Abstract
Although there is little doubt that the domestication of mammals was instrumental for the modernization of human societies, even basic features of the path towards domestication remain largely unresolved for many species. Reindeer are considered to be in the early phase of domestication with wild and domestic herds still coexisting widely across Eurasia. This provides a unique model system for understanding how the early domestication process may have taken place. We analysed mitochondrial sequences and nuclear microsatellites in domestic and wild herds throughout Eurasia to address the origin of reindeer herding and domestication history. Our data demonstrate independent origins of domestic reindeer in Russia and Fennoscandia. This implies that the Saami people of Fennoscandia domesticated their own reindeer independently of the indigenous cultures in western Russia. We also found that augmentation of local reindeer herds by crossing with wild animals has been common. However, some wild reindeer populations have not contributed to the domestic gene pool, suggesting variation in domestication potential among populations. These differences may explain why geographically isolated indigenous groups have been able to make the technological shift from mobile hunting to large-scale reindeer pastoralism independently.
Keywords: microsatellites, mitochondrial DNA, Rangifer tarandus, reindeer husbandry
1. Introduction
Reindeer (Rangifer tarandus) constitute a biological resource of vital importance to the physical and cultural survival of arctic residents, and have been exploited for food and other subsistence commodities for thousands of years (Kofinas et al. 2000; Huntington & Fox 2005). The species was probably essential for human immigration and colonization of the Eurasian arctic and subarctic following the retreat of ice after the last glacial period—The Weichselian (Beach 1990; Gordon 2003). More recently, domestication has allowed an even more specialized use of the species (Skjenneberg & Slagsvold 1968; Beach 1990). Just as sheep, goat, cattle and horse were important for the advance of agricultural societies (Zeder & Hesse 2000; Troy et al. 2001; Vilà et al. 2001; Diamond 2002; Bruford et al. 2003; Beja-Pereira et al. 2006), domestic reindeer were probably important for the development of many northern indigenous cultures (Vainshtein 1980; Aronsson 1991; Kofinas et al. 2000; Jernsletten & Klokov 2002). Indeed, a better knowledge of reindeer domestication could be the key to understanding the history of many arctic communities.
In contrast to most other livestock species where the wild forms are extinct (e.g. cattle and horse), threatened (donkeys, llama and alpaca) or geographically restricted (sheep and goat) wild populations of reindeer are still widely distributed across northern Eurasia and North America (caribou). Today, almost 50% of the approximate 3 000 000 reindeer in the Old World are wild animals, and wild and domestic herds are managed in close coexistence in many areas (Syroechkovskii 1995; Baskin 2005). This provides a unique opportunity to analyse the interaction between domestic and wild lineages. Reindeer are considered to be in the early phase of domestication (Baskin 2000; Reimers & Colman 2006) and could, therefore, serve as an excellent model species to understand how the early domestication processes may have taken place. Archaeological evidence suggests that livestock domestication of other species, such as cattle and goat, is likely to have initially involved the management of wild herds and the restocking of domestic herds with wild individuals (Clutton-Brock 1987; Troy et al. 2001; Zeder 2006; Zeder et al. 2006). The degree of separation between coexisting domestic and wild reindeer herds might illustrate how wild and domestic herds may have interacted in other livestock species during the initial stages of the domestication process.
Two competing hypotheses address the origin of reindeer herding (Mirov 1945; Nelleman 1961; Gordon 2003). According to the monocentric hypothesis, domestic reindeer first appeared a few thousand years ago east of the Urals in the southern part of the Siberian taiga from where they spread to other regions. The polycentric hypothesis, on the other hand, argues that the domestication of reindeer occurred independently multiple times in different parts of Eurasia. In order to address the origin of reindeer herding, the domestication history and the interaction between wild and domestic animals, we characterized mitochondrial and microsatellite DNA variation of wild and domestic herds throughout Eurasia.
