Abstract
Expression of p16Ink4a and p19Arf increases with age in both rodent and human tissues. However, whether these tumour suppressors are effectors of ageing remains unclear, mainly because knockout mice lacking p16Ink4a or p19Arf die early of tumours. Here, we show that skeletal muscle and fat, two tissues that develop early ageing-associated phenotypes in response to BubR1 insufficiency, have high levels of p16Ink4a and p19Arf. Inactivation of p16Ink4a in BubR1-insufficient mice attenuates both cellular senescence and premature ageing in these tissues. Conversely, p19Arf inactivation exacerbates senescence and ageing in BubR1 mutant mice. Thus, we identify BubR1 insufficiency as a trigger for activation of the Cdkn2a locus in certain mouse tissues, and demonstrate that p16Ink4a is an effector and p19Arf an attenuator of senescence and ageing in these tissues.
Cellular senescence is a state of irreversible growth arrest that can be induced by various cellular stressors1,2. The Cdkn2a locus encodes two separate tumour suppressors, p16Ink4a (A001711), a cyclin-dependent kinase (Cdk) inhibitor that can block G1–S progression when present above a certain level, and p19Arf (A001713), a positive regulator of the transcription factor p53 that integrates and responds to a wide variety of cellular stresses1,3–5. Both p16Ink4a and p19Arf are effectors of senescence in cultured cells6 and their levels increase with ageing in many tissues7,8. This has led to speculation that their induction is causally implicated in in vivo senescence and organismal ageing. However, rigorous testing of this notion has been difficult because mice that lack p16Ink4a or p19Arf die of cancer long before they reach the age at which normal mice start to develop age-related disorders1,2. Recent evidence in middle-aged p16Ink4a knockout mice indicates that the age-induced expression of p16Ink4a limits the proliferative and regenerative capacity of progenitor populations9–11. Yet, whether the increased stem-cell proliferation and tissue regeneration seen in p16Ink4a knockouts actually delay onset of age-related pathologies remains unknown because of the limited animal lifespan1,12.
One approach to study the role of p16Ink4a and p19Arf in ageing would be to determine whether their respective inactivation by single gene mutations, in mouse models that develop ageing-associated pathologies at an early age, would prevent or delay premature ageing. Mutant mice with low levels of the mitotic checkpoint protein BubR1 (called BubR1 hypomorphic or BubR1H/H mice, A003172) undergo premature separation of sister chromosomes and develop progressive aneuploidy along with various progeroid phenotypes that include short lifespan, cachectic dwarfism, lordokyphosis (abnormal curvature of the spine), sarcopaenia (age-related skeletal muscle atrophy), cataracts, craniofacial dysmorphisms, arterial stiffening, loss of (subcutaneous) fat, reduced stress tolerance and impaired wound healing13–15. During the course of natural ageing, several mouse tissues show a marked decline in BubR1 protein expression, which, combined with the observation that BubR1H/H mice age prematurely, suggests a possible role for BubR1 in regulating natural ageing13–15. Here we show that certain mouse tissues induce p16Ink4a and p19Arf in response to BubR1 hypomorphism. Using BubR1H/H mice in which these tumour suppressors are lacking, we have demonstrated that p16Ink4a is an effector of cellular senescence and ageing, whereas, p19Arf acts to suppress cellular senescence and ageing.
RESULTS
p16Ink4a inactivation increases the lifespan of BubR1H/H mice
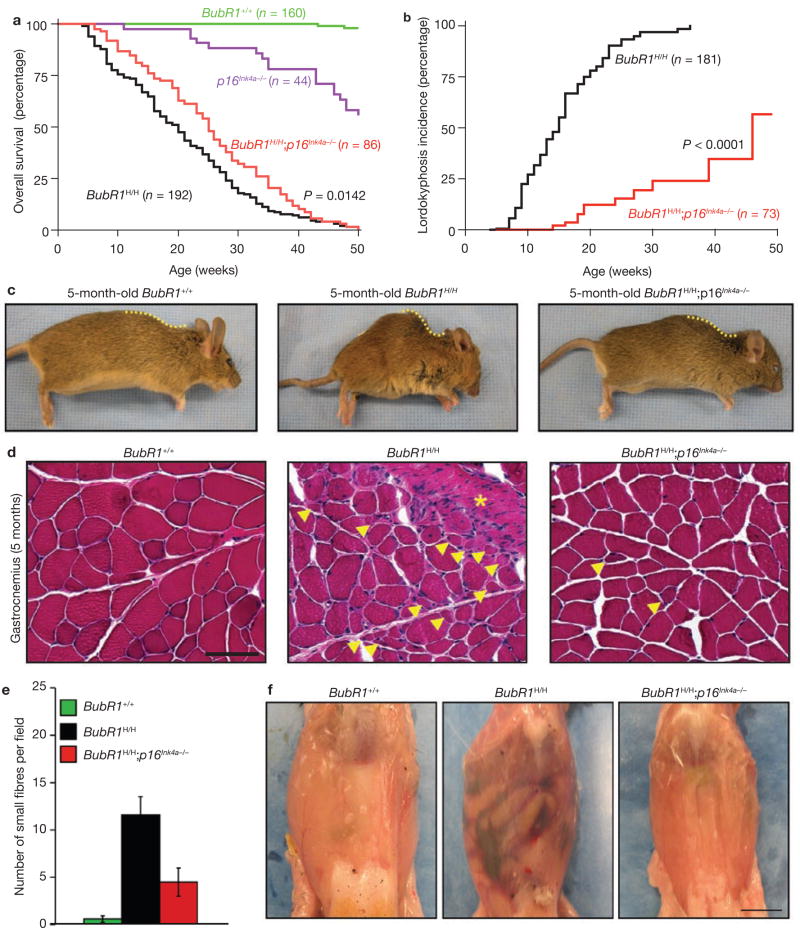
To determine the requirement for p16Ink4a in the development of progeroid phenotypes in BubR1-insufficient mice, we bred BubR1H/H mice on a p16Ink4a homozygous-null genetic background. In total, 86 BubR1H/H;p16Ink4a−/−, 192 BubR1H/H, 160 BubR1+/+ and 44 p16Ink4a−/− mice were generated and monitored for development of age-related phenotypes for a period of one year. Inactivation of p16Ink4a extended the lifespan of BubR1H/H mice by 25% (Fig. 1a). Although the median lifespan of BubR1H/H mice was extended in the absence of p16Ink4a, the maximum lifespan was not, suggesting that the condition(s) that cause(s) death was not rescued by p16Ink4a inactivation.
Figure 1.
Ablation of p16Ink4a in BubR1H/H mice extends lifespan and attenuates sarcopaenia. (a) Overall survival curves for wild-type, p16Ink4a−/−, BubR1H/H and BubR1H/H;p16Ink4a−/− mice. The median overall survival of combined BubR1H/H;p16Ink4a−/− mice is 25 weeks, a 25% extension in lifespan compared with BubR1H/H animals. We note that the p16Ink4a−/−, BubR1H/H and BubR1H/H;p16Ink4a−/− curves are all significantly different from the wild-type (BubR1+/+) curve (P < 0.0001, log-rank tests). Moreover, the BubR1H/H;p16Ink4a−/− curve is significantly different from the BubR1H/H curve (P = 0.0142). (b) Incidence and latency of lordokyphosis in BubR1H/H and BubR1H/H;p16Ink4a−/− mice. The curves are significantly different (P < 0.0001, log-rank test). We note that no wild-type or p16Ink4a−/− mice developed lordokyphosis during our one-year observation period (data not shown). (c) Images of 5-month-old wild-type, BubR1H/H and BubR1H/H;p16Ink4a−/− mice. Note the profound difference in the curvature of the spine in the BubR1H/H;p16Ink4a−/− mouse. (d) Cross-sections of gastrocnemius muscles from 5-month-old wild-type, BubR1H/H and BubR1H/H;p16Ink4a−/− mice. Arrowheads mark degenerated fibres and asterisks mark areas of connective tissue infiltration. Scale bar is 100 μm. (e) Quantification of the number of deteriorating (atrophic) muscle fibres in gastrocnemius muscles shown in d. Note that BubR1H/H;p16Ink4a−/− muscles have 3-fold less atrophic fibres than BubR1H/H muscles. Data are mean ± s.d. (n = 4). (f) Skinned 5-month-old wild-type, BubR1H/H and BubR1H/H;p16Ink4a−/− mice demonstrating that abdominal wall thickness is visually increased in BubR1H/H;p16Ink4a−/− mice when compared with BubR1H/H animals. Scale bar is 1 cm.
