Abstract
Although chromosome mis-segregation is a hallmark of cancer cells, its genetic basis and role in malignant transformation remain poorly understood. In recent years, several mouse models have been generated that harbor gene defects that perturb high-fidelity chromosome segregation. Analysis of these models has revealed that whole chromosome instability (W-CIN) can cause, inhibit or have no effect on tumorigenesis. Here we propose that the effect of W-CIN on tumor development depends on the particular W-CIN gene that is defective, including its other cellular functions, the extent or nature of the gene defect, the affected tissue or cell type and the context of other cancer gene mutations.
Aneuploidy and cancer – an overview
Aneuploidy, an alteration in the number of whole chromosomes, has been recognized as a trait of cancer cells for over a century [1,2]. Whether aneuploidy is a cause or consequence of cancer is one of the long-standing questions in cancer biology and a subject of debate. Some researchers assert that aneuploidy is the primary cause and driving force of tumorigenesis [3], whereas others argue that, whereas oncogenes and tumor suppressor genes propel malignant cell transformation, aneuploidy is simply a benign side effect of neoplastic growth [4]. Based on mathematical modeling, others propose that aneuploidy is an early event in tumor formation that precedes the inactivation of tumor suppressor genes [5].
Part of the difficulty in understanding the role of aneuploidy in cancer lies in the highly divergent nature of the chromosomal abnormalities in tumor cells, including cells within the same tumor [6,7]. Sporadic chromosome segregation errors represent one way in which diploid cells could become aneuploid [3]. These errors are thought to occur coincidentally or by exposure to certain chemical compounds. Aneuploidy might also be driven by genetic alterations that promote inaccurate chromosome segregation, thus increasing the rate with which whole chromosomes are lost or gained [8]. This condition is referred to as whole chromosomal instability (W-CIN) [9]. An important difference between these two scenarios is that cells that become aneuploid through a coincidental error are not necessarily chromosomally unstable, whereas cells that have a W-CIN gene defect continually scramble their aneuploid karyotypes. In addition to numerical chromosomal abnormalities, cancer cells often exhibit changes in chromosome structure, including reciprocal or nonreciprocal chromosomal translocations, deletions of chromosome arms and amplifications of large chromosome regions [10,11]. This condition is termed structural or segmental chromosomal instability (S-CIN) [9].
To better understand the mechanisms that cause chromosomal instability and their role in cancer development, it is crucially important to define the molecular basis of mitotic activity and progression [12]. In yeast, more than a hundred genes, when defective, can cause chromosomal instability [13,14]. These genes are implicated in mitotic checkpoint control, chromosome condensation, sister-chromatid cohesion, kinetochore assembly, spindle formation and several other mitotic events. Several hundred genes have been estimated to contribute to proper chromosome segregation in humans [5,15]. The application of gene knockout technology to determine the physiological relevance of known CIN genes has been instrumental in providing new details in the relationship between aneuploidy and cancer. Mice harboring defective mitotic checkpoint genes have been particularly helpful because they accumulate cells that exhibit numerical chromosomal changes in the absence of apparent structural abnormalities [16,17]. Although many mouse models with numerical chromosomal changes exhibit increased tumor susceptibility, the relationship between aneuploidy and cancer seems to be highly complex, as will be discussed below. The debate surrounding the role of aneuploidy in cancer has overshadowed two important questions: which genes that have been implicated in chromosome mis-segregation have the most prominent role in cancer prevention, and why? These two issues will also be considered in this review.
Many W-CIN mouse models are tumor prone
The mitotic checkpoint is a surveillance system that ensures high-fidelity chromosome segregation by delaying anaphase onset until the kinetochores of duplicated chromosomes are properly attached to microtubules from opposite spindle poles (Figure 1). Core components of this checkpoint are Bub1 (budding uninhibited by benzimidazoles 1 homolog), BubR1 (budding uninhibited by benzimidazoles 1 homolog β), Bub3 (budding uninhibited by benzimidazoles 3 homolog), Mad1 (mitotic arrest deficient-like 1) and Mad2 (mitotic arrest deficient-like 2) [12]. In certain types of human cancers, including breast, colorectal and gastric cancers, mutations have been observed in mitotic checkpoint genes, although at very low frequency [17]. Downregulation of mitotic checkpoint genes is, however, seen much more frequently in human tumors [17]. Several groups have tried to determine whether these alterations are sufficient to cause W-CIN and tumorigenesis by generating mitotic checkpoint-defective mice. Thus far, classical mouse knockouts of mitotic checkpoint genes have resulted in embryonic lethality [18–23]. Although heterozygotes have in some cases provided valuable information regarding the biologically relevant function of the gene, not all heterozygous-null mutations result in an overt phenotype. This can occur when gene expression remains above the threshold necessary to impair function. Therefore, the generation of hypomorphic (H) alleles has been extremely useful in determining the effect of reduced gene expression while maintaining organism viability. Hypomorphic alleles produce more functional protein than a knockout allele but significantly less than wild type, allowing for a substantial yet incomplete disruption of function.
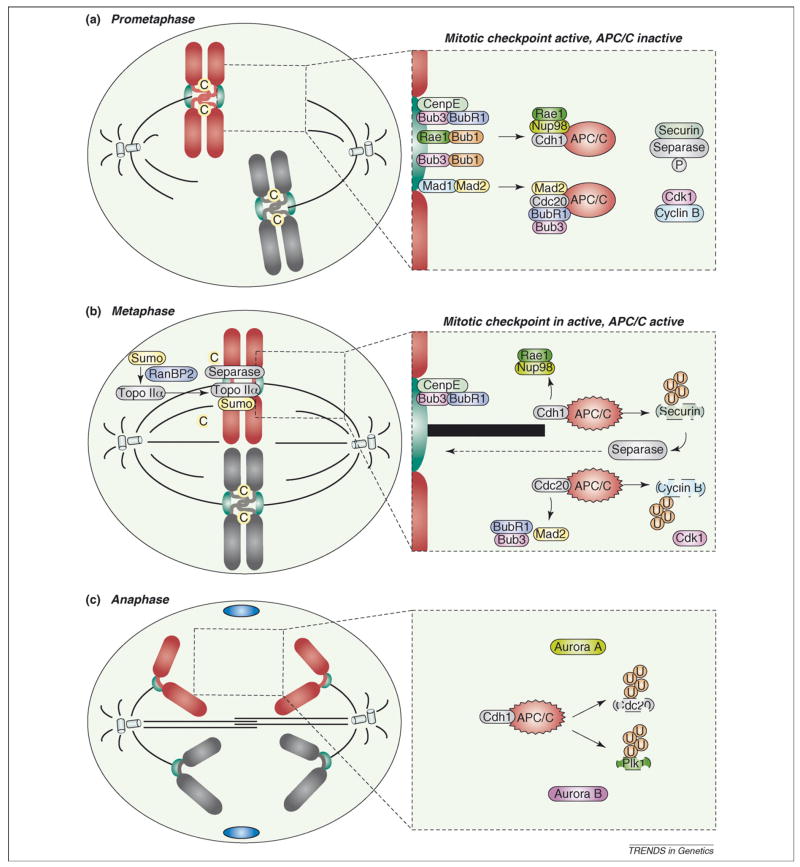
Figure 1.
