Summary
We explore the interplay between amino acid sequence, thermodynamic stability, and functional fitness in the M2 proton channel of influenza A virus. Electrophysiological measurements show that drug-resistant mutations have minimal effects on M2’s specific activity, and suggest that resistance is achieved by altering a binding site within the pore rather than a less direct allosteric mechanism. In parallel we measure the effects of these mutations on the free energy of assembling the homotetrameric transmembrane pore from monomeric helices in micelles and bilayers. Although there is no simple correlation between the evolutionary fitness of the mutants and their stability, all variants formed more stable tetramers in bilayers, and the least fit mutants showed the smallest increase in stability upon moving from a micelle to a bilayer environment. We speculate that the folding landscape of a micelle is rougher than that of a bilayer, and more accommodating of conformational variations in non-optimized mutants.
Keywords: thermodynamics, mutagenesis, disulfide, influenza A virus, M2, membrane protein
Introduction
Our understanding of the determinants of membrane protein structure is at a relatively primitive level when compared to that of water-soluble proteins. One reason for this disparity is the relative paucity of membrane proteins in the crystallographic database, but a second reason relates to the lack of methodical studies of how systematic changes in sequence affect the thermodynamics of folding, particularly in bilayers. While considerable progress has been made on this front for β-proteins (Tamm et al., 2004), the α-helical class of membrane proteins have been more refractory to analysis. While encouraging results have been obtained in micelles, the interpretation of these studies is complicated by the difficulty in characterizing the amount of residual structure in the unfolded state (Bowie, 2004). One exception to this problem is the case of glycophorin A; because the native state of this TM helix is dimeric, the unfolded state can be defined as a monomeric helix (Fisher et al., 1999) that is devoid of any interactions with its partner. The thermodynamics of dimerization of glycophorin A and other related peptides (Fleming et al., 1997; Fleming and Engelman, 2001; Fleming et al., 2004) are well correlated with their of association in bacterial membranes (Russ and Engelman, 1999; Russ and Engelman, 2000; Senes et al., 2000). However, extensive thermodynamic measurements in bilayers as compared to micelles have not been accomplished. Clearly, direct measurement in both systems on the same peptide system would be a great contribution to the current knowledge.
Here we examine the effects of mutations on the function and thermodynamics of folding in phospholipid bilayers of a series of mutations of the AM2 proton channel from the influenza A virus, some of which were previously characterized by equilibrium analytical ultracentrifugation (EAUC) in micelles. AM2 is a 97 amino acid protein from the influenza A virus that homotetramerizes to form a proton channel. This protein is one of the smallest ion channels known, yet it exhibits properties of selectivity (Chizhmakov et al., 1996; Lin and Schroeder, 2001; Mould et al., 2000; Shimbo et al., 1996; Venkataraman et al., 2005), activation (Chizhmakov et al., 2003; Wang et al., 1995), and gating (Okada et al., 2001; Tang et al., 2002a). AM2 has a single transmembrane helix of 25 residues (M2TM, Figure 1) that retains the ability to tetramerize, conduct protons, and bind the anti-influenza drug amantadine (Salom et al., 2000). The thermodynamic analysis of this ion channel is simplified because folding involves the homo-oligomerization of a single membrane spanning helix. However, due to AM2’s functional complexity, our findings may be extended to more complex membrane proteins.
Figure 1.
Sequence alignment of M2TM22–46 from the Udorn strain of influenza A virus used in EAUC studies and M219–62 used in thiol-disulfide exchange experiments. The positions of single site mutations and their identities are highlighted in cyan. At bottom is the sequence of the Weybridge strain of influenza A virus containing the L38F mutation (variations in magenta).
A significant motivation of this study has been to determine how different mutations allow the protein to escape inhibition by amantadine, and how overall stability relates to this process. Of the several amantadine-insensitive mutations observed in the last two decades, S31N is particularly fit (Abed et al., 2005; Holsinger et al., 1994). This mutation renders the virus highly resistant to amantadine inhibition, and the incidence of clinical isolates bearing this mutation has jumped from a few percent to approximately 97% in recent years (Deyde et al., 2007), prompting the centers for disease control to recommend discontinuing the use of this drug (Bright et al., 2005; Bright et al., 2006). This mutation is found in 99% of the H3N2 viruses, which dominated the 2005–2006 flu season. It also occurs with very high frequency in H5N1, and is being found at increasing rates in H1N1 viruses, where it also occurs even in the absence of amantadine usage (Hayden, 2006). Recent strains of the virus bearing this mutation are now called the N-lineage of the virus (Simonsen et al., 2007). In the crystal structure (Stouffer et al., 2008), solution NMR structure (Schnell and Chou, 2008), solid-state NMR structural model (Hu et al., 2007) and computational models of M2 (Chen et al., 2007; Nishimura et al., 2002; Pinto et al., 1997), this mutation lies near the conduction pore of the virus and was widely believed to escape inhibition by altering the shape of the amantadine-binding site observed in the crystal structure of the protein. However, a recent NMR investigation of the protein in the presence of 40 mM of a related drug, rimantadine, concluded that the drug instead binds “allosteric” sites on the outside of the closed state of the channel, and that amantadine-resistant mutations strongly stabilize the open conformation (Schnell and Chou, 2008). To address this question, we report here measurements of the specific activity of drug-resistant mutants expressed in oocytes, but fail to find evidence for strong stabilization of an open conformation.
Previously, we used the EAUC method to determine the effect of numerous sequence variations on the stability of the proton channel in detergent micelles (Howard et al., 2002; Stouffer et al., 2005). The pore lining residues and residues at the helix-helix interface were mutated to alanine and phenylalanine (Howard et al., 2002; Stouffer et al., 2005). We found that almost all of the mutations favored tetramerization over the wild type and only mutations involving the polar His 37 residue destabilized the tetramer. These findings constrast with the extreme conservation of the amino acid sequence found throughout AM2 proton channel strains, suggesting that the sequence requirements for stability of the tetramer are quite modest, while the requirements for function are quite stringent. Thus, the channel protein’s sequence does not appear to evolve towards thermodynamic stability, but for function – specifically, to allow formation and interchange of a number of functional conformations, each required for gating and transmission of protons. Here, we extend this analysis to determine the effect of a subset of these mutations on the channel stability in a bilayer environment. We also examine how these changes in sequence influence proton channel activities.
To determine the free energy of association in bilayers we employed the method of equilibrium thiol-disulfide interchange to examine some of the most and least stabilizing mutations as determined in detergent micelles. Additionally, the reversal voltage and specific activity of some AM2 mutants were measured in oocytes. We find that the all of the variants form more stable tetramers in phospholipid bilayers versus micelles. Moreover, there are functionally interesting differences in the degree of stabilization of the mutants in micelles versus bilayers; sequence variants from highly fit and virulent forms of the virus favor tetramerization in bilayers over mutations that have not been observed in field of isolates of the virus.
Results
Variants of M2TM
This study focuses on natural and unnatural mutations that have interesting effects on the thermodynamic stability and function of the channel in micelles. These variants include mutations whose stabilities vary by 3.7 kcal/mol tetramer (Table 1), some of which also perturb the binding of the anti-influenza drug amantadine to the proton pore. In our previous study (Howard et al., 2002; Stouffer et al., 2005), we found H37A to be the only mutation that strongly destabilizes the tetramer. It was also chosen for this study because of the importance of histidine in channel function. The four histidine residues that line the channel pore are involved in proton selectivity, which is eliminated by the H37A mutation (Czabotar et al., 2004; Gandhi et al., 1999; Venkataraman et al., 2005), and form transient interhelical interactions with Trp 41 in the gating process (Okada et al., 2001; Wu and Voth, 2005).
Table 1.
