Abstract
Background
With multiple strains of various pathogens being sequenced, it is necessary to develop high-throughput methods that can simultaneously process multiple bacterial or viral genomes to find common fingerprints as well as fingerprints that are unique to each individual genome. We present algorithmic enhancements to an existing single-genome pipeline that allows for efficient design of microarray probes common to groups of target genomes. The enhanced pipeline takes advantage of the similarities in the input genomes to narrow the search to short, nonredundant regions of the target genomes and, thereby, significantly reduces the computation time. The pipeline also computes a three-state hybridization matrix, which gives the expected hybridization of each probe with each target.
Results
Design of microarray probes for eight pathogenic Burkholderia genomes shows that the multiple-genome pipeline is nearly four-times faster than the single-genome pipeline for this application. The probes designed for these eight genomes were experimentally tested with one non-target and three target genomes. Hybridization experiments show that less than 10% of the designed probes cross hybridize with non-targets. Also, more than 65% of the probes designed to identify all Burkholderia mallei and B. pseudomallei strains successfully hybridize with a B. pseudomallei strain not used for probe design.
Conclusion
The savings in runtime suggest that the enhanced pipeline can be used to design fingerprints for tens or even hundreds of related genomes in a single run. Hybridization results with an unsequenced B. pseudomallei strain indicate that the designed probes might be useful in identifying unsequenced strains of B. mallei and B. pseudomallei.
Background
Sequence-based pathogen identification is an increasingly important tool for clinical diagnostics and environmental monitoring of biological threat agents. Developments in sequencing technology have led to the availability of many pathogen genome sequences. Many more pathogen genomes and near-neighbors are being sequenced due to initiatives by the National Institute of Allergy and Infectious Diseases and the U.S. Department of Defense. Availability of these genomic sequences has opened up opportunities for the development of whole-genome-based diagnostic assays, such as DNA microarrays and polymerase chain reaction (PCR) assays, which offer more flexibility than traditional methods based on a single gene or selected regions of a target genome [1]. Microarray-based pathogen diagnostic assays are gaining popularity due to their ability to test for hundreds, or even thousands, of pathogens in a single diagnostic test [2].
Oligonucleotide probes designed for pathogen diagnostic assays should be unique to the pathogen with respect to all other non-target genomes. Clinical and environmental samples may contain a multitude of non-target genomes, and hence probes designed for diagnostic assays must be unique with respect to all non-target genomes. As a result, the design of pathogen diagnostic assays entails the computationally expensive comparison of target genomes with all known non-target sequences. Many different methods have been developed to guide the design of pathogen diagnostic assays. Some methods [3-5] are intended for PCR-based assays, whereas others [6-12] are intended for microarray-based assays. Kaderali and Schliep [6] presented one of the first methods for designing microarrays for pathogen identification. Their approach is very similar to that of designing probes for gene expression analysis; they design a single probe for each target, with the probe being unique to the target with respect to all other input target sequences. However, the specificity of the probe with respect to other non-target genomes is not analyzed. A somewhat similar approach is that of host-blind probe design presented by Putonti et al. [13], in which the probes are unique only with respect to a host genome. With a few exceptions, however, most of these tools do not have the capability of testing for specificity against a large number of non-target genomes. This is clearly not adequate if the signatures are to be used to identify the pathogen from environmental/clinical samples containing any number of unanticipated non-target organisms.
Some tools for designing PCR assays, such as KPATH [5] and Insignia [4], perform in silico comparisons against all known non-target sequences. They also have the ability to design common signatures for multiple pathogen sequences. In KPATH, common PCR signatures are selected from a multiple sequence alignment of the target genomes. As described by Fitch et al. [3], this approach is inherently based on the assumption of collinearity within the target genomes, which may not hold true for bacterial genomes. Insignia, on the other hand, selects common signatures from shared sequences discovered through pairwise local alignments and, hence, does not assume collinearity.
Neither KPATH nor Insignia is applicable for designing microarray fingerprints, as the design and specificity requirements of microarray fingerprints are quite different from those of PCR signatures. The most commonly used PCR signatures consist of a probe and two primers and, due to their short length [18–25 base pairs (bp)] and constraints on the interprimer distance, inexact matches with non-target sequences can be tolerated without much degradation in specificity. Conversely, in addition to being characterized by only one DNA segment with no spacing constraints, microarray probes are generally longer and more susceptible to cross hybridization even in the absence of an exact match [14]. This requires more extensive searches, for both exact and inexact matches, against non-target sequences to identify highly specific fingerprints for microarrays.
