Abstract
The neural crest, a transient population of migratory cells, forms the craniofacial skeleton and peripheral nervous system, among other derivatives in vertebrate embryos. The transcriptional repressor Snail2 is thought to be crucial for the epithelial-to-mesenchymal transition (EMT) that promotes neural crest delamination from the neural tube; however, little is known about its downstream targets. To this end, we depleted avian Snail2 in the premigratory neural crest using morpholino antisense oligonucleotides and examined effects on potential targets by quantitative PCR. Several dorsal neural tube genes were upregulated by alleviating Snail2 repression; moreover, the cell adhesion molecule cadherin6B was derepressed within 30 minutes of blocking Snail2 translation. Examination of the chick cadherin6B genomic sequence reveals that the regulatory region contains three pairs of clustered E boxes, representing putative Snail2 binding sites. Furthermore, in vivo and in vitro biochemical analyses demonstrate that Snail2 directly binds to these sites and regulates cadherin6B transcription. These results are the first to describe a direct target of Snail2 repression in vivo and in the context of the EMT that characterizes neural crest development.
Keywords: Snail2 (Slug), cadherin6B, Neural crest, Epithelial-to-mesenchymal transitions, E boxes
INTRODUCTION
The neural crest is a transient embryonic cell type that arises within the dorsal neural tube but subsequently delaminates and initiates migration to multiple sites in the periphery of the embryo. Following this migratory phase, neural crest cells form many diverse derivatives, including much of the peripheral nervous system and craniofacial skeleton. The development of the neural crest is thought to critically rely upon the activity of Snail2 (formerly known as Slug) (Nieto et al., 1994; LaBonne and Bronner-Fraser, 2000), a zinc-finger transcription factor that plays a key role in regulating epithelial-to-mesenchymal transitions (EMTs), resulting in neural crest delamination from the neural tube.
Snail2 is expressed in premigratory and early migrating neural crest cells. Overexpression of Snail2 results in expansion of the neural crest domain in Xenopus and an increase in the number of migratory neural crest cells from the chick hindbrain (LaBonne and Bronner-Fraser, 1998; del Barrio and Nieto, 2002); conversely, knock-down using antisense Snail2 RNA in Xenopus results in inhibition of neural crest migration and a decrease or loss of many neural crest derivatives (Carl et al., 1999). As a member of the Snail superfamily of transcriptional repressors, Snail2 functions as an important regulator through its ability to modulate the activity of other genes. However, surprisingly little is known about the downstream targets of Snail2 during neural crest development.
Here, we have combined the techniques of morpholino antisense oligonucleotide (MO) knock-down and quantitative PCR to identify possible downstream target genes of Snail2 in avian embryos. In particular, we have assessed the response of several transcripts in order to better describe the molecular basis of Snail2-mediated neural crest EMT. Our results identify a number of genes that are upregulated upon alleviation of Snail2 repression in the premigratory chick neural crest cell population, thus representing potential targets of Snail2 repression during the process of EMT. We further show that one putative target of Snail2 is cadherin6B (Cad6B), which has been previously reported to be expressed in the premigratory neural crest and to be downregulated as neural crest cells delaminate and migrate away from the dorsal neural tube in the avian trunk (Nakagawa and Takeichi, 1995; Nakagawa and Takeichi, 1998). Through the identification of Snail2 binding sites (E boxes) within the Cad6B regulatory region using the available chick Cad6B genomic sequence, together with chromatin immunoprecipitation, electrophoretic mobility shift and luciferase assays, we demonstrate that Snail2 directly represses Cad6B transcription in a highly dynamic manner that is required for neural crest EMT. Furthermore, our studies show that the regulation of neural crest EMT by Snail2 occurs in a Cad6B-mediated manner. Our results are the first to reveal a direct downstream target of Snail2 repression during avian neural crest EMT.
MATERIALS AND METHODS
Design and electroporation of antisense morpholinos
A 3′ lissamine-labeled antisense Snail2 MO, 5′-TCTTGACCAGGAAGGAGC-3′, was designed according to the manufacturer’s criteria (Gene Tool, LLC). A 5-mismatch 3′ lissamine-labeled antisense Snail2 MO, 5′-TgTTcACCAcGAAcGAcC-3′, was designed as a negative control (mutated bases shown in lower case). A 3′ lissamine-labeled antisense Cad6B MO was also designed: 5′-ACCAGAAGCAATGGTAAGTTCTCAT-3′. Each MO was introduced into the developing chick embryo using a modified version of the electroporation technique (Itasaki et al., 1999). Briefly, the MO was injected into the lumen of the neural tube and two 25 volt, 25 msecond pulses were applied across the embryo. To determine the reduction in numbers of cells containing Snail2 protein on the electroporated compared with the non-electroporated side, we examined the presumptive neural crest domain in transverse sections through the dorsal neural tube of embryos unilaterally transfected with the Snail2 MO (five embryos by counting cells in ⩾four sections per embryo). We then took the ratio of the number of Snail2-positive cells on the electroporated side to the number of Snail2-positive cells on the control side. To determine the efficiency of the Snail2 MO to knock-down Snail2 protein translation in these same sections, we examined the number of Snail2 protein-positive cells (green) on the electroporated side that also contained the Snail2 MO (red) and thus were yellow. This number was then divided by the total number of Snail2 MO-positive (red) cells in the same sections, with the efficiency of reduction expressed as a percentage.
Chick embryo culture
Fertilized chicken eggs were obtained from AA Enterprises (Ramona, CA) and incubated on their sides at 38°C in a humidified incubator (Lyon Electric, Chula Vista, CA). Tissue was dissected in PB-1 standard medium. Chamber slides (LabTek) were coated with 10 μg/ml recombinant fibronectin (BD Biosciences) and 16.6 μg/ml poly-L-lysine (Sigma). Neural folds were explanted to slides and cultured in serum-free DMEM plus N2 supplement (Gibco-BRL) for 4 hours prior to fixation and imaging. To determine the number of cells that have undergone EMT in explanted tissue, the number of cells containing the MO (red) that have undergone EMT (i.e. mesenchymal in appearance) and have emigrated from the explants were counted, using DAPI staining and phase-contrast microscopy to identify individual cells. This number was divided by the total number of cells that have undergone EMT. Emigrating cells were counted in this manner from at least thirty explants over three independent experiments.
Whole-mount in situ hybridization
Linearized cDNAs were used to synthesize digoxigenin-labeled antisense RNA probes, and whole-mount in situ hybridizations were performed using ‘Protocol Four’ as previously described (Xu and Wilkinson, 1998). Stained embryos were imaged on a Zeiss Stemi SV11 microscope and processed using Photoshop 7.0 (Adobe Systems).
Immunohistochemistry
The distribution of Snail2 protein was assessed using anti-Snail2 antisera [Developmental Studies Hybridoma Bank (DSHB), Iowa University; clone 62.IE6]. Briefly, whole embryos were fixed for 15 minutes in 4% paraformaldehyde at room temperature and washed with PBS containing 0.1% Tween 20 (PBTw). Embryos were then blocked in 10% sheep serum in PBTw before adding the anti-Snail2 antibody (1:100, in 5% sheep serum in PBTw) and incubating at 4°C overnight. The primary antibody was washed off with PBTw, and replaced by an Alexa-Fluor 488 secondary antibody (Molecular Probes, 1:500, diluted in 5% sheep serum in PBTw) with incubation overnight at 4°C. The embryos were then washed, mounted and imaged. Immunohistochemical detection of Cad6B (DSHB, clone CCD8B-1; 1:100) was performed as described (Nakagawa and Takeichi, 1998) with an Alexa-Fluor 488 secondary antibody (Molecular Probes, 1:500). If required, embryos were cryostat-sectioned at 14 μm, and sections were DAPI-stained.