Free energies of dissociation (ΔGtet) for wild type and M2TM mutant peptides in the units of kcal/mole tetramer. The change of energy upon mutation is also expressed relative to wild type as ΔΔGtet. Curve fits for the M2TM S31N and H37A peptide is located in Supplementary Figure 1. Experimental error for ΔΔGtet in EAUC (micelles) was found in repeated measurements of WT to be ± 0.12 (Stouffer et al., 2006) With the thiol-disulfide interchange method used for vesicles, errors in absolute ΔGtet, estimated by fixing K2 to its upper and lower limits (determined from the WT fitting) and refitting each mutant data set, were between 0.5 and 0.8 kcal/mole. However, because we were interested in ΔΔGtet values for the different environments, ΔGtet values for the vesicle experiments were calculated from the least-squares fitting errors in K1 for each mutant and WT. We then used the RMS error for each mutant with WT to calculate the resulting errors for the ΔΔGtets. This was done because K2, whose value showed strong correlation with K1, was assumed to be the same as WT for all mutants.
| Micelles | Vesicles | |||
|---|---|---|---|---|
| ΔG tet (kcal/mol) | ΔΔG tet (kcal/mol) | ΔG tet (kcal/mol) | ΔΔG tet (kcal/mol) | |
| WT | *8.3 ± 0.12 | 11.2 ± 0.12 | ||
| L26F | *10.6 ± 0.12 | 2.3 ± 0.17 | 12.7 ± 0.27 | 1.5 ± 0.3 |
| S31N | 8.7 ± 0.12 | 0.4 ± 0.17 | 12.4 ± 0.27 | 1.2 ± 0.3 |
| I33A | *10.1 ± 0.12 | 1.8 ± 0.17 | 12.2 ± 0.59 | 1.0 ± 0.2 |
| H37A | 6.9 ± 0.12 | −1.4 ± 0.17 | 10.2 ± 0.16 | −1.0 ± 0.2 |
| L38F | *10.4 ± 0.12 | 2.1 ± 0.17 | 14.1 ± 0.16 | 2.9 ± 0.6 |
previously reported in (Stouffer et al., 2005).
We also studied L26F, I33A, and L38F, which were the three most stabilizing sequence variants previously studied (Stouffer et al., 2005). Each of these has a different phenotype of interest to this work. I33A was chosen as an amantadine-resistant mutant that, to our knowledge, has not been observed in naturally occurring strains of influenza A virus. L26F was investigated as an amantadine-resistant mutant that is not widely distributed in the absence of selective pressure from the presence of amantadine (Abed et al., 2005; Kitahori et al., 2006; Suzuki et al., 2003); viruses bearing this mutation have smaller plaque sizes than normal (Abed et al., 2005). Finally, L38F is a natural mutation that occurs in the Weybridge strain (Figure 1) and other strains of influenza A virus. Although L38F shows a decrease in its ability to bind amantadine (Stouffer et al., 2005), the Weybridge strain is vulnerable to the drug (Wang et al., 1993).
The peptides studied here, designated M2TM19–46, are slightly longer than the peptides previously studied, M2TM22–46, by EAUC (Figure 1). The peptides have been extended to Cys19, which forms an interchain disulfide (Holsinger and Lamb, 1991) in native M2, and enables measurement of stability through thiol-disulfide interchange in the present study. In the reduced free-thiol form, the additional residues do not significantly affect the free energy of tetramerization of the peptides in micelles using analytical ultracentrifugation (Cristian et al., 2003a). Mutants whose free energy of tetramerization were not previously reported have now been determined by the same method and their stabilities are reported in Table 1. For the functional assays, full length M2 protein mutants were used.
Measurement of stability using equilibrium thiol-disulfide interchange in phospholipids bilayers
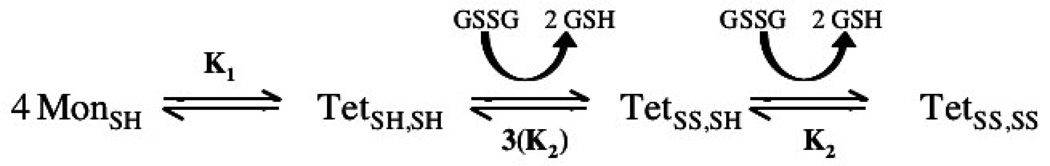
The method of thiol–disulfide interchange in lipid bilayers has been successfully developed to measure the tetramer dissociation constant of M2TM19–46 (Cristian et al., 2003a; Cristian et al., 2003b). The method involves measuring the equilibrium between the reduced and disulfide-bonded form of the protein under reversible redox conditions using a glutathione redox buffer at equilibrium. If the experiments are conducted under conditions in which the protein is primarily monomeric in the reduced form, but tetrameric when oxidized, one can analyze the peptide concentration dependence of the equilibrium to obtain the dissociation constant for tetramerization (Scheme 1).
Scheme 1.
The thermodynamic equilibrium between disulfide coupling and tetramer formation, showing thermodynamically significant species (Cristian et al., 2003b). The oxidation states of monomer (Mon) and tetramer (Tet) are denoted in the subscripts as “SH” for reduced and “SS” for oxidized. Oxidized and reduced glutathione is represented as GSSG and GSH, respectively. The first K2 value is given a coefficient of three because there are three potential disulfide partners in the reduced tetramer.
This method requires a substantial monomer population in equilibrium with the tetrameric form of the protein at experimentally accessible peptide to lipid ratios. Previously we found that this could be accomplished by incorporating the peptide into short-chain lipid bilayers formed from DLPC (1,2-dilauroyl-sn-glycero-3-phosphocholine), in which the tetrameric conformation is less stable than in longer chain lipid bilayers such as DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine) or POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) (Cristian et al., 2003b). Although a thicker bilayer is more biologically relevant, the tetramer was essentially fully formed in long chain lipids. Also, the head group and hydrocarbon chain length of DLPC are similar to that of the dodecyl phosphocholine (DPC) detergent used in the analytical centrifugation experiments allowing a more direct comparison between the two studies.
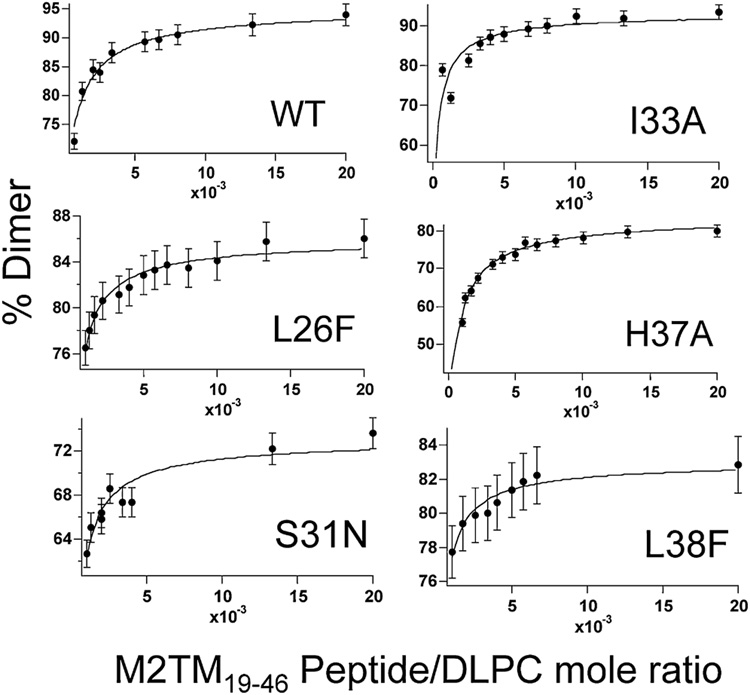
Once the system achieves equilibrium, analytical HPLC is used to quantitate the amount of monomer and disulfide bonded dimer (as a result of tetramerization) at equilibrium for various peptide to lipid ratios. The resultant dimer population is an attribute of the affinity and cooperativity of M2TM tetramerization (Cristian et al., 2003a). The dissociation constants and equilibria are fit to the experimental data as previously shown (Figure 2) using Scheme 1 (Cristian et al., 2003b). The free energy of tetramer dissociation (Ktet) is then calculated by ΔG = −RT lnKtet. To obtain the best estimate of Ktet, the experiments were conducted at a glutathione oxidized to reduced molar ratio of 1:2. This ratio allowed measurement of the least stable to the most stable variants under a single experimental condition.