Current model of the mitotic checkpoint. To prevent chromosome mis-segregation, cells have developed a multiprotein surveillance mechanism, called the mitotic checkpoint, that delays anaphase onset until all kinetochores are correctly attached to mitotic spindle microtubules and aligned in the metaphase plate (the central area between the two spindle poles). (a) Various mitotic checkpoint proteins, including Rae1 (green), Bub3 (pink), Bub1 (orange), BubR1 (purple), Mad1 (blue) and Mad2 (yellow), bind kinetochores (green) that lack attachment or tension and generate a ‘stop anaphase’ signal that diffuses into the mitotic cytosol. This signal is believed to consist of Bub3–BubR1–Mad2 and Rae1–Nup98 (light yellow) protein complexes, which bind and inhibit APCCdc20 (dark grey) and APCCdh1 (light grey), respectively. Once each pair of sister kinetochores attaches to microtubules (black), and microtubule motors generate tension that stretches them, the production of inhibitory ‘stop anaphase’ signals at those kinetochores quenches. (b) Silencing of the ‘stop anaphase’ signal at the last kinetochore triggers the release of Bub3–BubR1–Mad2 and Rae1–Nup98 from APCCdc20 and APCCdh1, respectively. This event allows APCCdc20-mediated cyclin B (blue) destruction and APCCdh1-mediated securin (brown) degradation. Separase (grey), which is inhibited through its association with securin and by cyclin B–Cdk1-mediated phosphorylation, triggers sister chromatid disjunction by cohesin cleavage. Sister chromatid separation is also dependent on Topo IIα (grey), which decatenates centromeric DNA. Topo IIα targeting to inner centromeres is dependent on RanBP2 (purple)-mediated sumoylation. (c) In anaphase, fully separated sister chromatids move to opposite poles. Proper progression through anaphase requires APC/CCdh1-mediated degradation of Cdc20 and Polo like kinase 1 (Plk1; green). AurkA (yellow) and AurkB (pink) remain intact until their degradation in late mitosis. Blue ovals in (c) represent portions of the contractile ring that mediates cell division. Encircled C, cohesin; encircled U, ubiquitin.
The effect of reduced expression of mitotic checkpoint genes, including Bub1, Bub1b (encoding BubR1 protein), Bub3, centromere protein E (Cenp-E), Mad1 and Mad2, has been examined [21–27]. In each case, haploinsufficiency or hypomorphism of these genes results in aneuploidy in both mouse embryonic fibroblasts (MEFs) and in tissues, albeit to varying degrees (Table 1). In addition to these canonical mitotic checkpoint factors, several proteins that mediate macromolecule transport through nuclear pores have recently been implicated in mitosis, including Ran binding protein 2 [RanBP2, also called nucleoporin 358 (Nup358)], nucleoporin 98 (Nup98) and the Bub3-related protein RNA export factor 1 (Rae1) [28,29] (Figure 1). Nup98 and Rae1 form an inhibitory complex that binds securin-bound APC/CCdh1 complexes [29], preventing securin degradation until the release of mitotic checkpoint inhibition. RanBP2, a SUMO E3 ligase, is essential for topoisomerase IIα (Topo IIα) localization to inner centromeres, thereby promoting disentanglement of sister chromatids and proper chromosome segregation [30]. Mice and MEFs that express decreased levels of these transport factors exhibit W-CIN and develop aneuploidy (Table 1). Compound heterozygous null mutations of Rae1 and Nup98 or Bub3 produced mouse models that accumulate very high percentages of aneuploid cells [28]. Homozygous inactivation of pituitary tumor-transforming gene 1 (Pttg1; also named securin) in mice causes mild aneuploidy and premature sister chromatid separation (PMSCS) (Table 1) [31], although no such defects were reported for an independently generated knockout strain [32]. Furthermore, mice lacking Chfr (checkpoint with forkhead and ring finger domains) display W-CIN and develop substantial aneuploidy (Table 1) [33]. Chfr is an E3 ubiquitin ligase that delays entry into mitosis after exposure to microtubule-depolymerizing agents by inhibiting cyclin B nuclear import, an event that is required for mitosis onset [33]. Chfr also controls mitotic checkpoint activity, presumably through its ability to regulate Mad2 and BubR1 functions [34].
Table 1.
Overview of W-CIN mouse models and their chromosome segregation and cancer phenotypes
| Mouse model | Aneuploidy
|
S-CIN | Mitotic checkpoint defect | Main segregation defect(s) | Carcinogen-induced tumorigenesis
|
Spontaneous tumorigenesis
|
Refs | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Mutant MEFs (control MEFs) | Mutant tissue (control tissue) | Tumor susceptibility | Tumor spectrum | Tumor susceptibility | Tumor spectrum | |||||
| MMTV-Aurka | N.D. | N.D. | N.D. | N.D. | N/A | N.D. | N/A | Up | AC | [70] |
| MMTV-Aurka;Trp53+/− | N.D. | N.D. | N.D. | N.D. | N/A | N.D. | N/A | Up | AC | [70] |
| CAG-CAT-Aurka; WAP-Cre | N.D. | N.D. | N.D. | N.D. | CMA/CMS | N.D. | N/A | Normal | N/A | [71] |
| Bub1+/− | 14% (7%) | 16% (1%) S | No (G) | Yes | LC/CF | Yes (D) | LG | Normal | N/A | [24] |
| Bub1H/H | 35% (7%) | 35% (1%) S | No (G) | Yes | LC/CF | N.D. | N/A | Up | HC, LA, SA | [24] |
| Bub1−/H | 36% (7%) | 39% (1%) S | No (G) | Yes | LC/CF | N.D. | N/A | Up | LA, HC, SA, LY | [24] |
| Bub1b+/− | 14% (9%) | 0% S | No (G) | Yes | LC | N.D. | N/A | Normal | N/A | [22] |
| 360% (17%) | N.D. | No (G) | Yes | N.D. | Yes (A) | MA, AD, AC | N.D. | N/A | [41,46] | |
| Bub1bH/H | 36% (9%) | 15% (0%) S | No (Sk) | Yes | LC | Yes (D) | LG | Normal | N/A | [22,36] |
| Bub1b+/−;APC+/Min | 366% (17%) | N.D. | N.D. | Yes | N.D. | N.D. | N/A | Up, Down | Colon up, SI down | [41] |
| Bub3+/− | 19% (9%) | 9% (1%) S | No (G) | Yes | LC | Yes (D) | LG | Normal | N/A | [28,36] |
| 42% (35%) | N.D. | N.D. | N.D. | N.D. | N.D. | N/A | Normal | N/A | [19,50] | |
| Bub3+/−;Trp53+/− | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N/A | Normal | N/A | [50] |
| Bub3+/−;Rae1+/− | 41% (9%) | 36% (0%) S | No (G) | Yes | LC | Yes (D) | LG | Normal | N/A | [36] |
| Bub3+/−;Rb1+/− | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N/A | Normal | N/A | [50] |
| Cenp-E+/− | 335% (20%) | 35% (10%) S | No (Sk) | Yes | CF | Protected (D) | LG | Up, Down | S, LG up | [25] |
| 335% (12%) PB | LV down | |||||||||
| 320% (5%) C | ||||||||||
| Cenp-E+/−;p19Arf−/− | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N/A | Down | LY | [25] |
