Colorectal carcinoma (CRC) is the third leading cause of cancer-related deaths in the United States. Currently, there are an estimated 150,000 new cases of CRC and over 50,000 deaths due to this disease annually in the U.S.(1) (2). While localized tumor growth may cause significant organ dysfunction and even death, metastases cause the vast majority (~90%) of human cancer deaths(3).The ability to metastasize is linked with the ability of cancer cells to invade adjacent tissues, to gain access to vascular or lymphatic channels and to survive transit through the bloodstream so that they may extravasate, then reside and colonize heterologous organs or tissues. Cancer cells acquire the capacity of invasion and metastasis through regulated processes related to tissue development and homeostasis. Identification of the key regulators of these processes may provide valuable prognostic information to patients with CRC as well as opportunities for enhanced intervention.
Developmental biology and carcinogenesis in intestinal epithelial cells
At least three major signaling pathways are critical for the development and maintenance of homeostasis in the gastrointestinal tract: the TGF-β superfamily, Wnt/β-catenin/TCF (T cell-specific transcription factor) and Notch signaling pathways. These three signaling pathways contribute to the development, differentiation and maintenance of homeostasis in intestinal epithelium, whereas mutations or aberrant regulation of these pathways contribute to tumor initiation and progression in the intestine (reviewed in (4)). In this review, we will emphasize the interactions of two of these pathways, TGF-β superfamily and Wnt, and their potential roles in colorectal cancer cell invasiveness and metastatic potential. It is well recognized that other genetic alterations, signaling pathways and processes such as angiogenesis are also important contributors of carcinogenesis, tumor progression and metastasis, but these will not be addressed in this brief review.
Adverse phenotypic behaviors have been linked to a process known as epithelial to mesenchymal transition (EMT). EMT is tightly controlled, reversible and required for embryonic development, tissue reorganization and wound healing. During EMT, cells lose epithelial polarity and acquire a mesenchymal phenotype with invasive characteristics(5). Cell-cell junctions are disrupted in EMT by mechanisms involving loss of expression of the adherens junction protein, E-Cadherin. Tight junctions lose polarity and function, and the cells undergoing EMT express mesenchymal markers such as vimentin and α-smooth muscle actin (α-SMA). In many cases, cellular transformation recapitulates a molecular environment favoring EMT, which in turn disrupts normal epithelial cell polarity and results in the acquisition of invasive and metastatic potential. Thus, a growing body of evidence implicates EMT in tumor cell migration, invasiveness and metastatic behavior(6–8).
Overview of TGF-β superfamily signaling
The TGF-β superfamily is made up of two subfamilies of cytokines: the TGF-β/Activin/Nodal subfamily and the Bone Morphogenic Protein (BMP)/Growth and Differentiation Factor (GDF)/Muellerian Inhibiting Substance (MIS) subfamily(9). For purposes of this review, we will not focus on the accessory receptors (i.e., Betaglycan, Cripto, or Endoglin), interactions with inhibin, GDFs, or the Mullerian subfamily (AMH/MIS) and their associations in the TGF-β superfamily. Specific TGF-β and BMP-related cytokines and their shared signaling intermediates function as critical developmental regulators, cell growth inhibitors and tumor suppressors in normal tissue. Germline loss of certain components of the signaling intermediates for these cytokines often results in embryonic lethality due to their critical roles in development. On the other hand, somatic cell mutations or acquired loss of function may contribute to the development or progression of cancer.
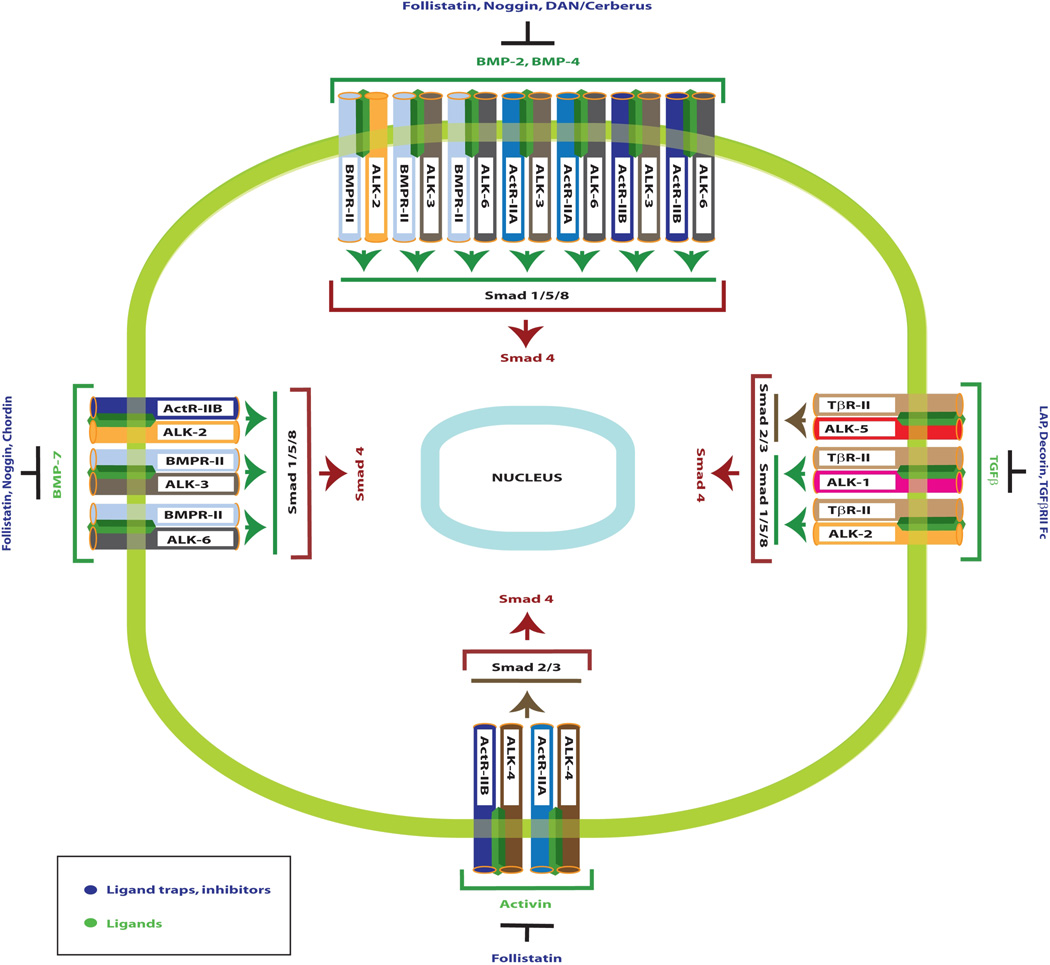
The TGF-β family of ligands bind and signal through a heteromeric complex of ligand-specific type I (Alk 1–7) and type II (ligand-specific) serine/threonine kinase receptors(9). Ligand access to the serine/threonine kinase receptors is regulated by a family of proteins known as ligand traps, proteins that selectively bind to specific TGF-β superfamily ligands, thereby blocking access to the receptors (10) (9) (Figure 1). For example, the ligand trap protein, Noggin, inhibits receptor activation of the BMP cytokines 2, 4, and 7. A type II receptor is necessary for specific ligand binding. Following binding of ligand to type II receptors, the ligand-bound type II receptor forms an oligomeric complex with the type I receptor, resulting in type I receptor phosphorylation. Information from the tissue microenvironment to the cell nucleus by these growth inhibitors is then transmitted by specific Smad proteins (R-Smads) associated with the growth factor (type II) receptors. Nine members of the vertebrate TGF-β family intracellular signaling pathways have been identified, and by consensus are now referred to as Smad1 through Smad9(9). As a general rule, Smads1, 5, and 8 are R-smads that function to transduce signals from the bone morphogenetic proteins (BMPs) and their specific type II (BMPR-II, ActR-II/B) and type I (Alk3, Alk6, Alk2) receptors, mediating cellular transition to an epithelial phenotype.
Figure 1. Signaling specificity of Smad proteins as they relate to TGF-β ligands, Ligand Binding Traps and the Type I and Type II receptors.
Important ligand traps, ligands, and receptor complexes in TGF-β superfamily signaling are noted. Downstream R-Smads 1, 2, 3, 5 and 8 are grouped based on their signaling specificity. Adapted from9.
Smads2 and 3 are important R-Smad substrates of Alk5 that may be activated by either activin (through the type II ActR-IIB receptor) or by TGF-β selective type II receptors. Inhibitory Smads 6 and 7 (I-Smads) inhibit the signaling function of the receptor-activated Smads. Smad6 preferentially inhibits BMP signaling, whereas Smad7 can inhibit both TGF-β and BMP signaling by preventing receptor-mediated phosphorylation of the receptor-activated Smad proteins(9). When phosphorylated by the type I receptor, R-Smads associate with the common signaling intermediate, Smad4, which translocates the entire R-Smad:Co-Smad complex to the nucleus, where it associates with one or more of a number of DNA-binding partners and activates the transcription of specific target genes important for both growth inhibition and EMT. Importantly, Smad4 translocates to the nucleus when complexed with the R-Smads(11). Steady-state levels of Smad proteins are regulated through the ubiquitin-proteasome degradation pathway.
TGF-β and Smad4 loss in Colorectal Cancer
The TGF-β family of peptides has a growth inhibitory role in intestinal epithelium. Kurokowa and colleagues(12) were the first to report that TGF-β was an inhibitor of cultured rat intestinal epithelial cells. We subsequently determined that inhibition of cultured intestinal cell proliferation after TGF-β treatment results from mid-to-late G1 cell cycle arrest associated with downregulation of cyclin D1(13) and inhibition of Cdk4-associated Rb kinase activity(14). In vivo, there is increased expression of both TGF-β1 and type II TGF-β receptor (TβRII) in intestinal epithelial cells as they migrate from the proliferative compartment toward the lumen in both the small intestine and the colon(15) (16) (17) (18). This pattern of expression is inversely correlated with the mitotic activity in the gut epithelium. Taken together, these findings suggest that TGF-β plays a regulatory role in intestinal cell proliferation, and perhaps differentiation.