2. Material and methods
We obtained 732 blood or tissue samples for wild and domestic reindeer from 26 localities throughout Eurasia (see table S1 in the electronic supplementary material). DNA was extracted according to a standard chloroform:phenol protocol. A subset of 407 (accession nos. EU653306–EU653712) samples were amplified and sequenced for a 470 bp region of the mitochondrial D-loop (see methods in Flagstad & Røed 2003). All samples (n=732) were analysed for 14 reindeer-specific microsatellites (Nvhrt-01, Nvhrt-03, Nvhrt-16, Nvhrt-21, Nvhrt-24, Nvhrt-31, Nvhrt-48, Nvhrt-73, Nvhrt-76 (Røed & Midthjell 1998) and Rt-1, Rt-5, Rt-6, Rt-9, Rt-27 (Wilson et al. 1997); see methods in Røed et al. 2002). We used the Micro-Checker software (Van Oosterhout et al. 2004) to assess the quality of our microsatellite scoring. Less than 5% (15 out of 364) of the microsatellite locus–population combinations gave evidence of scoring errors due to stutter bands, allelic dropout or null alleles, and we concluded that the scoring quality was satisfactory.
Mitochondrial sequences were aligned manually using the sequence editor program SeqScape v. 2.0 (AB Applied Biosystems). An appropriate model of nucleotide substitution was selected using the hierarchical test approach implemented in Modeltest v. 3.06 (Posada & Crandall 1998). Phylogenetic relationships among different haplotypes were estimated using the Bayesian approach implemented in MrBayes v. 3.1.2 (Ronquist & Huelsenbeck 2003) with one million Metropolis-coupled Markov chain Monte Carlo (MCMC) cycles and 50 000 burn-in cycles using two Markov chains. In addition, a reduced median-joining network was constructed using the software Network v. 4.1.1.2 (Bandelt et al. 1995, 1999).
We used Fstat (http://www2.unil.ch/popgen/softwares/fstat.htm) and Arlequin (Excoffier et al. 2005) to estimate the levels of genetic variability in microsatellites and mtDNA, respectively. Genetic distances between populations were calculated from the mtDNA data using Kimura's two-parameter model (Kimura 1980), and a population dendrogram was constructed from the NJ algorithm as implemented in the software Sendbs (http://www.kms.ac.jp/∼genomelb/takezaki.eng.html#software). For the microsatellite data, genetic distances between populations were estimated by Nei's Da-distance (Nei et al. 1983) and a population dendrogram was constructed using the software Poptree (http://www.kms.ac.jp/∼genomelb/genomelab.eng.html#software). Support values at the nodes were estimated from 500 bootstrap replicates for both trees. An analysis of molecular variance (AMOVA; Excoffier et al. 1992; Weir 1996) was used to examine the proportion of genetic variation that could be explained at group level, given one, two or three domestication origins. For each number of origins, populations were grouped in several constellations, determined from geography and wild or domestic status of the analysed herds.
Genetic structure at an individual level was analysed by the Bayesian assignment approach as implemented in the software Structure (Pritchard et al. 2000). The log likelihood of our data [ln Pr(X|K)] was estimated, given different numbers of genetic clusters (K∈[1,10]), using an admixture model with uniform priors (α=1, αmax=50), correlated allele frequencies, 20 000 burn-in cycles and 50 000 MCMC iterations. All analyses were run without prior population information and were repeated 10 times for each K value. Delta (K) was then calculated as m|L″(K)|/s[L(K)], where m and s are the mean and standard deviation, respectively. The modal value of this distribution was considered as the uppermost level of genetic structuring (Evanno et al. 2005). A factorial correspondence analysis using the computer program Genetix (Belkhir et al. 2004) was performed to visualize the distribution of genetic variation across individuals.