| Chfr−/− | 330% (10%) | N.D. | N.D. | N.D. | N.D. | Yes (D) | Skin | Up | LY, epithelial | [33] |
| Mad1+/− | N.D. | N.D. | N.D. | Yes | LC/CF | N.D. | N/A | Up | Lung tumors | [21] |
| Mad2+/− | 357% (16%) | 18% (0%) S | N.D. | Yes | LC | N.D. | N/A | Up | Lung tumors | [26,36] |
| Mad2+/TA | 340% (8%) | N.D. | Yes (G)a | Yes | LC/CF | N.D. | N/A | Up | LD, H | [36,39] |
| Nup98+/− | 12% (9%) | 0% S | No (G) | N.D. | N.D. | No (D) | N/A | Normal | N/A | [29,35] |
| Pttg1−/− | 315% (1%) | N.D. | Yes (G)b | N/A | N.D. | N.D. | N/A | Normal | N/A | [31,42] |
| N.D. | N.D. | N.D. | N.D. | No | N.D. | N.D. | N.D. | N.D. | [32] | |
| Pttg1−/−;Rb1+/− | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | Protected | PA | [42] |
| Rae1+/− | 20% (9%) | 9% (0%) S | No (G) | Yes | LC | Yes (D) | LG | Normal | N/A | [28] |
| Rae1+/−;Nup98+/− | 37% (9%) | 37% (0%) S | No (G) | Yes | LC | Yes (D) | LG | Normal, Down | LA down | [29,35] |
| RanBP2+/− | 13% (9%) | 1% (0%) S | No (G) | N.D. | AB | No (D) | N/A | N.D. | N/A | [30] |
| RanBP2H/H | 26% (9%) | 5% (0%) S | No (G) | N.D. | AB | Yes (D) | Skin, LG | Up | LA, HC | [30] |
| RanBP2−/H | 33% (9%) | 15% (0%) S | No (G,Sk) | N.D. | AB | Yes (D) | Skin, LG | Up | LA, HC, SA | [30] |
| Cdh1+/− | N.D. | N.D. | N.D. | N.D. | N.D. | Yes (D/T)c | Skin | Upc | B, LA, LV, T | [78] |
Abbreviations: (A), azoxymethane or AOM; AB, anaphase bridges; AC, Adenocarcinomas; AD, adenoma; B, breast; C, colon cells; CF, congression failure; (D), dimethylbenzanthrene or DMBA; (D/T), combined DMBA/12-O-tetradecanoylphorbol-13-acetate; FC, failed cytokinesis; (G), Giemsa staining of metaphase spreads did not provide evidence of chromosome breaks, gaps, fragmentation, or chromosome fusion events; H, hepatoma; HC, hepatocellular carcinoma; LA, lung adenocarcinoma; LC, lagging chromosomes; LD, lung adenoma; LG, lung; LV, liver; LY, lymphoma; MA, microadenoma; N.D., not determined; N/A, not applicable; PA, pituitary adenoma; PB, peripheral blood cells; S, splenocytes; SA, sarcoma; (Sk), spectral karyotype analysis on MEFs; T, testis; W-CIN, whole chromosome instability.
Mad2 overexpression resulted in a significant increase in the number of chromosomal breaks and fragments, chromosomal fusions, chromatid breaks and gaps.
Aberrant chromosome structures including quadriradials, triradials and breaks were observed in 4–6% of Pttg1−/− metaphase spreads but not in wild-type controls.
Spontaneous and carcinogen-induced tumor formation were, respectively, increased and decreased, but these differences were not reported to be statistically significant.
Are these W-CIN mouse models prone to tumor formation? When exposed to carcinogens, 10 of 13 W-CIN models were tumor prone; this is perhaps the most compelling evidence for a causal role of aneuploidy in tumor formation. Several W-CIN models also show clear evidence of increased spontaneous tumor susceptibility, including Bub1 and RanBP2 hypomorphic mice and Mad1, Mad2 and Cenp-E heterozygous mice. Despite the presence of vast amounts of aneuploid cells, other W-CIN models do not seem to be predisposed to spontaneous tumors, including Bub1, Bub1b, Bub3 and Rae1 heterozygous mice and Rae1;Bub3 and Rae1;Nup98 compound heterozygotes [24,28,35].
Remarkably, there is no direct correlation between the incidence of spontaneous tumor development and the level of aneuploidy. In particular, Bub1−/H and Bub1H/H mice show high aneuploidy in MEFs and splenocytes and are prone to spontaneous tumors [24], whereas Rae1;Bub3 and Rae1;Nup98 double heterozygous mice exhibit similar levels of aneuploidy but are not susceptible to spontaneous tumorigenesis [29,36]. W-CIN models that are prone to spontaneous tumors display a similar tumor spectrum as aged wild-type mice, which include lymphomas, lung adenocarcinomas, hepatocellular carcinomas and sarcomas. Of these, lung adenocarcinomas consistently develop at a much higher incidence in W-CIN models, implying that aneuploidy is particularly effective in driving tumor progression in lung epithelial cells. The underlying basis for this is currently unclear. Other tumor types are also enhanced, but in a more gene specific fashion. By example, sarcomagenesis is enhanced in Bub1−/H, RanBP2−/H and Mad1+/−, but not in Mad2+/− and Cenp-E+/− mice, whereas increased lymphomagenesis is observed only in Bub1−/H and Cenp-E+/−mice. Hepatocellular carcinomas are specifically increased in Bub1H/H mice; this tumor type is not increased in either Bub1+/− or in Bub1−/H mice, suggesting that neoplastic growth of hepatocytes requires an optimal level of Bub1 reduction.
It is unclear why some W-CIN gene defects promote tumorigenesis in certain tissues but not in others. One complication is that the aneuploidy levels of the tumor-prone cell types are unknown because of technical limitations. Preparation of metaphase spreads from tissues and organs other than spleen, thymus and bone marrow requires collagenase-treatment and long-term culturing, causing culture stress-induced aneuploidy. Therefore, aneuploidy levels from short-term splenocyte cultures might not be representative for other tissue and cell types. The observation that aneuploidy levels in germ cells of mitotic checkpoint-defective mice are much lower than in splenocytes supports this notion [37]. It is also unclear if chromosome mis-segregation elicits the same response in each cell type and W-CIN gene defect. Perhaps in some instances, chromosome mis-segregation results in continued proliferation, whereas in others, cell cycle arrest, senescence or apoptosis might occur.
Mad2 is activated by E2F transcription factor 1 (E2F1) and overexpressed in human tumors lacking a functional retinoblastoma (Rb) pathway [38]. Transgenic mice that ubiquitously overexpress Mad2 are highly susceptible to a wide variety of tumors, suggesting that Mad2 contributes to cancer development after inactivation of Rb activity [39]. However, in addition to aneuploidy, Mad2 transgenic mice show massive structural chromosome defects, including chromosome breaks, deletions, amplifications and fusions. Thus, whether Mad2-dependent tumorigenesis results from W-CIN, S-CIN or both remains unclear. It will be important to test whether overexpression of other mitotic (checkpoint) genes also causes chromosomal instability and cancer, particularly if these genes are controlled by E2F transcription factors and/or are upregulated in human tumors [40].