p16Ink4a loss blunts sarcopaenia induced by BubR1 insufficiency
A prominent ageing-associated phenotype of BubR1H/H mice is the development of lordokyphosis13. The incidence of this phenotype was markedly reduced in BubR1H/H;p16Ink4a−/− animals when compared with BubR1H/H mice (Fig. 1b, c). Furthermore, the median time to onset of lordokyphosis was three times longer in BubR1H/H;p16Ink4a−/− mice than in BubR1H/H mice (Fig. 1b). Lordokyphosis is associated with both osteoporosis and age-related degenerative loss of muscle mass and strength (sarcopaenia) in wild-type mice of extremely advanced age16. Histological evaluation of longitudinal femur sections from kyphotic BubR1H/H mice revealed no evidence for osteoporosis (Supplementary Information, Fig. S1a, b). Histopathology on gastrocnemius and paraspinal muscles of 5-month-old BubR1H/H mice, however, revealed clear signs of skeletal muscle atrophy and degeneration (Fig. 1d and data not shown). Muscle degeneration was greatly reduced in BubR1H/H muscles lacking p16Ink4a (Fig. 1d, e). In addition, abdominal muscles of BubR1H/H mice were poorly developed, as revealed by macroscopic analysis and magnetic resonance imaging (Fig. 1f; Supplementary Information, Fig. S1c). Depletion of p16Ink4a resulted in substantial correction of this defect. These data demonstrate that p16Ink4a has a major role in establishing sarcopaenia in BubR1H/H mice.
p16Ink4a limits the regenerative capacity of β cells and has been linked to pancreatic islet atrophy and development of diabetes9,17,18, which in turn can cause muscle atrophy through accelerated degradation of muscle protein19. This prompted us to test whether the sarcopaenia observed in BubR1H/H mice might be due to β cell failure. BubR1H/H mice showed highly efficient glucose clearance in a glucose-tolerance test (Supplementary Information, Fig. S2a). Complementary blood insulin measurements indicated that insulin sensitivity was not impaired in BubR1H/H mice and showed no evidence for insulin resistance (Supplementary Information, Fig. S2b). Furthermore, overall pancreatic morphology, as well as islet size, shape and abundance were similar in 12-month-old BubR1H/H and control mice, as verified by histology (Supplementary Information, Fig. S2c). Consistently, p16Ink4a expression in the pancreas was not significantly elevated in BubR1H/H mice, compared with BubR1+/+ counterparts. Thus, sarcopaenia in BubR1H/H mice is unlikely to be caused by p16Ink4a-mediated β cell degeneration or insulin resistance.
BubR1 and p16Ink4a levels are inversely linked in skeletal muscle
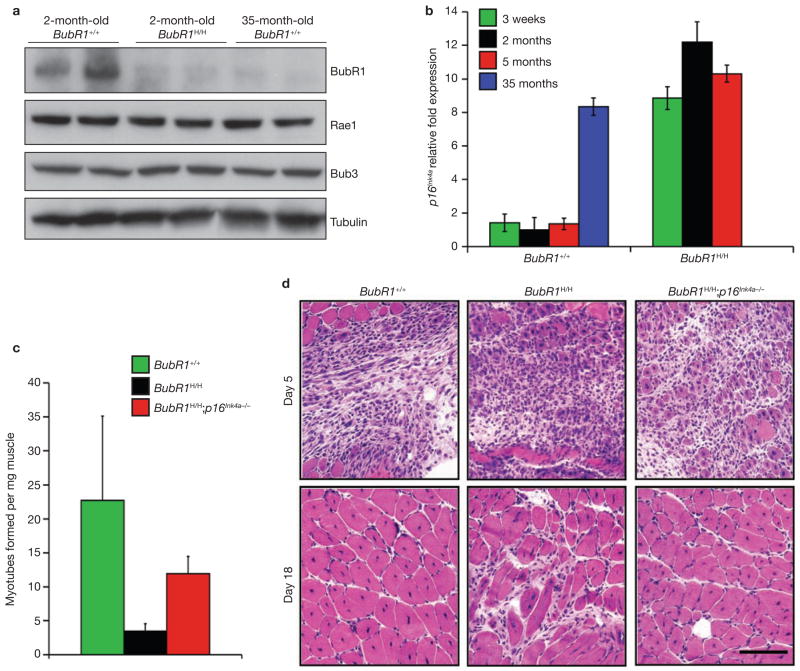
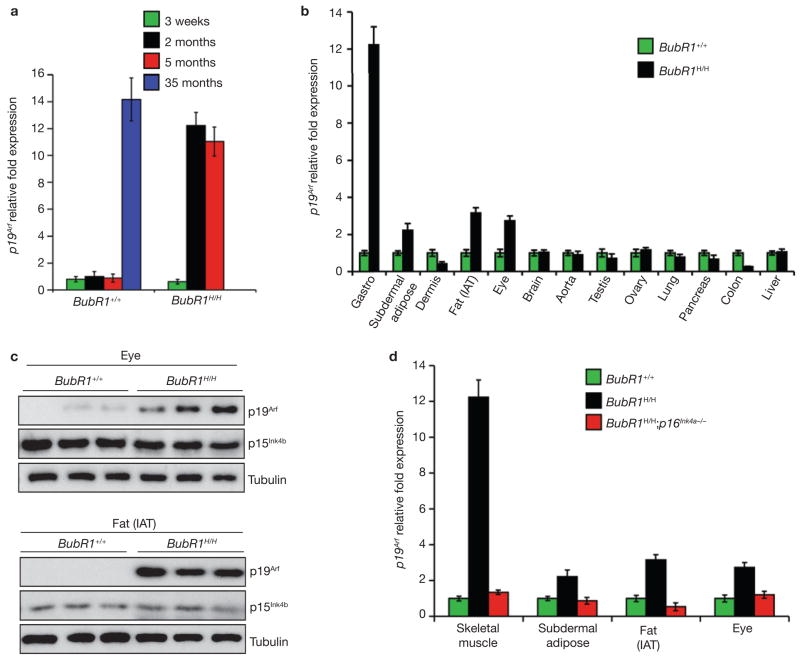
To determine whether BubR1 may have a role in normal skeletal muscle ageing, we measured BubR1 protein levels in skeletal muscle of young and old wild-type mice by western blot analysis. Gastrocnemius muscles had considerably higher levels of BubR1 protein at 2 months than at 35 months of age (Fig. 2a; Supplementary Information, Fig. S6a). BubR1 transcripts were undetectable by qRT–PCR in the gastrocnemius of 35-month-old mice but were readily present at 2 months (data not shown), suggesting that reduced BubR1 transcriptional activity contributes to the decline in BubR1 protein levels at advanced age. In contrast to BubR1 transcription, p16Ink4a transcription increased markedly with age in gastrocnemius muscles of old wild-type mice (Fig. 2b). Gastrocnemius of 2- and 5-month-old BubR1H/H mice also had high p16Ink4a transcript levels (Fig. 2b), providing evidence for an inverse relationship between BubR1 and p16Ink4a expression. To characterize this relationship further, we measured p16Ink4a expression in gastrocnemius of 3-week-old BubR1H/H mice, when skeletal muscle atrophy is histologically undetectable (Supplementary Information, Fig. S2d). Transcript levels of p16Ink4a were similarly elevated for 3-week-old, and 2- and 5-month-old mice (Fig. 2b), indicating that p16Ink4a induction is an early response to BubR1 hypomorphism that precedes histological signs of sarcopaenia.
Figure 2.