TGF-β regulation of epithelial cell proliferation is altered by cellular transformation. TGF-β inhibits growth of non-tumorigenic human colonic adenoma cells in culture; however, conversion of adenoma to a tumorigenic adenocarcinoma is associated with a decreased response to the inhibitory actions of TGF-β(19). Studies in human colon carcinoma cell lines have demonstrated a correlation between the differentiation state of tumors and sensitivity to the anti-proliferative and differentiation-promoting effects of TGF-β(20). Thus, loss of growth-inhibitory responses to TGF-β appears to be a common and important event that attends malignant transformation of epithelial cells.
One of the mechanisms by which tumor cells become resistant to the growth inhibitory actions of TGF-β may be through down-regulation or mutation of the TβRII. Several studies have suggested that a decrease in expression of TβRII is a key step for the neoplastic transformation of epithelial cells(21) (22) (23). Inactivation of the TβRII has been detected in a subgroup of colorectal carcinomas associated with the microsatellite instability or replication error phenotype found in approximately 13% of all colorectal cancers(24). Mutations of TβRII have also been identified in 15% of microsatellite stable colorectal cancers(25). Of potential importance are the observations that the subset of colorectal cancers that exhibit microsatellite instability (and TβRII mutations) tend to be proximal colon cancers and have a better prognosis (stage for stage) than the majority of sporadic colorectal cancers that do not share these genetic defects(26) (27). An important recent observation was made in a murine model of invasive mammary carcinoma wherein conditional loss of TβRII in breast cancer cells resulted in chemokine-mediated recruitment of myeloid cells into the tumor stroma and promotion of invasion and metastasis(28).
In addition to loss of the normal growth inhibitory processes mediated from mutations of the TβRII, mutations or epigenetic loss of expression of Smad signal transduction proteins also contributes to tumorigenesis and tumor progression in the GI tract(29). Mutations of Smad3 have not been identified in human cancers, but 37% of gastric carcinomas exhibit decreased Smad3 immunoreactivity and when Smad3 deficient gastric cancer cells are forced to express ectopic Smad3, TGF-β responsiveness is restored and xenograft growth in nude mice is suppressed in Smad3 transfected gastric cancer cells versus controls(30). Smad2 mutations have been identified in a small subset (under 10%) of colorectal cancers(31) (32).
The most commonly disrupted Smad mediator in cancers, including colorectal cancer, is Smad4. Mutations of Smad4 have been identified in 50% of pancreatic cancers(33), 20–30% of colorectal cancer(34) (35) (36) and a significant fraction of small bowel adenocarcinomas(37). Loss of Smad4 expression is correlated with loss of E-cadherin expression(38), liver metastasis and poor prognosis in colon cancer(39) (40). Loss of Smad4 expression and deletion of chromosome 18q have recently been associated with increased incidence of lymph node metastasis in colorectal cancer(41). Loss of Smad4-dependent signaling in T cells leads to de novo epithelial cancers in the murine gut(42). These reports indicate that loss of Smad4 expression is an important contributing factor for tumorigenesis in CRC.
BMP in the Intestinal Tract
The role of BMP subfamily ligands and receptors in the GI tract are less well known than that of the TGF-β subfamily, though recent experimental observations indicate their activity in growth, development and cancer. BMP2 and its receptors are expressed in the mouse and human colon, predominantly in mature colonocytes at the epithelial surface. BMP2 promotes apoptosis and growth inhibition in cultured colon cancer cells(43). Howe(44) and Sayed(45) have identified germline mutations in the gene encoding the BMP receptor 1A in Juvenile Polyposis Syndrome (JPS). Of note, these studies also implicated Smad4 mutations in a significant subset of JPS patients. Villin promoter-directed expression of Noggin, a natural BMP antagonist, in the intestine of mice as well as targeted disruption of the BMP signaling pathway through conditional inactivation of the Bmpr1a gene in mice resulted in similar histopathology as is seen in JPS(46) (47). Similarly, BMP2 expression was found to be decreased in microadenomas of familial adenomatous polyposis (FAP) patients. These studies indicate that BMP signaling is critical for homeostasis in the intestinal tract, and loss of this function leads to pre-cancerous conditions such as JPS and FAP.
EMT and TGF-β superfamily signaling
In contrast with the tumor suppressive effects of Smad4 intrinsic to TGF-β and BMP signaling, additional data demonstrate that Smad4 may also regulate EMT, perhaps through TGF-β and/or BMP mediated signaling interactions with Ras/MAP kinase, PI3 Kinase or Wnt signaling pathways(48). Others have reported that TGF-β and BMP ligands and their respective ligand trap peptides have homeostatic opposing effects on EMT in some systems with TGF-β promoting and BMPs inhibiting EMT(49) (reviewed in(10)). The balance between a differentiated epithelial phenotype and the more aggressive and invasive mesenchymal phenotype is also influenced by the abundance and activity of locally produced cytokines. Rees et al(50) recently reported evidence of EMT at the leading invasive front of esophageal adenocarcinomas that consisted of loss of membrane E-cadherin expression, increased α-SMA and vimentin, along with diffuse stromal TGF-β1 immunostaining. In contrast, BMP7 immunostaining was absent at the invasive front with occasional cells showing increased immunoreactivity in the central tumor areas.
Inhibitor of DNA binding/differentiation (Id) proteins (Id1–4) are downstream targets of TGF-β/BMP that regulate differentiation in osteoblasts, fibroblasts, epithelial cells and endothelial cells in a cell-type specific and contextual manner(51). Id proteins are helix loop helix proteins (HLH) that both antagonize basic helix-loop-helix (bHLH) transcription factors, such as Id2 binding to the pocket proteins of the Rb gene, and mediate mitogenic signals as they inhibit differentiation and play critical roles in development and cancer(52) (53) (reviewed in(54)). Id proteins also function to integrate Smad signals to create a permissive or refractory nuclear environment ordering cell fate and proliferation(55).
The regulation of Id expression and their roles in gene expression may help to explain how different cell types or similar cell types under diverse conditions may have markedly dissimilar responses to TGF-β or BMP. For example, some cells respond to TGF-β by undergoing EMT, while others are simply growth inhibited and/or undergo an apoptotic response. The mechanisms underlying these differential responses are not completely understood, but studies by Kowanetz et. al, indicate that Id proteins play an important role(55). In the Kowanetz studies, Smad4-dependent TGF-β induction of EMT was associated with sustained inhibition of Id2 and Id3 proteins, while Smad4-dependent BMP-7 treatment induced sustained upregulation of the same. Forced overexpression of Id2 or Id3 by adenoviral infection in this study was sufficient to block TGF-β1-mediated EMT whereas knockdown of Id2 induced EMT in mouse mammary epithelial cells following treatment with either TGF-β1 or BMP-7 (55). Thus, Id2 levels, along with combinations of transcriptional co-regulatory factors, may determine the cellular response to TGF-β-family signaling, thus dictating whether or not a cell will exhibit the EMT phenotype independent of ligand treatment. Accordingly, high levels of Id2 expression are correlated with favorable prognosis in patients with primary breast cancer, and overexpression of Id2 in MDA-MB-468 breast cancer cells results in decreased invasive capacity of the transfected cells(56), presumably due to a blockade of TGF-β mediated EMT. Interestingly, Id proteins 1, 2, and 3 are elevated in colon cancers when compared to normal tissue(57); however, the roles and the balance of the various Id proteins in gastrointestinal tumorigenesis are still unclear(51). In fact, loss of Id4 has been shown to correlate with increased differentiation and poor prognosis in CRC(58). Conversely, deletion of the Id2 gene in mice resulted in failure of terminal differentiation and expansion of the proliferative zone in the intestine of the developing fetus and neonatal mice; and, the Id2−/− mice develop intestinal epithelial dysplasia and multiple adenomas between the age of 2 and 13 months, thus defining a role for Id2 as a potential tumor suppressor in the gut (59).
Canonical Wnt signaling in intestinal growth and differentiation and in colorectal cancer
Wnt signaling is a critical regulator of early embryonic development. In adult organisms, Wnt signaling regulates maintenance of intestinal crypt progenitor cell compartments and the control of cell fate along the intestinal crypt-villus axis(4). This pathway is highly conserved from invertebrates to vertebrate organisms and intracellular levels of the key signaling mediator, β-catenin, are tightly controlled. The differentiated epithelial cell goes to great lengths to restrict the levels of cytosolic β-catenin. The Wnt pathway is normally activated by Wnt ligand and receptor interactions in an embryologic or stem cell microenvironment. “The defining event in canonical Wnt signaling is cytoplasmic accumulation of β-catenin and its subsequent nuclear translocation and activity”(60).