RNA and cDNA preparation
Total RNA was isolated using the RNAqueous Total RNA Isolation Kit (Ambion), and cDNA was synthesized using random hexamers and the Superscript II RT-PCR system (Invitrogen) according to the manufacturer’s instructions.
Quantitative polymerase chain reaction (QPCR)
QPCR was performed using the ABI 7000 in a TaqMan (Applied Biosciences) or SYBR Green (Bio-Rad) assay as described (Taneyhill and Bronner-Fraser, 2005). Briefly, 50 μl reactions were performed using the 2×TaqMan mix in the presence of cDNA, 100-300 nM of each primer, and 450 nM of each probe; 25 μl reactions were performed using the 2×iTaq SYBR Green mix in the presence of cDNA and 50–100 nM of each primer. After normalization to a standard (chick 18S rRNA), fold upregulation or downregulation was determined by dividing the relative expression value for the gene of interest in the presence of the Snail2 MO by that obtained for the gene of interest in the presence of the control MO.
Chicken Cad6B sequence identification, isolation and construct preparation
The genomic location of chick Cad6B was identified using the Ensembl (www.ensembl.org) web browser by searching with the complete chick Cad6B cDNA sequence (Genbank accession number D42149). Sequence reads were assembled using AssemblyLIGN and analysis was performed using MacVector 7.1.1.
The Cad6B genomic sequence was divided in silico into portions containing two E-box motifs. Fragments were amplified by PCR from BAC clone CH261-110K4 using Expand Long Template Polymerase (Roche) with appropriate primers:
E4_5′, 5′-TTCATCCACCAGTAACCCCTTG-3′;
E3_3′, 5′-GCCCTTTTCCCTTCATTCTCC-3′;
E2_5′, 5′-GCAAAGATACCCTGGACTTCGG-3′;
E1_3′, 5′-CCAACATCCCTCCTTGTAACCC-3′;
EA_5′, 5′-TTCTCCACCACACATACCAGTGC-3′; and
EB_3′, 5′-AGACACAGTCCCATTTGAGTTTGG-3′.
Fragments were inserted into the KpnI (5′) and XhoI (3′) cloning sites of the pGL2-Promoter Luciferase vector (Promega). All constructs were sequenced to confirm PCR and cloning accuracy.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation of Snail2 protein associated with the Cad6B regulatory region was carried out using described procedures and buffers (Upstate). Briefly, dorsal neural tube tissue dissected from the premigratory chick trunk or midbrain region was fixed in 1% formaldehyde and then subjected to sonication (ten times, 15-second pulse and 1 minute recovery on ice) using a microtip attachment on a Branson 450 Sonifier set at 50% output. After sonication, 20 μl of chromatin was taken as a standard curve template for QPCR. Equivalent amounts of chromatin were used in three different immunoprecipitations carried out at 4°C overnight: an anti-rabbit GFP antibody control (5 μg of GFP antibody; Molecular Probes), a no antibody control, and an anti-Snail2 antibody (150–250 μl of Snail2 antibody). QPCR was then performed on the immunoprecipitated chromatin as described above to detect association of Snail2 to each of the E-box pairs, as well as to the 3′ region of the Cad6B transcript in exon 11 (a region with no predicted E-box motifs) as a negative control. The results reported for the Snail2 antibody were corrected for any background immunoprecipitated chromatin by subtracting the averaged value obtained in the presence of the GFP (or IgG) and no antibody immunoprecipitates (both usually undetectable). Because the amount of chromatin obtained from the tissue prior to immunoprecipitation differs between experiments, the results are presented as a representative ChIP experiment, with the s.d. reported for association of Snail2 with each E-box pair.
Electrophoretic mobility shift assay (EMSA)
Snail2 or control nuclear extract was isolated from transiently transfected 293T cells according to standard techniques. 10 μg of nuclear extract was incubated with double-stranded E box-containing oligonucleotides end-labeled with 33P (GE Healthcare) using T4polynucleotide kinase (Promega). Protein-DNA binding reactions were incubated with or without unlabeled competitor DNA for 20 minutes at room temperature in the presence of an EMSA buffer, prior to addition of the labeled probe (Wu et al., 2005). Poly(dI-dC) (GE Healthcare) was used as a non-specific competitor. E1, 5′-TAGGTTCACGACAGGTATGCAGTTAGAT-3′; E2, 5′-ACAAATCCTGTCAGGTAGTTCTGCTCCA-3′; all mutated oligonucleotides had the E-box core consensus sequence (underlined) altered from CAGGTA to TTGGTA. Incubated reaction mixtures were separated on a high-ionic-strength non-denaturing 4% PAGE gel. Dried gels were exposed to phosphorimaging screens, and signal was detected using a STORM840 Phosphorimager (GE Healthcare).
Construction of mutant Cad6B E box-luciferase reporters and QPCR-based luciferase assays
Primers were designed to change the E-box core consensus sequence from CAGGTA to TTGGTA (for E boxes 1, 2, 3 and 4; sequences available on request). Site-directed mutagenesis was carried out using the QuikChange II Site-Directed Mutagenesis Kit according to the manufacturer’s instructions (Stratagene), utilizing the wild-type Cad6B E box-luciferase reporters as the template. All constructs were sequenced to ensure accuracy of mutagenesis.
200 ng of wild-type or mutant luciferase reporter construct [Cad6B E box-luciferase or control (pGL2-Promoter)] and 240 ng of either a Snail2 or empty vector control construct [Tag-2B-Snail2 or Tag-2B vector (Stratagene)] were transiently transfected into 293T cells using the Lipofectamine 2000 reagent and Opti-MEM I media (Gibco-BRL) according to the manufacturer’s instructions (Invitrogen). Cells were harvested 24 hours post-transfection and total RNA was isolated. SYBR Green QPCR was carried out using primers designed to amplify both the luciferase and ampicillin amplicons (sequence available upon request). A normalized value of luciferase expression for each construct was obtained by dividing the average luciferase quantity obtained for each construct by the corresponding amount of ampicillin. The effect of Snail2 on this normalized luciferase expression was then calculated by dividing the normalized value obtained in the presence of Snail2 by the normalized value obtained in the presence of the empty vector. The reported results are an average of at least three independent transfection experiments, with each QPCR reaction performed in triplicate. Experimental results for the mutant E-box reporters represent the mode value obtained from three independent transfection experiments.
Immunoprecipitation and immunoblotting
Cells utilized for immunoprecipitation and/or immunoblotting were lysed in Lysis Buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.5% NP-40, 1 mM EDTA, 1 mM DTT plus Complete Protease Inhibitors from Roche) by vortexing, followed by a 20 minute incubation on ice and 10 minute centrifugation at 20,800 g, 4°C. Total protein concentration (cytoplasmic and nuclear fractions) was determined by performing a Bradford assay (Bio-Rad). Immunoprecipitations and immunoblotting were carried out using a Flag-M2 peroxidase antibody (Sigma) and/or the Snail2 antibody using standard methods.