Figure 2.
The dissociation constant and equilibria from scheme 1 (curve) is fit to the percent dimer formation for each respective peptide to DLPC lipid mole ratio (●) for wild type M2TM19–46 and the five sequence variants.
Sequence Variants are More Stable in Lipids vs. Micelles yet the Effects of Mutations on Stability are Correlated
As expected, ΔGtet of dissociation for the wild type and each of the sequence variants is more favorable in DLPC vesicles than in the DPC micelles used in the EAUC experiments (Table 1). The tetramers are more stable in bilayers than micelles by up to 4.2 kcal/mol tetramer, depending on the sequence variant. Considering that the contribution to the overall stability of the additional N-terminal residues, is only 0.5 kcal/mol tetramer (Cristian et al., 2003a), these stabilizing effects are quite significant.
L26F and S31N mutant ion channel proteins retain activity and proton selectivity while amantadine sensitivity is lost
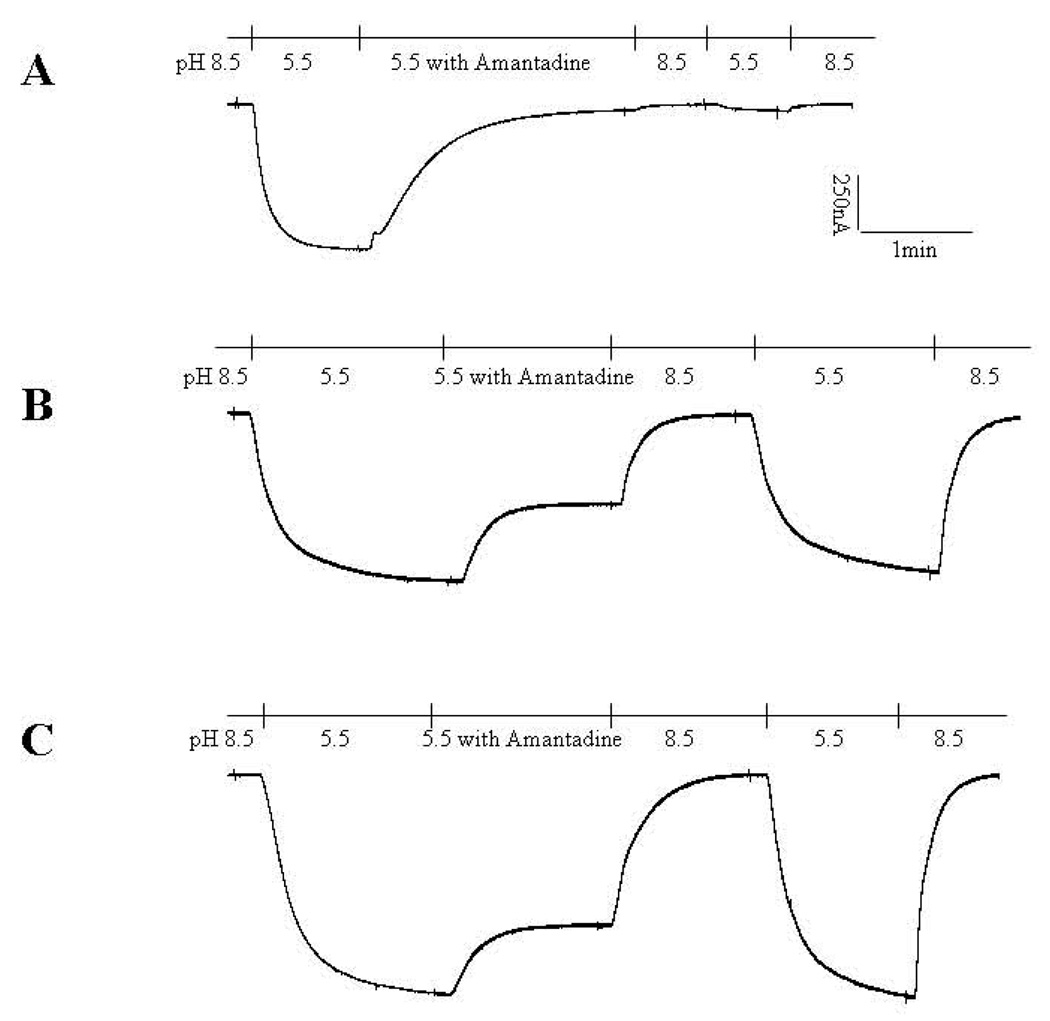
To gain an understanding of the function of these point mutations, we compared the properties of the mutants in the full-length proteins in Xenopus oocytes. The effects of mutating H37 to Gly and other small residues have already been very extensively studied; these mutations lead to channels with highly compromised selectivity(Venkataraman et al., 2005). The effects of mutating L38 to Phe has also been thoroughly investigated, and this mutation has little affect when made in the sequence studied here (Betakova et al., 2005; Chizhmakov et al., 2003; Wang et al., 1993). Thus, we focused on a comparison of WT, L26F and S31N, as these mutations eliminate amantadine binding and thermodynamically stabilize the formation of tetramers in the M2TM peptides (Stouffer et al., 2005). As depicted in Figure 3, both of L26F and S31N mutant channels displayed a robust pH-activated inward current, similar to that seen for the wild type ion channel. To allow the detection of weak amantadine-sensitivity expected for the mutants, measurements were conducted at a high (100 µM) drug concentration, which is approximately 100-fold greater than the IC50 for the WT protein. As expected, the wild type protein was essentially completely inhibited by treatment for 2–3 min with 100 µM amantadine, and this inhibition did not reverse over a five-minute period following removal of the drug (Figure 3A). By contrast, the ion channel activity of the S31N mutant was inhibited by only 30%, and this small inhibition was immediately reversed after the removal the drug (Figure 3C). The L26F mutant ion channel was also reversibly inhibited by amantadine (Figure 3B), although the inhibition (47%) was somewhat higher than for S31N (p < 0.001). Thus, S31N is more resistant to inhibition by amantadine than L26F.
Figure 3.
pH activation and amantadine sensitivity of L26F, S31N and WT AM2 channels. Oocytes expressing wt AM2 protein or either mutant protein displayed robust inward current (downward reflection), 48–72 hrs after mRNA injection when oocytes were bathed in pH5.5 Barth’s solution. Amantadine sensitivity of these AM2 variants was evaluated by bathing the oocytes in pH 5.5 Barth’s solution that contained 100 µM amantadine when the oocyte displayed maximal inward current. A representative trace is shown for each AM2 variant. (A) WT AM2. Amantadine inhibits 95.2 ± 1.5% (n = 5) of the inward current, and this inhibition is not reversible. (B) L26F AM2. Amantadine inhibits 46.8 ± 0.7% (n= 4) of the inward current, and this inhibition is readily reversible. (C) S31N AM2. Amantadine inhibits 30.7 ± 1.8% (n =4) of the inward current, and this inhibition is also highly reversible. Inhibition is greater for L26F than for S31N mutant protein (P < 0.001).