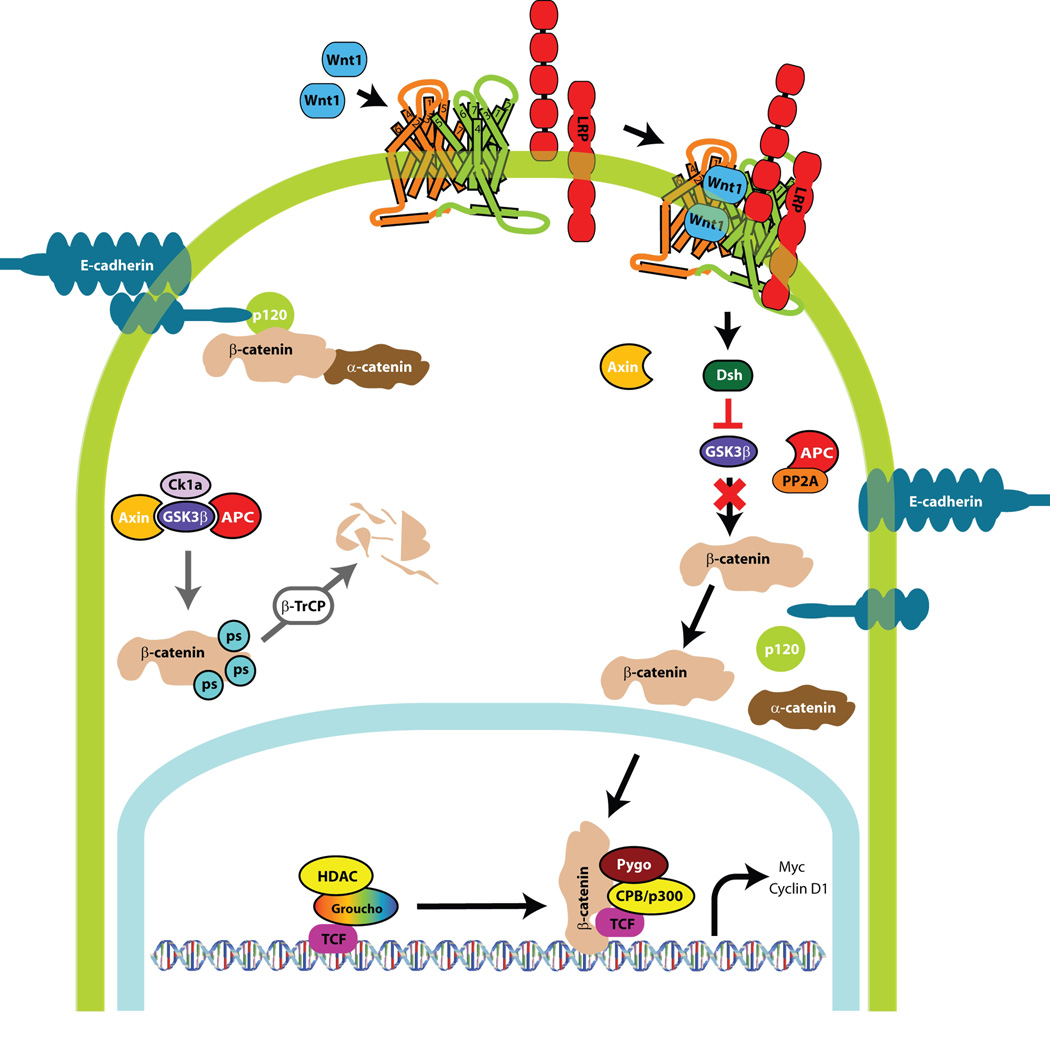
When Wnt signaling is activated, β-catenin levels transiently increase intracellularly and β-catenin translocates to the nucleus where it forms a transcriptional regulatory complex with the T cell-specific transcription factor/lymphoid enhancer-binding factor 1 (TCF/LEF) to activate transcription of cyclin D1, c-Myc and other downstream targets(61, 62). In the absence of Wnt signaling, β-catenin levels in the cytoplasm are exceedingly low and the β-catenin pools are localized to the adherens junctions in a complex with E-cadherin. Normally, in the absence of a Wnt ligand, low cytoplasmic concentrations of β-catenin are maintained by the scaffold protein Axin. Axin coordinates the formation of a β-catenin destruction complex that consists of casein kinase 1a (Ck1a), glycogen synthase kinase 3β (Gsk3β), the tumor suppressor adenomatous polyposis coli (APC) and protein phosphatase 2A (PP2A) (Figure 2). Together, components of this complex act to constitutively phosphorylate β-catenin, leading to its recognition by the β-TrCP subunit of the SCF ubiquitin ligase complex (SCFβ-TrCP)(63) (64) (65), catalytic transfer of polyubiquitin chains to β-catenin and its rapid degradation by the 26S proteasome(66)
Figure 2. Wnt Signaling Pathway.
Wnt ligand interaction with the LRP receptor complex activates the pathway and β-catenin levels increase intracellularly. β-catenin translocates to the nucleus where it forms a transcriptional regulatory complex with TCF/LEF to activate transcription of cyclin D1, c-Myc and other downstream targets. E-cadherin negatively regulates Wnt signaling. E-cadherin binds and sequesters β-catenin to the plasma cell membrane at the adherens junctions, rendering it unavailable for transcriptional activity. Maintenance of low β-catenin levels by axin occur in the absence of Wnt ligand. Axin coordinates the β-catenin destruction complex (casein kinase 1a (Ck1a), glycogen synthase kinase 3β (Gsk3β), the tumor suppressor adenomatous polyposis coli (APC) and protein phosphatase 2A (PP2A)). Together, these components lead to the sequential phosphorylation of β-catenin and eventual proteasomal degradation.
Inactivating mutations in the Apc (adenomatous polyposis coli) gene or activating mutations in the β-catenin gene result in failure to phosphorylate β-catenin with stabilization and accumulation of β-catenin in the cytoplasm and aberrant activation of Wnt signaling. Hypophosphorylation of β-catenin leads to accumulation in the cytoplasm and translocation to the nucleus with resultant regulation of target gene expression with the TCF/LEF family of transcription factors(60). The significance of the role of APC in colorectal cancer is revealed by the fact that over 80% of cases of sporadic CRC have truncating mutations in the Apc gene, which has often been considered the gatekeeper in the genesis of colorectal cancer (CRC) because it is the earliest detectable genetic lesion in most premalignant colorectal adenomas. Aberrant stabilization of β-catenin in CRC can result from inactivating APC mutation or stabilizing mutations in β-catenin (found in 10% of sporadic colon cancers).
Adherens junctions, Wnt and EMT
Adherens junctions are specialized forms of cadherin-based adhesive contacts important for tissue organization in developing and adult organisms. E-cadherin is an epithelium-specific cadherin that forms protein complexes with cytoplasmic proteins (catenins) that convert the specific, homophilic-binding capacity of the extracellular domain into stable cell adhesion. Critical proteins found at the adherens junction include E-cadherin, α-catenin, β-catenin, and p120 catenin. E-cadherin is a calcium-dependent adhesion protein. It is the major component of the adherens junction that facilitates cell-cell communication, and E-cadherin also functions as a tumor suppressor(67). Junctional proteins may also serve important roles other than as structural components of intercellular junctions. E-cadherin binds and sequesters cytoplasmic β-catenin, rendering β-catenin unavailable for signaling in the canonical Wnt/β-catenin/TCF signaling cascade(68). Therefore, E-cadherin like the APC protein is an important tumor suppressor protein. Both E-cadherin and APC have essential roles in preventing accumulation of cytoplasmic β-catenin, and thereby prevent inappropriate activation of the Wnt pathway(67).
We have recently found that forced expression of Smad4 in Smad4-deficient SW480 colon cancer cells induced E-cadherin expression and restored its membrane localization while reducing β-catenin levels. These effects were accompanied by a marked decrease in β-catenin/TCF activity(69). These data implicate Smad4 as a key modulator of both Wnt signaling and EMT in these colon cancer cells.
Decreased expression or loss of membrane localization of E-cadherin has been reported in breast(70), colon(71), esophageal(72) (73), gastric(74), pancreatic(75) and other carcinomas. Loss of E-cadherin from the junctional complex in epithelial malignancies is associated with invasion, metastasis, and worse prognosis(76) (77) (78) (79) (80) (81).
Loss of normal expression of E-cadherin may occur by any of several well-described mechanisms. The E-cadherin gene (CDH1) is located on chromosome 16q22.1. Germline mutations in CDH1 are found in families with hereditary diffuse gastric cancer(82) (83) (84). Somatic mutations in CDH1 have been identified in sporadic diffuse gastric carcinomas(85) (86) (87), lobular breast cancers(88) (89) and carcinomas of the endometrium and ovaries(90). Notwithstanding these examples, mutational loss of E-cadherin expression is a relatively rare event in colorectal carcinoma, but more commonly, expression of E-cadherin is extinguished by epigenetic mechanisms(91). These mechanisms may include regulation at the transcriptional level and post-translational regulation of E-cadherin membrane localization and stability.
Reynolds and colleagues have recently demonstrated that p120 catenin interaction with E-cadherin is essential for maintenance of E-cadherin stability and function as a tumor suppressor(92) (93). Transcriptional repression is also a prominent regulatory mechanism by which E-cadherin expression may be suppressed in tumors. The zinc finger transcriptional repressors of the Snail/Slug family, Sip1 and ZEB1, bind to two E-boxes in the E-cadherin promoter and interact with the transcriptional corepressor, CtBP, which recruits histone deacetylases to facilitate silencing of E-cadherin expression (94) (95) (96) (97). In addition, two types of basic-helix-loop-helix (bHLH) proteins, E12/E47(98) and Twist(99) have also been identified as interacting with the E-cadherin promoter E-box region to repress E-cadherin expression and to induce the EMT phenotype(100). In colorectal carcinoma, recent immunohistochemistry analysis of E-cadherin expression in a 577 tumor set tissue microarray with 10-years of clinical follow-up revealed that normal membrane localization of E-cadherin was present in only 38% of samples, while ~34% exhibited cytoplasmic immunolocalization of E-cadherin and 23% had complete absence of E-cadherin immunostaining(101). The 5-and 10-year survival rates were significantly lower for those patients whose tumors exhibited aberrant or absent E-cadherin expression.
Alteration of E-cadherin expression in experimental tumor models has profound effects on the invasive and metastatic potential of human cancer cells. Forced E-cadherin expression in E-cadherin-deficient breast cancer cells decreased invasiveness in collagen matrix(102), and forced expression of E-cadherin in human MDA MB 231 breast cancer cells suppressed the development of bone metastases in athymic (nude) mice(103). We have found that suppression of SLUG (or SNAI2) expression by siRNA restores E-cadherin expression and partially rescues the transformed phenotype in Ras transformed rat intestinal epithelial cells(104).