RESULTS
Knock-down of Snail2 expression reveals potential downstream target genes
To identify possible direct targets of Snail2 in presumptive neural crest cells about to undergo EMT, we utilized a morpholino antisense oligonucleotide (MO) to knock-down Snail2 protein levels in the premigratory neural crest region (see Fig. S1 in the supplementary material). To verify that cells inheriting the MO failed to express Snail2 protein, we determined the efficacy of the Snail2 MO by counting the number of cells that expressed Snail2 protein in at least three sections of five representative embryos receiving the Snail2 MO. On average, the Snail2 MO is 82% efficient in blocking Snail2 translation. Furthermore, by counting the number of Snail2-positive cells on the electroporated side versus the contralateral control side, we find that there is typically greater than a 33% decrease in detectable Snail2 protein. Although these numbers reflect the varying amount of MO received by each cell and the mosaic nature of the electroporation technique, the observed effects on candidate gene expression are nonetheless robust (see below).
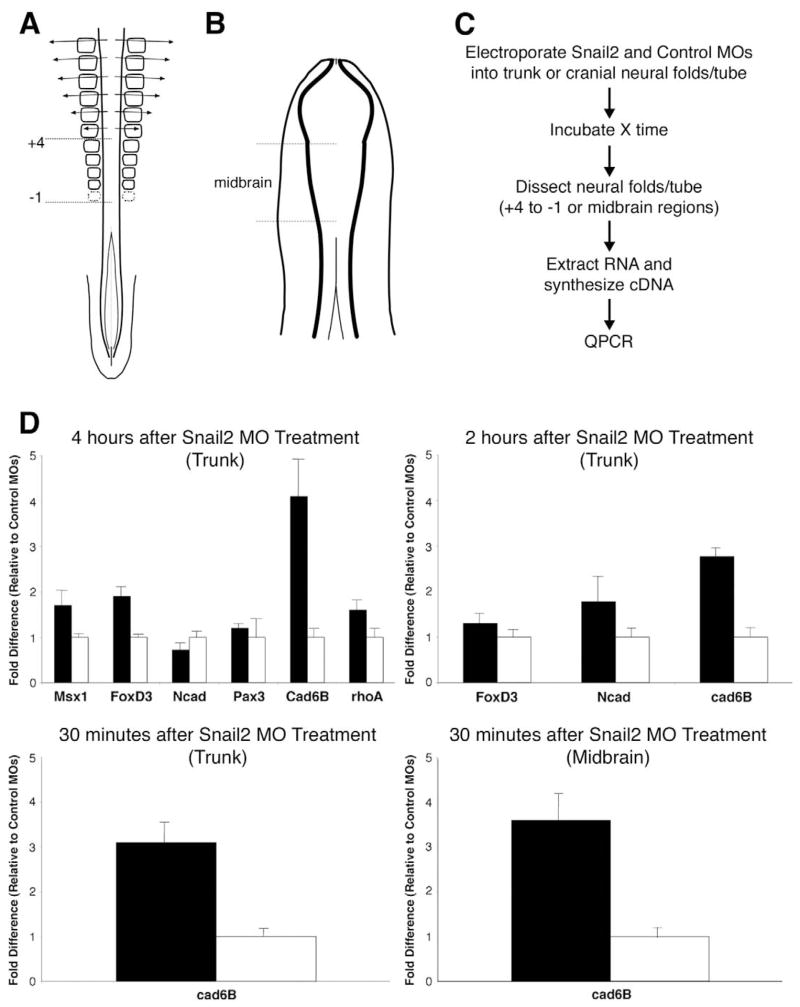
A single lissaminated MO to Snail2 was electroporated into neural tube cells at two different stages in order to target premigratory trunk neural crest (embryos with 21–23 somites) or premigratory midbrain neural crest (embryos with 5–7 somites). Importantly, the MO was introduced just at the onset of Snail2 expression at each axial level. At selected times after MO electroporation, the neural folds were dissected on the electroporated side at the level adjacent to somites +4 to −1 for the trunk region, or in the midbrain for the cranial region (Fig. 1A, B). The dissected tissue was then assayed by quantitative PCR (QPCR) using primers for candidate target genes (Fig. 1C, D).
Fig. 1. Depletion of Snail2 reveals changes in gene expression in premigratory avian neural crest cells.
(A, B) Depictions of avian embryos at developmental stages when premigratory neural crest cells are undergoing EMT in the dorsal neural tube: (A) in the trunk (between somites −1 and +4) of an embryo with 21–23 pairs of somites, and (B) in the midbrain of an embryo with approximately six pairs of somites. (C) Flow diagram of the assays performed to elucidate the effect of depleted levels of Snail2 on candidate gene expression levels during EMT in trunk and midbrain neural crest. (D) Responsiveness of various candidate genes to decreased Snail2 protein levels in the trunk 4 hours and 2 hours after treatment with the MO, as assessed by QPCR. Results for Cad6B expression in the trunk and midbrain are also shown after a 30 minute treatment with the Snail2 or control MO. Results are reported as fold difference relative to that obtained with the control MO. Black bars, Snail2 MO; white bars, control MO. Results presented are an average of at least two independent experiments performed with quadruple replicates for each condition (Snail2 or control MO).
Taking advantage of the known role of Snail2 as a transcriptional repressor in multiple systems (Nieto, 2002; Barrallo-Gimeno and Nieto, 2005), we examined genes whose transcript levels were increased as a result of knocking-down Snail2 translation. We selected a number of good candidates based on their relative expression patterns compared with Snail2. These included FoxD3, Msx1, Pax3, Ncad, Cad6B, and RhoB.
Four and 2 hours after electroporation of the Snail2 MO in the trunk, several candidate transcripts, including FoxD3 and Cad6B (2 and 4 hours), Ncad (2 hours), and Msx1 (4 hours) (Fig. 1D), were upregulated in Snail2 MO-treated compared with control MO-treated embryos. Further characterization revealed that Cad6B was upregulated 3-fold in the presence of the Snail2 MO as early as 30 minutes post-electroporation in the trunk (Fig. 1D). In addition, upregulation of at least 1.5-fold (mode value of 3.6) of Cad6B was observed 30 minutes after electroporation of the midbrain region with the Snail2 MO (Fig. 1D). Although detectable by QPCR, these changes in Cad6B transcript levels were below the threshold of detection possible by in situ hybridization and/or immunohistochemistry. Interestingly, no upregulation of a related cadherin, Ncad, was observed by QPCR after 30 minutes, showing the specificity of Snail2 for Cad6B (data not shown).
At later times (e.g. 15 hours) after MO electroporation in the trunk, some genes (Ncad, Cad6B, RhoA) exhibited decreased transcript levels in the presence of the Snail2 MO, whereas others were unchanged, making it likely that their repression is not mediated directly by Snail2 at this time point (see Fig. S2 in the supplementary material).
Snail2 and Cad6B have largely non-overlapping expression patterns during chick neural crest emigration
The rapid upregulation of Cad6B transcripts in the presence of the Snail2 MO after only 30 minutes raises the intriguing possibility that it may be a direct target of Snail2 repression. Consistent with this hypothesis, Cad6B has been shown to be downregulated upon neural crest emigration in the trunk (Nakagawa and Takeichi, 1995; Nakagawa and Takeichi, 1998). To gain further insight into the relationship between Snail2 and Cad6B at stages corresponding to EMT of the neural crest, we compared their mRNA and protein expression patterns at midbrain and trunk levels using whole-mount in situ hybridization and immunostaining (Figs 2, 3).