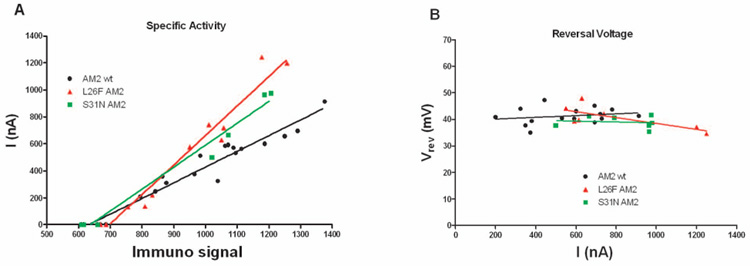
Given that these mutants are approximately 100-fold more resistant to amantadine than the WT, the allosteric model predicts (Schnell and Chou, 2008) a similarly large change in their conductance characteristics associated with a parallel increase in the stability of the open state. Furthermore, the effect should be more marked for S31N than L26F, because S31N is more resistant to amantadine. We therefore measured the surface expression, specific activity and proton selectivity for these AM2 variants (Figure 4A). The amount of protein expressed on the surface of the cell was approximately the same for the S3N1, L26F, and WT protein, indicating that the mutations did not affect the ability of the protein to be biosynthetically incorporated into the plasma membrane. The specific activity is defined as the current at pH 5.5 divided by the amount of protein expressed at the surface of the oocyte for each cell studied. In practice, it is computed from the slope of the current at pH 5.5 versus the surface expression of M2 obtained in 10 to 20 experiments. In contrast to the allosteric model (Schnell and Chou, 2008) the specific activity of S31N is very similar to wild type, showing only a 1.4-fold increase in specific activity. In further contradiction to the allosteric model, the specific of L26F is a 1.35-fold greater than the specific activity of S31N, despite the fact that S31N is more resistant to amantadine than L26F.
Figure 4.
Specific activity and reversal voltage (Vrev) of L26F, S31N and wt AM2 channel expressed in oocytes. (A) For each AM2 variant at least 4 oocytes expressing the M2 protein and 3 uninjected oocytes were measured. The current for each uninjected cell and for each cell expressing a given protein was plotted against the immunofluorescence signal intensity for that cell and a straight line was fitted to the data. The slope of this plot was taken as the relative specific activity of the protein. The resulting slopes were as follows: AM2 wt, slope = 1.164 ± 0.059; L26F, slope = 2.183 ± 0.143; and S31N, slope = 1.624 ± 0.116 (F =29.9, ΔFn = 35, P < 0.001) (B) Membrane conductance measurements were made at pH5.5; after membrane current reached the maximal value, a 100 mV ramp of voltage from −40 mV to 60 mV was applied. For each cell expressing a given M2 channel the reversal voltage was plotted against the steady inward current for that cell and a straight line was fitted to the plot. The Y-intercept of this plot was taken as the reversal voltage (Vrev) of the protein. The Vrev values were as follows: AM2 wt, Vrev = 41.0 ± 2.4 mV; L26F, Vrev = 49.4 ± 3.8 mV, and S31N, Vrev = 40.2 ± 4.5 mV (F = 0.47, ΔFn = 25, P = 0.63).
We also examined the ion selectivity of these two channel proteins by measuring the reversal voltage (Vrev) in normal Na+ Barth’s solution (Figure 4B). The reversal potentials of all the mutant ion channels were the same within experimental error, indicating that the L26F and S31N mutant M2 channels retain very high proton selectivity.
Discussion
Structural basis for modulation of stability
The M2 proton channel is a pH-gated channel that is activated at low pH. Very recently, structures of the tetrameric transmembrane region of the protein were solved at low and neutral pH by X-ray crystallography (Stouffer et al., 2008), and at high pH by NMR structure (Schnell and Chou, 2008). The structures are broadly similar, but show conformational changes consistent with the pH-dependent opening of the channel at low pH. Given that our measurements were conducted at high pH it is most appropriate to consider their effects in the context of the high pH-form of the channel, and the positions of the changes have been shown on the transmembrane domain in Fig. 5A and 5B. Both the NMR and crystal structures confirm that all of the mutations studied in this work lie along the helix-helix interface of the tetramer, with the exception of His37, which projects towards the center of the channel. His 37 engages in numerous stabilizing interactions with nearby Trp residues, explaining the destabilizing nature of mutations to this residue. The effects of the other mutations are also broadly consistent with the structure. For example, the mutation of L26 or L38 to Phe can easily be accommodated at the helix-helix interface, where the larger Phe sidechain can pack favorable against the neighboring helix. Interestingly, I33A occurs in a region that is rich in small residues, which are particularly favorable for helix-helix interactions in membranes (Senes et al., 2004; Senes et al., 2000); mutation of I33 to Ala might facilitate and extend the packing of a patch of small residues that includes Ser31 and G34 on one helix and A30 of a neighboring helix. Finally, the mutation S31N can be accommodated by placing the Asn sidechain within the aqueous pore (Stouffer et al., 2008).
Figure 5.
Positions of mutations in a side view (A) and top view (B) of the channel. Panel C shows the position of rimantadine inferred from the NMR study in pink near the bottom of the panel, versus the position of rimantadine (in orange) inferred from crystallographic and mutagenesis studies. The geometric complementarity of the amantadine docked into the pore of the channel is striking. Panels A and B show the positions of residues mutated in space-filling with the carbon atoms of each mutated residue in orange, except for L38, which is shown in cyan for contrast. The remaining carbon atoms are shown in green.
Thermodynamic stability in bilayers versus micelles
All five homotetrameric peptides have a higher stability in bilayers than micelles (Table 1), which can be related to intrinsic differences between the two hydrophobic environments. In this study we used short - chained lipid vesicles to facilitate the experiment, and to allow comparison with the measurements in DPC micelles. However, the protein is even more stable in membranes with longer chain lengths and/or in the presence of cholesterol (Cristian et al., 2003b). Thus, chain lengths that are more similar to physiological lipids would show much greater increases in the stability and have an even greater impact on the conformational specificity of the tetramer.