Epigenetics, E-cadherin, and EMT
Histone acetylation contributes to epigenetic regulation of E-cadherin. Chromatin structure has a profound impact on gene transcription. Genomic DNA is coiled around the nucleosome, which is comprised of histones. Specific modifications of histones such as acetylation or deacetylation influence how tightly the DNA is coiled around the nucleosomes. Acetylation of histones weakens the interactions between histones and DNA, thereby loosening the coils of DNA, which allows better access of transcription factors to the promoters of target genes. Histone acetylation is catalyzed by histone acetyltransferases. Conversely, histone deacetylation is catalyzed by histone deacetylases (HDACs). Histone deacetylation results in chromatin compaction and inactivation of gene expression(105). At least 11 HDACs have been identified in humans and these are classified into two general classes: class I (HDAC1, 2, 3, 8 and 11) and class II (HDAC4, 5, 6, 7, 9 and 10)(106) (107) (108). HDAC1 and 2 participate in Sin3A, NuRD and CoRest transcriptional co-repressor complexes(109), while HDAC3 complexes with NCoR and SMRT co-repressors(110). Co-repressor complexes are recruited to specific gene promoter regions by several target-specific transcription factors such as the Snail/Slug family, Sip1, and ZEB1, all known to be involved in the regulation of EMT. Inhibition of HDAC activity may restore E-cadherin expression in cancer cells in which it is epigenetically repressed. Ohira et. al(111) demonstrated that trichostatin A treatment could restore E-cadherin expression in E-cadherin-deficient H661 lung carcinoma cells. Witta, et. al,(112) also demonstrated that the HDAC inhibitor MS-275, could induce E-cadherin mRNA expression in three different lung cancer cell lines in which E-cadherin repression was associated with high level expression of the ZEB1 transcription factor. They also observed that restoration of E-cadherin expression in the E-cadherin-deficient lung cancer cells increased the sensitivity to the growth inhibitory and apoptotic effects of Epidermal Growth Factor receptor (EGFR) tyrosine kinase inhibitors such as gefitinib.
Tumor biology at the invasive front
Increased nuclear β-catenin localization and Wnt signaling have been demonstrated at the tumor-stromal interface of the leading invasive edge of tumors and is associated with localized evidence of EMT (loss of E-cadherin at the cell membrane) and disruption of cell polarity(81). This group recently showed that well-to-moderately differentiated pT3 rectal adenocarcinomas express basement membrane proteins, but breakdown of the basement membrane in many of the rectal cancers occurs in discrete regions adjacent to cancer cells at the invasive front of tumors. At the sites of breakdown of the basement membrane (the invasive front), these investigators found localized expression of the transcription factor ZEB1 and decreased expression of one of its targets, Lama3 (a component of laminin 5 in the basement membrane)(113). A provocative observation from the Spaderna report was that the basement membrane was rebuilt in both lymph node and distant metastases, accompanied by the same glandular morphology observed in the majority of the primary tumor, including the predominance of the epithelial phenotype, but that the entire process was recapitulated in the invasive regions of the metastatic lesions. These observations support the notion that in most cases, EMT is a dynamic, regulated and reversible process.
The exact mechanisms by which ZEB1 and the transient EMT phenotype are induced and regulated at the invasive front remain under investigation. It is likely that local environmental factors have a major influence on this important change in tumor cell phenotype. Such factors may result from interactions between tumor cells and nearby stromal cells, changes in local intercellular signaling, nutrient availability, pH or oxygen tension. Dissolution of the basement membrane potentially brings epithelial and stroma cells into direct contact. Also, ZEB1 cooperates with the NADH+ sensor, CtBP, which is recruited to the E-cadherin promoter under hypoxic conditions to repress expression of E-cadherin, and the same hypoxic conditions increase cell motility of lung cancer H1299 cells(114).
Evidence continues to accumulate implicating localized hypoxic stress and anaerobic metabolism in colorectal and other cancers in association with poor prognosis(115) (116) (117). Futhermore, proliferating fibroblasts at the invading tumor front exhibit endogenous markers of hypoxia, acidity and oxidative stress(118). Interplay between tumor epithelial cells and stromal cells appear to provide a symbiotic environment that may promote tumorigenesis. Balance between a differentiated epithelial phenotype and the more aggressive and invasive mesenchymal phenotype is also influenced by abundance and activity of locally produced cytokines.
Tight junctions (TJs) and EMT, Invasion and Metastasis
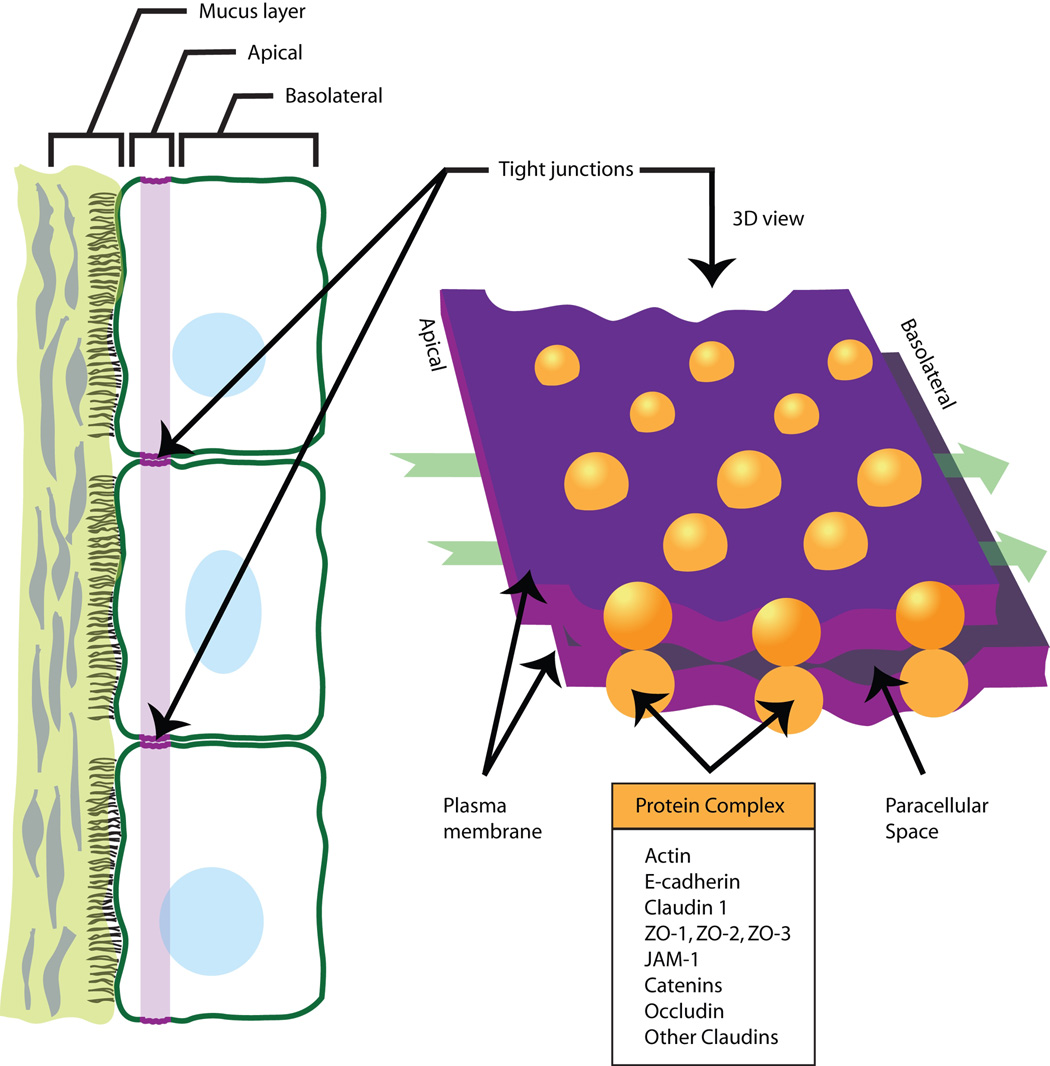
TJs are the most apical cell-cell contacts in epithelial and many endothelial cell sheets(119). These junctions are important for their barrier function and serve as a gate for paracellular transport as well as a fence modulating the lateral diffusion of proteins within the cell membrane (Figure 3). Loss of cellular polarity resulting from EMT requires disruption of tight junctions. Recent studies have demonstrated frequently mislocalized expression of TJ proteins from their normal apical membrane localization to the cell cytoplasm during pathological conditions and tumorigenesis, indicating roles other than those known to be related to TJs. TJ components have been suggested to be potential players in the regulation of gene expression. For example, proteins such as ASH1 can translocate from TJs to the nucleus and bind to specific transcriptional factors or function as transcriptional factors themselves(120) (121).
Figure 3. Tight junctions.
Depicted are two-and three-dimensional renditions of a tight junction with associated proteins. Tight junctions are closely associated areas of two cells whose membranes join forming a virtually impermeable barrier to fluid. It is a type of junctional complex only present in vertebrates. Similar junctions occurring in invertebrates are called septate junctions. Adapted from http://www.tier-guide.com and illustrations via personal communication with Punita Dhawan, Ph.D.