3. Results and discussion
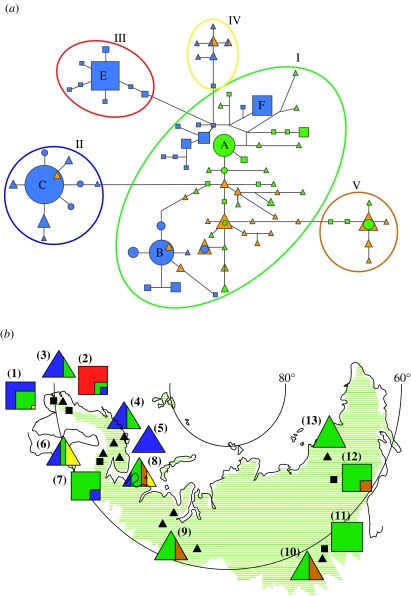
Ninety-five different mitochondrial haplotypes were identified. A median-joining network (figure 1a) reveals an internal assemblage of haplotypes (I), which is connected to a few terminal star-like clusters (II, III, IV and V), corresponding to four highly supported clades in a phylogenetic tree (see figure S1 in the electronic supplementary material). It is likely that the large but poorly structured haplotype assemblage I represents the large reindeer population that was present across Beringia during the last glacial period (Guthrie & Matthews 1971; Elias et al. 1996; Flagstad & Røed 2003). The terminal clusters may have arisen in smaller glacial refugia, or alternatively by random haplotype sorting and subsequent population expansion during postglacial re-colonization of the Eurasian arctic. The Beringian haplotype cluster is by far the most common in all Russian herds, whereas clusters II and III dominate in Fennoscandia (Norway, Sweden and Finland; figure 1b). Haplotypes from cluster II are found in high frequencies in all Fennoscandian domestic herds and in the southwestern wild population in Norway. Notably, cluster III is restricted to wild herds in central Norway and was never observed in any of the 107 domestic reindeer sampled in Fennoscandia (figure 1b).
Figure 1.
(a) Median-joining network for mitochondrial DNA haplotypes found in Eurasian reindeer. Haplotypes found only in wild herds are represented by squares, whereas the triangles represent domestic haplotypes. Circles represent haplotypes found in both wild and domestic herds. Blue haplotypes are found in Fennoscandia, whereas the orange and green haplotypes represent western and eastern Russia, respectively. Five different haplotype clusters (haplogroups, I–V) are encircled with different colours. Haplotypes that are directly mentioned in the text are labelled with capital letters. (b) Haplogroup frequencies in wild (squares) and domestic (triangles) reindeer herds. Populations are pooled according to geography and whether they are wild or domestic herds: (1) southwestern Norway wild (Nor-Wild 6–8), (2) central Norway wild (Nor-Wild 1–5), (3) central Norway domestic (Nor-Dom 1–3), (4) northern Norway domestic (Nor-Dom 4–6), (5) northern Finland domestic (Fin-Dom 2), (6) eastern Finland domestic (Fin-Dom 1), (7) eastern Finland wild (Fin-Wild 1), (8) northwestern Russia domestic (Rus-Dom 6 and 7), (9) north–central Russia domestic (Rus-Dom 3–5), (10) southeastern Russia domestic (Rus-Dom 2), (11) southeastern Russia wild (Rus-Wild 2), (12) northeastern Russia wild (Rus-Wild 1), (13) northeastern Russia domestic (Rus-Dom 1; see table S1 in the electronic supplementary material). Colours representing the different haplogroups are the same that are used to mark them in (a). Green lines in the map indicate the distribution range of Eurasian reindeer.
Haplotype sharing is very limited between Russia and Fennoscandia (figure 1a), suggesting separate origins of domestic reindeer in the two regions. This implies limited exchange of animals between the reindeer herding people of Fennoscandia and the indigenous cultures in western Russia. This is particularly remarkable for the two Russian domestic herds sampled on the Kola peninsula (Rus-Dom 6 and Rus-Dom 7), which are located very close to the northernmost parts of Norway and Finland (figure 1b). By sharp contrast, reindeer herds show high levels of haplotype sharing within regions, suggesting not only a common origin for the herds within each of these regions but also extensive intra-regional exchange and trade of animals. This interpretation is further supported by the high levels of genetic diversity in all domestic herds, which in some areas is even higher than that found in local wild herds (see table S1 in the electronic supplementary material). High levels of genetic diversity in domestic herds might further suggest that augmentation of domestic herds with animals from local wild herds has been common (Vilà et al. 2005), which is also compatible with the extensive haplotype sharing observed between neighbouring wild and domestic herds, e.g. haplotype A in eastern Russia and haplotypes B and C in Fennoscandia (figure 1a). Nevertheless, the most common haplotype in the central Norwegian wild population (E) and the wild Finnish population (F) are not found in any domestic herd (figure 1) strongly suggesting that neither of these wild populations have been sources for domestic reindeer in Fennoscandia.