We have previously developed a software tool [9,12] for designing microarray probes that identify fingerprints for a single target genome. The software, named TOFI (Tool for Oligonucleotide Fingerprint Identification), is an integrated, scalable, high-performance-computing pipeline, which combines genome comparison tools, probe design software, and sequence alignment programs to design highly specific microarray probes for pathogen identification. To our knowledge, TOFI is the only software that has the ability to design microarray probes that are specific to the target with respect to all sequenced non-target genomes.
In this paper, we extend the TOFI pipeline to design microarray probes for multiple, related, bacterial and viral pathogens. Our aim is to efficiently identify all probes in the input target sequences that are unique with respect to all available non-target sequences. The major contributions of this paper include: (1) an efficient algorithm that pre-processes the input target sequences to take advantage of the similarities among them and reduce their effective size, which results in considerable speedup (nearly four-fold for eight targets and greater for larger numbers) when compared to the previous, single-target version of the pipeline, (2) a novel three-state hybridization matrix computation strategy, which identifies probes that can be used to characterize the possible combinations of the input target genomes, and (3) a set of experimental results with multiple Burkholderia genomes, which allow us to validate the new algorithms and the specificity criteria that were recently introduced (but not experimentally validated) in [12]. The improved pipeline scales well with increase in the number of target genomes, and can potentially design fingerprints for hundreds of related target genomes in a single run.
Methods
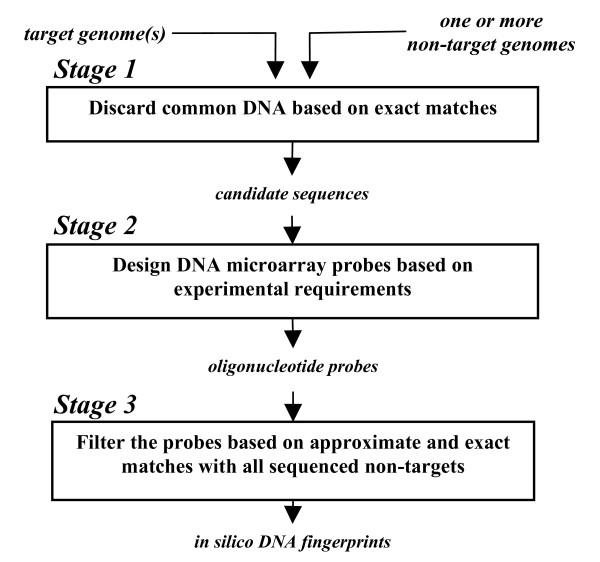
In the following, we briefly describe the TOFI pipeline for a single genome, and then present the algorithmic improvements implemented to accommodate multiple genomes. The TOFI pipeline consists of the three main stages illustrated in Figure 1. The stages are designed so that large portions of the target genome are eliminated in the less-expensive two initial stages, and the computationally more expensive searches for specific fingerprints are performed over smaller regions of the target genome in the final stage. The reader should refer to [12] for a detailed description of the TOFI pipeline.
Figure 1.
Overview of the TOFI pipeline. Stage 1 and Stage 3 of the TOFI pipeline have been improved to handle multiple genomes. In stage 1, the target genomes are compared with each other to eliminate redundant sequences. In Stage3, an in silico hybridization matrix is computed, which indicates which probes hybridize to which targets.
Overview of TOFI pipeline
The first stage of TOFI uses the suffix-tree-based MUMmer [15] program to perform pairwise comparisons of the target genome with each non-target genome and eliminate regions in the target genome that have exact matches with any of the non-target genomes. Given a pair of sequences, MUMmer finds all maximal matches that are at least as long as a threshold (termed minmatch) between the two sequences. TOFI uses MUMmer to find these maximal matches and eliminate regions in the target genome that are covered by them. The selection of minmatch is based on the specificity parameters supplied by the user. This ensures that every segment of the target genome that satisfies the restrictions on probe length and specificity parameters is part of the surviving regions of the target genome. These surviving regions, referred to as candidate sequences, are then passed on to the second stage of the pipeline.
In the second stage, TOFI identifies oligonucleotides of desired length from the candidate sequences that satisfy experimental conditions, such as melting temperature (Tm) and GC content. TOFI uses the Oligonucleotide Modeling Platform (OMP) software to identify these oligonucleotides, also referred to here as probes. OMP uses the nearest-neighbor hybridization model [16] to calculate Tm and to estimate if a probe forms any secondary structures that may prevent it from hybridizing to the intended target.