Claudins encode integral tight junction proteins with molecular masses ranging from ~20 to 27 kDa, which have four transmembrane domains and cytoplasmic N-and C-terminal ends(122). Each claudin exhibits a distinct tissue-specific pattern of expression. The C-terminal domains of claudin proteins serve as a binding site for a complex set of proteins including a number of PDZ-domain proteins (ZO-1, ZO-2, ZO-3)(123). Growth factor stimulation (EGF and TGF-β) induces diiferential change in claudin expression and cellular localization(124) (125). Claudins can also recruit and promote the activation of pro-matrix metalloproteinase-2 (MMP-2), a key molecule involved in tumor invasion and metastasis(126)
Claudin-1 regulates EMT and metastasis
Based on our growing awareness of EMT and the need for cells to dissolve cell-cell junctions in order to invade, we investigated the relationships of the tight junction proteins of the claudin family and EMT in colorectal cancer. We were surprised to find that claudin-1 expression is increased in human colon carcinoma and that nuclear claudin-1 localization is a frequent occurrence in metastatic lesions(127). Genetic manipulation of claudin-1 expression in colon cancer cell lines induces changes in cellular phenotype with structural and functional changes in markers of EMT. To determine if claudin-1 protein expression has a causal role in colon tumor progression and invasion, we manipulated the levels of claudin-1 expression in primary (SW480) and metastatic (SW620) colorectal cancer cell lines derived from a single patient(128). Increased expression of claudin-1 in the SW480 cells resulted in no changes in the rate of cell proliferation in culture, but increased invasiveness through extracellular matrix in transwell in vitro assays. Overexpression of claudin-1 in the SW480 cells to levels similar to those expressed in SW620 cells also resulted in decreased apoptosis in response to anoikis (detachment in cell culture), and increased expression of matrix metalloproteinases. Claudin-1 overexpression in the SW480 cells also resulted in enhanced xenograft subcutaneous tumor growth and increased hepatic metastases when cells were injected into the splenic circulation. In contrast, RNA interference-mediated inhibition of claudin-1 levels in metastatic SW620 cells resulted in opposite effects, including decreased invasiveness in culture, increased apoptosis in response to anoikis, decreased xenograft tumor growth and a significant decrease in hepatic metastases in the splenic injection assay(127). Increased claudin-1 expression was associated with increased Wnt signaling activity, while inhibition of claudin-1 expression resulted in decreased Wnt signaling activity in both SW480 and SW620 cell lines. Of interest, claudin-1 has been identified as a probable target of β-catenin/TCF signaling, supporting a potential role for claudin-1 dysregulation in colorectal carcinogenesis(129). Taken together, the data suggest that Wnt/β-catenin/TCF not only regulates claudin-1 expression, but that conversely, claudin-1 expression in colon cancer cells positively impacts Wnt/β-catenin/TCF signaling.
During the course of our examination of the role of claudin-1 in EMT and metastasis, we noted an inverse correlation of claudin-1 and Smad4 expression amongst several colon cancer cell lines. For example, Smad4 null lines SW480, SW620 and HT29 all expressed abundant levels of claudin-1, whereas the Smad4 expressing lines HCT116, HCT115, the immortalized rat small intestinal (RIE) cells and the immortalized mouse colonocyte (YAMC) line did not express detectable levels of claudin-1. Further, we found an inverse relationship between claudin-1 levels and Smad4 levels in tumor lysates assessed by immunoblotting. We hypothesized that Smad4 may regulate the expression of claudin-1. We tested this hypothesis by expressing Smad4 in claudin-1 expressing colon cancer cells that were Smad4 deficient, and found that expression of Smad4 was sufficient to silence claudin-1 expression in both SW480 and HT29 cells. Smad4 expression in the SW480 cells results in a more epithelial phenotype, increased expression of E-cadherin, redistribution of β-catenin to the cell membrane, and decreased invasiveness(69). Consistent with these findings, Schwarte-Waldhoff and Muller(130) (131), noted that stable Smad4 expression in SW480 cells is correlated with decreased xenograft tumorigenicity and enhanced E-cadherin expression. To reiterate, if Smad4 is re-expressed in SW480 cells that are both Smad4 and E-cadherin deficient, this results in the induction and membrane localization of E-cadherin, suppression of Wnt signaling and suppression of invasiveness and xenograft growth in nude mice.
SUMMARY
Metastases account for the vast majority of fatalities due to colon cancer. The metastatic phenotype, or the ability of normally polarized epithelial cells to separate from their tissue of origin, invade basement membrane, survive transit through the bloodstream and re-colonize at distal sites recapitulates a developmental process known as EMT. Regulators of EMT in adult epithelial cells, including the TGF-β and Wnt signal transduction systems, are important mediators of embryonic tissue development, tissue homeostasis, wound healing and they have important roles in tumor cell behaviors that are still being defined. Research into the potential epigenetic and reversible nature of EMT, and the downstream effector signaling pathways leading to EMT may enable the identification of novel therapeutic interventions to disrupt the metastatic process. Through a better understanding of the biological mechanisms that contribute to metastasis lies the hope of translating these discoveries into new treatments that will improve the survival of cancer patients.
Acknowledgments
This work was supported by the following grants from the National Institutes of Health:
CA69457
DK52334
CA084239
DK07673
CA106183
CA95103
CA068485
CA46413
CA77839
Abbreviations
- CRC
Colorectal Carcinoma
- TGF-β
Transforming Growth Factor-βeta
- BMP
Bone Morphogenetic Protein
- TβRII
type II TGF-β receptor
- EMT
Epithelial to Mesenchymal transition
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007 Jan–Feb;57(1):43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008 Mar–Apr;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000 Jan 7;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Radtke F, Clevers H. Self-renewal and cancer of the gut: two sides of a coin. Science. 2005 Mar 25;307(5717):1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- 5.Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003 Dec;15(6):740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Cui W, Fowlis DJ, Bryson S, Duffie E, Ireland H, Balmain A, et al. TGFbeta1 inhibits the formation of benign skin tumors, but enhances progression to invasive spindle carcinomas in transgenic mice. Cell. 1996 Aug 23;86(4):531–542. doi: 10.1016/s0092-8674(00)80127-0. [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi A. [Genetic changes in liver metastasis of colorectal cancer and their clinical application] Nippon Geka Gakkai Zasshi. 2001 May;102(5):370–375. [PubMed] [Google Scholar]
- 8.Janda E, Lehmann K, Killisch I, Jechlinger M, Herzig M, Downward J, et al. Ras and TGF[beta] cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. J Cell Biol. 2002 Jan 21;156(2):299–313. doi: 10.1083/jcb.200109037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003 Jun 13;113(6):685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 10.Neilson EG. Setting a trap for tissue fibrosis. Nat Med. 2005 Apr;11(4):373–374. doi: 10.1038/nm0405-373. [DOI] [PubMed] [Google Scholar]
- 11.de Caestecker MP, Piek E, Roberts AB. Role of transforming growth factor-beta signaling in cancer. J Natl Cancer Inst. 2000 Sep 6;92(17):1388–1402. doi: 10.1093/jnci/92.17.1388. [DOI] [PubMed] [Google Scholar]
- 12.Kurokowa M, Lynch K, Podolsky DK. Effects of growth factors on an intestinal epithelial cell line: transforming growth factor beta inhibits proliferation and stimulates differentiation. Biochem Biophys Res Commun. 1987 Feb 13;142(3):775–782. doi: 10.1016/0006-291x(87)91481-1. [DOI] [PubMed] [Google Scholar]
- 13.Ko TC, Sheng HM, Reisman D, Thompson EA, Beauchamp RD. Transforming growth factor-beta 1 inhibits cyclin D1 expression in intestinal epithelial cells. Oncogene. 1995 Jan 5;10(1):177–184. [PubMed] [Google Scholar]
- 14.Ko TC, Yu W, Sakai T, Sheng H, Shao J, Beauchamp RD, et al. TGF-beta1 effects on proliferation of rat intestinal epithelial cells are due to inhibition of cyclin D1 expression. Oncogene. 1998 Jul 2;16(26):3445–3454. doi: 10.1038/sj.onc.1201902. [DOI] [PubMed] [Google Scholar]
- 15.Barnard JA, Beauchamp RD, Coffey RJ, Moses HL. Regulation of intestinal epithelial cell growth by transforming growth factor type beta. Proc Natl Acad Sci U S A. 1989 Mar;86(5):1578–1582. doi: 10.1073/pnas.86.5.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnard JA, Warwick GJ, Gold LI. Localization of transforming growth factor beta isoforms in the normal murine small intestine and colon. Gastroenterology. 1993 Jul;105(1):67–73. doi: 10.1016/0016-5085(93)90011-z. [DOI] [PubMed] [Google Scholar]
- 17.Winesett MP, Ramsey GW, Barnard JA. Type II TGF(beta) receptor expression in intestinal cell lines and in the intestinal tract. Carcinogenesis. 1996 May;17(5):989–995. doi: 10.1093/carcin/17.5.989. [DOI] [PubMed] [Google Scholar]