Inverse correlation between BubR1 and p16Ink4a expression levels with ageing. (a) Western blot analysis of gastrocnemius muscle in young wild-type and BubR1H/H mice and old wild-type mice. Blots were probed with antibodies against BubR1, Bub3 and Rae1. Anti-tubulin was used as a loading control. Note that the mitotic checkpoint proteins Bub3 and Rae1 remain highly expressed as wild-type mice age. Uncropped images of the scans are shown in Supplementary Information, Fig. S6a. (b) p16Ink4a expression in wild-type and BubR1H/H gastrocnemius muscles at various ages analysed by qRT–PCR. Data are mean ± s.d. (n = 3 males per genotype and age group, with triplicate measurements taken). Values were normalized to GAPDH. Relative fold expression is to 2-month-old wild-type values. (c) Myotube formation potential of gastrocnemius muscles from 5-month-old mice of the indicated genotypes analysed by a well-standardized in vitro assay. Data are mean ± s.d. (n = 4). (d) Cardiotoxin-treated gastrocnemius muscle of 5-month-old wild-type, BubR1H/H and BubR1H/H;p16Ink4a−/− mice at 5 or 18 days after injection. Note that all gastrocnemius muscles show an extensive hypercellular response to cardiotoxin injection by day 5 regardless of genotype. Wild-type and BubR1H/H;p16Ink4a−/− mice have complete restoration of muscle architecture by myofibres with central nuclei by day 18, whereas BubR1H/H mice have been unable to restore normal tissue structure. Scale bar is 100 μm.
Increased expression of p16Ink4a with age in adult stem cells is associated with reduced tissue repair and regeneration in several mouse tissues9–12. To explore whether p16Ink4a-mediated exhaustion of myogenic stem-cell potential might contribute to premature sarcopaenia in BubR1H/H mice, in vitro myoblast-to-myofibre differentiation assays were performed on gastrocnemius muscles from 5-month-old wild-type, BubR1H/H and BubR1H/H;p16Ink4a−/− mice. In these assays, the average number of myotubes obtained per milligram of muscle tissue was about 7-fold lower in BubR1H/H mice than in wild-type mice (Fig. 2c). By contrast, only a 2-fold reduction in myotube formation was observed in BubR1H/H mice lacking p16Ink4a. To confirm these data, 5-month-old wild-type, BubR1H/H and BubR1H/H;p16Ink4a−/− mice were challenged to regenerate muscle fibres by injection of cardiotoxin, a 60-amino-acid polypeptide that causes acute injury by rapidly destroying muscle fibres20. Consistent with our in vitro data, muscle regeneration was overtly delayed in BubR1H/H mice, but not in BubR1H/H;p16Ink4a−/− counterparts (Fig. 2d). Collectively, these data indicate that p16Ink4a promotes sarcopaenia in BubR1H/H mice, at least in part, by impairing muscle regeneration.
p16Ink4a loss attenuates ageing in selective BubR1 hypomorphic tissues
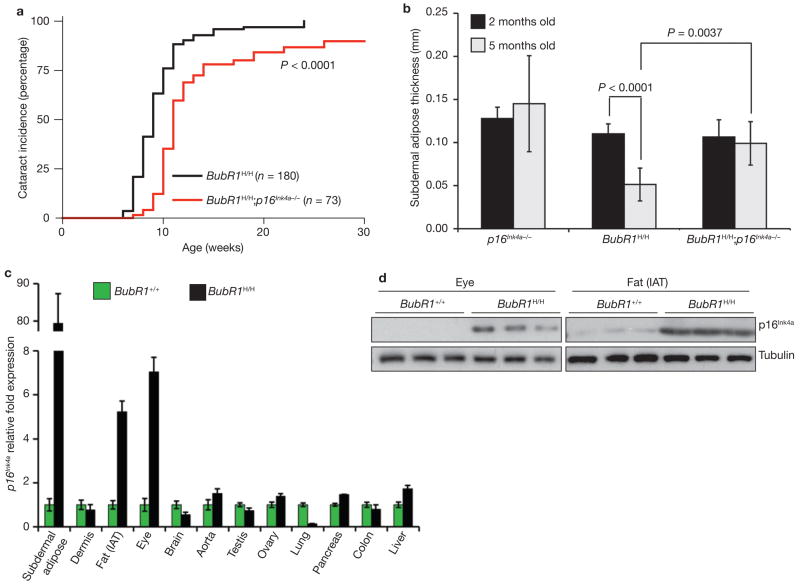
Loss of p16Ink4a caused a modest, yet significant, delay in the latency of cataract formation in BubR1H/H mice (Fig. 3a). Aged skin is characterized by reduced dermal thickness and subcutaneous adipose tissue, both of which are observed in BubR1H/H mice at young ages13. At 2 months of age, BubR1H/H, BubR1H/H;p16Ink4a−/− and p16Ink4a−/− mice had similar amounts of subdermal adipose tissue (Fig. 3b). As expected, the mean thickness of the subcutaneous adipose layer decreased notably in 5-month-old BubR1H/H mice. This decline was not accompanied by increased fat storage in liver tissue (Supplementary Information, Fig. S2e). The decrease in subcutaneous fat was much less severe in age-matched BubR1H/H;p16Ink4a−/− mice (Fig. 3b), indicating that p16Ink4a is, at least in part, responsible for loss of subcutaneous adipose tissue in BubR1H/H mice. Tolerance of anaesthetic stress was also greatly improved in BubR1H/H;p16Ink4a−/− mice (Supplementary Information, Table S1), as was adipose tissue deposition (Supplementary Information, Fig. S2f). However, several progeroid symptoms seen in BubR1H/H mice remained unchanged following loss of p16Ink4a, including dwarfism, dermal thinning, arterial wall stiffening and infertility (Supplementary Information, Table S1 and data not shown). No progeroid phenotypes of BubR1H/H mice were aggravated by p16Ink4a loss.
Figure 3. p16Ink4a disruption attenuates selective progeroid features of BubR1 hypomorphic mice.
(a) Incidence and latency of cataract formation in BubR1H/H and BubR1H/H;p16Ink4a−/− mice as detected by the use of slit light after dilation of eyes. The curves are significantly different (P < 0.0001, log-rank test). We note that no wild-type or p16Ink4a−/− mice developed cataracts during this observation period. (b) Subcutaneous adipose layer thickness of p16Ink4a−/−, BubR1H/H and BubR1H/H;p16Ink4a−/− mice at 2 and 5 months of age. Data are mean ± s.d. (n = 4 male mice for each age per genotype). A two-tailed Mann-Whitney test was used for statistical analysis. (c) qRT–PCR analysis for relative expression of p16Ink4a in a variety of 2-month-old tissues from BubR1H/H and wild-type mice. Values were normalized to GAPDH, and relative fold is to 2-month-old wild-type samples. Data are mean ± s.d. (n = 3 male mice for each tissue, with triplicate measurements taken). (d) Western blots of eye and fat extracts from 2-month-old BubR1+/+ and BubR1H/H mice probed with anti-p16Ink4a antibody. Anti-tubulin antibody served as loading control. Uncropped images of the scans are shown in Supplementary Information, Fig. S6b, c.
The differential corrective effects of p16Ink4a disruption on individual progeroid phenotypes suggest tissue-specific differences in engagement of the p16Ink4a pathway in the cellular response to BubR1 deficiency. BubR1H/H tissues in which p16Ink4a loss causes a significant delay of premature ageing, such as eye and (subdermal) adipose tissue, showed strong induction of p16Ink4a expression in response to BubR1 hypomorphism (Fig. 3c, d; Supplementary Information, Fig. S6b, c). BubR1H/H tissues in which p16Ink4a inactivation has no discernible corrective effect, such as dermis, brain, aorta, testis and ovary, did not exhibit significant p16Ink4a induction (Fig. 3c and data not shown). Furthermore, mutant tissues that are not subjected to premature ageing, including lung, pancreas, colon and liver13, maintained low p16Ink4a expression levels. Together, these data demonstrate that p16Ink4a is activated in a subset of tissues in BubR1H/H mice, where it contributes to progeroid phenotypes.