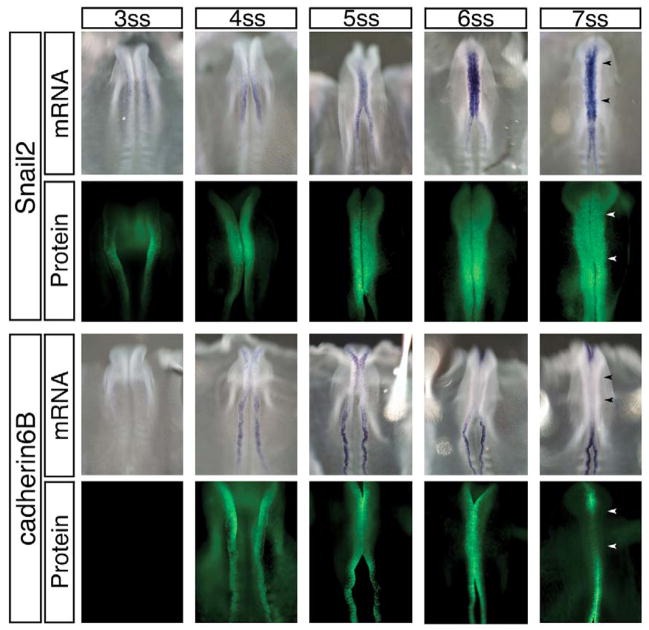
Fig. 2. In the midbrain, increasing levels of Snail2 expression coincide with a decrease in Cad6B transcript and protein prior to neural crest EMT.
Time-course of the transcript (whole-mount in situ hybridization) and protein (immunohistochemistry) distribution of Snail2 and Cad6B in the midbrain region during the period of EMT, just prior to the onset of neural crest cell emigration. Cad6B transcripts are initially detected in the midbrain region at the 4–5 somite stage (ss), but are diminished as both Snail2 transcripts and protein are upregulated. Interestingly, Cad6B protein is present after its transcripts are no longer detected up until 6 ss, and is eliminated at 7 ss, just one stage before the onset of neural crest cell migration from this region. Arrowheads identify midbrain boundaries.
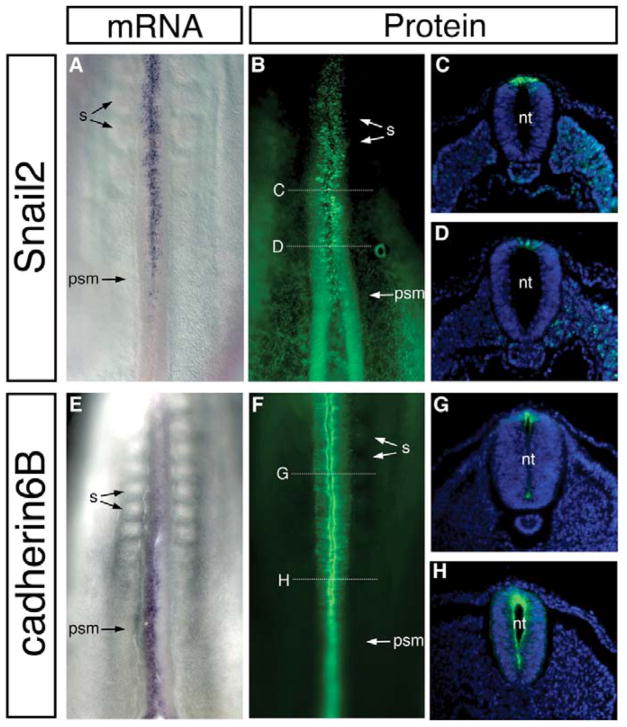
Fig. 3. Cad6B and Snail2 have largely non-overlapping expression patterns in the trunk.
Whole-mount in situ hybridizations (A, E) and immunohistochemistry (B–D, F–H) of 21- to 23-somite stage chick embryos for Snail2 (A–D) and Cad6B (E–H) expression. High levels of Snail2 mRNA and protein are detected in the dorsal neural tube as somites form, at axial levels that coincide with neural crest EMT. Conversely, Cad6B mRNA and protein are high in the caudal region of the embryo and decrease at more rostral regions at levels where neural crest EMT is occurring and migration is about to commence. nt, neural tube; psm, presomitic mesoderm; s, somite.
At midbrain levels, both Snail2 and Cad6B transcripts and proteins were found to be expressed initially in a small population of cells (Fig. 2). Snail2 transcripts and protein accumulated when the embryo had 6–7 somites, just prior to neural crest emigration. Cad6B transcripts were initially expressed in the neural tube but were subsequently downregulated at the 5–6 somite stage (ss), prior to initiation of neural crest emigration. However, Cad6B protein persisted until the 6–7 ss, just one stage prior to the onset of neural crest cell emigration from the midbrain (8 ss). After this stage, Cad6B protein was downregulated. Thus, in the midbrain region, we initially observed somewhat overlapping expression of Snail2 and Cad6B, but this expression became largely reciprocal (high Snail2 protein, low Cad6B mRNA) immediately prior to and during neural crest EMT at the 8 ss. A similar correlation was noted at truncal levels, with Snail2 protein levels high in the premigratory neural crest when Cad6B levels were low (Fig. 3). Thus, the expression profiles of Snail2 protein and Cad6B transcripts in the midbrain and trunk regions are consistent with a possible regulatory relationship between Snail2 and Cad6B.
Snail2 binds to the Cad6B regulatory region via E boxes
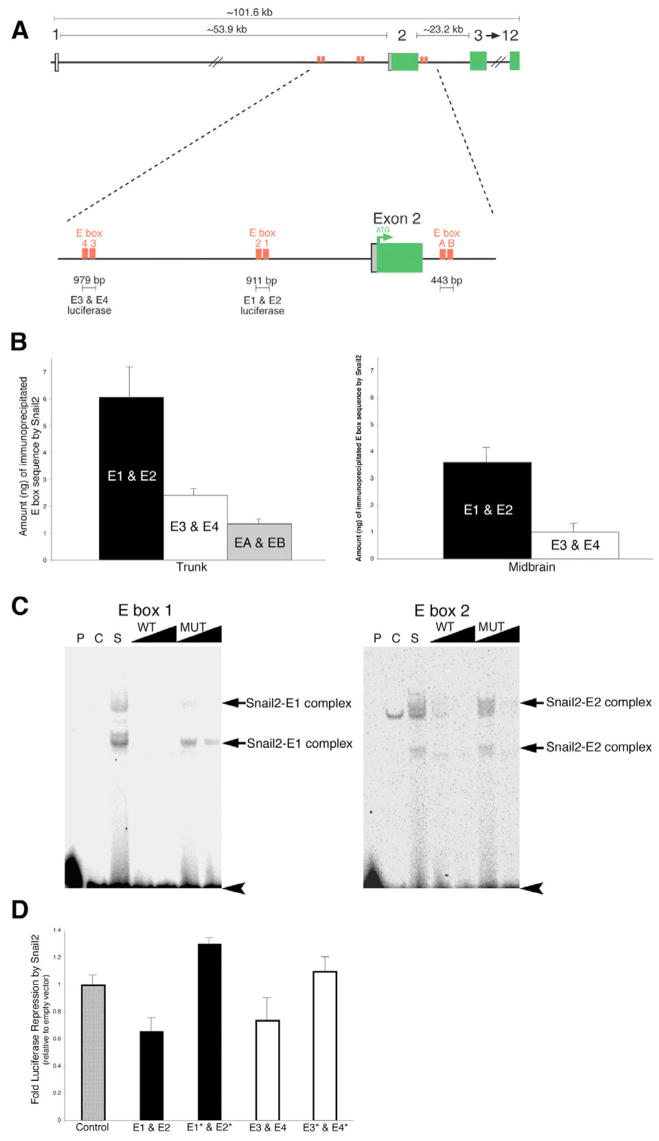
The Snail family of transcriptional repressors interacts with a core E-box motif CANNTA/G to mediate repression of their target genes (Giroldi et al., 1997; Nieto, 2002). To determine if the Cad6B sequence contains E-box motifs, we analyzed the genomic locus 130 kb upstream and downstream of the coding region in silico. Chick Cad6B is composed of 12 exons that are spread over ~102 kb on chromosome 2 (Ensembl Gene ID; ENSGALG00000012917). Sequence analysis identified a number of clustered E boxes containing the sequence CAGGTA surrounding the second exon of Cad6B, the first coding exon (Fig. 4A). To confirm the accuracy of the deposited sequence, the sequence was compared with a BAC clone (CH261-110K4) containing the region of interest. Approximately 8 kb of the region surrounding the ATG of Cad6B was sequenced in both directions, and six E-box motifs were found to be clustered in this putative regulatory region. The E-box motifs are grouped in three distinct regions and have been termed E4 and E3 (265 bp apart and 6.5 kb upstream of exon 2), E2 and E1 (305 bp apart and 2343 bp upstream of exon 2), and EA and EB (133 bp apart and 1.1 kb downstream of exon 2) (Fig. 4A). It should be noted that we identified several errors in the assembly of the deposited genomic sequence.