In the third and final stage of the pipeline, TOFI performs a BLAST [17] search for each probe against a comprehensive sequence database, such as the nt database provided by the National Center for Biotechnology Information (NCBI). The BLAST comparisons are performed in parallel on multiple processors using the blastn program of mpiBLAST [18]. Probes with significant alignments to non-target genomes are eliminated, and the surviving probes become the in silico DNA fingerprints for the target genomes. These probes are then subjected to experimental validation to test their sensitivity and specificity.
Multiple Genomes
Given a set of target genomes, our aim is to find microarray fingerprints that are unique to any subset of the target genomes with respect to all sequenced non-target genomes. The input consists of a set of K target genomes T = {t1, t2, ..., tK}, where each target tk, 1 ≤ k ≤ K, is a collection of all the sequences (a FASTA file containing chromosomes, unassembled contigs, etc.) from the kth target genome. The aim is to select a set of N probes P = {p1, p2, ..., pN}, where each probe pn of length |pn| is a substring of some sequence in T, with Lmin ≤ |pn| ≤ Lmax, and Lmin and Lmax representing the minimum and maximum probe length constraints, respectively. In addition, a probe pn should satisfy experimental constraints like GC content and melting temperature, and should not have significant sequence similarity with any known genomic sequence not in T.
The multiple-genome pipeline differs from the single-genome pipeline in Stage 1 and Stage 3. The major enhancements include: (1) comparison of each target genome with all other target sequences to eliminate redundant sequence segments from further consideration in Stage 1, and (2) computation of an in silico hybridization matrix of patterns in Stage 3, where each pattern identifies the input target sequences that can be characterized by a probe.
Preprocessing the target genomes
A brute-force approach for designing fingerprints for multiple genomes would entail the design of fingerprints for each genome separately. For K target genomes, this approach would take approximately K times the computation time necessary for designing fingerprints for a single genome. Given the recent sequence availability of multiple bacterial strains of interest (ranging from tens to hundreds) and the large computation time to identify fingerprints for a typical bacterial genome (~5 hours on 74 processors), such brute-force approach would be impractical.
In general, there is significant sequence similarity among closely related genomes. Our approach takes maximum advantage of such similarities by eliminating (redundant) sequence segments that are shared by any two input target genomes. This is done as the first step in Stage 1, where TOFI compares the targets within themselves to construct a set of nonredundant target sequences. The fingerprints for the multiple targets are then designed from these nonredundant target sequences. Comparison of the input target sequences is performed using an iterative process that effectively compares each target genome with all other target genomes.
We start with the set of target genomes T and a set of nonredundant target sequences S, which is initially empty. All sequences in the first target genome t1 are added to S. Next, we find all exact matches between the sequences in t2 and S using MUMmer. All exact matches that are longer than an input threshold are removed from t2, and the remaining sequences are added to S. This process is sequentially repeated for all other target genomes. When processing the kth genome, all nonredundant sequences from the previous k-1 sequences are already included in S. Hence, only the nonredundant sequences in tk are added to S. In this process, minmatch, the threshold for minimum exact matches, should be equal to Lmin. If minmatch is larger than Lmin, shared sequences of length ≥Lmin not reported by MUMmer will be included in S, leading to highly similar segments being added to the list of nonredundant sequences. The nonredundant sequences at the end of this preprocessing step are then subjected to comparisons with non-target sequences to remove exact matches with non-target sequences. The candidate sequences at the end of Stage 1 are submitted to Stage 2, which selects probes satisfying the experimental constraints. These probes are then subjected to extensive specificity analysis in Stage 3, which includes hierarchical BLAST comparisons against increasingly larger databases of non-target sequences [12].
In silico Hybridization Expectations
As the probes are designed to identify multiple target genomes, any given probe is not necessarily a substring of every target. As a result, it is necessary to explicitly compare each probe against each target sequence to identify the targets for which the probe can serve as a fingerprint. We consider multiple criteria in determining the specificity of a probe. Many measures, such as overall sequence identity, contiguous matches, and predicted free energy, have all been previously shown to be important measures of the potential for cross hybridization [19,20]. In addition to these measures, we use several measures of near-contiguous matches introduced in a previous paper on TOFI [12]. To incorporate contiguous and near-contiguous matches in determining probe specificity and estimate hybridization expectations, we use a series of thresholds, M0, M1, M2, and M3, where Mi is the maximum length of a contiguous region in which the alignment between a probe and a genome sequence has (Mi - i) matches and i mismatches/insertions/deletions. Accordingly, M0 is the length of the longest stretch of contiguous matches between a probe and a genome sequence. Identity, contiguous matches, and near-contiguous matches between a probe and a target/non-target genome are computed from the BLAST alignments between the two.
