquid nitrogen and dissolved in buffer RLT (QIAGEN) followed by RNA purification according to manufacturer's protocol. Two-and-a-half micrograms of total RNA was used for first-strand cDNA synthesis with SuperScript II RT and oligo dT primer (Invitrogen). Real-time PCR was performed with SYBR Green PCR Master Mix (Applied Biosystems). All reactions were performed at least in duplicate. Primer sequences are provided in the Supplemental Experimental Procedures.
Transfection and RNA Interference
RPE-myrAkt-ER cells were transfected using Fugene (Roche) according to manufacturer's instructions. Twenty-four hours post transfection, cells were placed into medium containing 1% LPDS and induced with 100 nM 4-OHT as indicated.
For gene-silencing experiments, cells were transfected using DharmaFECT reagent 1 (Dharmacon) following a reverse transfection protocol. Twenty-four hours post transfection, cells were split and divided into parallel cultures for different treatments. SiRNA sequences are provided in the Supplemental Experimental Procedures.
For depletion experiments in Kc167 cells, dsRNAs were generated by in vitro transcription using the MEGASCRIPT T7 transcription kit (Ambion). Primer sequences are provided in the Supplemental Experimental Procedures. Silencing was performed using 25 μg of dsRNA, and cells were analyzed after 5 days.
Transgenic Fly Lines
A sequence comprising 700 bp of the 5′ end of the dSREBP gene was amplified by PCR from genomic DNA and inserted as an inverted repeat into pWIZ-UAST (Lee and Carthew, 2003). Transgenic flies were generated on a yw background. Single transgene insertions on chromosomes 2 and 3 were combined to increase silencing efficiency. dSREBPRNAi-S1: one insertion, dSREBPRNAi-S2 and dSREBPRNAi-S3: two insertions.
UAS-Dp110WT, UAS-Dp110[KD], and dpp-GAL4/UAS-Dp110[KD] fly lines were described previously (Weinkove et al., 1999). MS1096-GAL4/UAS-Dp110WT flies were a gift from Ernst Hafen (ETH, Zuerich). dSREBP189 flies were a gift from Robert B. Rawson (University of Texas Southwestern Medical Center, Dallas). UAS-dSREBP fly lines were obtained from the Bloomington Stock Center. The dSREBP RNAi line T1 was obtained form the Vienna Drosophila RNAi Centre. Genotypes of all fly lines are provided in the Supplemental Experimental Procedures.
Analysis of Wing Area and Cell Number
Wings of male flies were dehydrated in ethanol, mounted in Euparal (Agar Scientific), and photographed. Wing area was determined by measuring pixel number. Cell number was determined by counting wing hairs in an area of fixed size in anterior or posterior wing compartments. Unless stated, data represent values generated from at least 20 wings of each genotype.
Statistical Analysis
P values were calculated using a two-tailed student's t test assuming equal variance.
Acknowledgments
We wish to thank S. Marygold, B. Thompson, and N. Tapon for helpful discussions. We also thank J. Downward for advice and discussion. We are very grateful to E. Hafen, H. Stocker, and R. Rawson for providing fly lines. We also thank J. Ericsson for providing material. We would like to thank S. Basu for providing cell lines. We thank T. Gilbank for generating transgenic flies, A. Schmukle for help with the analysis, and the LRI Equipment Park and FACS Laboratory for valuable assistance. C.R.S. was supported by an EMBO Long-Term Fellowship. This work was funded by Cancer Research UK.
Published: September 2, 2008
Footnotes
Supplemental Data include 14 figures and Supplemental Experimental Procedures and can be found with this article online at http://www.cellmetabolism.org/cgi/content/full/8/3/224/DC1/.