Fig. 4. Snail2 binds specifically to E boxes surrounding the ATG codon of Cad6B.
(A) Chick Cad6B is encoded by 12 exons on chromosome 2. Sequence analysis identified six E boxes with the motif CAGGTA clustered upstream and downstream of the ATG located in exon 2 of Cad6B. Primers and probes for chromatin immunoprecipitation (ChIP) and QPCR assays were designed to anneal to the region between E boxes within each clustered E-box pair. For luciferase experiments, the region of sequence spanning each clustered pair of E boxes that was cloned into the luciferase reporter vector is indicated. (B) ChIP experiments identify the preferential association of Snail2 with E boxes in the premigratory neural crest region of the chick trunk and midbrain, with E1 and E2>E3 and E4>EA and EB. At each axial level, the experiment was repeated at least three times, and the results presented are representative of a typical experiment. No association of Snail2 to distal sequences lacking E-box motifs in exon 11 of the Cad6B coding region was observed, and no E-box motifs were amplified by QPCR performed on samples immunoprecipitated with control antibodies (GFP, IgG) or with no antibodies added. (C) Electrophoretic mobility shift assay (EMSA) identifying the specific interaction of Snail2 with E box 1 and E box 2 in the Cad6B regulatory region (see also Fig. S3 in the supplementary material). Double-stranded 33P-end-labeled E-box oligonucleotide probes were incubated with control or Snail2 nuclear extract in the presence or absence of various competitor DNA oligonucleotide probes. P, no protein; C, 10 μg control nuclear extract. In the remaining lanes, 10 μg Snail2 nuclear extract was added to each binding reaction. S, no competitors added; WT and MUT, 10-fold and 100-fold molar excess of either unlabeled wild-type or unlabeled mutant E-box probes added, respectively. Retarded Snail2–E-box complexes are identified by arrows, and unbound probes are indicated by arrowheads. Two bands corresponding to E-box complexes were observed in the presence of the Snail2 nuclear extract. These bands were competed by the addition of wild-type E-box oligonucleotides, but to a much reduced efficiency with mutated E-box oligonucleotides. (D) Luciferase assays demonstrate that Snail2 represses luciferase expression from wild-type Cad6B E box-luciferase reporters but not from reporters with mutated E boxes. E1* and E2*, E1 and E2 double mutant; E3* and E4*, E3 and E4 double mutant. Results are reported as fold repression (relative to empty vector), and are an average of at least three independent QPCR experiments.
To define a direct molecular interaction between Snail2 and Cad6B, we employed a combination of in vivo (ChIP) and in vitro (EMSA and luciferase experiments) biochemical assays, as detailed below.
In vivo ChIP assay
To demonstrate in vivo association of Snail2 protein with the putative Cad6B regulatory region, we performed ChIP experiments with an antibody to Snail2 in conjunction with QPCR, using primers and probes designed to amplify genomic sequence between the E boxes comprising an E-box pair, on tissue isolated from the premigratory region of the chick trunk (+4 to −1 somite region) and the chick midbrain. The nature of this experiment allowed us to quantitatively determine the amount of Cad6B genomic sequence (represented by each clustered pair of E boxes) with which Snail2 potentially associates, generating a profile that describes the affinity of Snail2 for different E-box pairs (Fig. 4B). In the premigratory region of the avian trunk, Snail2 was found to preferentially associate with E1 and E2, with the overall interaction affinity described as E1 and E2>E3 and E4>EA and EB. This preferential association also held true for the midbrain, except that little to no Snail2 was found to associate with EA and EB at this axial level. No association of Snail2 was observed in a control QPCR reaction using Cad6B sequence from exon 11 that lacks any predicted E-box motifs, and no E boxes were immunoprecipitated in the absence of antibody or in the presence of a non-specific antibody, such as GFP or IgG. These in vivo results are the first to describe Snail2 association with the regulatory region of a bona fide target gene in a biological system.
EMSAs
Our ChIP data suggested that Snail2 associates predominantly with E1 and E2. To further validate the robustness with which Snail2 associates with these E boxes, EMSAs were performed using nuclear extract prepared from 293T cells transiently transfected with a Snail2 or empty vector construct. The Snail2-containing nuclear extract was able to interact directly with E1 and E2, forming two shifted complexes in this assay (Fig. 4C). These interactions were shown to be specific by out-competing the two Snail2–E-box complexes with unlabeled probes containing the wild-type E-box motif. By contrast, unlabeled probes containing E boxes with two point mutations in the E-box core motif were unable to compete for the Snail2 protein. In addition, the specificity of this interaction was confirmed by the ability of a Snail2 antibody to associate with a complex formed by E box 1 and affinity-purified recombinant GST-tagged Snail2 protein, thus further retarding the migration (i.e. ‘supershifting’) the complex in the gel (see Fig. S3 in the supplementary material). These results are similar to those describing the binding of mammalian Snail2 to E boxes in the E-cadherin promoter (Bolós et al., 2003).
Luciferase assays
QPCR-based luciferase assays were performed to assess the ability of Snail2 to regulate luciferase expression driven by various Cad6B E box-luciferase reporters (Fig. 4A, D). Co-transfection of Snail2 and Cad6B E box-luciferase reporters into 293T cells resulted in a statistically significant decrease in luciferase expression in the presence of E1 and E2, and E3 and E4, when compared with co-transfection of these reporters with an empty vector control (Fig. 4D). These results are in good agreement with those shown previously for the regulation of the human (Batlle et al., 2000) and mouse (Bolós et al., 2003; Conacci-Sorrell et al., 2003; Peinado et al., 2004) E-cadherin promoters by Snail1 and Snail2. Furthermore, we mutated the E boxes in these luciferase reporters (making the same two point mutations in the E-box core motif as in the EMSA experiments) to assess effects on luciferase expression in the presence or absence of Snail2. Our results showed that mutation of these E boxes abrogates the ability of Snail2 to repress luciferase expression. Our combined biochemical results suggest that the binding of Snail2 to E boxes in the Cad6B regulatory region mediates Snail2 repression of Cad6B transcription.