We use two sets of thresholds to compute in silico hybridization expectations. The first set of design thresholds, denoted by CU, indicates the minimum value of each parameter necessary for hybridization. The second set of thresholds, denoted by CL, indicates the maximum value of each parameter permissible for avoiding hybridization. CU is used to identify probes that will potentially hybridize to a genome, and CL is used to identify probes that will not hybridize to a genome. The individual thresholds in CU are denoted by IdentityU, M0U, M1U, M2U, and M3U, and the individual thresholds in CL are denoted by IdentityL, M0L, M1L, M2L, and M3L. For a probe to be considered as an in silico fingerprint for a target, all specificity measures between the probe and the target sequence must be greater than the corresponding thresholds in CU and all specificity measures between the probe and any non-target sequence must be less than or equal to the corresponding thresholds in CL. Note that the set of thresholds CU is only employed to compute the in silico hybridization expectations, which are used to identify the targets for which each probe can serve as a fingerprint. Accordingly, the number of probes reported by TOFI is solely controlled by the thresholds in CL.
We use the pairwise BLAST program bl2seq to compare each probe with each target sequence. Based on the alignments of the N probes with the K targets, we build an N × K hybridization matrix H, where each entry Hnk indicates whether probe pn hybridizes to target tk. Unlike earlier representations [21-23], which use a binary matrix to represent the hybridization expectations, we use a three-state matrix. Each Hnk ∈ {-1,0,1}, where Hnk = 1 indicates that fingerprint pn hybridizes with genome tk, Hnk = -1 indicates that pn does not hybridize to tk, and Hnk = 0 indicates that pn may or may not hybridize to tk. We opt for a three-state representation to more accurately represent the expected behavior of the probes. In many situations, probes may have some sequence similarity with a given target, but this identity may not be high enough to guarantee hybridization or low enough to rule out the possibility of hybridization.
The hybridization matrix is constructed based on the highest scoring alignment between a probe pn and a target genome tk, as follows:
i. Hnk = -1 if all the specificity measures are less than or equal to the corresponding thresholds in CL;
ii. Hnk = 1 if all the specificity measures are greater than the corresponding thresholds in CU; and
iii. Hnk = 0 if neither (i) nor (ii) is satisfied.
A probe pn is considered unique to a target tk if Hnk = 1, and Hnj = -1 ∀ j ≠ k. A probe pn is common to a set of targets Ts if:
i. Hnj = 1 ∀ j | tj ∈ Ts, and
ii. Hnj = -1 ∀ j | tj ∉ Ts.
According to this definition, any probe pn with Hnk = 0 for any target k can neither be a probe unique to a target nor a probe common to a set of targets. However, as these are only in silico expectations, some of these probes may prove to be useful after experimental validation.
Results
In this section, we present the results for the identification of in silico fingerprints (i.e., probe design) and their associated experimental evaluation. For the probe design process, we used probe length parameters Lmin = 35 and Lmax = 40, and optimal melting temperature of 70°C. Probe lengths of 35–40 bases were chosen to ensure compatibility of the probe sequences with microarrays available from various vendors, some of which are limited to the in situ synthesis of probes that are 40 bases or less. We used the entire NCBI nt database to estimate probe specificity.
Probe design
We designed probes for four strains of Burkholderia mallei and four strains of B. pseudomallei, employing B. thailandensis as the non-target near-neighbor genome. Table 1 shows the details of the eight target genomes and the near-neighbor genome, each consisting of two chromosomes. The table shows the combined sizes of the two chromosomes.
Table 1.