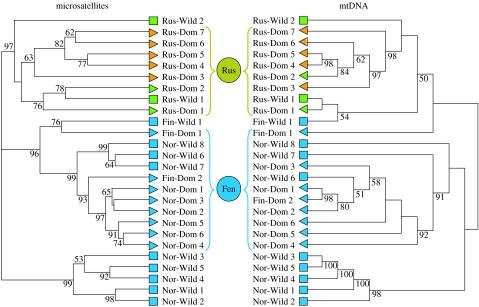
A clustering analysis at population level confirms that Russian and Scandinavian herds are strongly differentiated (figure 2). Three highly supported clades are evident in the microsatellite tree comprising (i) all Russian herds, (ii) the central Norwegian wild herds, and (iii) the rest of the Fennoscandian herds, wild as well as domestic. Virtually the same pattern appears from the mtDNA data, pointing towards the same origin for both sexes in the initial domestic herds. The division of the data into three main groups is also supported by an AMOVA (table 1) where there is a marked increase in the amount of variation explained at group level when separating herds in Russia and Fennoscandia and a further increase when separating the wild populations in central Norway from the rest of Fennoscandia (model G). Further division of the data gives only a marginal increase in the amount of variation explained at group level.
Figure 2.
Population dendrograms as inferred from microsatellite and mtDNA data. Wild and domestic populations are represented by squares and triangles, respectively. Geographical origins are represented by different colours. Blue, Fennoscandia; Orange, western Russia; Green, eastern Russia. Two putative origins for domestication (Russia and Fennoscandia) are indicated by filled circles between the trees.
Table 1.
AMOVA for wild (W) and domestic (D) reindeer herds in Eurasia. (The percentage of the total variation that can be explained at group level is given for one, two or three origins of domestication. Origins of domestication are based on the number of groups containing one or more domestic herds, putatively representing a separate domestication origin.)
| model | origins of domestication | no. of groups | groups | variation among groups | |
|---|---|---|---|---|---|
| mtDNA | microsatellites | ||||
| A | 1 | 2 | [domestic] [wild] | 9.13 | 2.29 |
| B | 1 | 2 | [Fennoscandia-D-W+ Russia-D] [Russia-W] | 7.12 | 1.40 |
| C | 1 | 2 | [Fennoscandia-D+Russia-D-W] [Fennoscandia-W] | 11.67 | 2.54 |
| D | 1 | 3 | [domestic] [Fennoscandia-W] [Russia-W] | 12.10 | 2.66 |
| E | 2 | 2 | [Fennoscandia-D-W] [Russia-D-W] | 20.97 | 3.73 |
| F | 2 | 4 | [Fennoscandia-D] [Russia-D] [Fennoscandia-W] [Russia-W] | 24.93 | 4.50 |
| G | 2 | 3 | [Fennoscandia-D+Norway-W1a+Finland-W] [Russia-D-W] [Norway-W2b] | 36.64 | 5.77 |
| H | 3 | 4 | Fennoscandia-D+Norway-W1a] [Norway-W2b] [west Russia-D+Finland-W] [east Russia-D-W] | 38.27 | 5.70 |
| I | 3 | 5 | [Fennoscandia-D+Norway-W1a] [Norway-W2b] [west Russia-D] [east Russia D-W] [Finland-W] | 41.50 | 6.02 |
Norway-W1 denotes the natural population in southwestern Norway.
Norway-W2 denotes the natural population in central Norway.