Supplemental Data
References
- Amemiya-Kudo M., Shimano H., Yoshikawa T., Yahagi N., Hasty A.H., Okazaki H., Tamura Y., Shionoiri F., Iizuka Y., Ohashi K. Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J. Biol. Chem. 2000;275:31078–31085. doi: 10.1074/jbc.M005353200. [DOI] [PubMed] [Google Scholar]
- Azzout-Marniche D., Becard D., Guichard C., Foretz M., Ferre P., Foufelle F. Insulin effects on sterol regulatory-element-binding protein-1c (SREBP-1c) transcriptional activity in rat hepatocytes. Biochem. J. 2000;350:389–393. [PMC free article] [PubMed] [Google Scholar]
- Bauer D.E., Hatzivassiliou G., Zhao F., Andreadis C., Thompson C.B. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- Bennett M.K., Toth J.I., Osborne T.F. Selective association of sterol regulatory element-binding protein isoforms with target promoters in vivo. J. Biol. Chem. 2004;279:37360–37367. doi: 10.1074/jbc.M404693200. [DOI] [PubMed] [Google Scholar]
- Berwick D.C., Hers I., Heesom K.J., Moule S.K., Tavare J.M. The identification of ATP-citrate lyase as a protein kinase B (Akt) substrate in primary adipocytes. J. Biol. Chem. 2002;277:33895–33900. doi: 10.1074/jbc.M204681200. [DOI] [PubMed] [Google Scholar]
- Drenan R.M., Liu X., Bertram P.G., Zheng X.F. FKBP12-rapamycin-associated protein or mammalian target of rapamycin (FRAP/mTOR) localization in the endoplasmic reticulum and the Golgi apparatus. J. Biol. Chem. 2004;279:772–778. doi: 10.1074/jbc.M305912200. [DOI] [PubMed] [Google Scholar]
- Du X., Kristiana I., Wong J., Brown A.J. Involvement of Akt in ER-to-Golgi Transport of SCAP/SREBP: A Link between a Key Cell Proliferative Pathway and Membrane Synthesis. Mol. Biol. Cell. 2006;17:2735–2745. doi: 10.1091/mbc.E05-11-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle D., Hegarty B., Bossard P., Ferre P., Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86:839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Edinger A.L., Thompson C.B. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol. Biol. Cell. 2002;13:2276–2288. doi: 10.1091/mbc.01-12-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchinetti V., Ouyang W., Wei H., Soto N., Lazorchak A., Gould C., Lowry C., Newton A.C., Mao Y., Miao R.Q. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 2008;27:1932–1943. doi: 10.1038/emboj.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann M., Iynedjian P.B. Regulation of sterol regulatory-element binding protein 1 gene expression in liver: role of insulin and protein kinase B/cAkt. Biochem. J. 2000;349:13–17. doi: 10.1042/0264-6021:3490013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin D.A., Stevens D.M., Thoreen C.C., Burds A.A., Kalaany N.Y., Moffat J., Brown M., Fitzgerald K.J., Sabatini D.M. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Gutierrez E., Wiggins D., Fielding B., Gould A.P. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature. 2007;445:275–280. doi: 10.1038/nature05382. [DOI] [PubMed] [Google Scholar]
- Hardie D.G., Carling D. The AMP-activated protein kinase–fuel gauge of the mammalian cell? Eur. J. Biochem. 1997;246:259–273. doi: 10.1111/j.1432-1033.1997.00259.x. [DOI] [PubMed] [Google Scholar]
- Hatzivassiliou G., Zhao F., Bauer D.E., Andreadis C., Shaw A.N., Dhanak D., Hingorani S.R., Tuveson D.A., Thompson C.B. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Hirano Y., Yoshida M., Shimizu M., Sato R. Direct demonstration of rapid degradation of nuclear sterol regulatory element-binding proteins by the ubiquitin-proteasome pathway. J. Biol. Chem. 2001;276:36431–36437. doi: 10.1074/jbc.M105200200. [DOI] [PubMed] [Google Scholar]