p16Ink4a loss attenuates in vivo senescence
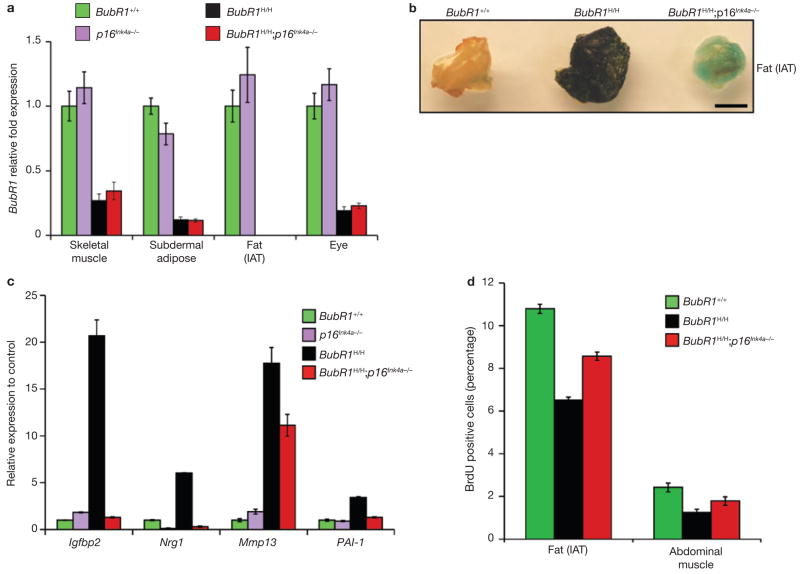
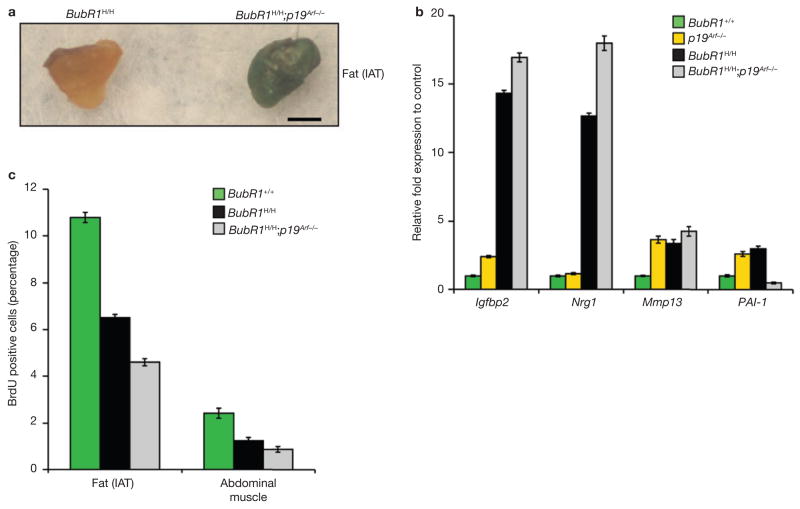
BubR1 is a putative E2F-regulated gene21 and loss of p16Ink4a leads to increased E2F transcriptional activity22. Accordingly, attenuation of ageing in skeletal muscle, fat and eye may be the result of increased BubR1 gene expression. However, this is unlikely as BubR1 transcript levels in these tissues were not affected by loss of p16Ink4a (Fig. 4a). As p16Ink4a is an effector of cellular senescence, p16Ink4a deletion may delay ageing in hypomorphic mice by decreasing senescence. As shown in Fig. 4b, BubR1H/H adipose tissue expresses high levels of senescence-associated (SA)-β-galactosidase, a marker of cellular senescence23. SA-β-galactosidase staining was much lower in adipose tissue of BubR1H/H;p16Ink4a−/− mice (Fig. 4b). Skeletal muscles of 2-month-old BubR1H/H mice did not stain positive for SA-β-galactosidase but expressed high levels of several other senescence-associated genes, including Igfbp2, Nrg1, Mmp13 and PAI-124–27 (Fig. 4c). Expression of these markers was decreased markedly in skeletal muscles of age-matched BubR1H/H;p16Ink4a−/− mice. A key feature of senescence is loss of proliferative potential. In vivo 5-bromo-2-de-oxyuridine (BrdU) labelling showed that 2-month-old BubR1H/H mice had much lower percentages of ycling cells in skeletal muscle and fat than wild-type mice (Fig. 4d). These reductions were less profound in BubR1H/H;p16Ink4a−/− mice. Collectively, these data suggest that BubR1 hypomorphism causes cellular senescence in adipose tissue and skeletal muscle through a p16Ink4a-dependent mechanism. As p16Ink4a inactivation attenuates both senescence and ageing in these tissues, the mechanism by which BubR1 hypomorphism accelerates the ageing phenotypes may involve p16Ink4a-induced senescence.
Figure 4. p16Ink4a induction in BubR1H/H mice promotes cellular senescence.
(a) Relative expression of BubR1 in gastrocnemius, subdermal adipose, fat deposits and eyes from 2-month-old wild-type, p16Ink4a−/−, BubR1H/H and BubR1H/H;p16Ink4a−/− mice as determined by qRT–PCR. Values were normalized to GAPDH. Relative fold is to 2-month-old wild-type samples. Data are mean ± s.d. (n = 3 male mice per genotype, with triplicate measurements taken per sample). We note that ablation of p16Ink4a was unable to increase the amount of BubR1 present in either wild-type or BubR1 hypomorphic mice. (b) IAT of 5-month-old wild-type, BubR1H/H and BubR1H/H;p16Ink4a−/− mice stained for SA-β-galactosidase activity. Scale bar is 2 mm. (c) Relative expression of senescence markers in gastrocnemius muscles of 2-month-old wild-type, p16Ink4a−/−, BubR1H/H and BubR1H/H;p16Ink4a−/− mice analysed by qRT–PCR. Data are mean ± s.d. (n = 3 male mice per genotype). Values were normalized to GAPDH. Relative fold expression is to 2-month-old wild-type muscle. (d) Analysis of replicative senescence in skeletal muscle and fat of 2-month-old wild-type, BubR1H/H and BubR1H/H;p16Ink4a−/− mice by analysing in vivo BrdU incorporation. Data are mean ± s.d. (n = 3 males per genotype).
p19Arf is elevated in BubR1H/H tissues with high levels of p16Ink4a
Besides p16Ink4a, p19Arf is expressed at increased levels in many tissues of wild-type mice with advanced age7, including skeletal muscle (Fig. 5a). Although p19Arf is an established effector of senescence in cultured mouse embryonic fibroblasts (MEFs), its role in senescence and ageing in the context of the whole organism has not been clarified1,2. To explore the role of p19Arf in BubR1-mediated ageing, we analysed its relative expression in tissues from 2-month-old BubR1H/H and BubR1+/+ mice. Increased p19Arf expression was consistently observed in BubR1H/H tissues that were subjected to premature ageing and had high p16Ink4a levels, including skeletal muscle, (subdermal) adipose tissue and eye, but not in tissues that developed age-related pathology in a p16Ink4a-independent fashion or had no age-related phenotypes (Fig. 5b, c; Supplementary Information, Fig. S6b, c and data not shown). Skeletal muscle, (subdermal) adipose tissue and eye from BubR1H/H mice lacking p16Ink4a had normal p19Arf transcript levels (Fig. 5d), suggesting that the observed increase in p19Arf expression is dependent on high p16Ink4a levels. p15Ink4b, which encodes a Cdk inhibitor that has been linked to ageing in some tissues7, was neither increased in BubR1H/H tissues with increased p16Ink4a and p19Arf expression, nor in any other tissue of BubR1H/H mice (Supplementary Information, Figs S3a, S6b, c). In contrast to p16Ink4a and p19Arf, p15Ink4b was not expressed at increased levels in skeletal muscles of aged wild-type mice (Supplementary Information, Fig. S3b).