- 18.Zhang T, Nanney LB, Peeler MO, Williams CS, Lamps L, Heppner KJ, et al. Decreased transforming growth factor beta type II receptor expression in intestinal adenomas from Min/+ mice is associated with increased cyclin D1 and cyclin-dependent kinase 4 expression. Cancer Res. 1997 May 1;57(9):1638–1643. [PubMed] [Google Scholar]
- 19.Manning AM, Williams AC, Game SM, Paraskeva C. Differential sensitivity of human colonic adenoma and carcinoma cells to transforming growth factor beta (TGF-beta): conversion of an adenoma cell line to a tumorigenic phenotype is accompanied by a reduced response to the inhibitory effects of TGF-beta. Oncogene. 1991 Aug;6(8):1471–1476. [PubMed] [Google Scholar]
- 20.Hoosein NM, McKnight MK, Levine AE, Mulder KM, Childress KE, Brattain DE, et al. Differential sensitivity of subclasses of human colon carcinoma cell lines to the growth inhibitory effects of transforming growth factor-beta 1. Exp Cell Res. 1989 Apr;181(2):442–453. doi: 10.1016/0014-4827(89)90101-8. [DOI] [PubMed] [Google Scholar]
- 21.Arteaga CL, Tandon AK, Von Hoff DD, Osborne CK. Transforming growth factor beta: potential autocrine growth inhibitor of estrogen receptor-negative human breast cancer cells. Cancer Res. 1988 Jul 15;48(14):3898–3904. [PubMed] [Google Scholar]
- 22.Kimchi A, Wang XF, Weinberg RA, Cheifetz S, Massague J. Absence of TGF-beta receptors and growth inhibitory responses in retinoblastoma cells. Science. 1988 Apr 8;240(4849):196–199. doi: 10.1126/science.2895499. [DOI] [PubMed] [Google Scholar]
- 23.Sun L, Wu G, Willson JK, Zborowska E, Yang J, Rajkarunanayake I, et al. Expression of transforming growth factor beta type II receptor leads to reduced malignancy in human breast cancer MCF-7 cells. J Biol Chem. 1994 Oct 21;269(42):26449–26455. [PubMed] [Google Scholar]
- 24.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995 Jun 2;268(5215):1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 25.Grady WM, Myeroff LL, Swinler SE, Rajput A, Thiagalingam S, Lutterbaugh JD, et al. Mutational inactivation of transforming growth factor beta receptor type II in microsatellite stable colon cancers. Cancer Res. 1999 Jan 15;59(2):320–324. [PubMed] [Google Scholar]
- 26.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993 May 7;260(5109):816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 27.Gryfe R, Kim H, Hsieh ET, Aronson MD, Holowaty EJ, Bull SB, et al. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N Engl J Med. 2000 Jan 13;342(2):69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 28.Yang L, Huang J, Ren X, Gorska AE, Chytil A, Aakre M, et al. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell. 2008 Jan;13(1):23–35. doi: 10.1016/j.ccr.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elliott RL, Blobe GC. Role of transforming growth factor Beta in human cancer. J Clin Oncol. 2005 Mar 20;23(9):2078–2093. doi: 10.1200/JCO.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 30.Han SU, Kim HT, Seong DH, Kim YS, Park YS, Bang YJ, et al. Loss of the Smad3 expression increases susceptibility to tumorigenicity in human gastric cancer. Oncogene. 2004 Feb 19;23(7):1333–1341. doi: 10.1038/sj.onc.1207259. [DOI] [PubMed] [Google Scholar]
- 31.Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, et al. MADR2 maps to 18q21 and encodes a TGFbeta-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell. 1996 Aug 23;86(4):543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 32.Ohtaki N, Yamaguchi A, Goi T, Fukaya T, Takeuchi K, Katayama K, et al. Somatic alterations of the DPC4 and Madr2 genes in colorectal cancers and relationship to metastasis. Int J Oncol. 2001 Feb;18(2):265–270. doi: 10.3892/ijo.18.2.265. [DOI] [PubMed] [Google Scholar]
- 33.Hahn SA, Hoque AT, Moskaluk CA, da Costa LT, Schutte M, Rozenblum E, et al. Homozygous deletion map at 18q21.1 in pancreatic cancer. Cancer Res. 1996 Feb 1;56(3):490–494. [PubMed] [Google Scholar]
- 34.Thiagalingam S, Lengauer C, Leach FS, Schutte M, Hahn SA, Overhauser J, et al. Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat Genet. 1996 Jul;13(3):343–346. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- 35.Riggins GJ, Thiagalingam S, Rozenblum E, Weinstein CL, Kern SE, Hamilton SR, et al. Mad-related genes in the human. Nat Genet. 1996 Jul;13(3):347–349. doi: 10.1038/ng0796-347. [DOI] [PubMed] [Google Scholar]
- 36.Riggins GJ, Kinzler KW, Vogelstein B, Thiagalingam S. Frequency of Smad gene mutations in human cancers. Cancer Res. 1997 Jul 1;57(13):2578–2580. [PubMed] [Google Scholar]
- 37.Blaker H, Aulmann S, Helmchen B, Otto HF, Rieker RJ, Penzel R. Loss of SMAD4 function in small intestinal adenocarcinomas: comparison of genetic and immunohistochemical findings. Pathol Res Pract. 2004;200(1):1–7. doi: 10.1016/j.prp.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 38.Reinacher-Schick A, Baldus SE, Romdhana B, Landsberg S, Zapatka M, Monig SP, et al. Loss of Smad4 correlates with loss of the invasion suppressor E-cadherin in advanced colorectal carcinomas. J Pathol. 2004 Apr;202(4):412–420. doi: 10.1002/path.1516. [DOI] [PubMed] [Google Scholar]
- 39.Alazzouzi H, Alhopuro P, Salovaara R, Sammalkorpi H, Jarvinen H, Mecklin JP, et al. SMAD4 as a prognostic marker in colorectal cancer. Clin Cancer Res. 2005 Apr 1;11(7):2606–2611. doi: 10.1158/1078-0432.CCR-04-1458. [DOI] [PubMed] [Google Scholar]
- 40.Miyaki M, Iijima T, Konishi M, Sakai K, Ishii A, Yasuno M, et al. Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene. 1999 May 20;18(20):3098–3103. doi: 10.1038/sj.onc.1202642. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka T, Watanabe T, Kazama Y, Tanaka J, Kanazawa T, Kazama S, et al. Loss of Smad4 protein expression and 18qLOH as molecular markers indicating lymph node metastasis in colorectal cancer--a study matched for tumor depth and pathology. J Surg Oncol. 2008 Jan 1;97(1):69–73. doi: 10.1002/jso.20896. [DOI] [PubMed] [Google Scholar]
- 42.Kim BG, Li C, Qiao W, Mamura M, Kasprzak B, Anver M, et al. Smad4 signalling in T cells is required for suppression of gastrointestinal cancer. Nature. 2006 Jun 22;441(7096):1015–1019. doi: 10.1038/nature04846. [DOI] [PubMed] [Google Scholar]
- 43.Hardwick JC, Van Den Brink GR, Bleuming SA, Ballester I, Van Den Brande JM, Keller JJ, et al. Bone morphogenetic protein 2 is expressed by, and acts upon, mature epithelial cells in the colon. Gastroenterology. 2004 Jan;126(1):111–121. doi: 10.1053/j.gastro.2003.10.067. [DOI] [PubMed] [Google Scholar]
- 44.Howe JR, Bair JL, Sayed MG, Anderson ME, Mitros FA, Petersen GM, et al. Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nat Genet. 2001 Jun;28(2):184–187. doi: 10.1038/88919. [DOI] [PubMed] [Google Scholar]
- 45.Sayed MG, Ahmed AF, Ringold JR, Anderson ME, Bair JL, Mitros FA, et al. Germline SMAD4 or BMPR1A mutations and phenotype of juvenile polyposis. Ann Surg Oncol. 2002 Nov;9(9):901–906. doi: 10.1007/BF02557528. [DOI] [PubMed] [Google Scholar]
- 46.Haramis AP, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus GJ, et al. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science. 2004 Mar 12;303(5664):1684–1686. doi: 10.1126/science.1093587. [DOI] [PubMed] [Google Scholar]
- 47.He XC, Zhang J, Tong WG, Tawfik O, Ross J, Scoville DH, et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat Genet. 2004 Oct;36(10):1117–1121. doi: 10.1038/ng1430. [DOI] [PubMed] [Google Scholar]
- 48.Valcourt U, Kowanetz M, Niimi H, Heldin CH, Moustakas A. TGF-beta and the Smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol Biol Cell. 2005 Apr;16(4):1987–2002. doi: 10.1091/mbc.E04-08-0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeisberg M, Hanai J, Sugimoto H, Mammoto T, Charytan D, Strutz F, et al. BMP-7 counteracts TGF-beta1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat Med. 2003 Jul;9(7):964–968. doi: 10.1038/nm888. [DOI] [PubMed] [Google Scholar]
- 50.Rees JR, Onwuegbusi BA, Save VE, Alderson D, Fitzgerald RC. In vivo and in vitro evidence for transforming growth factor-beta1-mediated epithelial to mesenchymal transition in esophageal adenocarcinoma. Cancer Res. 2006 Oct 1;66(19):9583–9590. doi: 10.1158/0008-5472.CAN-06-1842. [DOI] [PubMed] [Google Scholar]
- 51.Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005 Aug;5(8):603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- 52.Iavarone A, Garg P, Lasorella A, Hsu J, Israel MA. The helix-loop-helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev. 1994 Jun 1;8(11):1270–1284. doi: 10.1101/gad.8.11.1270. [DOI] [PubMed] [Google Scholar]
- 53.Lasorella A, Noseda M, Beyna M, Yokota Y, Iavarone A. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 2000 Oct 5;407(6804):592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- 54.Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003 Aug;13(8):410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 55.Kowanetz M, Valcourt U, Bergstrom R, Heldin CH, Moustakas A. Id2 and Id3 define the potency of cell proliferation and differentiation responses to transforming growth factor beta and bone morphogenetic protein. Mol Cell Biol. 2004 May;24(10):4241–4254. doi: 10.1128/MCB.24.10.4241-4254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stighall M, Manetopoulos C, Axelson H, Landberg G. High ID2 protein expression correlates with a favourable prognosis in patients with primary breast cancer and reduces cellular invasiveness of breast cancer cells. Int J Cancer. 2005 Jun 20;115(3):403–411. doi: 10.1002/ijc.20875. [DOI] [PubMed] [Google Scholar]