Snail2 affects neural crest EMT through a Cad6B-dependent mechanism
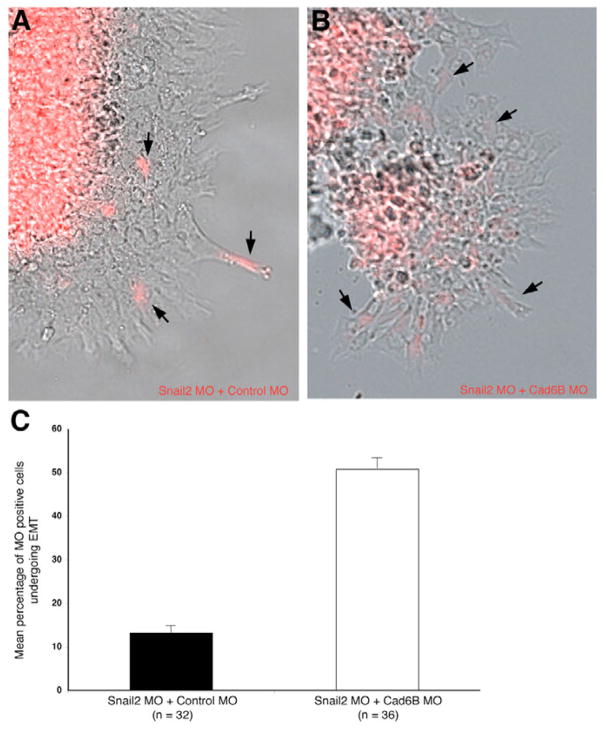
To examine whether Snail2 affects neural crest EMT in a Cad6B-mediated manner, we designed a morpholino to target the 5′ coding region of Cad6B in order to knock-down Cad6B translation. Embryos were electroporated with an equimolar mixture of either the Snail2 MO plus a control MO, or the Snail2 MO plus the Cad6B MO. Dissected neural folds from the electroporated side were placed in tissue culture, fixed 4 hours after explantation, and imaged. In the presence of the Snail2 MO alone, we observed only a small percentage of MO-positive neural crest cells (13%; n=32 explants) emigrating from the explanted neural folds (Fig. 5A), in keeping with previous work in chick (Nieto et al., 1994). The rescue of this decreased emigration, however, was partially achieved by inhibiting Cad6B translation concomitantly with the knock-down of Snail2 translation. Neural crest cells electroporated with an equimolar mixture of the Snail2 and Cad6B MOs were able to emigrate from cultured explants (51%; n=36 explants; P<0.0001; Fig. 5B, C). Collectively, these results demonstrate that Snail2 affects neural crest EMT, in part, via a Cad6B-dependent mechanism (Fig. 6).
Fig. 5. Snail2 modulates neural crest EMT in a Cad6B-dependent fashion.
Explantation and 4-hour culture of chick neural folds electroporated in vivo with an equimolar mixture of either (A) the Snail2 MO plus control MO or (B) the Snail2 MO plus Cad6B MO. (C) Bar chart showing s.e.m. of explantation results. Statistical analysis identifies a significant reduction in the number of neural crest cells undergoing EMT in the presence of Snail2 MO when compared with the Snail2 MO plus Cad6B MO (P<0.0001; unpaired Student’s t-test). 13% of emigrating neural crest cells from Snail2 MO-treated explants are Snail2 MO-positive, suggesting the requirement of Snail2 to ensure proper EMT (n=32 explants). 51% of emigrating neural crest cells contain both the Snail2 and Cad6B MOs, demonstrating that neural crest EMT occurs via a mechanism that involves repression of Cad6B by Snail2 (n=36 explants).
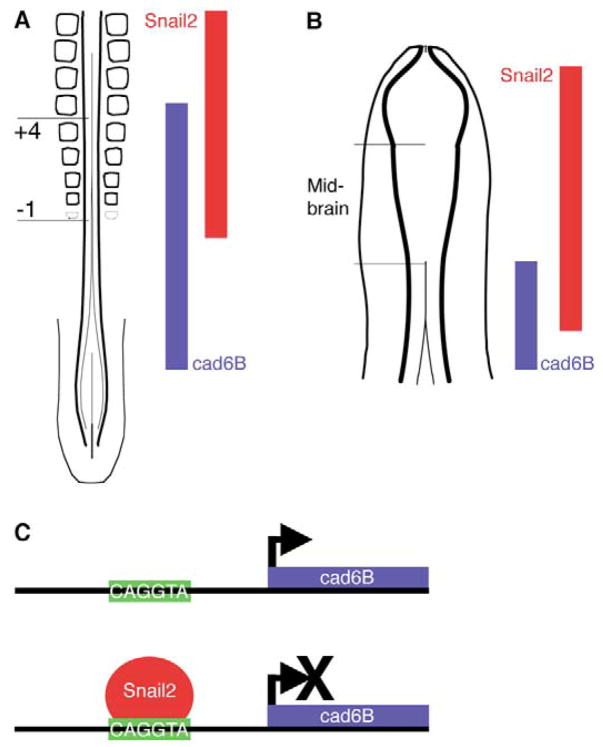
Fig. 6. Model illustrating the dynamic regulatory relationship between Snail2 and Cad6B in the developing avian embryo.
(A) In the premigratory region of the chick trunk, Snail2 protein is required to downregulate Cad6B transcripts along the rostro-caudal axis of the embryo such that, more rostrally, higher levels of Snail2 protein lead to lower levels of Cad6B transcripts, promoting neural crest EMT. More caudally, the absence of Snail2 protein results in the presence of Cad6B. (B) A similar pattern of expression is also observed in the midbrain, where, as neural crest cells undergo EMT (and just prior to neural crest migration), high Snail2 protein levels foster the repression of Cad6B transcripts. (C) The repression of Cad6B transcription is mediated by the direct binding of Snail2 to E-box motifs in the Cad6B regulatory region.
DISCUSSION
The role of Snail proteins in epithelial-to-mesenchymal transitions
The molecular events that govern EMTs coordinate the transformation of non-motile epithelial cells into motile mesenchymal cells, and are requisite for correct tissue assembly during embryogenesis. Aberrant activation of EMT can lead to cancer progression, endowing carcinoma cells with invasive and metastatic properties (Thiery, 2002). Members of the Snail superfamily (Sefton et al., 1998; Hemavathy et al., 2000b; Nieto, 2002; Barrallo-Gimeno and Nieto, 2005) have been shown to control genes whose products function in mediating cell movement both during metastasis (Hemavathy et al., 2000a; De Craene et al., 2005) and embryonic cell migration (Barrallo-Gimeno and Nieto, 2005). During oncogenesis, Snail1 and Snail2 play key roles in facilitating tumor metastasis through their ability to directly repress transcription of the adhesion molecule E-cadherin via an interaction with the E-cadherin promoter (Cano et al., 2000; Bolós et al., 2003; Côme et al., 2004).
A crucial developmental role for Snail family members is highlighted by the mouse knockout of Snail1, which is embryonic lethal owing to mesodermal defects during gastrulation (Carver et al., 2001). This process appears to be conserved during evolution because the mouse knockout phenotype is highly reminiscent of the abnormal migration of mesodermal precursor cells that occurs in Snail-deficient Drosophila gastrulae (Grau et al., 1984; Boulay et al., 1987). Previous work, however, has shown that Snail2 is not essential for mesoderm formation or neural crest development in mouse (Jiang et al., 1998). To address potential redundancies that might exist between mouse Snail family members, a double knockout of Snail1 and Snail2 in the mouse was recently generated; however, this mutant showed no defects in neural crest development (up to E9.5) (Murray and Gridley, 2006). These results raise the intriguing possibility that there are distinct functional roles, both conserved and non-conserved, for Snail family members in different organisms. For example, during mesoderm formation, the function of Snail1 in mouse is highly conserved with Xenopus Snail and chick Snail1 and Snail2.