- Horton J.D., Shah N.A., Warrington J.A., Anderson N.N., Park S.W., Brown M.S., Goldstein J.L. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenoue T., Inoki K., Yang Q., Zhou X., Guan K.L. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signaling. EMBO J. 2008;27:1919–1931. doi: 10.1038/emboj.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K., Li Y., Zhu T., Wu J., Guan K.L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Inoki K., Zhu T., Guan K.L. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- Inoue N., Shimano H., Nakakuki M., Matsuzaka T., Nakagawa Y., Yamamoto T., Sato R., Takahashi A., Sone H., Yahagi N. Lipid synthetic transcription factor SREBP-1a activates p21WAF1/CIP1, a universal cyclin-dependent kinase inhibitor. Mol. Cell. Biol. 2005;25:8938–8947. doi: 10.1128/MCB.25.20.8938-8947.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn A.D., Summers S.A., Birnbaum M.J., Roth R.A. Expression of a constitutively active Akt Ser/Thr kinase in 3T3–L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation. J. Biol. Chem. 1996;271:31372–31378. doi: 10.1074/jbc.271.49.31372. [DOI] [PubMed] [Google Scholar]
- Kunte A.S., Matthews K.A., Rawson R.B. Fatty acid auxotrophy in Drosophila larvae lacking SREBP. Cell Metab. 2006;3:439–448. doi: 10.1016/j.cmet.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Carthew R.W. Making a better RNAi vector for Drosophila: use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Leevers S.J., Weinkove D., MacDougall L.K., Hafen E., Waterfield M.D. The Drosophila phosphoinositide 3-kinase Dp110 promotes cell growth. EMBO J. 1996;15:6584–6594. [PMC free article] [PubMed] [Google Scholar]
- Liang G., Yang J., Horton J.D., Hammer R.E., Goldstein J.L., Brown M.S. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J. Biol. Chem. 2002;277:9520–9528. doi: 10.1074/jbc.M111421200. [DOI] [PubMed] [Google Scholar]
- Manning B.D., Cantley L.C. AKT/PKB Signaling: Navigating Downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakakuki M., Shimano H., Inoue N., Tamura M., Matsuzaka T., Nakagawa Y., Yahagi N., Toyoshima H., Sato R., Yamada N. A transcription factor of lipid synthesis, sterol regulatory element-binding protein (SREBP)-1a causes G(1) cell-cycle arrest after accumulation of cyclin-dependent kinase (cdk) inhibitors. FEBS J. 2007;274:4440–4452. doi: 10.1111/j.1742-4658.2007.05973.x. [DOI] [PubMed] [Google Scholar]
- Neufeld T.P. TOR Regulation: Sorting out the Answers. Cell Metab. 2007;5:3–5. doi: 10.1016/j.cmet.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Oshiro N., Takahashi R., Yoshino K.I., Tanimura K., Nakashima A., Eguchi S., Miyamoto T., Hara K., Takehana K., Avruch J. The proline-Rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mTOR complex 1. J. Biol. Chem. 2007;282:20329–20339. doi: 10.1074/jbc.M702636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende M., Um S.H., Mieulet V., Sticker M., Goss V.L., Mestan J., Mueller M., Fumagalli S., Kozma S.C., Thomas G. S6K1(−/−)/S6K2(−/−) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X.D., Xu P.Z., Chen M.L., Hahn-Windgassen A., Skeen J., Jacobs J., Sundararajan D., Chen W.S., Crawford S.E., Coleman K.G. Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking Akt1 and Akt2. Genes Dev. 2003;17:1352–1365. doi: 10.1101/gad.1089403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plas D.R., Thompson C.B. Akt-dependent transformation: there is more to growth than just surviving. Oncogene. 2005;24:7435–7442. doi: 10.1038/sj.onc.1209097. [DOI] [PubMed] [Google Scholar]
- Porstmann T., Griffiths B., Chung Y.L., Delpuech O., Griffiths J.R., Downward J., Schulze A. PKB/Akt induces transcription of enzymes involved in cholesterol and fatty acid biosynthesis via activation of SREBP. Oncogene. 2005;24:6465–6481. doi: 10.1038/sj.onc.1208802. [DOI] [PubMed] [Google Scholar]
- Potter C.J., Pedraza L.G., Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell Biol. 2002;4:658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- Radimerski T., Montagne J., Rintelen F., Stocker H., van der Kaay J., Downes C.P., Hafen E., Thomas G. dS6K-regulated cell growth is dPKB/dPI(3)K-independent, but requires dPDK1. Nat. Cell Biol. 2002;4:251–255. doi: 10.1038/ncb763. [DOI] [PubMed] [Google Scholar]
- Rawson R.B. The SREBP pathway–insights from Insigs and insects. Nat. Rev. Mol. Cell Biol. 2003;4:631–640. doi: 10.1038/nrm1174. [DOI] [PubMed] [Google Scholar]