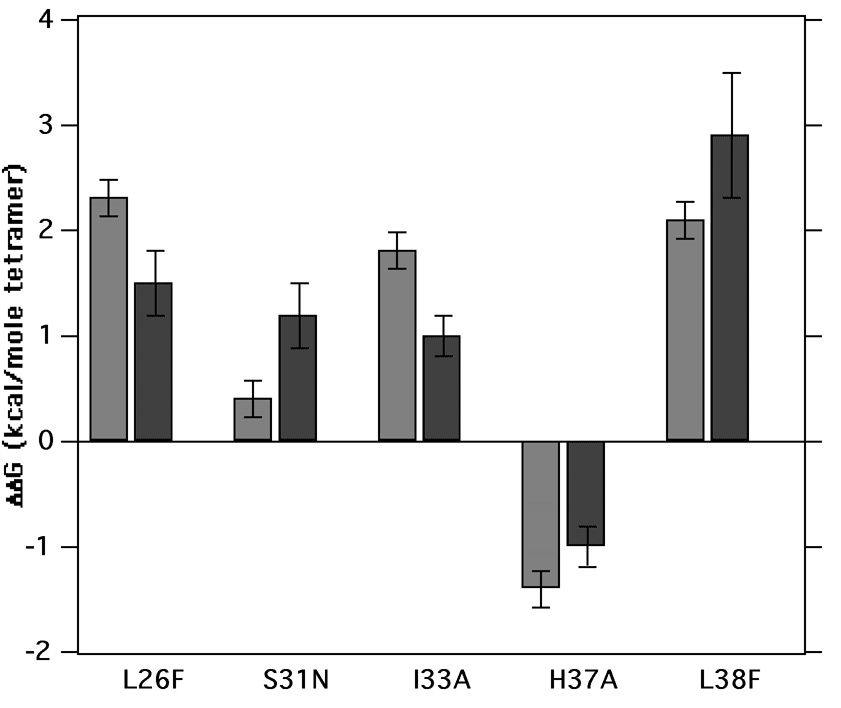
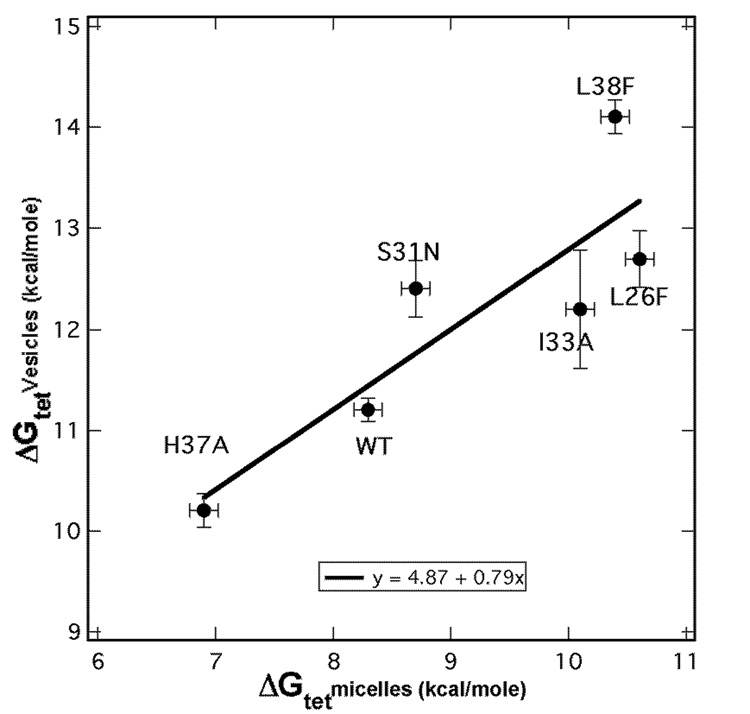
We observe a reasonable correlation between the stabilities in the two environments (Figure 6); mutations that are stabilizing in micelles remain stabilizing in bilayers, and the destabilizing mutation was also destabilizing in both environments. The probability that all five mutations would have the same effect in the two environments is approximately 0.55 = 0.03, indicating that this finding is significant. Regression analysis of the tetramerization free energies (Figure 7) gave a near unitary slope and a good correlation (R = 0.86) between the stability of the mutants in the two environments. In general the mutations are several kcal/mol more stable in bilayers versus micelles over the experimental range examined. There are, however, deviations of individual points from the regression line that are beyond the error bars, suggesting that there could be differential effects of the environment on each mutant.
Figure 6.
Change in free energy of tetramer dissociation upon mutation in micelles (grey) and vesicles (black). L26F, S31N, I33A and L38F mutations stabilize tetramer formation in both environments whereas H37A is destabilizing
Figure 7.
Correlation plot of the effect of single site mutations on free energy of tetramer formation (−ΔGtet) in vesicles and the corresponding effects in micelles. Points that fall above the trend line indicate that mutations are more stabilizing in vesicles, whereas points that fall below the line indicate that mutations are more stabilizing in micelles.
In micelles, the detergent molecules decorate themselves around a transmembrane protein, which allows for considerable structural flexibility. On the other hand, bilayers align the charged and the hydrophobic sectors of integral membrane proteins in a more geometrically restricted manner. Thus, folding within a micelle might occur on a more rugged energetic landscape, with many more potential minima than encountered in the more restricted conformational space available in a bilayer. In this scenario, mutations could have a net stabilizing effect in a micelle environment – not by lowering the energy level of the native conformations, but rather by stabilizing one or more non-native states that are accessible only in micelles. Also, these mutations might upset a delicate energetic balance by stabilizing only one of multiple conformations required for function. One might expect, then, that naturally occurring, highly fit mutations should be highly stabilizing to the full ensemble of native structures, whereas the unnatural or less fit mutations might induce non-native conformations in the more malleable environment of a micelle than in a bilayer. The data do suggest this could be the case; S31N and L38F lie above the trend line in Figure 7, while the less fit L26F and I33A (not observed in natural viruses) mutations lie below the line. The selective increase in stability of S31N over WT is also consistent with previous studies showing that Asn is able to stabilize TM helical bundles (Choma et al., 2000; Gratkowski et al., 2001; Lear et al., 2003). These observations, suggest a possible explanation for previous observations concerning the effects of mutations in membrane proteins (Bowie, 2001): it is often surprisingly easy to discover mutations that increase stability without showing major effects on a membrane protein’s function measured in vitro. We speculate that such mutations affect the energetic landscape, allowing misfolding or other events that might be apparent only after extended times in vivo.
Implications to the structure of the amantadine-M2 complex and the development of drug resistance
It is interesting to consider our results in light of the recent dramatic increase in the resistance of viruses to amantadine and rimantadine. In the crystal structure of the amantadine-M2TM complex, the drug appears to be bound in the pore surrounded by residues that are mutated in amantadine-resistant viruses (Stouffer et al., 2008). Mutating Ser31 to Asn is expected to decrease the radius and/or increase the polarity of the pore, thereby interfering with binding. The mutation L26F also decreases the amantadine-sensitivity, although not to the same extent as S31N. Its proximity to the crystallographically defined amantadine-binding site suggests that it might alter the geometry of the pore sufficiently to decrease the affinity for amantadine. This possibility is consistent with its location near N-terminus of the helix where the bundle is least restrained and hence might undergo conformational changes in response to even subtle mutations, particularly when the changes are repeated at each monomer of the subunit.
While, our results are consistent with the crystallographic structure of the amantadine-M2 complex, they are less consistent with the surface location of four drug-biding sites observed in the NMR structure (Schnell and Chou, 2008). Other facts that are difficult to reconcile with the NMR model include: 1) the drug is known to bind in a stoichiometry of one drug/tetramer with a Hill coefficient of 1.0 (Czabotar et al., 2004; Wang et al., 1993); 2) amantadine-resistant mutations cluster in a region surrounding the N-terminal region of the pore, rather than at the proposed surface sites near the C-terminus of the channel (Stouffer et al., 2008); 3) mutations to each of the residues predicted to comprise the NMR-defined drug-binding sites (residues 40–45) had little affect on the ability of amantadine to block the channel (Pinto et al., 1997; Tang et al., 2002b); 4) amantadine and rimantadine bind to the channel very slowly and dissociate even more slowly, which is inconsistent with the rapid kinetics expected for a surface binding site; 5) the NMR structure contains an unfilled site in the pore of the channel that is surrounded by residues important for amantadine binding and that could sterically accommodate the drug (Figure 5C); 6) the residues lining the site in the pore where the drug was expected to bind show no NOEs with water molecules, despite the fact the channel is 7 Å wide at this point, and hence would be expected to be filled with water (unless the drug was actually there but not detected due to motional averaging).
These contradictions were rationalized by suggesting that the drug inhibits the channel allosterically by binding to the surface site, which predicts that mutations strongly stabilize the open state of the channel. However, the electrophysiological studies of L26F and S31N were not consistent with this expectation. Instead, they lend support to a large body of data indicating that biologically relevant mutants escape amantadine inhibition by alterations to the amantadine-binding site in the pore. Thus, the binding sites observed on the outside of the channel in the NMR structure appear to be secondary sites, possibly related to the amine-binding sites observed under some conditions in vesicle assays (Lin et al., 1997). The observation of a secondary site might be a result of the experimental conditions used in the NMR structure determination (Schnell and Chou, 2008); the drug was present at 40 mM concentration, the detergent at 300 mM and the tetramer at 0.2 mM (0.75 mM in monomer). Given that rimantidine partitions almost entirely into the micelle, it constituted 13% of the micellar components (Schnell and Chou, 2008). A protein with a cross-sectional area similar to the M2 tetramer has been shown to bind at least 90 molecules of dihexanoylphosphatidylcholine (the same detergent as in the M2 work) (Fernandez et al., 2002; Hilty et al., 2004), indicating that there were at least 12 drug molecules per micelle compartment in which the channel resides. Furthermore, remarkably selective NOEs between residues on the surface of a protein and the detergents in a micelle are often observed (Fernandez et al., 2002; Hilty et al., 2004), although they do not necessarily reflect pharmacologically relevant binding events.