The absence of a neural crest phenotype in the mouse Snail1/Snail2 double knockout indicates that significant differences exist with respect to the function of Snail family members in the neural crest of different vertebrates. Xenopus Snail (Carl et al., 1999; LaBonne and Bronner-Fraser, 2000) and chick Snail2 (Nieto et al., 1994) (present study) have been shown to have important functions in neural crest development. In particular, knock-down of Xenopus Snail not only affects neural crest emigration and migration, but also the formation of some neural crest derivatives (Carl et al., 1999). It is possible that neural crest development in the mouse proceeds through a redundant pathway that compensates for the loss of both Snail family members, perhaps reflecting a change in placental mammals. In this scenario, additional neural crest specifier genes may play a more dominant role in mouse versus chick or Xenopus. Consistent with species-specific roles for different transcription factors, the expression patterns of mouse and chick Snail1 and Snail2 genes appear to be ‘swapped’ during neural crest development (Sefton et al., 1998). Taken together, these results suggest that there is conservation of some, but not all, functions of Snail family members in the vertebrate lineage. Therefore, the molecular mechanisms controlling neural crest EMT in mouse and chick are likely to differ based on changes in the expression of both Snail and cadherin family members in these organisms.
Snail2 regulates the expression of many genes, including the cell adhesion molecule cadherin6B, during the EMT that characterizes neural crest development
The process of EMT is elegantly exemplified during embryonic development by the delamination of neural crest cells from the dorsal neural tube. Using this model system, we delineate the molecular role of Snail proteins during chick neural crest development by identifying target genes regulated by Snail2, the avian functional homolog of Snail1 (Sefton et al., 1998; del Barrio and Nieto, 2002). To this end, we have knocked-down the expression of Snail2 in the developing neural crest region and have identified a number of genes that are sensitive to depleted levels of Snail2 protein. We provide the first quantitative insight into the dynamic regulation of gene transcription during neural crest EMT in vivo. We find that depletion of Snail2 causes rapid and robust changes in the transcript levels of many genes in the premigratory neural crest (Msx1, FoxD3, Ncad, Cad6B) at many different time points.
Of particular interest is the observation that Cad6B expression is elevated within 30 minutes of Snail2 depletion, suggesting a potentially direct regulatory relationship between Snail2 and Cad6B. Support for this possibility came from examination of the putative regulatory region of chick Cad6B, in which we identified three pairs of clustered E boxes. This is a very similar E-box motif to that which Snail proteins bind in the regulation of E-cadherin expression. Furthermore, the expression patterns of Snail2 and Cad6B in the trunk and midbrain region of the developing chick embryo are consistent with this putative regulatory relationship. In these regions, Snail2 and Cad6B expression are somewhat overlapping, but, prior to neural crest EMT, Snail2 protein accumulates and Cad6B transcripts decrease. Thus, we find a correlation between the time of neural crest EMT and the time that Snail2 protein levels are high and Cad6B transcripts are low in the developing embryo. We do observe a slight delay in the downregulation of Cad6B transcripts once Snail2 protein is detected, particularly evident in the midbrain region at early stages of development. This is likely to be due to the biochemical nature of the Snail2 protein, the half-life of which is approximately 19 minutes (see Fig. S4 in the supplementary material); this is slightly shorter than that reported for Snail1 (half-life of 25 minutes) (Zhou et al., 2004). Snail1 undergoes extensive phosphorylation that promotes its degradation and results in its localization to the cytoplasm, thus abrogating its function as a transcriptional repressor (Zhou et al., 2004). Given these biochemical properties that characterize the Snail superfamily, it is probable that complete repression of Cad6B in these regions cannot occur until a sufficient level of Snail2 protein has accumulated and has been dephosphorylated.
Snail2 represses Cad6B transcription in vitro and in vivo
Our biochemical analyses demonstrate that Snail2 directly binds to E boxes in the Cad6B regulatory region to repress Cad6B transcription. In vitro gel shift assays verified that chick Snail2 protein binding to E box 1 and 2 in the Cad6B regulatory region is mediated by the presence of functional E boxes, as mutations in the E-box core motif prohibit Snail2 binding. Luciferase assays also demonstrate that Snail2 represses luciferase expression driven by Cad6B E boxes, and that the repression is dependent upon the integrity of the E-box motif. Furthermore, in vivo chromatin immunoprecipitation experiments performed in the chick midbrain and trunk prior to the onset of crest emigration demonstrate an association of Snail2 protein with the E boxes identified in the Cad6B regulatory region, with E1 and E2 having the highest affinity for Snail2, followed by E3 and E4 and then EA and EB. Taken together, these results strongly suggest that Snail2 directly represses Cad6B transcription through specific E boxes in the Cad6B regulatory region, in a highly dynamic fashion and at multiple axial levels in the developing chick embryo. Previous reports of Snail2-mediated repression of target genes, such as E-cadherin, have relied primarily on experiments performed in tissue culture cell lines (Behrens et al., 1991; Cano et al., 2000; Bolós et al., 2003). In chick, E-cadherin is not expressed in the neural tube at the stages under consideration, ruling it out as a possible Snail2 target in the neural crest. Furthermore, our in vivo explantation results suggest that proper neural crest EMT in the avian embryo proceeds in a Cad6B-dependent manner, with repression of Cad6B achieved by Snail2 prior to neural crest cell emigration. Our combined results are the first to be obtained in vivo that describe a direct target for Snail2 repression during avian neural crest EMT (Fig. 6).
The important developmental function of Snail2 has been well characterized during gastrulation (Nieto et al., 1994; Ciruna et al., 1997), somitogenesis (Dale et al., 2006) and neural crest cell development (Nieto et al., 1994; LaBonne and Bronner-Fraser, 1998; LaBonne and Bronner-Fraser, 2000; Sakai et al., 2006). Increasingly, a number of genes regulated by Snail family members have been identified with roles in development and disease in the control of key EMT processes (De Craene et al., 2005). The EMT that characterizes neural crest cell emigration from the dorsal neural tube is a highly coordinated event that involves the upregulation of mesenchymal markers and molecules that promote morphological changes and migration (such as Bmp4, Msx1/2, RhoA/B and Cad7), as well as the repression of genes that function to maintain the epithelial, non-motile state of the cell (such as Cad6B). Indeed, alterations in the expression of some of these same molecular EMT players also render tumor cells invasive, further emphasizing the shared molecular properties ascribed to the neural crest and metastasizing cancer cells (Hemavathy et al., 2000a; Côme et al., 2004; De Craene et al., 2005). Collectively, our results demonstrate that repression of Cad6B by Snail2 is highly dynamic and one of the significant events during the epithelial-to-mesenchymal conversion of a non-motile dorsal neural tube cell into a migratory neural crest cell.
Acknowledgments
We thank Tatjana Sauka-Spengler and Titus Brown for assistance with genomic analysis, Peter Farlie for help with the Snail2 immunohistochemistry protocol, Dorota Skowronska-Krawczyk for advice on the chromatin immunoprecipitations, and David Arce for excellent technical assistance. This work was supported by grants from the NIH-NICHD (NRSA-F32 HD43535 to L.A.T.), American Heart Association (0525037Y to E.G.C.) and NIH (NS36585 to M.B.-F.).