- 57.Wilson JW, Deed RW, Inoue T, Balzi M, Becciolini A, Faraoni P, et al. Expression of Id helix-loop-helix proteins in colorectal adenocarcinoma correlates with p53 expression and mitotic index. Cancer Res. 2001 Dec 15;61(24):8803–8810. [PubMed] [Google Scholar]
- 58.Umetani N, Takeuchi H, Fujimoto A, Shinozaki M, Bilchik AJ, Hoon DS. Epigenetic inactivation of ID4 in colorectal carcinomas correlates with poor differentiation and unfavorable prognosis. Clin Cancer Res. 2004 Nov 15;10(22):7475–7483. doi: 10.1158/1078-0432.CCR-04-0689. [DOI] [PubMed] [Google Scholar]
- 59.Russell RG, Lasorella A, Dettin LE, Iavarone A. Id2 drives differentiation and suppresses tumor formation in the intestinal epithelium. Cancer Res. 2004 Oct 15;64(20):7220–7225. doi: 10.1158/0008-5472.CAN-04-2095. [DOI] [PubMed] [Google Scholar]
- 60.Gordon MD, Nusse R. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem. 2006 Aug 11;281(32):22429–22433. doi: 10.1074/jbc.R600015200. [DOI] [PubMed] [Google Scholar]
- 61.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999 Apr 1;398(6726):422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 62.Gottardi CJ, Gumbiner BM. Adhesion signaling: how beta-catenin interacts with its partners. Curr Biol. 2001 Oct 2;11(19):R792–R794. doi: 10.1016/s0960-9822(01)00473-0. [DOI] [PubMed] [Google Scholar]
- 63.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. Embo J. 1998 Mar 2;17(5):1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kishida S, Yamamoto H, Ikeda S, Kishida M, Sakamoto I, Koyama S, et al. Axin, a negative regulator of the wnt signaling pathway, directly interacts with adenomatous polyposis coli and regulates the stabilization of beta-catenin. J Biol Chem. 1998 May 1;273(18):10823–10826. doi: 10.1074/jbc.273.18.10823. [DOI] [PubMed] [Google Scholar]
- 65.Maniatis T. A ubiquitin ligase complex essential for the NF-kappaB, Wnt/Wingless, and Hedgehog signaling pathways. Genes Dev. 1999 Mar 1;13(5):505–510. doi: 10.1101/gad.13.5.505. [DOI] [PubMed] [Google Scholar]
- 66.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. Embo J. 1997 Jul 1;16(13):3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Christofori G, Semb H. The role of the cell-adhesion molecule E-cadherin as a tumour-suppressor gene. Trends Biochem Sci. 1999 Feb;24(2):73–76. doi: 10.1016/s0968-0004(98)01343-7. [DOI] [PubMed] [Google Scholar]
- 68.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004 Mar 5;303(5663):1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shiou SR, Singh AB, Moorthy K, Datta PK, Washington MK, Beauchamp RD, et al. Smad4 regulates claudin-1 expression in a transforming growth factor-beta-independent manner in colon cancer cells. Cancer Res. 2007 Feb 15;67(4):1571–1579. doi: 10.1158/0008-5472.CAN-06-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002 Mar 15;62(6):1613–1618. [PubMed] [Google Scholar]
- 71.Ikeguchi M, Makino M, Kaibara N. Clinical significance of E-cadherin-catenin complex expression in metastatic foci of colorectal carcinoma. J Surg Oncol. 2001 Jul;77(3):201–207. doi: 10.1002/jso.1095. [DOI] [PubMed] [Google Scholar]
- 72.Krishnadath KK, Tilanus HW, van Blankenstein M, Hop WC, Kremers ED, Dinjens WN, et al. Reduced expression of the cadherin-catenin complex in oesophageal adenocarcinoma correlates with poor prognosis. J Pathol. 1997 Jul;182(3):331–338. doi: 10.1002/(SICI)1096-9896(199707)182:3<331::AID-PATH860>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 73.Tamura S, Shiozaki H, Miyata M, Kadowaki T, Inoue M, Matsui S, et al. Decreased E-cadherin expression is associated with haematogenous recurrence and poor prognosis in patients with squamous cell carcinoma of the oesophagus. Br J Surg. 1996 Nov;83(11):1608–1614. doi: 10.1002/bjs.1800831138. [DOI] [PubMed] [Google Scholar]
- 74.Oda T, Kanai Y, Oyama T, Yoshiura K, Shimoyama Y, Birchmeier W, et al. E-cadherin gene mutations in human gastric carcinoma cell lines. Proc Natl Acad Sci U S A. 1994 Mar 1;91(5):1858–1862. doi: 10.1073/pnas.91.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pignatelli M, Ansari TW, Gunter P, Liu D, Hirano S, Takeichi M, et al. Loss of membranous E-cadherin expression in pancreatic cancer: correlation with lymph node metastasis, high grade, and advanced stage. J Pathol. 1994 Dec;174(4):243–248. doi: 10.1002/path.1711740403. [DOI] [PubMed] [Google Scholar]
- 76.Vleminckx K, Vakaet L, Jr, Mareel M, Fiers W, van Roy F. Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell. 1991 Jul 12;66(1):107–119. doi: 10.1016/0092-8674(91)90143-m. [DOI] [PubMed] [Google Scholar]
- 77.Ikeguchi M, Taniguchi T, Makino M, Kaibara N. Reduced E-cadherin expression and enlargement of cancer nuclei strongly correlate with hematogenic metastasis in colorectal adenocarcinoma. Scand J Gastroenterol. 2000 Aug;35(8):839–846. doi: 10.1080/003655200750023219. [DOI] [PubMed] [Google Scholar]
- 78.Kimura K, Endo Y, Yonemura Y, Heizmann CW, Schafer BW, Watanabe Y, et al. Clinical significance of S100A4 and E-cadherin-related adhesion molecules in non-small cell lung cancer. Int J Oncol. 2000 Jun;16(6):1125–1131. doi: 10.3892/ijo.16.6.1125. [DOI] [PubMed] [Google Scholar]
- 79.Kanazawa N, Oda T, Gunji N, Nozue M, Kawamoto T, Todoroki T, et al. E-cadherin expression in the primary tumors and metastatic lymph nodes of poorly differentiated types of rectal cancer. Surg Today. 2002;32(2):123–128. doi: 10.1007/s005950200004. [DOI] [PubMed] [Google Scholar]
- 80.Kanazawa T, Watanabe T, Kazama S, Tada T, Koketsu S, Nagawa H. Poorly differentiated adenocarcinoma and mucinous carcinoma of the colon and rectum show higher rates of loss of heterozygosity and loss of E-cadherin expression due to methylation of promoter region. Int J Cancer. 2002 Nov 20;102(3):225–229. doi: 10.1002/ijc.10690. [DOI] [PubMed] [Google Scholar]
- 81.Brabletz T, Jung A, Reu S, Porzner M, Hlubek F, Kunz-Schughart LA, et al. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci U S A. 2001 Aug 28;98(18):10356–10361. doi: 10.1073/pnas.171610498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guilford P, Hopkins J, Harraway J, McLeod M, McLeod N, Harawira P, et al. E-cadherin germline mutations in familial gastric cancer. Nature. 1998 Mar 26;392(6674):402–405. doi: 10.1038/32918. [DOI] [PubMed] [Google Scholar]
- 83.Guilford PJ, Hopkins JB, Grady WM, Markowitz SD, Willis J, Lynch H, et al. E-cadherin germline mutations define an inherited cancer syndrome dominated by diffuse gastric cancer. Hum Mutat. 1999;14(3):249–255. doi: 10.1002/(SICI)1098-1004(1999)14:3<249::AID-HUMU8>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 84.Huntsman DG, Carneiro F, Lewis FR, MacLeod PM, Hayashi A, Monaghan KG, et al. Early gastric cancer in young, asymptomatic carriers of germ-line E-cadherin mutations. N Engl J Med. 2001 Jun 21;344(25):1904–1909. doi: 10.1056/NEJM200106213442504. [DOI] [PubMed] [Google Scholar]
- 85.Tamura G, Sakata K, Nishizuka S, Maesawa C, Suzuki Y, Iwaya T, et al. Inactivation of the E-cadherin gene in primary gastric carcinomas and gastric carcinoma cell lines. Jpn J Cancer Res. 1996 Nov;87(11):1153–1159. doi: 10.1111/j.1349-7006.1996.tb03125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Becker KF, Atkinson MJ, Reich U, Becker I, Nekarda H, Siewert JR, et al. E-cadherin gene mutations provide clues to diffuse type gastric carcinomas. Cancer Res. 1994 Jul 15;54(14):3845–3852. [PubMed] [Google Scholar]
- 87.Muta H, Noguchi M, Kanai Y, Ochiai A, Nawata H, Hirohashi S. E-cadherin gene mutations in signet ring cell carcinoma of the stomach. Jpn J Cancer Res. 1996 Aug;87(8):843–848. doi: 10.1111/j.1349-7006.1996.tb02109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Berx G, Cleton-Jansen AM, Nollet F, de Leeuw WJ, van de Vijver M, Cornelisse C, et al. E-cadherin is a tumour/invasion suppressor gene mutated in human lobular breast cancers. Embo J. 1995 Dec 15;14(24):6107–6115. doi: 10.1002/j.1460-2075.1995.tb00301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Berx G, Cleton-Jansen AM, Strumane K, de Leeuw WJ, Nollet F, van Roy F, et al. E-cadherin is inactivated in a majority of invasive human lobular breast cancers by truncation mutations throughout its extracellular domain. Oncogene. 1996 Nov 7;13(9):1919–1925. [PubMed] [Google Scholar]
- 90.Risinger JI, Berchuck A, Kohler MF, Boyd J. Mutations of the E-cadherin gene in human gynecologic cancers. Nat Genet. 1994 May;7(1):98–102. doi: 10.1038/ng0594-98. [DOI] [PubMed] [Google Scholar]
- 91.Nollet F, Berx G, van Roy F. The role of the E-cadherin/catenin adhesion complex in the development and progression of cancer. Mol Cell Biol Res Commun. 1999 Aug;2(2):77–85. doi: 10.1006/mcbr.1999.0155. [DOI] [PubMed] [Google Scholar]
- 92.Ireton RC, Davis MA, van Hengel J, Mariner DJ, Barnes K, Thoreson MA, et al. A novel role for p120 catenin in E-cadherin function. J Cell Biol. 2002 Nov 11;159(3):465–476. doi: 10.1083/jcb.200205115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Davis MA, Ireton RC, Reynolds AB. A core function for p120-catenin in cadherin turnover. J Cell Biol. 2003 Nov 10;163(3):525–534. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Remacle JE, Kraft H, Lerchner W, Wuytens G, Collart C, Verschueren K, et al. New mode of DNA binding of multi-zinc finger transcription factors: deltaEF1 family members bind with two hands to two target sites. Embo J. 1999 Sep 15;18(18):5073–5084. doi: 10.1093/emboj/18.18.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chinnadurai G. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol Cell. 2002 Feb;9(2):213–224. doi: 10.1016/s1097-2765(02)00443-4. [DOI] [PubMed] [Google Scholar]