Finally, it is interesting to ask why the S31N mutation has only recently become dominant, even in areas where there is not selective pressure from drug use. This mutation had appeared many times over the last decade, but until now it never showed a selective advantage in the absence of the drug. Thus, the dominance of this lineage must arise from interactions with fitness-enhancing mutations unrelated to drug resistant at other genomic sites (Simonsen et al., 2007). A given variant of M2 must work in conjunction with the viral hemagglutinin (HA) (Steinhauer et al., 1991), which is responsible for the pH-dependent fusion of the viral envelope with the endosomal membrane. When in the Golgi network the proton channel activity of M2 is important for preventing a premature conformational change of the HA of some subtypes (Ciampor et al., 1992). Thus, the activity of the M2 must be sufficient to ensure proper transit of HA to the cell surface, which depends to a significant extent on the pH-sensitivity of the HA molecules in the predominant strain of virus at a given time (Grambas et al., 1992; Lamb, 1994; Pinto and Lamb, 2006).
We hypothesize that optimal proton flux and pH-sensitivity of M2 will drift with time in response to changes in the predominant strain of HA. S31N is better matched to the requirements of the HA proteins in the currently circulating viruses than the previous wild type with Ser31. Thus, this mutation might be seen to enjoy multiple advantages including enhanced thermodynamic stability, almost complete resistance to amantadine, and improved functional matching to the requirements of the host and virus. On the other hand, excessive channel activity might be deleterious to the host cell, which could contribute to the small-plaque phenotype seen for L26F. Thus, the increased specific activity of this mutant together with its potential loss of conformational specificity might contribute to its low frequency of occurrence in the absence of a strong selective pressure from the presence of amantadine.
Experimental Procedures
Peptide Synthesis and Purification
N-terminally acetylated sequence variants of M2TM22–46 and M2TM19–46 peptides (Figure 1) were synthesized using an Applied Biosystems 430A peptide synthesizer on a 0.25 mmole scale, using Rink-amide resin with a 0.35 mmole/g substitution level as in (Stouffer et al., 2005). Fmoc-arginine-(Pbf)-OH and Fmoc-glycine-OH were doubly coupled. A cocktail of TFA/ thioanisole/1,2-ethanedithiol/anisole (90:5:3:3 v/v) under nitrogen for 2 h was used to cleave the peptides from the resin and deprotect the amino acid side chains. Crude peptide was precipitated and washed three times with ether/hexanes (50:50 v/v) and dried under reduced pressure overnight. The peptide was then purified on a semi-preparative reverse phase HPLC (Vydac C-4 column) using a linear gradient of buffer A (0.1% aqueous TFA) and buffer B 2-propanol/acetonitrile/water (6:3:1 containing 0.1% TFA) running from 60% to 100% buffer B. Elution of pure peptide occurred at ~82% buffer B. The purity of the peptides was assessed on an analytical C4 HPLC column using a linear buffer A/B gradient and molecular weights were verified by MALDI-TOF (PerSeptive Biosystems) mass spectrometry.
Site-specific mutagenesis, mRNA synthesis, microinjection and culture of oocytes
M2L26F and M2S31N, full length protein from the Udorn strain of influenza A virus, were prepared employing the QuikChange site-directed mutagenesis method. Wild type, M2L26F and M2S31N cDNAs were linearized before mRNA synthesis using their respective restriction sites. In vitro synthesis of mRNA was performed using the mMessage mMachine T7 transcription Kit (Ambion, Austin, TX). Ovarian lobules from female Xenopus laevis were removed by surgery and treated with collagenase B (~ 1mg/ml; Roche Diagnostics) in Ca2+ -free OR-II solution (82.5 mM NaCl, 2 mM KCl, 1 mM MgCl2, 5 mM HEPES-NaOH, pH7.5) at room temperature for 1 to 1.5 hr to free oocytes from follicle cells. These oocytes were washed with OR-II for a few times and maintained in ND96 solution (96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 2.5 mM sodium pyruvate, 0.1 mg/ml gentamycin, 5 mM HEPES-NaOH, pH 8.5). Oocytes of good size, color and shape were injected 50 nl mRNA. The injected oocytes were still maintained in ND96 solution.
Equilibrium Analytical Ultracentrifugation
Centrifugation of M2TM22–46 S31N and H37A sequence variants were performed and analyzed according to (Stouffer et al., 2005).
Thiol-disulfide Exchange Sample Preparation and Redox Equilibration Reaction
M2TM19–46 peptide from a 2-propanol/water (1:1) stock was added to DLPC from an ethanol stock. The peptide concentration was kept constant (20 µM) and the peptide to detergent mole ratio was varied between samples from 1:50 to 1:2000. Stock solvents were evaporated under a stream of nitrogen and the resultant films were left overnight under high vacuum. Sample vials with air-tight caps were evacuated with argon gas. The film was then hydrated with a degassed 100 mM Tris-HCl, 0.2 M KCl, 1 mM EDTA pH 8.6 buffer solution. This thiol-disulfide interchange reaction was initiated by adding oxidized (GSSG) and reduced (GSH) glutathione so that their final concentrations were 0.66 mM and 1.33 mM respectively.
Samples were vigorously vortexed and sonicated until the peptide/detergent film was completely incorporated into the glutathione containing buffer. Vesicles were equilibrated using the freeze-thaw method every 2 hrs for 6 hrs, using a dry-ice acetone bath, followed by sonication until the samples became clear (Cristian et al., 2003b). After equilibration, the reaction was quenched by lowering the pH with HCl. The equilibrated samples were analyzed by reverse-phase HPLC using an analytical C-4 column with a linear buffer A/ buffer B gradient. Elutant species consisted of DLPC, followed by a mixed disulfide of peptide with GSH, the thiol free monomer, and the disulfide bonded peptide. The latter three were identified by MALDI-TOF mass spectrometry. HPLC peaks were integrated to determine the fraction of covalently bound dimer.
Data were fit to a function defined by the equilibria of Scheme 3 using Igor Pro as described previously. Fitting variables for WT were K1, K2, and a baseline correction required to account for irreversible aggregation. The WT value of K2 was used as a fixed parameter in fitting the mutant data, reasoning that the similar structures and identical buffer conditions should result in the same cysteine reactivities. Previously, we found that a baseline correction was necessary to account for a fraction aggregated protein that was not in thermodynamic equilibrium. This value showed no systematic trend between the mutants, and varied by approximately 10% (standard deviation of the difference from the mean).
Electrophysiological recording
Whole-cell currents were recorded 48–72 hrs after mRNA injection with a two-electrode voltage-clamp apparatus consisting a Dagan TEV 200A amplifier (Axon Instruments, Union City, CA) at room temperature using two electrodes filling with 3 mM KCl. Individual oocytes were held at −20mV and oocytes current were measured at standard Barth’s solution (88 mM NaCl, 1.0 mM KCl, 2.4 mM NaHCO3, 0.3 mM NaNO3, 0.71 mM CaCl2, 0.82 mM MgSO4, 15 mM HEPES (pH 8.5), or 15 mM MES (pH 5.5). Data were collected using pClamp10.0 software. 100 µM amantadine hydrochloride was dissolved in pH 5.5 Barth’s solution, and the final pH was checked and adjusted to 5.5.