W-CIN and tumor suppression
Weaver et al. [25] recently introduced the concept that aneuploidy can prevent or delay the formation of certain kinds of tumors. Three observations in Cenp-E heterozygous mice led to this idea. First, loss of one Cenp-E gene copy caused a slight delay in tumorigenesis in mice lacking the gene that encodes p19Arf (alternative reading frame; cyclin-dependent kinase inhibitor 2a isoform 3). Second, spontaneous liver tumors developed at much lower rates in Cenp-E+/− mice than in controls and were typically much smaller. Third, Cenp-E+/− mice were less prone to 7,12-dimethylbenz[a]anthracene (DMBA)-induced tumors. Although the first finding was statistically significant, the other two were not, which led to some controversy about the concept. However, in addition to Cenp-E, a second mitotic checkpoint gene, Bub1b, has shown evidence of haploinsufficient tumor suppression. Specifically, when the Min (multiple intestinal neoplasia) point mutation in the murine APC (adenomatous polyposis coli) gene was bred onto a Bub1b+/− genetic background, mice formed twofold fewer small intestinal tumors than APC+/Min mice containing two wild-type Bub1b copies [41] (Table 1). Another example of a W-CIN model with tumor suppression is Nup98;Rae1 compound heterozygous mice [35], which show a twofold reduction lung tumor incidence. Chesnokova et al. [42] reported that Rb1+/− mice develop nearly threefold fewer pituitary adenomas on a Pttg1−/− than on a Pttg1+/+ genetic background. However, in this instance, it remains unclear whether tumor suppression results from aneuploidy. In each of the above models, the tumor suppressive effect seems to be tissue restricted. It is important to note that several W-CIN mouse models with substantial aneuploidy were comprehensively screened for tumor development, including Bub1b−/H, Bub3+/−;Rae1+/− and RanBP2−/H mice [24,30,36] and did not present any evidence of reduced tumorigenesis. These findings suggest that certain W-CIN genes lack antitumor activity, although it remains possible that these genes suppress tumorigenesis only in particular genetic contexts.
How might W-CIN gene defects suppress tumor formation? Although the exact mechanism remains unclear, one thought is that Cenp-E and Bub1b haploin-sufficiency perturb chromosome segregation so severely that the cell dies as a consequence [41,43]. Apoptosis is greatly increased in small intestinal tissue of Bub1b+/−;APC+/Min mice, providing compelling evidence that increased cell death can prevent tumor formation [41]. In addition, aneuploidy is substantially increased in Bub1b MEFs carrying the APCMin mutation and in Cenp-E+/− MEFs treated with DMBA or lacking p19Arf 43. Although these data are encouraging, further work is needed to determine whether aneuploidy is also increased in the specific tissues in which Cenp-E and Bub1b haploinsufficiency suppress tumorigenesis. Moreover, most W-CIN models with substantial aneuploidy show increased tumor susceptibility after carcinogen exposure (Table 1). Of note, Nup98+/−;Rae1+/− and Bub3+/−;Rae1+/− mutant mice display similar aneuploidy levels as Cenp-E+/− mice. Therefore, they might also be expected to show reduced DMBA-induced tumorigenesis, similar to Cenp-E+/− mice, if cell death resulting from mitotic failure was indeed the mechanism of tumor suppression. Further testing to elucidate the role of aneuploidy level in tumor suppression is required. Notably, recent findings show that the gain of a single or several chromosomes in budding yeast causes a proliferative disadvantage; this finding reveals another potential mechanism by which aneuploidy might suppress tumorigenesis [44].
Genetic context: a crucial modulator of W-CIN pathology
One idea regarding cancer initiation posits that it starts with a coincidental mis-segregation of one or a few chromosomes, changing the copy numbers of thousands of genes including some of the hundreds of genes thought to regulate proper chromosome segregation [3,45]. This event causes chromosomal instability and allows progeny cells to further scramble their chromosomal content until rare chromosome combinations that provide malignant growth properties eventually emerge through successive rounds of clonal expansion. The finding that some mice with W-CIN accumulate vast amounts of aneuploid cells, but have little-or-no predisposition to spontaneous tumors, does not support this idea. Rather than initiating cancer, it seems that certain W-CIN gene mutations contribute to tumor progression in instances where specific mutations in crucial cancer genes have already increased the risk for neoplastic growth [24,28,30,35,46].
A key question emerges: which types of cancer genes cooperate with W-CIN in tumor formation? A candidate gene approach in which known cancer gene mutations are crossed into W-CIN mouse strains is a practicable means of identifying cooperating genes. Using this strategy, Baker et al. [47] demonstrated that the incidence of lung adenocarcinomagenesis in Bub1b hypomorphic mice increases nearly 20-fold in mice lacking the gene that encodes p16Ink4a (cyclin-dependent kinase inhibitor 2a isoforms 1/2) (Figure 2). Tissues other than lung were unaffected, indicating that BubR1 hypomorphism specifically synergizes with p16Ink4a loss in lung tumorigenesis. Interestingly, the absence of p16Ink4a is expected to inactivate Rb1 and lead to E2F hyperactivity. As a result, Mad2 might be overexpressed in Bub1bH/H; p16Ink4 mutant mice and accelerate lung tumorigenesis [39]. When p19Arf instead of p16Ink4a is absent in Bub1bH/H mice, tumorigenesis does not accelerate in any tissue, underscoring the crucial nature of gene context in modulating the effect of W-CIN on tumor formation (Figure 2). Using a similar approach, Rao et al. [41] showed that APC+/Min mice develop on average ten times more colon tumors on a Bub1b+/− background than on a Bub1b+/+ background [41], suggesting that Bub1b haploinsufficiency contributes to tumorigenesis in various genetic contexts (Figure 2). It is tempting to speculate that W-CIN resulting from Bub1b haploinsuficiency accelerates loss of heterozygosity (LOH) of the remaining wild-type APC allele. However, proof for this mechanism will not be easy to obtain, because the APCMin mutation itself causes W-CIN [48,49].
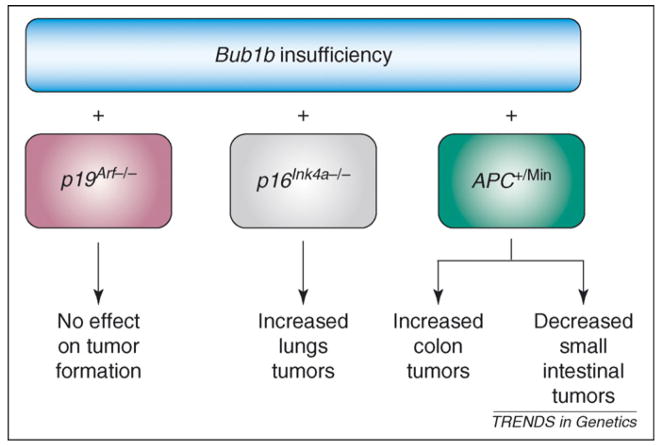
Figure 2.
Genetic context of whole chromosome instability (W-CIN) genes modulates tumorigenesis propensity. The genetic context of W-CIN defects dramatically alters the tumorigenetic potential outcome of a given tissue type. As a specific example, Bub1b hypomorphism has been analyzed in mice lacking either p16Ink4a or p19Arf 47. Tumorigenesis was accelerated only in the context of p16Ink4a, but not p19Arf, loss. Similarly, APC+/Min mice develop more colon tumors, and fewer small intestine tumors, on a Bub1b+/− background than on a Bub1b+/+ background.