Figure 5.
p19Arf is elevated in BubR1 hypomorphic tissues with high p16Ink4a. (a) Skeletal muscles of wild-type and BubR1H/H mice of various ages were analysed for p19Arf expression by qRT–PCR. All values were normalized to GAPDH. Data are mean ± s.d. (n = 3 mice were used per genotype and age group). (b) Relative expression of p19Arf in various tissues of 2-month-old BubR1H/H and BubR1+/+ mice as measured by qRT–PCR. Data are mean ± s.d. (n = 3 males per genotype). All values were normalized to GAPDH. Relative expression is to wild-type samples. (c) Western blots of eye and fat extracts from 2-month-old BubR1+/+ and BubR1H/H mice probed with anti-p19Arf and p15Ink4b antibodies. Anti-tubulin antibody was used as a loading control. Uncropped images of the scans are shown in Supplementary Information, Fig. S6b, c. (d) Relative expression of p19Arf in skeletal muscle (gastrocnemius), subdermal adipose, fat deposits and eyes of 2-month-old wild-type, BubR1H/H and BubR1H/H;p16Ink4a−/− mice as determined by qRT–PCR. Data are mean ± s.d. (n = 3 males per genotype). All values were normalized to GAPDH. Relative expression is to wild-type samples.
p19Arf disruption accelerates ageing in BubR1H/H mice
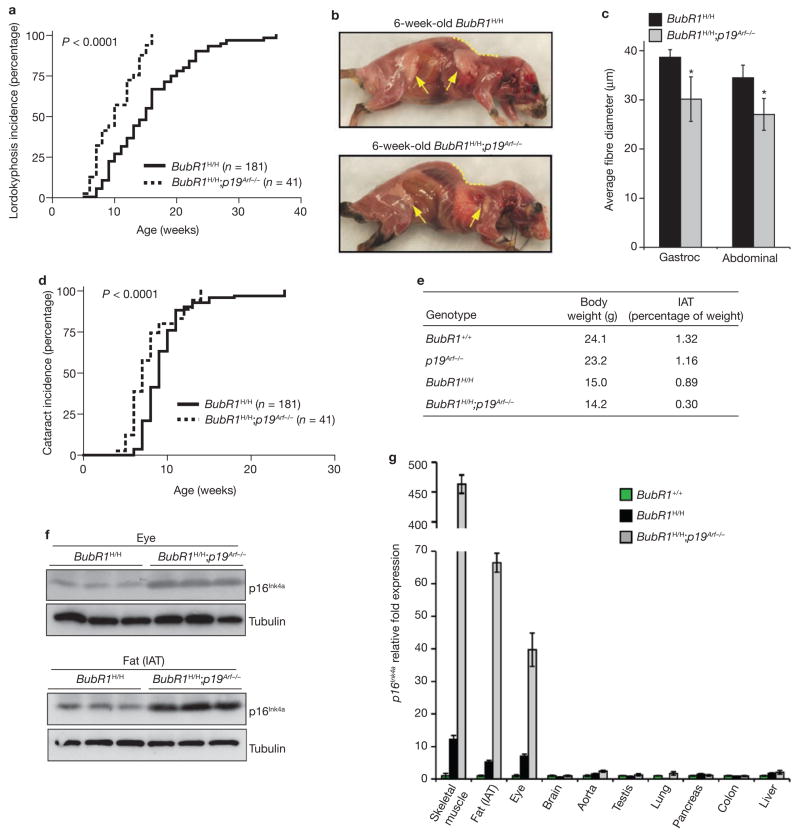
To determine whether p19Arf also acts as an effector of ageing in BubR1H/H mice, 41 BubR1H/H;p19Arf−/− mice were generated and monitored for development of age-related phenotypes. Surprisingly, lordokyphosis developed at a significantly faster rate in BubR1H/H;p19Arf−/− mice than in BubR1H/H mice (Fig. 6a, b). Six-week-old BubR1H/H;p19Arf−/− mice had significantly smaller fibres in gastrocnemius and abdominal muscles than age-matched BubR1H/H mice (Fig. 6c; Supplementary Information, Fig. S4a), indicating that muscle wasting was accelerated in the absence of p19Arf. Cataract formation was also significantly accelerated when p19Arf was absent (Fig. 6d). Skinned 6-week-old BubR1H/H;p19Arf−/− mice showed overt reductions in deposition of adipose tissue (Fig. 6b). This was confirmed by weighing inguinal adipose tissue (IAT, Fig. 6e). Furthermore, the mean thickness of the subcutaneous adipose layer was significantly smaller in BubR1H/H;p19Arf−/− mice than in BubR1H/H mice (0.07 versus 0.11 mm; P < 0.0001, two-tailed Mann-Whitney test). Other progeroid features of BubR1H/H mice seemed to be unchanged by p19Arf inactivation (Supplementary Information, Fig. S4b and Table S1). Taken together, these data indicate that p19Arf acts to delay ageing in response to BubR1 hypomorphism.
Figure 6.
Accelerated ageing in BubR1H/H mouse tissues with increased p16Ink4a expression when p19Arf is lacking. (a) Incidence and latency of lordokyphosis in BubR1H/H and BubR1H/H;p19Arf−/− mice. The curves are significantly different (P < 0.0001, log-rank test). (b) Skinned 6-week-old BubR1H/H and BubR1H/H;p19Arf−/− males. Note that the BubR1H/H;p19Arf−/− mouse has more profound lordokyphosis (dotted line) and reduced subcutaneous fat deposits (arrows). (c) Average muscle fibre size of gastrocnemius (Gastroc) and abdominal muscles of BubR1H/H and BubR1H/H;p19Arf−/− males. Data are mean ± s.d. (n = 3 mice per genotype). A two-tailed Mann-Whitney test was used for statistics. For both comparisons, P < 0.0001. (d) Incidence and latency of cataract formation in BubR1H/H and BubR1H/H;p19Arf−/− mice. The curves are significantly different (P < 0.0001, log-rank test). (e) Amount of inguinal adipose tissue in 6-week-old mice of the indicated genotypes. IAT is expressed as percentage of total body weight. Three male mice of each genotype were used. (f) Western blots of eye and fat extracts from 2-month-old BubR1H/H and BubR1H/H;p19Arf−/− mice probed with anti-p16Ink4a and anti-tubulin antibody. Uncropped images of the scans are shown in Supplementary Information, Fig. S6d. (g) Relative expression of p16Ink4a in various tissues of 2-month-old BubR1+/+, BubR1H/H and BubR1H/H;p19Arf−/− mice as measured by qRT–PCR. Data are mean ± s.d. (n = 3 males per genotype). All values were normalized to GAPDH. Relative expression is to wild-type samples.
One possible explanation for the pro-ageing effect of p19Arf inactivation in BubR1H/H mice may involve increased expression of p16Ink4a. Consistent with this idea, p16Ink4a levels in skeletal muscle, fat and eye increased markedly when p19Arf was knocked out in BubR1H/H mice (Fig. 6f, g; Supplementary Information, Fig. S6d). A potential explanation for these results may be that genetic manipulation of p19Arf sequences changes the normal regulatory balance in the Cdkn2 locus, thereby increasing p16Ink4a expression. However, disruption of p19Arf had no appreciable effect on p16Ink4a levels in tissues undergoing p16Ink4a-independent ageing or lacking age-related pathologies (Fig. 6g), arguing against this possibility.
Inactivation of p19Arf increases cellular senescence
Next, we investigated whether p19Arf loss accelerates ageing in BubR1H/H mice through increased senescence. Indeed adipose tissue of 6-week-old BubR1H/H;p19Arf−/− mice showed much higher SA-β-galactosidase activity than that of corresponding BubR1H/H mice (Fig. 7a). Furthermore, two senescence-associated genes, Igfbp2 and Nrg1, which are expressed at increased levels in gastrocnemius muscles of 6-week-old BubR1H/H mice, were further elevated in corresponding muscles of age-matched BubR1H/H;p19Arf−/− mice (Fig. 7b). In vivo BrdU incorporation showed that cell proliferation in skeletal muscle and fat was considerably lower in BubR1H/H;p19Arf−/− mice than in BubR1H/H mice (Fig. 7c). Collectively, these data indicate that p19Arf induction in BubR1H/H mice functions to prevent or delay senescence and provide further evidence for the notion that in vivo senescence promotes ageing.
Figure 7.