Immunofluorescence of living oocytes
Individual oocytes were fixed immediately after recording in ND96 solution containing 2% formaldehyde in a 96-well plate for measurement of relative surface expression levels. Oocytes were then incubated in ND96 solution with 2% nonfat milk for 1 hr, for 30 min with 14C2 mouse monoclonal antibody (1:500 dilution, in ND96 solution with 2% milk), then washed (3X) with 2% milk containing ND96 solution (10 min each), and finally incubated with goat anti-mouse IgG1 (g1) labeled with Alexa Fluor® 546 (A21123, Molecular Probes, Medford, OR; 40 mg/ml in 2% milk containing ND96 solution) for 30 min. Free fluorophore was removed by five more washes (10 min each) with ND96 solution. All steps were carried out at 4°C, and the ND96 solution used in this immunofluorescence study lacked CaCl2, gentamycin and pyruvate. Fluorescence was quantified using a PTI image master microfluorometer (Photon Technologies) with a 20 X 0.5 NA objective as described previously (Venkataraman et al., 2005). The brightest part of the each oocyte’s surface was imaged using a CCD camera when exciting the AlexaFluor® with a 540 nm wavelength of light. Intensity quantifications were made using Image Master Version 5.0 software (Photon Technologies). Uninjected oocytes were subjected to the above conditions as a control for the autofluorescence from the yolk (Beumer et al., 1995).
Supplementary Material
Acknowledgement
ALS thanks Kathleen Howard for providing resin containing M2TM H37A22–46. Work supported by NIH grants GM56423 (WFD) and AI-31882 (LHP).
Abbreviations
- TM
transmembrane
- EAUC
equilibrium analytical ultracentrifugation
- DLPC
1,2-dilauroyl-sn-glycero-3-phophocholine
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- DPC
dodecyl phosphocholine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abed Y, Goyette N, Boivin G. Generation and characterization of recombinant influenza A (H1N1) viruses harboring amantadine resistance mutations. Antimicrob Agents Chemother. 2005;49:556–559. doi: 10.1128/AAC.49.2.556-559.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betakova T, Ciampor F, Hay AJ. Influence of residue 44 on the activity of the M2 proton channel of influenza A virus. J Gen Virol. 2005;86:181–184. doi: 10.1099/vir.0.80358-0. [DOI] [PubMed] [Google Scholar]
- Beumer TL, Veenstra GJ, Hage WJ, Destree OH. Whole-mount immunohistochemistry on Xenopus embryos using far-red fluorescent dyes. Trends Genet. 1995;11:9. doi: 10.1016/s0168-9525(00)88978-7. [DOI] [PubMed] [Google Scholar]
- Bowie JU. Stabilizing membrane proteins. Curr Opin Struct Biol. 2001;11:397–402. doi: 10.1016/s0959-440x(00)00223-2. [DOI] [PubMed] [Google Scholar]
- Bowie JU. Membrane proteins: a new method enters the fold. Proc Natl Acad Sci U S A. 2004;101:3995–3996. doi: 10.1073/pnas.0400671101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright RA, Medina MJ, Xu XY, Perez-Oronoz G, Wallis TR, Davis XHM, Povinelli L, Cox NJ, Klimov AI. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet. 2005;366:1175–1181. doi: 10.1016/S0140-6736(05)67338-2. [DOI] [PubMed] [Google Scholar]
- Bright RA, Shay DK, Shu B, Cox NJ, Klimov AI. Adamantane resistance among influenza A viruses isolated early during the 2005–2006 influenza season in the United States. Jama. 2006;295:891–894. doi: 10.1001/jama.295.8.joc60020. [DOI] [PubMed] [Google Scholar]
- Chen H, Wu Y, Voth GA. Proton transport behavior through the influenza A M2 channel: insights from molecular simulation. Biophys J. 2007;93:3470–3479. doi: 10.1529/biophysj.107.105742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizhmakov IV, Geraghty FM, Ogden DC, Hayhurst A, Antoniou M, Hay AJ. Selective proton permeability and pH regulation of the influenza virus M2 channel expressed in mouse erythroleukaemia cells. J Physiol. 1996;494(Pt 2):329–336. doi: 10.1113/jphysiol.1996.sp021495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizhmakov IV, Ogden DC, Geraghty FM, Hayhurst A, Skinner A, Betakova T, Hay AJ. Differences in conductance of M2 proton channels of two influenza viruses at low and high pH. J Physiol. 2003;546:427–438. doi: 10.1113/jphysiol.2002.028910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choma C, Gratkowski H, Lear JD, DeGrado WF. Asparagine-mediated self-association of a model transmembrane helix. Nature Struct Biol. 2000;7:161–166. doi: 10.1038/72440. [DOI] [PubMed] [Google Scholar]
- Ciampor F, Thompson CA, Grambas S, Hay AJ. Regulation of pH by the M2 protein of influenza A viruses. Virus Res. 1992;22:247–258. doi: 10.1016/0168-1702(92)90056-f. [DOI] [PubMed] [Google Scholar]
- Cristian L, Lear JD, DeGrado WF. Determination of membrane protein stability via thermodynamic coupling of folding to thiol-disulfide interchange. Protein Sci. 2003a;12:1732–1740. doi: 10.1110/ps.0378603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristian L, Lear JD, DeGrado WF. Use of thiol-disulfide equilibria to measure the energetics of assembly of transmembrane helices in phospholipid bilayers. Proc Natl Acad Sci U S A. 2003b;100:14772–14777. doi: 10.1073/pnas.2536751100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czabotar PE, Martin SR, Hay AJ. Studies of structural changes in the M2 proton channel of influenza A virus by tryptophan fluorescence. Virus Res. 2004;99:57–61. doi: 10.1016/j.virusres.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Deyde VM, Xu X, Bright RA, Shaw M, Smith CB, Zhang Y, Shu Y, Gubareva LV, Cox NJ, Klimov AI. Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J Infect Dis. 2007;196:249–257. doi: 10.1086/518936. [DOI] [PubMed] [Google Scholar]
- Fernandez C, Hilty C, Wider G, Wuthrich K. Lipid-protein interactions in DHPC micelles containing the integral membrane protein OmpX investigated by NMR spectroscopy. Proc Natl Acad Sci U S A. 2002;99:13533–13537. doi: 10.1073/pnas.212515099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher LE, Engelman DM, Sturgis JN. Detergents modulate dimerization, but not helicity, of the glycophorin A transmembrane domain. J Mol Biol. 1999;293:639–651. doi: 10.1006/jmbi.1999.3126. [DOI] [PubMed] [Google Scholar]
- Fleming KG, Ackerman AL, Engelman DM. The effect of point mutations on the free energy of transmembrane alpha- helix dimerization. J Mol Biol. 1997;272:266–275. doi: 10.1006/jmbi.1997.1236. [DOI] [PubMed] [Google Scholar]
- Fleming KG, Engelman DM. Specificity in transmembrane helix-helix interactions can define a hierarchy of stability for sequence variants. Proc Natl Acad Sci U S A. 2001;98:14340–14344. doi: 10.1073/pnas.251367498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming KG, Ren CC, Doura AK, Eisley ME, Kobus FJ, Stanley AM. Thermodynamics of glycophorin A transmembrane helix dimerization in C14 betaine micelles. Biophys Chem. 2004;108:43–49. doi: 10.1016/j.bpc.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Gandhi CS, Shuck K, Lear JD, Dieckmann GR, DeGrado WF, Lamb RA, Pinto LH. Cu(II) inhibition of the proton translocation machinery of the influenza A virus M2 protein. J Biol Chem. 1999;274:5474–5482. doi: 10.1074/jbc.274.9.5474. [DOI] [PubMed] [Google Scholar]
- Grambas S, Bennett MS, Hay AJ. Influence of amantadine resistance mutations on the pH regulatory function of the M2 protein of influenza A viruses. Virology. 1992;191:541–549. doi: 10.1016/0042-6822(92)90229-i. [DOI] [PubMed] [Google Scholar]
- Gratkowski H, Lear JD, DeGrado WF. Polar sidechains drive the association of model, transmembrane peptides. Proc Natl Acad Sci USA. 2001;98:880–885. doi: 10.1073/pnas.98.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden FG. Antiviral resistance in influenza viruses--implications for management and pandemic response. N Engl J Med. 2006;354:785–788. doi: 10.1056/NEJMp068030. [DOI] [PubMed] [Google Scholar]