Although it has been postulated that W-CIN might drive tumorigenesis by promoting LOH of tumor suppressor genes, experimental evidence for this idea is still lacking. In one study, this idea was tested by breeding Bub3+/− mice onto Trp53 (transformation related protein 53; encoding p53) or Rb1 heterozygous null backgrounds [50]. In both cases, Bub3 haploinsufficiency failed to accelerate tumorigenesis, implying that W-CIN does not accelerate LOH of either tumor suppressor gene. It is possible that the level of W-CIN resulting from Bub3 deficiency mutant mice is too low to be effective in these mouse models. To settle this issue, it will be necessary to test additional W-CIN models for their ability to promote tumorigenesis through LOH of tumor suppressor genes. Finally, it should be noted that there are fundamental differences between the mechanisms of tumorigenesis in mice and humans [51]; therefore, the impact of W-CIN on tumorigenesis might be different in the two species.
W-CIN gene hierarchy
In recent years, studies using W-CIN mouse models have demonstrated that tumor formation is a possible, but not absolute, consequence of W-CIN and the ensuing aneuploidy. However, the reasons why certain W-CIN models that have similar levels of aneuploidy display such distinct spontaneous tumor predispositions remain puzzling. This phenomenon is not unique for W-CIN models; indeed, it has also been observed in mouse models for other genome maintenance systems. In particular, xeroderma pigmentosum, complementation group A (Xpa)- and xeroderma pigmentosum, complementation group E (Xpe)-deficient mice both have defective nucleotide excision repair (NER) functions and accumulate DNA damage at increased rates, but only Xpe-null animals are prone to a wide variety of spontaneous tumors [52]. By contrast, Xpa-null mice are highly susceptible to carcinogen-induced cancer, which mirrors the carcinogen sensitivity of some W-CIN models that are not prone to spontaneous tumors (Table 1).
What could explain the differences in tumorigenicity among W-CIN gene defects? One possibility is that the most potent tumorigenesis inhibitors among the W-CIN genes perform multiple tumor suppressive activities, similar to Trp53, whose protein product performs several functions linked to tumor suppression [e.g. DNA repair, cell cycle arrest, senescence, apoptosis and genomic stability (prevention of endoreduplication)] [53,54] (Figure 3). Indeed, APC is an example of a W-CIN gene that performs multiple tumor suppressive functions. APC disruption not only causes chromosome mis-segregation through weakened mitotic checkpoint control [48], destabilized kinetochore–microtubule attachment and cytokinesis failure [55], but also promotes uncontrolled proliferation through deregulated β-catenin signaling [56]. Bub1 also fulfills this criterion: aside from controlling accurate chromosome segregation, kinetochore–microtubule attachment and sister-chromatid cohesion [24,27], this mitotic checkpoint protein can also regulate cell death pathways, particularly in response to chromosome mis-segregation [24,57]. By contrast, W-CIN gene defects that display little or no tumor susceptibility might cause aneuploidy and also activate pathways that have antitumor activities (Figure 3). One such example might be Bub1b hypomorphism, which can impair cell proliferation and induce senescence [22,58]. Thus, it is important to determine which W-CIN genes are most relevant to human cancer. To address this question, it will be important to understand the entire spectrum of functions that individual W-CIN genes might have. Furthermore, the tumor suppressive potential of each W-CIN gene family member must be measured in mice.
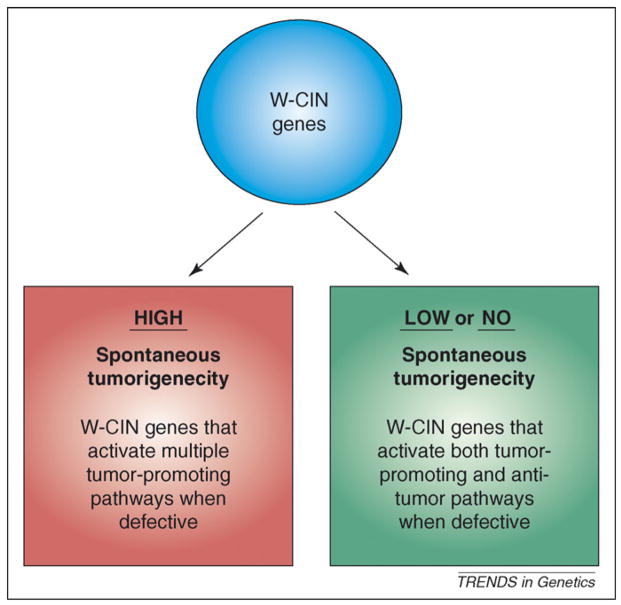
Figure 3.
Differences among whole chromosome instability (W-CIN) genes in their tumor suppressive activities. Defects in some W-CIN genes are more probable than others to promote transformation. Based on the currently available data, we hypothesize that two broad classes of W-CIN genes exist. Defects in W-CIN genes that result in high spontaneous tumorigenecity (red box) might activate multiple tumor-promoting activities. Examples of such defects are: Bub1 hypomorphism (causes aneuploidy and promotes cell survival); RanBP2 hypomorphism (causes aneuploidy and perhaps aberrant nucleocytoplasmic transport) and APC inactivation (causes aneuploidy and uncontrolled cell proliferation through β-catenin signaling). We also suggest that defects in W-CIN genes that have either little or no effect on spontaneous tumorigenecity (green box) might, when deregulated, activate both tumor-promoting and anti-tumor activities. For example, Bub1b hypomorphism causes aneuploidy but also induces senescence, a cellular mechanism that protects against tumorigenesis.
Tetraploidization: genetic basis and cancer relevance
Based on the nature of their numerical chromosomal changes, human cancers can be subdivided into two broad groups: (i) cancers with minor imbalances in chromosome copy number resulting in near-diploid DNA content (copy number abnormalities involving one or a few chromosomes) and (ii) cancers with vast changes in chromosome numbers (near tetraploid). The latter cancers are thought to originate from cells that incidentally become tetraploid [59]. These cells initially display poor growth properties but seem to improve their proliferative capacity by scrambling their chromosomes. Typically, fast growing progeny cells not only contain substantial amounts of extra chromosomes but also a multitude of structural chromosomal defects, including translocations, amplifications and deletions [60–62]. Whether numerical or structural chromosome instability, or both, drives malignant transformation of cells that become tetraploid is currently unknown. Indeed, it is a question that is difficult to address. We note that tetraploid cells can restore diploidy through a process of concerted chromosome loss, also referred to as reduction mitosis [59]. Thus, tetraploidization might only transiently perturb the euploid status in certain cells.
Two types of mitotic defects might cause cancer-related tetraploidization: failure of karyokinesis or cytokinesis (Figure 4). Karyokinesis failure might occur when cells encounter a serious mitotic spindle formation defect. Initially, the mitotic checkpoint will delay mitosis in pro-metaphase to provide time to correct the defect [12]. However, this checkpoint is active for only a limited period of time, and, when this time expires, (pro)metaphase cells are forced into G1 phase with 4N DNA content [60]. Because mitotic checkpoint gene defects substantially shorten the time of prometaphase arrest in response to aberrant kinetochore–microtubules attachment, they are expected to facilitate tetraploidization in rare situations where a substantial delay of anaphase onset would be necessary to properly repair aberrant kinetochore–microtubule attachments [63]. Whether mice with a defective mitotic checkpoint indeed accumulate higher rates of tetraploid cells has not been studied in great detail and should be considered in future studies.