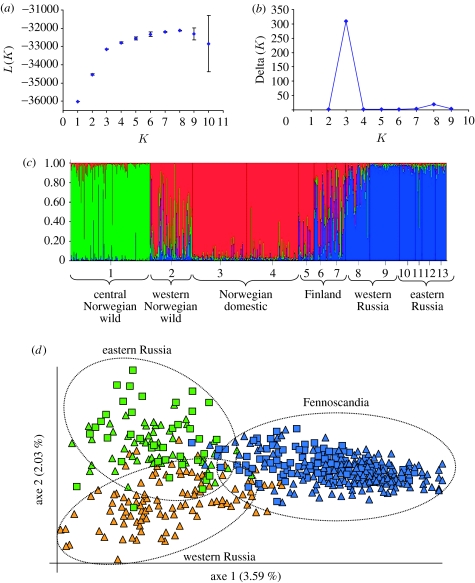
Similar to the analysis at population level, genetic variability at an individual level shows a partition of the sample into three main groups (figure 3a–c), supporting independent origins of domestic reindeer in Fennoscandia and Russia. Reindeer herding in Fennoscandia, and particularly in the northern part, has traditionally been connected to the Saami culture. Thus, our analyses strongly point towards an independent origin of Saami reindeer herding. Notably, the domestic gene pools in Fennoscandia and Russia seem to meet in eastern Finland, where the examined herds appear as a mixture of the two origins (figure 3c). This may reflect the frequent trade and transport of animals that occurred in the eighteenth century between the reindeer herders in eastern Finland (traditionally of Finnish origin) and the indigenous reindeer herding people towards the east as well as the north (Nieminen 2006). In contrast to the sharp genetic boundary between Russia and areas inhabited by the Saami people, the Russian domestic gene pool appears remarkably homogeneous across a vast region (figure 1b). However, when a factorial correspondence analysis is performed after removing the central Norwegian wild population, which clearly has not contributed to the domestic gene pool, a marked difference also appears between western and eastern Russian herds (figure 3d). Domestic and wild reindeer individuals from each of these three geographical regions appear mixed within the three clusters, demonstrating little differentiation between domestic and wild herds within areas and that the main differentiation is found between the following geographical areas: Fennoscandia; western Russia; and eastern Russia. This division is supported by the very limited mtDNA haplotype sharing between these three regions (figure 1a), and may suggest not only two but three different centres of domestication in Eurasian reindeer. Alternatively, local augmentation of domestic herds in western and eastern Russia could be partly or entirely responsible for this pattern.
Figure 3.
Clustering analysis in Eurasian reindeer herds, using (a–c) Bayesian assignment and (d) a factorial correspondence analysis. (a) Mean likelihood (L(K) (±s.d.)) over 10 runs dividing the entire dataset into K populations, for K values between 1 and 10. (b) Delta (K) where the modal value of the distribution is considered as the highest level of structuring, in our case three clusters. (c) Individual assignment to each of the three clusters, where the numbers refer to the same populations as given in figure legend 1b. Each individual is represented by a line and the proportion of each colour indicates proportion of ancestry from each group. (d) Factorial correspondence analysis of Eurasian reindeer, after excluding wild reindeer from central Norway. The graphics shows the two first axes. Wild and domestic individuals are symbolized by squares and triangles, respectively, and the colours are the same as given in figure legend 2.
Although we often observe a similar genetic composition of wild and domestic herds within areas, our data reveal some striking exceptions. As discussed previously, the wild populations in Finland and central Norway have contributed little or nothing to the domestic gene pool. In addition, the wild reindeer residing in the mountain taiga in southeastern Russia (Rus-Wild 2) show a genetic composition that is markedly different from that of the local domestic herds. In fact, all of the 10 wild individuals analysed carried herd-specific mtDNA haplotypes whereas domestic reindeer from the same area (Rus-Dom 2) have a much stronger genetic affinity towards more westerly and northerly distributed herds (figure 2). These differences may indicate variation in domestication potential among different wild populations, possibly due to the behavioural differences or variation in herd structure and size. In fact, our data indicate that the animals used in the domestication process probably derived from the large tundra herds instead of the smaller herds residing in the forest. Among the reindeer analysed, the two populations with the most characteristic wild forest ecotype, the Finnish (Fin-Wild 1) and the southeastern Russian (Rus-Wild-2), seem to have contributed little or nothing to the domestic gene pool. The tundra type that inhabits open areas is more gregarious than the forest dwelling types and has evolved a more sophisticated social organization (Geist 2003). This could represent an advantage for their exploitation by humans (Clutton-Brock 1987), which was likely to be especially important during the course of the transition from transport reindeer herding (mobile hunting) to managing larger herds of reindeer for food and skins (large-scale reindeer pastoralism).