Senescence increases in BubR1H/H tissues with high p16Ink4a when p19Arf is lacking. (a) IAT of 2-month-old BubR1H/H and BubR1H/H;p19Arf−/− mice stained for SA-β-galactosidase activity. Scale bar is 2 mm. (b) Relative expression of senescence markers in gastrocnemius muscles of 6-week-old wild-type, p19Arf−/−, BubR1H/H and BubR1H/H;p19Arf−/− mice. All values were normalized to GAPDH. Relative fold expression is to wild-type gastrocnemius. Data are mean ± s.d. (n = 3 male mice were evaluated per genotype). (c) Replicative senescence in skeletal muscle and fat of 2-month-old wild-type, BubR1H/H and BubR1H/H;p19Arf−/− mice as analysed by in vivo BrdU incorporation. Data are mean ± s.d. (n = 3 male mice per genotype were used for this experiment).
Distinct in vivo and in vitro effects of p16Ink4a and 19Arf inactivation on senescence
Previously, we have shown that BubR1H/H MEFs express high levels of p16Ink4a and p19Arf and age prematurely13. To determine the effects p16Ink4a and p19Arf on cellular senescence in these MEFs, we stained BubR1H/H;p16Ink4a−/− and BubR1H/H;p19Arf−/− MEFs for SA-β-galactosidase. Inactivation of p19Arf caused a marked decrease in senescence in BubR1H/H MEFs, whereas inactivation of p16Ink4a had no effect (Supplementary Information, Fig. S5a, b). Consistently, the percentage of cycling cells was greatly increased in BubR1H/H;p19Arf−/− MEFs, but not in BubR1H/H;p16Ink4a−/− MEFs (Supplementary Information, Fig. S5c, d). Furthermore, BubR1H/H;p19Arf−/− MEFs grew considerably faster than BubR1H/H MEFs, but BubR1H/H;p16Ink4a−/− MEFs did not (Supplementary Information, Fig. S5e, f). Immunoblotting showed that the p19Arf–53 pathway remained highly active in BubR1H/H;p16Ink4a−/− MEFs, similarly to BubR1H/H MEFs, whereas it was inactive in BubR1H/H;p19Arf−/− MEFs (Supplementary Information, Fig. S5h, g). Together, these data demonstrate that the effects of p16Ink4a and p19Arf ablation on in vivo senescence in skeletal muscle and fat of BubR1H/H mice are not recapitulated by their effects on in vitro senescence in BubR1H/H MEFs.
p16Ink4a loss synergizes with BubR1 insufficiency in lung tumorigenesis
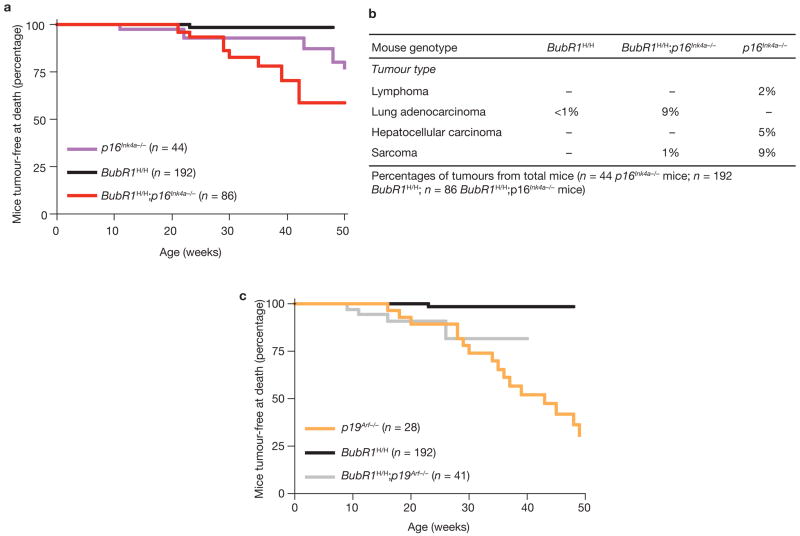
BubR1H/H mice show progressive and severe aneuploidy but rarely develop tumours13. Activation of p16Ink4a or p19Arf in response to BubR1 hypomorphism may act to suppress tumorigenesis. To test for this possibility, BubR1H/H mice lacking p16Ink4a or p19Arf were monitored for tumour formation. Live BubR1H/H;p16Ink4a−/− and BubR1H/H mice showed no overt tumours, but biopsy of moribund or dead animals revealed that BubR1H/H;p16Ink4a−/− mice had significantly more tumours than BubR1H/H mice (Fig. 8a). Eight out of nine BubR1H/H;p16Ink4a−/− tumours were lung adenocarcinomas, a type of tumour observed in only one BubR1H/H mouse and none of the p16Ink4a−/− mice (Fig. 8b). Sarcomas, the most prevalent tumour type in p16Ink4a−/− mice, were rare in BubR1H/H;p16Ink4a−/− mice and not present in BubR1H/H mice. Thus, the effect of BubR1 insufficiency is synergistic with that of p16Ink4a loss during tumorigenesis in lung epithelial cells, but not in other cell types (see Supplementary Discussion). BubR1H/H;p19Arf−/− and p19Arf−/− mice, however, had overlapping tumour-free survival curves (Fig. 8c), indicating that BubR1 insufficiency and p19Arf loss do not synergize in tumorigenesis.
Figure 8.
Ablation of p16Ink4a accelerates lung tumorigenesis in BubR1 insufficient mice. (a) Percentage of mice with tumours at time of death as a function of time for p16Ink4a−/−, BubR1H/H and BubR1H/H;p16Ink4a−/− mice. Biopsies were performed on moribund animals and all tissues were screened for tumours. Tumour tissues were collected and processed for histological confirmation. The BubR1H/H;p16Ink4a−/− curve is significantly different from the BubR1H/H curve with P = 0.0027 (calculated using a log-rank test). (b) Tumour spectra of p16Ink4a−/−, BubR1H/H and BubR1H/H;p16Ink4a−/− mice. (c) As in a but for p19Arf−/−, BubR1H/H and BubR1H/H;p19Arf−/− mice. There is no significant difference between the curves of BubR1H/H;p19Arf−/− and p19Arf−/− mice using a log-rank test.
DISCUSSION
Here, we report that inactivation of p16Ink4a attenuates the development of age-related pathologies in BubR1H/H tissues with elevated p16Ink4a. This shows that induction of p16Ink4a by cellular stress resulting from BubR1-insufficiency drives the development of ageing-associated phenotypes, and provides direct evidence for a causal involvement of this tumour suppressor in organismal ageing. Importantly, skeletal muscle and fat, two tissues that are subject to p16Ink4a-dependent ageing when BubR1 levels are low, have higher numbers of replicating cells and show decreased expression of senescence-associated proteins in the absence of p16Ink4a. These observations support the notion that p16Ink4a contributes to ageing-associated pathologies through accumulation of senescent cells. This is a significant finding as evidence that cellular senescence promotes ageing has thus far been largely circumstantial2,6.
BubR1H/H mouse tissues, in which p16Ink4a is elevated, also have increased p19Arf. However, p19Arf inactivation accelerates rather than delays ageing in these tissues, indicating that this tumour suppressor provides anti-ageing activity. This seems surprising, considering that p19Arf is an effector of senescence in cultured cells2,6. However, the recent observation that transgenic mice carrying an extra copy of both p19Arf and p53 are protected from ageing-associated damage and live longer than normal mice28, is consistent with our finding that p19Arf has anti-ageing activity in BubR1H/H mice. It has been proposed that the p19Arf–p53 pathway in response to low, chronic stress, may primarily induce genes that promote cell survival and repair, thereby extending lifespan5,28,29. High, acute types of stress, on the other hand, may accelerate ageing by triggering a more robust p53 response, causing irreversible cell-cycle arrest and/or apoptosis5,29–31. Therefore, one possibility is that p19Arf may elicit a p53 transcriptional response that provides protection against cellular stress resulting from BubR1 hypomorphism, thus delaying the onset of cellular senescence. The observation that skeletal muscle and fat from BubR1H/H mice lacking p19Arf accumulate more senescent cells is consistent with this idea. Strong additional support for the conclusion that p19Arf has anti-ageing activity is provided by our unpublished observations indicating that BubR1H/H mice lacking p53 phenocopy those lacking p19Arf. Two observations reported here suggest that p19Arf may exert its anti-ageing effect, at least in part, through negative regulation of p16Ink4a expression. First, inactivation of p19Arf in BubR1H/H mice resulted in increased p16Ink4a expression in skeletal muscle, fat and eye, three tissues that have high p19Arf levels and are subjected to accelerated ageing. Second, inactivation of p16Ink4a prevented the induction of p19Arf in these BubR1H/H tissues. How p19Arf attenuates p16Ink4a expression remains to be addressed.