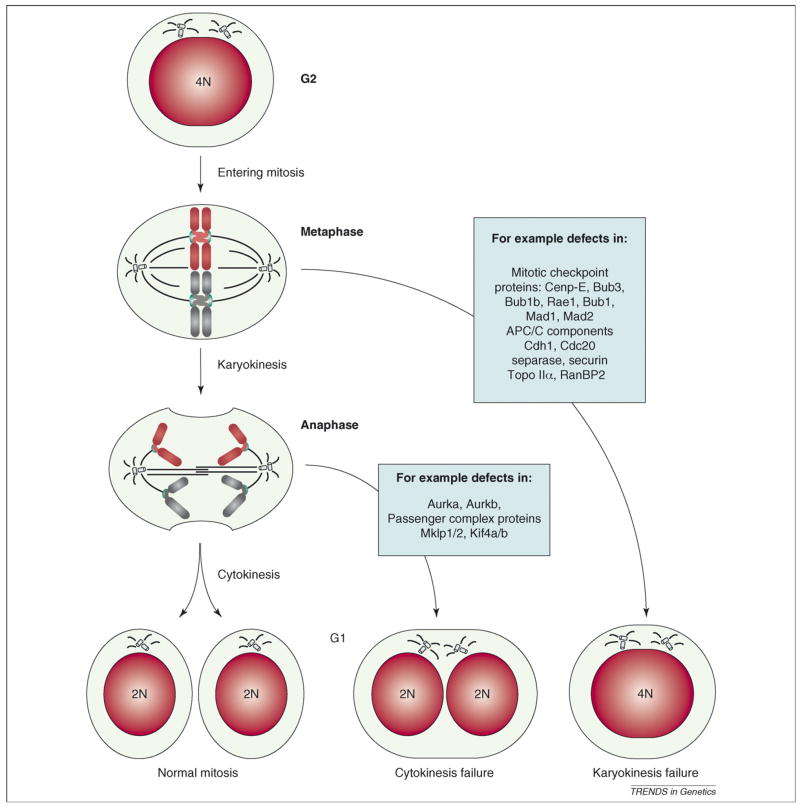
Figure 4.
Tetraploidization by mitotic failure. Normally a 4N cell in G2 enters mitosis, aligns its chromosomes in the metaphase plate and equally distributes the DNA over two nuclei (karyokinesis) and subsequently two daughter cells (cytokinesis). When karyokinesis fails, cells with 4N content are observed. When cytokinesis fails, the DNA is divided into two nuclei that remain within one cell. Mitotic genes that could have a role in failure of karyokinesis and cytokinesis are indicated (blue boxes). Note that cells undergoing karyokinesis and cytokinesis both inherit two centrosomes that could lead to abnormal spindles and chromosome mis-segregation during the next round of division (Box 1).
A second mechanism of karyokinesis failure involves the separation of sister chromatids [64]. Separation of duplicated chromosomes, which is required for anaphase onset, is an intricate process requiring APC/C activity [65], cohesin cleavage [64] and DNA decatenation [30]. Thus, defects in proteins that mediate key steps in this process, including APC/C components, separase [66] and Topo IIα30, might promote tetraploidization through karyokinesis failure.
Like mitosis, cytokinesis is a complex process whose underlying molecular mechanisms are just beginning to become understood [67]. Currently, ~50 genes are known to cause cytokinesis failure when defective [68]. These genes encode proteins that make up or are associated with actin filaments or microtubules or have known functions in karyokinesis or vesicular transport. One crucial next step toward understanding the role of cytokinesis failure in tumorigenesis will be to screen human tumors with massive chromosomal instability for alterations in genes known to regulate cytokinesis.
Several studies have provided direct evidence for the idea that tetraploidization plays a causal role in tumorigenesis. Fujiwara et al. [61] published an elegant experiment in which Trp53−/− mouse mammary gland epithelial cells, tetraploidized in culture by dihydrocytochalasin B treatment, developed mammary tumors in nude mice, whereas their nontreated diploid counterparts did not. Moreover, aurora kinase A (Aurka) overexpression, which causes tetraploidization through cytokinesis failure, is frequently observed in particular tumor types, including breast tumors [69]. A recent study showed that transgenic mice that overexpress Aurka in mammary glands are prone to breast tumors with high proportions of (near) tetraploid cells [70,71] (Table 1). The finding that tetraploid epithelial cells accumulate in transgenic mammary glands before breast tumors emerge supports the notion that tetraploidization is a cause of neoplastic growth. It will be important to create additional models of karyokinesis and cytokinesis failure to further define the role of tetraploidization in tumorigenesis and to identify the most crucial tumor suppressors among genes that guard against tetraploidization.
Cells undergoing tetraploidization inherit double the normal centrosome amount. Centrosome amplification is a hallmark of solid tumors and a known cause of mitotic spindle defects. Such defects might contribute to chromosome mis-segregation and expedite aneuploidization of tetraploid cells (Box 1).
Box 1. Centrosome amplification and aneuploidy development
In addition to chromosome number abnormalities, human tumors often also display centrosome amplification [72,73]. This finding led to the idea that centrosome amplification drives tumorigenesis by causing mitotic spindle abnormalities, inaccurate chromosome segregation and aneuploidy [74]. Centrosomes are microtubule organizing centers from which bipolar spindles emanate during mitosis [74]. At the beginning of G1 phase, cells contain a single centrosome that is composed of two centrioles, which duplicate in S phase. The two centriole pairs remain interconnected until mitosis onset, when they split and start to move apart and form a bipolar spindle.
The molecular basis of centrosome amplification remains poorly defined, but it is thought to occur via one of two distinct mechanisms [74]. The first scenario is that a cell acquires two centrosomes after failure to undergo karyokinesis or cytokinesis. In these instances, there is no partitioning of spindle poles or DNA into daughter cells. During the next cell cycle, these centrosomes again duplicate to form four spindle poles. The biological consequence of this could be a multipolar spindle, which generally is antagonistic to cell viability and growth. However, a rare cell might survive with an aberrant chromosomal complement capable of supporting growth [75]. The second model proposes that deregulation of kinases or tumor suppressors that contribute to centriole replication disrupt the normally once per cell cycle duplication of centrosomes. These include the Plk1, Aurka and Nek2 kinases and the p53, Brca1 and Bcra2 tumor suppressors [74].
Although centrosome amplification has been linked to aneuploidy and multipolar spindles, not all cells with supranumerary centrosomes will form multipolar spindles. In flies, the presence of extra centrosomes in the majority of somatic cells is compatible with normal development and survival [76]. However, extra centrosomes in the larval neural cells, which undergo asymmetric division, promotes metastatic tumorigenesis [76]. The observation that most cells can proliferate with little difficulty with extra centrosomes is consistent with a report demonstrating that extra centrosomes in nontransformed cells cluster during mitosis to prevent spindle multipolarity. At least one way to overcome this clustering mechanism to is disrupt the microtubule motor dynein [77]. Therefore, in addition to acquiring additional centrosomes, cells must seemingly also acquire a means to deregulate centrosome coalescence to generate multipolar spindles.