- Hilty C, Wider G, Fernandez C, Wuthrich K. Membrane protein-lipid interactions in mixed micelles studied by NMR spectroscopy with the use of paramagnetic reagents. Chembiochem. 2004;5:467–473. doi: 10.1002/cbic.200300815. [DOI] [PubMed] [Google Scholar]
- Holsinger LJ, Lamb RA. Influenza Virus M2 Integral Membrane Protein Is a Homotetramer Stabilized by Formation of Disulfide Bonds. Virology. 1991;183:32–43. doi: 10.1016/0042-6822(91)90115-r. [DOI] [PubMed] [Google Scholar]
- Holsinger LJ, Nichani D, Pinto LH, Lamb RA. Influenza A virus M2 ion channel protein: a structure-function analysis. J Virol. 1994;68:1551–1563. doi: 10.1128/jvi.68.3.1551-1563.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard KP, Lear JD, DeGrado WF. Sequence determinants of the energetics of folding of a transmembrane four-helix-bundle protein. Proc Natl Acad Sci U S A. 2002;99:8568–8572. doi: 10.1073/pnas.132266099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Asbury T, Achuthan S, Li C, Bertram R, Quine JR, Fu R, Cross TA. Backbone structure of the amantadine-blocked trans-membrane domain M2 proton channel from Influenza A virus. Biophys J. 2007;92:4335–4343. doi: 10.1529/biophysj.106.090183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitahori Y, Nakano M, Inoue Y. Frequency of amantadine-resistant influenza A virus isolated from 2001–02 to 2004–05 in Nara Prefecture. Jpn J Infect Dis. 2006;59:197–199. [PubMed] [Google Scholar]
- Lamb RA, Holsinger LJ, Pinto LH. The influenza A virus M2 ion channel protein and its role in the influenza virus life cycle. In: Wimmer E, editor. Cellular Receptors and Animal Viruses. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 303–321. [Google Scholar]
- Lear JD, Gratkowski H, Adamian L, Liang J, DeGrado WF. Position-dependence of stabilizing polar interactions of asparagine in transmembrane helical bundles. Biochemistry. 2003;42:6400–6407. doi: 10.1021/bi020573j. [DOI] [PubMed] [Google Scholar]
- Lin T-I, Heider H, Schroeder C. Different modes of inhibition by adamantane amine derivatives and natural polyamines of the functionally reconstituted influenza virus M2 proton channel protein. J Gen Virol. 1997;78:767–774. doi: 10.1099/0022-1317-78-4-767. [DOI] [PubMed] [Google Scholar]
- Lin TI, Schroeder C. Definitive assignment of proton selectivity and attoampere unitary current to the M2 ion channel protein of influenza A virus. J Virol. 2001;75:3647–3656. doi: 10.1128/JVI.75.8.3647-3656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mould JA, Drury JE, Frings SM, Kaupp UB, Pekosz A, Lamb RA, Pinto LH. Permeation and activation of the M2 ion channel of influenza A virus. J Biol Chem. 2000;275:31038–31050. doi: 10.1074/jbc.M003663200. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Kim S, Zhang L, Cross TA. The closed state of a H+ channel helical bundle combining precise orientational and distance restraints from solid state NMR. Biochemistry. 2002;41:13170–13177. doi: 10.1021/bi0262799. [DOI] [PubMed] [Google Scholar]
- Okada A, Miura T, Takeuchi H. Protonation of histidine and histidine-tryptophan interaction in the activation of the M2 ion channel from influenza a virus. Biochemistry. 2001;40:6053–6060. doi: 10.1021/bi0028441. [DOI] [PubMed] [Google Scholar]
- Pinto LH, Dieckmann GR, Gandhi CS, Papworth CG, Braman J, Shaughnessy MA, Lear JD, Lamb RA, DeGrado WF. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc Natl Acad Sci U S A. 1997;94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto LH, Lamb RA. The M2 proton channels of influenza A and B viruses. J Biol Chem. 2006;281:8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- Russ WP, Engelman DM. TOXCAT: a measure of transmembrane helix association in a biological membrane. Proc Natl Acad Sci U S A. 1999;96:863–868. doi: 10.1073/pnas.96.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- Salom D, Hill BR, Lear JD, DeGrado WF. pH-dependent tetramerization and amantadine binding of the transmembrane helix of M2 from the influenza A virus. Biochemistry. 2000;39:14160–14170. doi: 10.1021/bi001799u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senes A, Engel DE, DeGrado WF. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol. 2004;14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Senes A, Gerstein M, Engelman DM. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J Mol Biol. 2000;296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- Shimbo K, Brassard DL, Lamb RA, Pinto LH. Ion selectivity and activation of the M2 ion channel of influenza virus. Biophys J. 1996;70:1335–1346. doi: 10.1016/S0006-3495(96)79690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen L, Viboud C, Grenfell BT, Dushoff J, Jennings L, Smit M, Macken C, Hata M, Gog J, Miller MA, Holmes EC. The genesis and spread of reassortment human influenza A/H3N2 viruses conferring adamantane resistance. Mol Biol Evol. 2007;24:1811–1820. doi: 10.1093/molbev/msm103. [DOI] [PubMed] [Google Scholar]
- Steinhauer DA, Wharton SA, Skehel JJ, Wiley DC, Hay AJ. Amantadine selection of a mutant influenza virus containing an acid-stable hemagglutinin glycoprotein: evidence for virus-specific regulation of the pH of glycoprotein transport vesicles. Proc Natl Acad Sci U S A. 1991;88:11525–11529. doi: 10.1073/pnas.88.24.11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, Soto CS, Tereshko V, Nanda V, Stayrook S, DeGrado WF. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffer AL, DeGrado WF, Lear JD. Analytical Ultracentrifugation Studies of the Influenza M2 Homotetramerization Equilibrium in Detergent Solutions. Progr Colloid Polym Sci. 2006;131:108–115. [Google Scholar]
- Stouffer AL, Nanda V, Lear JD, DeGrado WF. Sequence determinants of a transmembrane proton channel: an inverse relationship between stability and function. J Mol Biol. 2005;347:169–179. doi: 10.1016/j.jmb.2005.01.023. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Saito R, Masuda H, Oshitani H, Sato M, Sato I. Emergence of amantadine-resistant influenza A viruses: epidemiological study. J Infect Chemother. 2003;9:195–200. doi: 10.1007/s10156-003-0262-6. [DOI] [PubMed] [Google Scholar]
- Tamm LK, Hong H, Liang B. Folding and assembly of beta-barrel membrane proteins. Biochim Biophys Acta. 2004;1666:250–263. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Tang Y, Zaitseva F, Lamb RA, Pinto LH. The gate of the influenza virus M2 proton channel is formed by a single tryptophan residue. J Biol Chem. 2002a;277:39880–39886. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- Tang Y, Zaitseva F, Lamb RA, Pinto LH. The gate of the influenza virus M2 proton channel is formed by a single tryptophan residue. J Biol Chem. 2002b;14:14. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- Venkataraman P, Lamb RA, Pinto LH. Chemical rescue of histidine selectivity filter mutants of the M2 ion channel of influenza A virus. J Biol Chem. 2005;280:21463–21472. doi: 10.1074/jbc.M412406200. [DOI] [PubMed] [Google Scholar]
- Wang C, Lamb RA, Pinto LH. Activation of the M2 ion channel of influenza virus: a role for the transmembrane domain histidine residue. Biophys J. 1995;69:1363–1371. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Takeuchi K, Pinto LH, Lamb RA. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J Virol. 1993;67:5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Voth GA. A computational study of the closed and open states of the influenza a M2 proton channel. Biophys J. 2005;89:2402–2411. doi: 10.1529/biophysj.105.066647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.