- 96.Light W, Vernon AE, Lasorella A, Iavarone A, LaBonne C. Xenopus Id3 is required downstream of Myc for the formation of multipotent neural crest progenitor cells. Development. 2005 Apr;132(8):1831–1841. doi: 10.1242/dev.01734. [DOI] [PubMed] [Google Scholar]
- 97.Vernon AE, LaBonne C. Tumor metastasis: a new twist on epithelial-mesenchymal transitions. Curr Biol. 2004 Sep 7;14(17):R719–R721. doi: 10.1016/j.cub.2004.08.048. [DOI] [PubMed] [Google Scholar]
- 98.Perez-Moreno MA, Locascio A, Rodrigo I, Dhondt G, Portillo F, Nieto MA, et al. A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J Biol Chem. 2001 Jul 20;276(29):27424–27431. doi: 10.1074/jbc.M100827200. [DOI] [PubMed] [Google Scholar]
- 99.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004 Jun 25;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 100.Kang Y, Massague J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004 Aug 6;118(3):277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 101.Bellovin DI, Bates RC, Muzikansky A, Rimm DL, Mercurio AM. Altered localization of p120 catenin during epithelial to mesenchymal transition of colon carcinoma is prognostic for aggressive disease. Cancer Res. 2005 Dec 1;65(23):10938–10945. doi: 10.1158/0008-5472.CAN-05-1947. [DOI] [PubMed] [Google Scholar]
- 102.Frixen UH, Behrens J, Sachs M, Eberle G, Voss B, Warda A, et al. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J Cell Biol. 1991 Apr;113(1):173–185. doi: 10.1083/jcb.113.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mbalaviele G, Dunstan CR, Sasaki A, Williams PJ, Mundy GR, Yoneda T. E-cadherin expression in human breast cancer cells suppresses the development of osteolytic bone metastases in an experimental metastasis model. Cancer Res. 1996 Sep 1;56(17):4063–4070. [PubMed] [Google Scholar]
- 104.Schmidt CR, Gi YJ, Patel TA, Coffey RJ, Beauchamp RD, Pearson AS. E-cadherin is regulated by the transcriptional repressor SLUG during Ras-mediated transformation of intestinal epithelial cells. Surgery. 2005 Aug;138(2):306–312. doi: 10.1016/j.surg.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 105.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006 Jan;5(1):37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 106.de Ruijter AJ, Meinsma RJ, Bosma P, Kemp S, Caron HN, van Kuilenburg AB. Gene expression profiling in response to the histone deacetylase inhibitor BL1521 in neuroblastoma. Exp Cell Res. 2005 Oct 1;309(2):451–467. doi: 10.1016/j.yexcr.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 107.Hu E, Dul E, Sung CM, Chen Z, Kirkpatrick R, Zhang GF, et al. Identification of novel isoform-selective inhibitors within class I histone deacetylases. J Pharmacol Exp Ther. 2003 Nov;307(2):720–728. doi: 10.1124/jpet.103.055541. [DOI] [PubMed] [Google Scholar]
- 108.Gray SG, Ekstrom TJ. The human histone deacetylase family. Exp Cell Res. 2001 Jan 15;262(2):75–83. doi: 10.1006/excr.2000.5080. [DOI] [PubMed] [Google Scholar]
- 109.Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002 Jan;9(1):45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 110.Jepsen K, Rosenfeld MG. Biological roles and mechanistic actions of co-repressor complexes. J Cell Sci. 2002 Feb 15;115(Pt 4):689–698. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- 111.Ohira T, Gemmill RM, Ferguson K, Kusy S, Roche J, Brambilla E, et al. WNT7a induces E-cadherin in lung cancer cells. Proc Natl Acad Sci U S A. 2003 Sep 2;100(18):10429–10434. doi: 10.1073/pnas.1734137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Witta SE, Gemmill RM, Hirsch FR, Coldren CD, Hedman K, Ravdel L, et al. Restoring E-cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Res. 2006 Jan 15;66(2):944–950. doi: 10.1158/0008-5472.CAN-05-1988. [DOI] [PubMed] [Google Scholar]
- 113.Spaderna S, Schmalhofer O, Hlubek F, Berx G, Eger A, Merkel S, et al. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology. 2006 Sep;131(3):830–840. doi: 10.1053/j.gastro.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 114.Zhang Q, Wang SY, Nottke AC, Rocheleau JV, Piston DW, Goodman RH. Redox sensor CtBP mediates hypoxia-induced tumor cell migration. Proc Natl Acad Sci U S A. 2006 Jun 13;103(24):9029–9033. doi: 10.1073/pnas.0603269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Koukourakis MI, Giatromanolaki A, Harris AL, Sivridis E. Comparison of metabolic pathways between cancer cells and stromal cells in colorectal carcinomas: a metabolic survival role for tumor-associated stroma. Cancer Res. 2006 Jan 15;66(2):632–637. doi: 10.1158/0008-5472.CAN-05-3260. [DOI] [PubMed] [Google Scholar]
- 116.Koukourakis MI, Giatromanolaki A, Polychronidis A, Simopoulos C, Gatter KC, Harris AL, et al. Endogenous markers of hypoxia/anaerobic metabolism and anemia in primary colorectal cancer. Cancer Sci. 2006 Jul;97(7):582–588. doi: 10.1111/j.1349-7006.2006.00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Koukourakis MI, Giatromanolaki A, Sivridis E, Gatter KC, Harris AL. Lactate dehydrogenase 5 expression in operable colorectal cancer: strong association with survival and activated vascular endothelial growth factor pathway--a report of the Tumour Angiogenesis Research Group. J Clin Oncol. 2006 Sep 10;24(26):4301–4308. doi: 10.1200/JCO.2006.05.9501. [DOI] [PubMed] [Google Scholar]
- 118.Sivridis E, Giatromanolaki A, Koukourakis MI. Proliferating fibroblasts at the invading tumour edge of colorectal adenocarcinomas are associated with endogenous markers of hypoxia, acidity, and oxidative stress. J Clin Pathol. 2005 Oct;58(10):1033–1038. doi: 10.1136/jcp.2005.026260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Furuse M, Hata M, Furuse K, Yoshida Y, Haratake A, Sugitani Y, et al. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: a lesson from claudin-1-deficient mice. J Cell Biol. 2002 Mar 18;156(6):1099–1111. doi: 10.1083/jcb.200110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nakamura T, Blechman J, Tada S, Rozovskaia T, Itoyama T, Bullrich F, et al. huASH1 protein, a putative transcription factor encoded by a human homologue of the Drosophila ash1 gene, localizes to both nuclei and cell-cell tight junctions. Proc Natl Acad Sci U S A. 2000 Jun 20;97(13):7284–7289. doi: 10.1073/pnas.97.13.7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Balda MS, Garrett MD, Matter K. The ZO-1-associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. J Cell Biol. 2003 Feb 3;160(3):423–432. doi: 10.1083/jcb.200210020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Morita K, Furuse M, Fujimoto K, Tsukita S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci U S A. 1999 Jan 19;93(2):511–516. doi: 10.1073/pnas.96.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Itoh M, Furuse M, Morita K, Kubota K, Saitou M, Tsukita S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J Cell Biol. 1999 Dec 13;147(6):1351–1363. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Singh AB, Harris RC. Epidermal growth factor receptor activation differentially regulates claudin expression and enhances transepithelial resistance in Madin-Darby canine kidney cells. J Biol Chem. 2004 Jan 30;279(5):3543–3552. doi: 10.1074/jbc.M308682200. [DOI] [PubMed] [Google Scholar]
- 125.Grande M, Franzen A, Karlsson JO, Ericson LE, Heldin NE, Nilsson M. Transforming growth factor-beta and epidermal growth factor synergistically stimulate epithelial to mesenchymal transition (EMT) through a MEK-dependent mechanism in primary cultured pig thyrocytes. J Cell Sci. 2002 Nov 15;115(Pt 22):4227–4236. doi: 10.1242/jcs.00091. [DOI] [PubMed] [Google Scholar]
- 126.Miyamori H, Takino T, Kobayashi Y, Tokai H, Itoh Y, Seiki M, et al. Claudin promotes activation of pro-matrix metalloproteinase-2 mediated by membrane-type matrix metalloproteinases. J Biol Chem. 2001 Jul 27;276(30):28204–28211. doi: 10.1074/jbc.M103083200. [DOI] [PubMed] [Google Scholar]
- 127.Dhawan P, Singh AB, Deane NG, No Y, Shiou SR, Schmidt C, et al. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest. 2005 Jul;115(7):1765–1776. doi: 10.1172/JCI24543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hewitt RE, McMarlin A, Kleiner D, Wersto R, Martin P, Tsokos M, et al. Validation of a model of colon cancer progression. J Pathol. 2000 Dec;192(4):446–454. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH775>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 129.Miwa N, Furuse M, Tsukita S, Niikawa N, Nakamura Y, Furukawa Y. Involvement of claudin-1 in the beta-catenin/Tcf signaling pathway and its frequent upregulation in human colorectal cancers. Oncol Res. 2001;12(11–12):469–476. doi: 10.3727/096504001108747477. [DOI] [PubMed] [Google Scholar]
- 130.Schwarte-Waldhoff I, Klein S, Blass-Kampmann S, Hintelmann A, Eilert C, Dreschers S, et al. DPC4/SMAD4 mediated tumor suppression of colon carcinoma cells is associated with reduced urokinase expression. Oncogene. 1999 May 20;18(20):3152–3158. doi: 10.1038/sj.onc.1202641. [DOI] [PubMed] [Google Scholar]
- 131.Muller N, Reinacher-Schick A, Baldus S, van Hengel J, Berx G, Baar A, et al. Smad4 induces the tumor suppressor E-cadherin and P-cadherin in colon carcinoma cells. Oncogene. 2002 Sep 5;21(39):6049–6058. doi: 10.1038/sj.onc.1205766. [DOI] [PubMed] [Google Scholar]