Although inactivation of p16Ink4a significantly delays the development of certain ageing-associated phenotypes in BubR1H/H mice, it does not completely prevent them. Furthermore, other progeroid phenotypes are not affected by loss of p16Ink4a. These findings suggest that BubR1 hypomorphism engages other progeroid effectors in addition to p16Ink4a. The identity of these effectors is currently unclear and remains to be established. Striking similarities exist between the progeroid phenotypes of BmalI knockout and BubR1H/H mice32. The molecular basis of this similarity is unclear, although, based on the known roles of each protein, it is unlikely that BubR1 and BmalI are functionally connected. However, it is possible that downstream pathways that respond to stress resulting from Bmal1 loss and BubR1 hypomorphism are shared.
METHODS
Generation of compound mutant mice
BubR1H/H mice were generated as described previously13. p16Ink4a and p19Arf knockout mice have been generated previously and were acquired from the Mouse Models of Human Cancers Consortium located at the National Cancer Institute, Frederick33,34. All mice were on a mixed 129 × C57BL/6 genetic background. They were housed in a pathogen-free barrier environment for the duration of the study. Experimental procedures involving the use of these laboratory mice were reviewed and approved by the Institutional Animal Care and Use Committee of the Mayo Clinic. Prism software (GraphPad Software) was used for generation of all survival curves and for statistical analyses.
Collection and analysis of tumours
Moribund mice were killed and all major organs were screened for overt tumours using a dissection microscope. Tumours that were collected were processed by standard procedures for histopathology. A Fisher’s exact test was used to compare tumour incidence proportions across the genotypes for mice that developed tumours. Board-certified pathologists assisted in the histological evaluation of tumour sections.
Analysis of progeroid phenotypes
Bi-weekly, mice were screened for the development of overt cataracts by examining dilated eyes with a slit-light. Incidence of lordokyphosis was checked bi-weekly. Mice that showed lordokyphosis for three consecutive monitoring periods were determined to have this condition. Various skeletal muscles were collected and processed for histology as described previously35,36. Fibre diameter measurements were performed on cross sections of gastrocnemius and abdominal muscles from 6-week-old male mice (n = 3 mice per genotype). A total of 50 fibres were measured per muscle using a calibrated computer program (Olympus MicroSuite Five). Dissection, histology and measurements of dermal and adipose layers of dorsal skin were performed as described previously16. Values represent an average of four males. Analysis of arterial wall stiffening was performed as described previously14. Measurements of body weight and IAT were performed on 6-week-old males (n = 3 per genotype). Oral glucose tolerance tests were performed on 5-month-old male mice (n = 5 per genotype) as described by the Jackson Laboratory (www.jax.org). Insulin measurements were performed as described previously37.
Quantitative real-time PCR
Total RNA was extracted from tissues using a Qiagen RNeasy RNA isolation kit according to the manufacturer’s protocol. Transcription into cDNA was performed using random hexamers and SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. All PCR reactions used SYBR green PCR Master Mix (Applied Biosystems) to a final volume of 12 μl, with each cDNA sample performed in triplicate in the ABI PRISM 7900 Sequence Detection System (Applied Biosystems) according to the protocol of the manufacturer. All experiments were performed on organs/tissues from at least three different animals in each age group and genotype. The expression of genes was normalized to GAPDH. Sequences of primers used for qRT–PCR of p15Ink4b (ref. 38), p16Ink4a (ref. 39), p19Arf (ref. 28), BubR1 (ref. 40), Mmp-13 (ref. 41), PAI-1 (ref. 42), Igfbp-2 (ref. 43), and GAPDH (ref. 44) were as published. Additionally, sequences for Nrg1 were: forward, CATGGTGAACATAGCGAATGGCC; reverse, CCACAATATGCTCACTGGAGATG A. Statistical differences were determined using an unpaired two-tailed t test.
Analysis of satellite-cell function
Analysis of in vivo satellite-cell function were carried out as described previously20. Briefly, mice anaesthetized with avertin (200 mg kg−1, i.p.) were given a single 50-μl injection of cardiotoxin (10 μM; Calbiochem) into the gastrocnemius muscle. After this injection, the skin incision was closed with a nylon suture. Mice were allowed to recover and then were analysed at both 5 and 18 days post-injection by routine histology. Isolation and culture of skeletal muscle satellite cells were performed as described previously45. Briefly, hindlimb muscles of 5-month-old mice were removed and trimmed of excess connective tissue and fat. Minced muscles were subjected to several 15-min rounds of digestion at 37 °C in incubation medium (50% DMEM high glucose (GIBCO)/50% F-12K (CellGro)/168 U ml−1 collagenase type II (Worthington)/0.04% Trypsin (GIBCO)). Once fully digested, cells were successively filtered through 70- and 40-μm strainers, collected by centrifugation at 300g for 5 min and resuspended in propagation medium (DMEM high glucose/15% FCS (GIBCO)/glutamine (CellGro)/penicillin-streptomycin (CellGro)). After seven days in culture, differentiation medium (propagation medium with 2% FCS) was applied and cells were fixed seven days after transfer to the medium. Myotube formation was quantified using the total number of myotubes for each sample normalized to the muscle mass extracted.
Magnetic resonance imaging
Magnetic resonance images with a 7-tesla scanner (Bruker) were obtained in 2%-isofluorane anaesthetized mice using a spinecho method as described previously46. Digital images were analysed with the Metamorph software (Visitron, Universal Imaging). The ratio of muscle area (paraspinal and chest or abdominal wall muscles) to total body cross-section was measured at the distal thorax and mid-abdomen levels.
In vivo BrdU incorporation
At 24 and 6 h before tissue collection, male mice of various genotypes were injected intraperitoneally with 200 μl of BrdU (10 mg ml−1; Sigma) in PBS. Mice were anaesthetized (avertin, 200 mg kg−1, i.p.) and successively perfused (transcardially) with PBS and 10% formalin. Organs were collected and embedded in paraffin. Five-μm sections were prepared and stained for BrdU according to the manufacturer’s protocol (BD Pharmingen). The percentage of BrdU-positive cells was determined by counting total and BrdU-positive nuclei in 10 non-overlapping fields at ×40 magnification (n =3 mice per genotype).
Generation and culture of MEFs
MEFs were generated as described previously47. BrdU incorporation assays on MEFs were performed according to the manufacturer’s protocol (BD Bioscience). Growth curves were generated as described previously13. Three independent MEF lines for each genotype were analysed in both experiments.
SA-β-galactosidase staining
Adherent MEFs were stained with a SA-β-galactosidase activity kit according to manufacturer’s protocol (Cell Signaling). Nuclei were stained with Hoechst to determine percentages of cells positive for SA-β-galactosidase activity. The percentage of SA-β-galactosidase-positive cells was the total number of cells positive for SA- β-galactosidase activity divided by the total number of cells (n = 3 independent MEF lines for each genotype at each passage). Adipose tissue depositions were stained for SA-β-galactosidase activity, as described previously13.
Western blot analyses
Western blot analyses were carried out as described previously48. Antibodies for senescence-associated proteins were as described13,16. The antibody for p15Ink4b was a gift from M. Barbacid49.
Acknowledgments
We thank Paul Galardy, Rick Bram, Randy Faustino, Amy Tang, Robin Ricke and Jim Kirkland for critical reading of the manuscript or helpful discussions. We would like to thank Mariano Barbacid for the generous gift of anti-p15Ink4b antibody. This work was supported by grants from the National Institutes of Health, the Ted Nash Foundation and the Ellison Medical Foundation to J.v.D.
Footnotes
Accession codes. USCD-Nature Signaling Gateway (http://www.signaling-gateway.org): A001711, A001713 and A003172
Note: Supplementary Information is available on the Nature Cell Biology website.
