Figure 4.
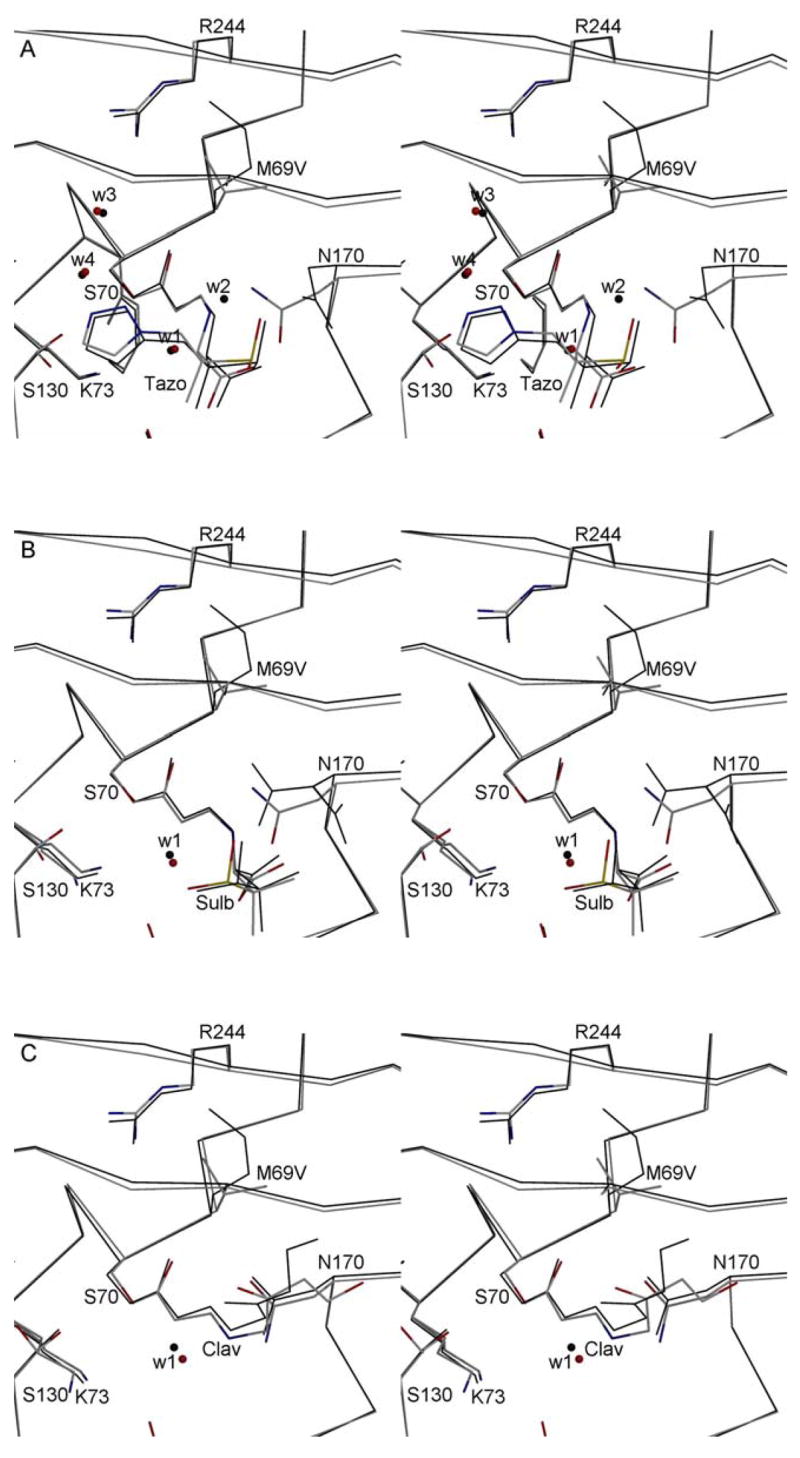
Stereo diagrams depicting the active sites of the inhibitor bound structures of the E166A and inhibitors resistant M69V/E166A variants. For each of the inhibitors, the M69V/E166A double mutant (grey with colored N, O, and S atoms) and the single E166A mutant (black lines) are superimposed. Tazobactam (tazo, 3A), sulbactam (sulb, 3B), and clavulanic acid (clav, 3C) are shown covalently attached to S70. The E166A SHV-1 β-lactamase structures used for the super positions have PDB identifiers 1RCJ (complexed with tazobactam), 2A49 (complexed with sulbactam), and 2A3U (complexed with clavulanic acid) with E166A) are used for comparison of the respective inhibitor bound double mutant structures. Water molecules present near the inhibitor are shown as small spheres. The catalytic water involved in deacylation (W1) is highlighted. For tazobactam in the single mutant structure, a ligand induced shift causes N170 to reorient thereby shifting the position of catalytic water to a new position (W2). Two additional waters are depicted in the tazobactam bound structures (W3 & W4) which are key for interacting with either tazobactam and K234 and/or R244.