The domestication of mammals is a slow process, which in its early phases may involve the management and control of wild herds rather than the capture of a few individuals and their subsequent breeding in captivity (Troy et al. 2001; Zeder 2006; Zeder et al. 2006). Archaeological evidence also suggests that these initial stages probably included the occasional augmentation of managed herds by adding wild individuals (Zeder 2006; Zeder et al. 2006). This is supported by a simulation study demonstrating that such backcrosses may have been common for several domestic species, contributing to their high genetic diversity (Vilà et al. 2005). Active management of reindeer herds—for example, the use of leading fences or enclosures and corrals for handling animals—can be tracked for a few thousand years (Mirov 1945; Aronsson 1991). This could explain the high genetic diversity observed in domestic herds and their similarity to the local wild populations. The ancestors of many livestock species, such as horse and cattle, probably lived in herds similar to the wild reindeer herds (small forest herds and larger tundra herds) that are present across the Eurasian arctic today. However, since their wild ancestors have disappeared or are greatly reduced, there is little or no information about the distribution of genetic diversity at the time of domestication. Thus, it is difficult to be conclusive about the early domestication history of these species. However, given the ecological and behavioural similarity between reindeer and the wild ancestors of these livestock species, the patterns observed for reindeer and reported herein may well be representative to how the initial stages of domestication of other livestock species took place.
In recent years, there has been increased attention towards the history and cultural traditions of indigenous people within the framework of laws and regulations of land exploitation. Our data contribute to a more detailed understanding of an important part of this cultural history. Indeed, understanding the long-lasting interaction between humans and reindeer is a crucial element for understanding the recent history of our own species in northern latitudes.
Acknowledgments
The present research adheres to the Norwegian Animal Welfare Act.
We thank the many hunters and herders and their organizers for their help in obtaining samples used in this study. The sampling effort of Svein D. Mathisen, Anti Oksanen and the Norwegian National Veterinary Institute are specially acknowledged. We also thank Liv Midthjell for skilful laboratory analyses and Reidar Andersen for useful comments on an earlier version of this paper. This work was partly supported by the Norwegian Directorate for Nature Management, the Norwegian Reindeer Husbandry Research Council and the Finnish Game and Fisheries Research Institute.
Supplementary Material
Sampling details and levels of genetic variability for 26 herds of Eurasian reindeer. Each population is given a code according to country origin (Nor=Norway, Fin=Finland and Rus=Russia) and status (Wild or Domestic). Listed are number of individuals analysed (n), mean number of allelic richness per locus (A), gene diversity (H), number of mtDNA haplotypes (nh), mtDNA haplotype diversity (h) and nucleotide diversity (π)
Bayesian phylogenetic tree for the 92 different control region haplotypes found in the 26 Eurasian reindeer herds. Posterior probabilities above 50 are indicated at the nodes. Haplotypes belonging to haplotype clusters II, III, IV, V are indicated. All other haplotypes belong to haplogroup I as given in figure 1. The North American woodland caribou (Rangifer tarandus caribou) is used as outgroup
References
- Aronsson K.-Å. Forest reindeer herding A.D. 1–1800. An archaeological and palaeoecological study in Northern Sweden. Archaeol. Environ. 1991;10:1–125. [Google Scholar]