Footnotes
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/134/8/1481/DC1
References
- Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor Snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- Behrens J, Lowrick O, Klein-Hitpass L, Birchmeier W. The E-cadherin promoter: functional analysis of a G.C-rich region and an epithelial cell-specific palindromic regulatory element. Proc Natl Acad Sci USA. 1991;88:11495–11499. doi: 10.1073/pnas.88.24.11495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolós V, Peinado H, Perez-Moreno M, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- Boulay JL, Dennefeld C, Alberga A. The Drosophila developmental gene Snail encodes a protein with nucleic acid binding fingers. Nature. 1987;330:395–398. doi: 10.1038/330395a0. [DOI] [PubMed] [Google Scholar]
- Cano A, Perez-Moreno M, Rodrigo I, Locascio A, Blanco M, del Barrio MG, Portillo F, Nieto MA. The transcription factor Snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Carl TF, Dufton C, Hanken J, Klymkowsky MW. Inhibition of neural crest migration in Xenopus using antisense Slug RNA. Dev Biol. 1999;213:101–115. doi: 10.1006/dbio.1999.9320. [DOI] [PubMed] [Google Scholar]
- Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21:8184–8188. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciruna BG, Schwartz L, Harpal K, Yamaguchi TP, Rossant J. Chimeric analysis of fibroblast growth factor receptor-1 (Fgfr1) function: a role for FGFR1 in morphogenetic movement through the primitive streak. Development. 1997;124:2829–2841. doi: 10.1242/dev.124.14.2829. [DOI] [PubMed] [Google Scholar]
- Côme C, Arnoux V, Bibeau F, Savagner P. Roles of the transcription factors Snail and Slug during mammary morphogenesis and breast carcinoma progression. J Mammary Gland Biol Neoplasia. 2004;9:183–193. doi: 10.1023/B:JOMG.0000037161.91969.de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conacci-Sorrell M, Simcha I, Ben-Yedidia T, Blechman J, Savagner P, Ben-Ze’ev A. Autoregulation of E-cadherin expression by cadherin-cadherin interactions: the roles of beta-catenin signaling, Slug, and MAPK. J Cell Biol. 2003;163:847–857. doi: 10.1083/jcb.200308162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale JK, Malapert P, Chal J, Vilhais-Neto G, Maroto M, Johnson T, Jayasinghe S, Trainor P, Herrmann B, Pourquie O. Oscillations of the Snail genes in the presomitic mesoderm coordinate segmental patterning and morphogenesis in vertebrate somitogenesis. Dev Cell. 2006;10:355–366. doi: 10.1016/j.devcel.2006.02.011. [DOI] [PubMed] [Google Scholar]
- De Craene B, van Roy F, Berx G. Unraveling signalling cascades for the Snail family of transcription factors. Cell Signal. 2005;17:535–547. doi: 10.1016/j.cellsig.2004.10.011. [DOI] [PubMed] [Google Scholar]
- del Barrio MG, Nieto NA. Overexpression of Snail family members highlights their ability to promote chick neural crest formation. Development. 2002;129:1583–1593. doi: 10.1242/dev.129.7.1583. [DOI] [PubMed] [Google Scholar]
- Giroldi LA, Bringuier PP, de Weijert M, Jansen C, van Bokhoven A, Schalken JA. Role of E boxes in the repression of E-cadherin expression. Biochem Biophys Res Commun. 1997;241:453–458. doi: 10.1006/bbrc.1997.7831. [DOI] [PubMed] [Google Scholar]
- Grau Y, Carteret C, Simpson P. Mutations and chromosomal rearrangements affecting the expression of snail, a gene involved in embryonic patterning in Drosophila melanogaster. Genetics. 1984;108:347–360. doi: 10.1093/genetics/108.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemavathy K, Ashraf SI, Ip YT. Snail/Slug family of repressors: slowly going into the fast lane of development and cancer. Gene. 2000a;257:1–12. doi: 10.1016/s0378-1119(00)00371-1. [DOI] [PubMed] [Google Scholar]
- Hemavathy K, Guru SC, Harris J, Chen JD, Ip YT. Human Slug is a repressor that localizes to sites of active transcription. Mol Cell Biol. 2000b;26:5087–5095. doi: 10.1128/mcb.20.14.5087-5095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itasaki N, Bel-Vialar S, Krumlauf R. ‘Shocking’ developments in chick embryology: electroporation and in ovo gene expression. Nat Cell Biol. 1999;1:E203–E207. doi: 10.1038/70231. [DOI] [PubMed] [Google Scholar]
- Jiang R, Lan Y, Norton CR, Sundberg JP, Gridley T. The Slug gene is not essential for mesoderm or neural crest development in mice. Dev Biol. 1998;198:277–285. [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Neural crest induction in Xenopus: evidence for a two-signal model. Development. 1998;125:2403–2414. doi: 10.1242/dev.125.13.2403. [DOI] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Snail-related transcriptional repressors are required in Xenopus for both the induction of the neural crest and its subsequent migration. Dev Biol. 2000;221:195–2005. doi: 10.1006/dbio.2000.9609. [DOI] [PubMed] [Google Scholar]
- Murray SA, Gridley T. Snail family genes are required for left-right asymmetry determination, but not neural crest formation, in mice. Proc Natl Acad Sci USA. 2006;103:10300–10304. doi: 10.1073/pnas.0602234103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Takeichi M. Neural crest cell-cell adhesion controlled by sequential and subpopulation-specific expression of novel cadherins. Development. 1995;121:1321–1332. doi: 10.1242/dev.121.5.1321. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Takeichi M. Neural crest emigration from the neural tube depends on regulated cadherin expression. Development. 1998;125:2963–2971. doi: 10.1242/dev.125.15.2963. [DOI] [PubMed] [Google Scholar]
- Nieto NA. The Snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- Nieto MA, Sargent MG, Wilkinson DG, Cooke J. Control of cell behavior during vertebrate development by Slug, a zinc finger gene. Science. 1994;264:835–839. doi: 10.1126/science.7513443. [DOI] [PubMed] [Google Scholar]
- Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/Histone Deacetylase 1 (HDAC1)/HDAC2 complexes. Mol Cell Biol. 2004;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai D, Suzuki T, Osumi N, Wakamatsu Y. Cooperative action of Sox9, Snail2, and PKA signaling in early neural crest development. Development. 2006;133:1323–1333. doi: 10.1242/dev.02297. [DOI] [PubMed] [Google Scholar]
- Sefton M, Sanchez S, Nieto MA. Conserved and divergent roles for members of the Snail family of transcription factors in the chick and mouse embryo. Development. 1998;125:3111–3121. doi: 10.1242/dev.125.16.3111. [DOI] [PubMed] [Google Scholar]
- Taneyhill LA, Bronner-Fraser M. Dynamic alterations in gene expression after Wnt-mediated induction of avian neural crest. Mol Biol Cell. 2005;16:5283–5293. doi: 10.1091/mbc.E05-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- Wu WS, Heinrichs S, Xu D, Garrison SP, Zambetti GP, Adams JM, Look AT. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 2005;123:641–653. doi: 10.1016/j.cell.2005.09.029. [DOI] [PubMed] [Google Scholar]
- Xu Q, Wilkinson DG. In situ hybridisation of mRNA with hapten labeled probes. In: Wilkinson DG, editor. In Situ Hybridisation: A Practical Approach. Oxford: Oxford University Press; 1998. pp. 87–106. [Google Scholar]
- Zhou BP, Deng J, Xia W, Xu J, Li YM, Gunduz M, Hung MC. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6:931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]