Concluding remarks and future perspectives
Mutant mice carrying gene defects that cause whole chromosome instability (W-CIN) are frequently prone to tumorigenesis, indicating that aneuploidy increases the organism’s risk of neoplastic transformation. Certain W-CIN gene defects pose a greater risk than others, although this difference does not seem to correlate with the degree of aneuploidy. We propose that W-CIN gene defects that perturb not only high-fidelity chromosome segregation, but also other tumor suppressive functions, constitute a greater cancer risk than those that engage antitumor mechanisms in addition to causing aneuploidy. Furthermore, in tissue where antitumor effects dominate tumor-promoting effects, W-CIN gene mutations might suppress tumor development.
Because defects in genes that prevent W-CIN do not phenocopy each other, a thorough analysis of all known W-CIN genes is required to determine the contribution of each to tumorigenesis. It will also be important to dissect the totality of tumor suppressive functions of W-CIN genes as well as the hierarchical relationship among various W-CIN gene defects. DNA mutagens cooperate with most W-CIN gene defects in tumorigenesis, but the underlying cancer gene mutations remain largely unknown. A major challenge for the future will therefore be to discover the genetic contexts in which W-CIN gene defects accelerate malignant transformation. Mouse modeling has mainly focused on the downregulation of mitotic checkpoint genes, but we might also consider investigating the effects of overexpressing W-CIN genes. Indeed, Mad2 transgenic mice are tumor prone. Thus, it is important to determine whether the consequence of overexpression of other mitotic checkpoint genes similarly results in a Mad2-like phenotype or whether, as observed for downregulation, phenotypes vary among different family members.
Acknowledgments
We apologize for omitting citations of numerous primary papers because of space limitations. We thank Darren Baker, Meelad Dawlaty, Scott Kaufmann and Liviu Malureanu for comments on the manuscript and critical discussion. We acknowledge funding from the National Institutes of Health (CA-96985; CA-126828 and CA-77262).
Glossary
- Aneuploidy
designates a karyotype that differs from diploid because of changes in the normal copy numbers of chromosomes
- Loss of heterozygosity (LOH)
loss of the normal functioning allele of a gene when the other allele has already been inactivated
- Mitotic checkpoint
molecular surveillance mechanism that guards against the inaccurate segregation of chromosomes during cell division by delaying the anaphase transition until all kinetochores are aligned at the metaphase plate and properly attached to the mitotic spindle
- Neoplastic growth
cell growth that is uncontrolled and uncoordinated with the growth of surrounding tissue
- Structural (or segmental) chromosomal instability (S-CIN)
the acquisition of chromosomes that have undergone changes to the structure of the chromosome, including translocations, deletions and amplifications of large portions of chromosomes
- Whole chromosomal instability (W-CIN)
referring to a condition in which cells are unable to segregate duplicated chromosomes with high accuracy during mitosis and, as a result, gain or lose whole chromosomes at high frequency
References
- 1.Boveri T. Zur Frage der Entstehung Maligner Tumoren. Gustav Fisher; 1914. [Google Scholar]
- 2.Boveri T. Uber mehrpolige Mitosen als Mittel zur Analyse der Zelkerns. Vehr d phys Med Ges zu Wurzburg Neu Folge. 1902;35:67–90. [Google Scholar]
- 3.Duesberg P, Li R. Multistep carcinogenesis: a chain reaction of aneuploidizations. Cell Cycle. 2003;2:202–210. [PubMed] [Google Scholar]
- 4.Zimonjic D, et al. Derivation of human tumor cells in vitro without widespread genomic instability. Cancer Res. 2001;61:8838–8844. [PubMed] [Google Scholar]
- 5.Michor F, et al. Can chromosomal instability initiate tumorigenesis? Semin Cancer Biol. 2005;15:43–49. doi: 10.1016/j.semcancer.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- 7.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 8.Lengauer C, et al. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 9.Geigl JB, et al. Defining ‘chromosomal instability’. Trends Genet. 2008;24:64–69. doi: 10.1016/j.tig.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 10.Maser RS, DePinho RA. Take care of your chromosomes lest cancer take care of you. Cancer Cell. 2003;3:4–6. doi: 10.1016/s1535-6108(02)00243-x. [DOI] [PubMed] [Google Scholar]
- 11.Rajagopalan H, et al. The significance of unstable chromosomes in colorectal cancer. Nat Rev Cancer. 2003;3:695–701. doi: 10.1038/nrc1165. [DOI] [PubMed] [Google Scholar]
- 12.Kops GJ, et al. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 13.Kolodner RD, et al. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- 14.Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- 15.Barber TD, et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci U S A. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker DJ, et al. The mitotic checkpoint in cancer and aging: what have mice taught us? Curr Opin Cell Biol. 2005;17:583–589. doi: 10.1016/j.ceb.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 17.Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr, Opin Cell Biol. 2006;18:658–667. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Putkey FR, et al. Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev Cell. 2002;3:351–365. doi: 10.1016/s1534-5807(02)00255-1. [DOI] [PubMed] [Google Scholar]
- 19.Kalitsis P, et al. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 2000;14:2277–2282. doi: 10.1101/gad.827500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobles M, et al. Chromosome mis-segregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–645. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- 21.Iwanaga Y, et al. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 2007;67:160–166. doi: 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- 22.Baker DJ, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 23.Wang Q, et al. BUBR1 deficiency results in abnormal megakaryopoiesis. Blood. 2004;103:1278–1285. doi: 10.1182/blood-2003-06-2158. [DOI] [PubMed] [Google Scholar]
- 24.Jeganathan K, et al. Bub1 mediates cell death in response to chromosome mis-segregation and acts to suppress spontaneous tumorigenesis. J Cell Biol. 2007;179:255–267. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weaver BA, et al. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Michel LS, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 27.Perera D, et al. Bub1 maintains centromeric cohesion by activation of the spindle checkpoint. Dev Cell. 2007;13:566–579. doi: 10.1016/j.devcel.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 28.Babu JR, et al. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome mis-segregation. J Cell Biol. 2003;160:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeganathan KB, et al. The Rae1-Nup98 complex prevents aneuploidy by inhibiting securin degradation. Nature. 2005;438:1036–1039. doi: 10.1038/nature04221. [DOI] [PubMed] [Google Scholar]
- 30.Dawlaty MM, et al. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–115. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, et al. Mice lacking pituitary tumor transforming gene show testicular and splenic hypoplasia, thymic hyperplasia, thrombocytopenia, aberrant cell cycle progression, and premature centromere division. Mol Endocrinol. 2001;15:1870–1879. doi: 10.1210/mend.15.11.0729. [DOI] [PubMed] [Google Scholar]