- Repa J.J., Liang G., Ou J., Bashmakov Y., Lobaccaro J.M., Shimomura I., Shan B., Brown M.S., Goldstein J.L., Mangelsdorf D.J. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robey R.B., Hay N. Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene. 2006;25:4683–4696. doi: 10.1038/sj.onc.1209595. [DOI] [PubMed] [Google Scholar]
- Sancak Y., Thoreen C.C., Peterson T.R., Lindquist R.A., Kang S.A., Spooner E., Carr S.A., Sabatini D.M. PRAS40 Is an Insulin-Regulated Inhibitor of the mTORC1 Protein Kinase. Mol. Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Sarbassov D.D., Ali S.M., Sabatini D.M. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Sarbassov D.D., Ali S.M., Sengupta S., Sheen J.H., Hsu P.P., Bagley A.F., Markhard A.L., Sabatini D.M. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Sato R., Yang J., Wang X., Evans M.J., Ho Y.K., Goldstein J.L., Brown M.S. Assignment of the membrane attachment, DNA binding, and transcriptional activation domains of sterol regulatory element-binding protein-1 (SREBP-1) J. Biol. Chem. 1994;269:17267–17273. [PubMed] [Google Scholar]
- Seegmiller A.C., Dobrosotskaya I., Goldstein J.L., Ho Y.K., Brown M.S., Rawson R.B. The SREBP pathway in Drosophila: regulation by palmitate, not sterols. Dev. Cell. 2002;2:229–238. doi: 10.1016/s1534-5807(01)00119-8. [DOI] [PubMed] [Google Scholar]
- Semenza G.L., Jiang B.H., Leung S.W., Passantino R., Concordet J.P., Maire P., Giallongo A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- Sitter B., Sonnewald U., Spraul M., Fjosne H.E., Gribbestad I.S. High-resolution magic angle spinning MRS of breast cancer tissue. NMR Biomed. 2002;15:327–337. doi: 10.1002/nbm.775. [DOI] [PubMed] [Google Scholar]
- Sun L.P., Li L., Goldstein J.L., Brown M.S. Insig required for sterol-mediated inhibition of Scap/SREBP binding to COPII proteins in vitro. J. Biol. Chem. 2005;280:26483–26490. doi: 10.1074/jbc.M504041200. [DOI] [PubMed] [Google Scholar]
- Sundqvist A., Bengoechea-Alonso M.T., Ye X., Lukiyanchuk V., Jin J., Harper J.W., Ericsson J. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7) Cell Metab. 2005;1:379–391. doi: 10.1016/j.cmet.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Sze D.Y., Jardetzky O. Characterization of lipid composition in stimulated human lymphocytes by 1H-NMR. Biochim. Biophys. Acta. 1990;1054:198–206. doi: 10.1016/0167-4889(90)90241-5. [DOI] [PubMed] [Google Scholar]
- Tyagi R.K., Azrad A., Degani H., Salomon Y. Simultaneous extraction of cellular lipids and water-soluble metabolites: evaluation by NMR spectroscopy. Magn. Reson. Med. 1996;35:194–200. doi: 10.1002/mrm.1910350210. [DOI] [PubMed] [Google Scholar]
- Vander Haar E., Lee S.I., Bandhakavi S., Griffin T.J., Kim D.H. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- Weinkove D., Neufeld T.P., Twardzik T., Waterfield M.D., Leevers S.J. Regulation of imaginal disc cell size, cell number and organ size by Drosophila class I(A) phosphoinositide 3-kinase and its adaptor. Curr. Biol. 1999;9:1019–1029. doi: 10.1016/s0960-9822(99)80450-3. [DOI] [PubMed] [Google Scholar]
- Welsh G.I., Hers I., Berwick D.C., Dell G., Wherlock M., Birkin R., Leney S., Tavare J.M. Role of protein kinase B in insulin-regulated glucose uptake. Biochem. Soc. Trans. 2005;33:346–349. doi: 10.1042/BST0330346. [DOI] [PubMed] [Google Scholar]
- Whiteman E.L., Cho H., Birnbaum M.J. Role of Akt/protein kinase B in metabolism. Trends Endocrinol. Metab. 2002;13:444–451. doi: 10.1016/s1043-2760(02)00662-8. [DOI] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M.N. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yokoyama C., Wang X., Briggs M.R., Admon A., Wu J., Hua X., Goldstein J.L., Brown M.S. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75:187–197. [PubMed] [Google Scholar]
- Yoon S., Lee M.Y., Park S.W., Moon J.S., Koh Y.K., Ahn Y.H., Park B.W., Kim K.S. Upregulation of acetyl-CoA carboxylase alpha and fatty acid synthase by HER2 at translational level in breast cancer cells. J. Biol. Chem. 2007;36:26,122–26,131. doi: 10.1074/jbc.M702854200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.