AUTHOR CONTRIBUTIONS
D.J.B., C.P.T., F.J., K.P., N.J.N., K.J., S.Y., S.R., L.R., H.J.H. and N.L.E. conducted experiments, prepared the figures and analysed the data; D.J.B., A.T. and J.M.v.D. planned the project and wrote the manuscript; J.M.v.D. supervised the project.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nature Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 3.Sharpless NE, DePinho RA. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9:22–30. doi: 10.1016/s0959-437x(99)80004-5. [DOI] [PubMed] [Google Scholar]
- 4.Sherr CJ, Weber JD. The ARF/p53 pathway. Curr Opin Genet Dev. 2000;10:94–99. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- 5.Vousden KH, Lane D. P p53 in health and disease. Nature Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 6.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Krishnamurthy J, et al. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zindy F, Quelle DE, Roussel MF, Sherr CJ. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
- 9.Krishnamurthy J, et al. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- 10.Janzen V, et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 11.Molofsky AV, et al. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beausejour CM, Campisi J. Ageing: balancing regeneration and cancer. Nature. 2006;443:404–405. doi: 10.1038/nature05221. [DOI] [PubMed] [Google Scholar]
- 13.Baker DJ, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nature Genet. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto T, et al. Aging-associated vascular phenotype in mutant mice with low levels of BubR1. Stroke. 2007;38:1050–1056. doi: 10.1161/01.STR.0000257967.86132.01. [DOI] [PubMed] [Google Scholar]
- 15.Hartman TK, Wengenack TM, Poduslo JF, van Deursen JM. Mutant mice with small amounts of BubR1 display accelerated age-related gliosis. Neurobiol Aging. 2007;28:921–927. doi: 10.1016/j.neurobiolaging.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 16.Baker DJ, et al. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J Cell Biol. 2006;172:529–540. doi: 10.1083/jcb.200507081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rane SG, et al. Loss of Cdk4 expression causes insulin-deficient diabetes and Cdk4 activation results in β-islet cell hyperplasia. Nature Genet. 1999;22:44–52. doi: 10.1038/8751. [DOI] [PubMed] [Google Scholar]
- 18.Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nature Rev Mol Cell Biol. 2007;8:703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- 19.Price SR, Mitch WE. Mechanisms stimulating protein degradation to cause muscle atrophy. Curr Opin Clin Nutr Metab Care. 1998;1:79–83. doi: 10.1097/00075197-199801000-00013. [DOI] [PubMed] [Google Scholar]
- 20.Koh TJ, Bryer SC, Pucci AM, Sisson TH. Mice deficient in plasminogen activator inhibitor-1 have improved skeletal muscle regeneration. Am J Physiol Cell Physiol. 2005;289:C217–C223. doi: 10.1152/ajpcell.00555.2004. [DOI] [PubMed] [Google Scholar]
- 21.Fridlyand J, et al. Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer. 2006;6:96. doi: 10.1186/1471-2407-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hengstschlager M, et al. Loss of the p16/MTS1 tumor suppressor gene causes E2F-mediated deregulation of essential enzymes of the DNA precursor metabolism. DNA Cell Biol. 1996;15:41–51. doi: 10.1089/dna.1996.15.41. [DOI] [PubMed] [Google Scholar]
- 23.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.West MD, Pereira-Smith OM, Smith JR. Replicative senescence of human skin fibroblasts correlates with a loss of regulation and overexpression of collagenase activity. Exp Cell Res. 1989;184:138–147. doi: 10.1016/0014-4827(89)90372-8. [DOI] [PubMed] [Google Scholar]
- 25.Wang S, Moerman EJ, Jones RA, Thweatt R, Goldstein S. Characterization of IGFBP-3, PAI-1 and SPARC mRNA expression in senescent fibroblasts. Mech Ageing Dev. 1996;92:121–132. doi: 10.1016/s0047-6374(96)01814-3. [DOI] [PubMed] [Google Scholar]
- 26.Shelton DN, Chang E, Whittier PS, Choi D, Funk WD. Microarray analysis of replicative senescence. Curr Biol. 1999;9:939–945. doi: 10.1016/s0960-9822(99)80420-5. [DOI] [PubMed] [Google Scholar]
- 27.Linskens MH, et al. Cataloging altered gene expression in young and senescent cells using enhanced differential display. Nucleic Acids Res. 1995;23:3244–3251. doi: 10.1093/nar/23.16.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matheu A, et al. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- 29.Serrano M, Blasco MA. Cancer and ageing: convergent and divergent mechanisms. Nature Rev Mol Cell Biol. 2007;8:715–722. doi: 10.1038/nrm2242. [DOI] [PubMed] [Google Scholar]
- 30.Varela I, et al. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature. 2005;437:564–568. doi: 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- 31.Cao L, Li W, Kim S, Brodie SG, Deng CX. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev. 2003;17:201–213. doi: 10.1101/gad.1050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharpless NE, et al. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 34.Sharpless NE, Ramsey MR, Balasubramanian P, Castrillon DH, DePinho RA. The differential impact of p16(INK4a) or p19(ARF) deficiency on cell growth and tumorigenesis. Oncogene. 2004;23:379–385. doi: 10.1038/sj.onc.1207074. [DOI] [PubMed] [Google Scholar]
- 35.Engel WK, Cunningham GG. Rapid examination of muscle tissue, an improved trichrome method for fresh-frozen biopsy sections. Neurology. 1963;13:919–923. doi: 10.1212/wnl.13.11.919. [DOI] [PubMed] [Google Scholar]
- 36.Kane GC, et al. ATP-sensitive K+ channel knockout compromises the metabolic benefit of exercise training, resulting in cardiac deficits. Diabetes. 2004;53(suppl 3):S169–S175. doi: 10.2337/diabetes.53.suppl_3.s169. [DOI] [PubMed] [Google Scholar]
- 37.Baur JA, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krimpenfort P, et al. p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a. Nature. 2007;448:943–946. doi: 10.1038/nature06084. [DOI] [PubMed] [Google Scholar]
- 39.Edwards MG, et al. Gene expression profiling of aging reveals activation of a p53-mediated transcriptional program. BMC Genomics. 2007;8:80. doi: 10.1186/1471-2164-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan B, et al. Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clin Cancer Res. 2006;12:405–410. doi: 10.1158/1078-0432.CCR-05-0903. [DOI] [PubMed] [Google Scholar]
- 41.Maes C, et al. Soluble VEGF isoforms are essential for establishing epiphyseal vascularization and regulating chondrocyte development and survival. J Clin Invest. 2004;113:188–199. doi: 10.1172/JCI19383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asahi M, et al. Protective effects of statins involving both eNOS and tPA in focal cerebral ischemia. J Cereb Blood Flow Metab. 2005;25:722–729. doi: 10.1038/sj.jcbfm.9600070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohlson N, Bergh A, Persson ML, Wikstrom P. Castration rapidly decreases local insulin-like growth factor-1 levels and inhibits its effects in the ventral prostate in mice. Prostate. 2006;66:1687–1697. doi: 10.1002/pros.20368. [DOI] [PubMed] [Google Scholar]
- 44.Jeong YJ, et al. Optimization of real time RT–PCR methods for the analysis of gene expression in mouse eggs and preimplantation embryos. Mol Reprod Dev. 2005;71:284–289. doi: 10.1002/mrd.20269. [DOI] [PubMed] [Google Scholar]
- 45.Beauchamp JR, Morgan JE, Pagel CN, Partridge TA. Dynamics of myoblast transplantation reveal a discrete minority of precursors with stem cell-like properties as the myogenic source. J Cell Biol. 1999;144:1113–1122. doi: 10.1083/jcb.144.6.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamada S, et al. Protection conferred by myocardial ATP-sensitive K+ channels in pressure overload-induced congestive heart failure revealed in KCNJ11 Kir6.2-null mutant. J Physiol. 2006;577:1053–1065. doi: 10.1113/jphysiol.2006.119511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Babu JR, et al. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol. 2003;160:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kasper LH, et al. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol. 1999;19:764–776. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Latres E, et al. Limited overlapping roles of P15(INK4b) and P18(INK4c) cell cycle inhibitors in proliferation and tumorigenesis. EMBO J. 2000;19:3496–3506. doi: 10.1093/emboj/19.13.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]