- 32.Mei J, et al. Securin is not required for cellular viability, but is required for normal growth of mouse embryonic fibroblasts. Curr Biol. 2001;11:1197–1201. doi: 10.1016/s0960-9822(01)00325-6. [DOI] [PubMed] [Google Scholar]
- 33.Yu X, et al. Chfr is required for tumor suppression and Aurora A regulation. Nat Genet. 2005;37:401–406. doi: 10.1038/ng1538. [DOI] [PubMed] [Google Scholar]
- 34.Privette LM, et al. Loss of CHFR in human mammary epithelial cells causes genomic instability by disrupting the mitotic spindle assembly checkpoint. Neoplasia. 2008;10:643–652. doi: 10.1593/neo.08176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeganathan KB, et al. Securin associates with APC(Cdh1) in prometaphase but its destruction is delayed by Rae1 and Nup98 until the metaphase/anaphase transition. Cell Cycle. 2006;5:366–370. doi: 10.4161/cc.5.4.2483. [DOI] [PubMed] [Google Scholar]
- 36.Baker DJ, et al. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J Cell Biol. 2006;172:529–540. doi: 10.1083/jcb.200507081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeganathan KB, van Deursen JM. Differential mitotic checkpoint protein requirements in somatic and germ cells. Biochem Soc Trans. 2006;34:583–586. doi: 10.1042/BST0340583. [DOI] [PubMed] [Google Scholar]
- 38.Hernando E, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- 39.Sotillo R, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren B, et al. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao CV, et al. Colonic tumorigenesis in BubR1+/− ApcMin/+ compound mutant mice is linked to premature separation of sister chromatids and enhanced genomic instability. Proc Natl Acad Sci U S A. 2005;102:4365–4370. doi: 10.1073/pnas.0407822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chesnokova V, et al. Pituitary hypoplasia in Pttg−/− mice is protective for Rb+/− pituitary tumorigenesis. Mol Endocrinol. 2005;19:2371–2379. doi: 10.1210/me.2005-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weaver BA, Cleveland DW. Aneuploidy: instigator and inhibitor of tumorigenesis. Cancer Res. 2007;67:10103–10105. doi: 10.1158/0008-5472.CAN-07-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 45.Duesberg P, et al. The chromosomal basis of cancer. Cell Oncol. 2005;27:293–318. doi: 10.1155/2005/951598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai W, et al. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res. 2004;64:440–445. doi: 10.1158/0008-5472.can-03-3119. [DOI] [PubMed] [Google Scholar]
- 47.Baker DJ, et al. Opposing roles for p16 Ink4a and p19 Arf in senescence and ageing caused by BubR1 insufficiency. Nat Cell Biol. 2008;10:825–836. doi: 10.1038/ncb1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dikovskaya D, et al. Loss of APC induces polyploidy as a result of a combination of defects in mitosis and apoptosis. J Cell Biol. 2007;176:183–195. doi: 10.1083/jcb.200610099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaplan KB, et al. A role for the Adenomatous Polyposis Coli protein in chromosome segregation. Nat Cell Biol. 2001;3:429–432. doi: 10.1038/35070123. [DOI] [PubMed] [Google Scholar]
- 50.Kalitsis P, et al. Increased chromosome instability but not cancer predisposition in haploinsufficient Bub3 mice. Genes Chromosomes Cancer. 2005;44:29–36. doi: 10.1002/gcc.20215. [DOI] [PubMed] [Google Scholar]
- 51.Rangarajan A, Weinberg RA. Opinion: comparative biology of mouse versus human cells: modelling human cancer in mice. Nat Rev Cancer. 2003;3:952–959. doi: 10.1038/nrc1235. [DOI] [PubMed] [Google Scholar]
- 52.Wijnhoven SW, et al. Tissue specific mutagenic and carcinogenic responses in NER defective mouse models. Mutat Res. 2007;614:77–94. doi: 10.1016/j.mrfmmm.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 53.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 54.Cross SM, et al. A p53-dependent mouse spindle checkpoint. Science. 1995;267:1353–1356. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- 55.Caldwell CM, et al. APC mutations lead to cytokinetic failures in vitro and tetraploid genotypes in Min mice. J Cell Biol. 2007;178:1109–1120. doi: 10.1083/jcb.200703186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aoki K, Taketo MM. Adenomatous polyposis coli (APC): a multi-functional tumor suppressor gene. J Cell Sci. 2007;120:3327–3335. doi: 10.1242/jcs.03485. [DOI] [PubMed] [Google Scholar]
- 57.Niikura Y, et al. BUB1 mediation of caspase-independent mitotic death determines cell fate. J Cell Biol. 2007;178:283–296. doi: 10.1083/jcb.200702134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baker DJ, et al. Opposing roles for p16 Ink4a and p19 Arf in senescence and aging caused by BubR1 insufficiency. Nat Cell Biol. 2008;10:825–836. doi: 10.1038/ncb1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol. 2004;5:45–54. doi: 10.1038/nrm1276. [DOI] [PubMed] [Google Scholar]
- 60.Rieder CL, Maiato H. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev Cell. 2004;7:637–651. doi: 10.1016/j.devcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 61.Fujiwara T, et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 62.Ganem NJ, et al. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 63.Baker DJ, et al. Mitotic regulation of the anaphase-promoting complex. Cell Mol Life Sci. 2007;64:589–600. doi: 10.1007/s00018-007-6443-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- 65.Pesin JA, Orr-Weaver TL. Regulation of APC/C activators in mitosis and meiosis. Annu Rev Cell Dev Biol. 2008;24:475–499. doi: 10.1146/annurev.cellbio.041408.115949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wirth KG, et al. Separase: a universal trigger for sister chromatid disjunction but not chromosome cycle progression. J Cell Biol. 2006;172:847–860. doi: 10.1083/jcb.200506119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131:847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 68.Eggert US, et al. Animal cytokinesis: from parts list to mechanisms. Annu Rev Biochem. 2006;75:543–566. doi: 10.1146/annurev.biochem.74.082803.133425. [DOI] [PubMed] [Google Scholar]
- 69.Giet R, et al. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol. 2005;15:241–250. doi: 10.1016/j.tcb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 70.Wang X, et al. Overexpression of aurora kinase A in mouse mammary epithelium induces genetic instability preceding mammary tumor formation. Oncogene. 2006;25:7148–7158. doi: 10.1038/sj.onc.1209707. [DOI] [PubMed] [Google Scholar]
- 71.Zhang D, et al. Cre-loxP-controlled periodic Aurora-A overexpression induces mitotic abnormalities and hyperplasia in mammary glands of mouse models. Oncogene. 2004;23:8720–8730. doi: 10.1038/sj.onc.1208153. [DOI] [PubMed] [Google Scholar]
- 72.Lingle WL, et al. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci U S A. 2002;99:1978–1983. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.D’Assoro AB, et al. Centrosome amplification and the development of cancer. Oncogene. 2002;21:6146–6153. doi: 10.1038/sj.onc.1205772. [DOI] [PubMed] [Google Scholar]
- 74.Nigg EA. Origins and consequences of centrosome aberrations in human cancers. Int J Cancer. 2006;119:2717–2723. doi: 10.1002/ijc.22245. [DOI] [PubMed] [Google Scholar]
- 75.Brinkley BR. Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol. 2001;11:18–21. doi: 10.1016/s0962-8924(00)01872-9. [DOI] [PubMed] [Google Scholar]
- 76.Basto R, et al. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quintyne NJ, et al. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–129. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- 78.Garci-Higuera I, et al. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat Cell Biol. 2008;10:802–811. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]