- Bandelt H.J, Forster P, Sykes B.C, Richards M.B. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H.J, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Baskin L.M. Reindeer husbandry/hunting in Russia in the past, present and future. Polar Res. 2000;19:23–30. doi:10.1111/j.1751-8369.2000.tb00324.x [Google Scholar]
- Baskin L.M. Number of wild and domestic reindeer in Russia in the late 20th century. Rangifer. 2005;25:51–58. [Google Scholar]
- Beach H. Comparative systems of reindeer herding. In: Galaty J.G, Johson D.L, editors. The world of pastoralism. The Guildford Press; New York, NY: 1990. pp. 255–298. [Google Scholar]
- Beja-Pereira A, et al. The origin of European cattle: evidence from modern and ancient DNA. Proc. Natl Acad. Sci. USA. 2006;103:8113–8118. doi: 10.1073/pnas.0509210103. doi:10.1073/pnas.0509210103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II; Montpellier, France: 2004. Genetix, logiciel sous Windows TM pour la génétique des populations. [Google Scholar]
- Bruford M.W, Bradley D.G, Luikart G. DNA markers reveal the complexity of livestock domestication. Nat. Rev. Genet. 2003;4:900–910. doi: 10.1038/nrg1203. doi:10.1038/nrg1203 [DOI] [PubMed] [Google Scholar]
- Clutton-Brock J. British Museum (Natural History), Cambridge University Press; London, UK: 1987. A natural history of domesticated animals. [Google Scholar]
- Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418:700–707. doi: 10.1038/nature01019. doi:10.1038/nature01019 [DOI] [PubMed] [Google Scholar]
- Elias S.A, Short S.K, Nelson C.H, Birks H.H. Life and times of the Bering land bridge. Nature. 1996;382:60–63. doi:10.1038/382060a0 [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software Structure: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. doi:10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse P.E, Quattro J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes—application to human mitochondrial-DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (v. 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Flagstad O, Røed K.H. Refugial origins of reindeer (Rangifer tarandus L.) inferred from mitochondrial DNA sequences. Evolution. 2003;57:658–670. doi: 10.1111/j.0014-3820.2003.tb01557.x. doi:10.1554/0014-3820(2003)057[0658;R00RRT]2.0.co;2 [DOI] [PubMed] [Google Scholar]
- Geist, V. 2003 Of reindeer and man, modern and Neanderthal: a creation story founded on a historic perspective on how to conserve wildlife, woodland caribou in particular. Rangifer (Special Issue) 14, 57–63.
- Gordon, B. 2003 Rangifer and man: an ancient relationship. Rangifer (Special Issue) 14, 15–27.
- Guthrie R.D, Matthews J.V., Jr The Cape Deceit fauna—Early Pleistocene mammalian assemblage from the Alaskan arctic. Quart. Res. 1971;1:474–510. doi:10.1016/0033-5894(71)90060-3 [Google Scholar]
- Huntington H, Fox H. The changing Arctic: indigenous perspectives. In: Symon C, Arris L, Heal B, editors. Arctic climate impact assessment. Cambridge University Press; Cambridge, UK: 2005. pp. 61–98. [Google Scholar]
- Jernsletten K.B, Klokov K.B. University of Tromsø; Tromsø, Norway: 2002. Sustainable reindeer husbandry. [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide-sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. doi:10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- Kofinas G, Osherenko G, Klein D, Forbes B. Research planning in the face of change: the human role in reindeer/caribou systems. Polar Res. 2000;19:3–21. doi:10.1111/j.1751-8369.2000.tb00323.x [Google Scholar]
- Mirov N.T. Notes on the domestication of reindeer. Am. Anthropol. 1945;47:393–408. doi:10.1525/aa.1945.47.3.02a00030 [Google Scholar]
- Nei M, Tajima F, Tateno Y. Accuracy of estimated phylogenetic trees from molecular-data. 2. Gene-frequency data. J. Mol. Evol. 1983;19:153–170. doi: 10.1007/BF02300753. doi:10.1007/BF02300753 [DOI] [PubMed] [Google Scholar]
- Nelleman G. Theories on reindeer breeding. Folk. 1961;3:91–103. [Google Scholar]
- Nieminen M. History and development of Finnish reindeer husbandry (in Finnish) Poromies. 2006;5:26–29. [Google Scholar]
- Posada D, Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Pritchard J.K, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimers E, Colman J.E. Reindeer and caribou (Rangifer tarandus) response towards human activities. Rangifer. 2006;26:56–71. [Google Scholar]
- Røed K.H, Midthjell L. Microsatellites in reindeer, Rangifer tarandus, and their use in other cervids. Mol. Ecol. 1998;7:1773–1776. doi: 10.1046/j.1365-294x.1998.00514.x. [DOI] [PubMed] [Google Scholar]
- Røed K.H, Holand Ø, Smith M.E, Gjøstein H, Kumpula J, Nieminen M. Reproductive success in reindeer males in a herd with varying sex ratio. Mol. Ecol. 2002;11:1239–1243. doi: 10.1046/j.1365-294x.2002.01509.x. doi:10.1046/j.1365-294X.2002.01509.x [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. doi:10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Skjenneberg S, Slagsvold L. Universitetsforlaget; Oslo, Norway: 1968. Reindriften og dens Naturgrunnlag. [Google Scholar]
- Syroechkovskii E.E. Smithsonian Institution Libraries; Washington, DC: 1995. Wild reindeer. [Google Scholar]
- Troy C.S, MacHugh D.E, Bailey J.F, Magee D.A, Loftus R.T, Cunningham P, Chamberlain A.T, Sykes B.C, Bradley D.G. Genetic evidence for Near-Eastern origins of European cattle. Nature. 2001;410:1088–1091. doi: 10.1038/35074088. doi:10.1038/35074088 [DOI] [PubMed] [Google Scholar]
- Vainshtein S. Nomads of south Siberia. The pastoral economies of Tuva. Cambridge University Press; Cambridge, UK: 1980. [Google Scholar]
- Van Oosterhout C, Hutchinson W.F, Wills D.P.M, Shipley P. Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 2004;4:535–538. doi:10.1111/j.1471-8286.2004.00684.x [Google Scholar]
- Vilà C, Leonard J.A, Götherstrom A, Marklund S, Sandberg K, Liden K, Wayne R.K, Ellegren H. Widespread origins of domestic horse lineages. Science. 2001;291:474–477. doi: 10.1126/science.291.5503.474. doi:10.1126/science.291.5503.474 [DOI] [PubMed] [Google Scholar]
- Vilà C, Seddon J, Ellegren H. Genes of domestic mammals augmented by backcrossing with wild ancestors. Trends Genet. 2005;21:214–218. doi: 10.1016/j.tig.2005.02.004. doi:10.1016/j.tig.2005.02.004 [DOI] [PubMed] [Google Scholar]
- Weir B.S. Sinauer Associates, Inc; Sunderland, MA: 1996. Data analysis. II. Methods for discrete population genetic data. [Google Scholar]
- Wilson G.A, Strobeck C, Wu L, Coffin J.W. Characterization of microsatellite loci in caribou, Rangifer tarandus, and their use in other artiodactyls. Mol. Ecol. 1997;6:697–699. doi: 10.1046/j.1365-294x.1997.00237.x. doi:10.1046/j.1365-294X.1997.00237.x [DOI] [PubMed] [Google Scholar]
- Zeder M.A. Central questions in the domestication of plants and animals. Evol. Anthropol. 2006;15:105–117. doi:10.1002/evan.20101 [Google Scholar]
- Zeder M.A, Hesse B. The initial domestication of goats (Capra hircus) in the Zagros mountains 10,000 years ago. Science. 2000;287:2254–2257. doi: 10.1126/science.287.5461.2254. doi:10.1126/science.287.5461.2254 [DOI] [PubMed] [Google Scholar]
- Zeder M.A, Emshwiller E, Smith B.D, Bradley D.G. Documenting domestication: the intersection of genetics and archaeology. Trends Genet. 2006;22:139–155. doi: 10.1016/j.tig.2006.01.007. doi:10.1016/j.tig.2006.01.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sampling details and levels of genetic variability for 26 herds of Eurasian reindeer. Each population is given a code according to country origin (Nor=Norway, Fin=Finland and Rus=Russia) and status (Wild or Domestic). Listed are number of individuals analysed (n), mean number of allelic richness per locus (A), gene diversity (H), number of mtDNA haplotypes (nh), mtDNA haplotype diversity (h) and nucleotide diversity (π)
Bayesian phylogenetic tree for the 92 different control region haplotypes found in the 26 Eurasian reindeer herds. Posterior probabilities above 50 are indicated at the nodes. Haplotypes belonging to haplotype clusters II, III, IV, V are indicated. All other haplotypes belong to haplogroup I as given in figure 1. The North American woodland caribou (Rangifer tarandus caribou) is used as outgroup