NCBI accession numbers and sizes of the Burkholderia genomes used for probe design
| Strain | Accession no./version | Size (bp) | |
| 1 | B. pseudomallei 1106a | NC_009076.1, NC_009078.1 | 7089249 |
| 2 | B. pseudomallei 1710b | NC_007434.1, NC_007435.1 | 7308054 |
| 3 | B. pseudomallei 668 | NC_009074.1, NC_009075.1 | 7040403 |
| 4 | B. pseudomallei K96243 | NC_006350.1, NC_006351.1 | 7247547 |
| 5 | B. mallei ATCC 23344 | NC_006348.1, NC_006349.1 | 5835527 |
| 6 | B. mallei NCTC 10229 | NC_008835.1, NC_008836.1 | 5742303 |
| 7 | B. mallei NCTC 10247 | NC_009079.1, NC_009080.1 | 5848380 |
| 8 | B. mallei SAVP1 | NC_008784.1, NC_008785.1 | 5232401 |
| 9 | B. thailandensis E264 (near-neighbor) | NC_007651.1, NC_007650.1 | 6723972 |
As expected, there is significant sequence similarity among the eight genomes. The four strains of B. pseudomallei are significantly different from each other. As a result, the combined nonredundant sequence size increased as each new B. pseudomallei sequence was processed. Conversely, the four strains of B. mallei are very similar to each other and to the four B. pseudomallei genomes, therefore only minimally increasing the combined nonredundant size as these genomes were processed. The combined size of the target genomes is 51343862 bp. However, the combined size of all nonredundant target sequences after the preprocessing step in Stage 1 was just 12011005 bp, a reduction of more than 75%. These nonredundant target sequences were further compared against the entire nt database retrieved from NCBI in July 2007. This version of the nt database consists of more than 5 million sequences with combined size greater than 21 Gbp.
The strategy of identifying and eliminating redundant portions of the target sequences considerably reduced the overall computation time. The total time to design fingerprints for the eight Burkholderia genomes with TOFI on a 74-processor Linux cluster with distributed memory was 9 hours and 41 minutes. Using the same number of processors, it took approximately 4 hours and 30 minutes to design fingerprints for each genome, for a total of 36 hours for processing the eight genomes. Therefore, for the eight Burkholderia genomes tested, the strategy used in the multiple-genome pipeline yielded a nearly four-fold reduction in the computation time in comparison with the single-genome pipeline. The savings in computation time would be even greater with larger number of target genomes.
Table 2 shows the values for the specificity thresholds CL and CU used for specificity computation with non-targets and targets, respectively. The thresholds for M0L and IdentityL were selected based on those suggested in the literature [14,19,24], and making the necessary adjustments to obtain a reasonably large number of fingerprints. The relaxation of these thresholds and the selection of other thresholds in CL and CU were based on empirical analyses of free energy computations previously presented [12]. Based on the CL thresholds, a total of 5015 probes were expected to be free of cross hybridization with non-targets. Table 3 shows the number of probes expected to identify each target. The third column in the table shows the number of probes that passed the design thresholds CU for each target strain; meaning that these are the number of probes that should hybridize with each strain. The fourth column indicates the number of probes that are unique to each target; meaning that these probes have matches ≤CL with all other genomes, including the other seven target strains. Column five shows the number of probes common to each subgroup, and the last column shows the number of probes common to all eight target strains.
Table 2.
The specificity thresholds used for probe design
| CL | CU |
| IdentityL = 85% | IdentityU = 90% |
| M0L = 18 | M0U = 27 |
| M1L = 21 | M1U = 29 |
| M2L = 24 | M2U = 32 |
| M3L = 27 | M3U = 34 |
Table 3.
Expected behavior of the 5015 designed probes
| Target | Total | Unique | Group | Common | |
| 1 | B. pseudomallei 1106a | 2710 | 259 | 504 | 981 |
| 2 | B. pseudomallei 1710b | 3346 | 739 | 504 | 981 |
| 3 | B. pseudomallei 668 | 2597 | 601 | 504 | 981 |
| 4 | B. pseudomallei K96243 | 3084 | 613 | 504 | 981 |
| 5 | B. mallei ATCC 23344 | 1373 | 0 | 31 | 981 |
| 6 | B. mallei NCTC 10229 | 1339 | 0 | 31 | 981 |
| 7 | B. mallei NCTC 10247 | 1567 | 0 | 31 | 981 |
| 8 | B. mallei SAVP1 | 1164 | 0 | 31 | 981 |
In all, 981 probes out of the total 5015 are expected to identify all eight strains. A total of 504 probes are unique to the B. pseudomallei subgroup, meaning that these probes have matches >CU with all the four B. pseudomallei strains and matches ≤CL with all other organisms, including the four B. mallei strains. Similarly, a total of 31 probes are unique to the B. mallei subgroup. There are hundreds of unique probes for each individual B. pseudomallei strain. However, because of the high similarity between the B. mallei genomes, none of the 5015 probes are unique to any individual B. mallei strain.
Hybridization experiments
Efficient hybridization of bacterial DNA requires that long genomic DNA molecules be fragmented to shorter lengths for optimal hybridization. In our experiments, we used restriction endonuclease digestion to fragment bacterial DNA prior to labeling and hybridization. Therefore, all probes that overlapped with restriction site positions corresponding to restriction enzymes used to prepare the DNA for hybridization were not included. This reduced the number of probes from 5015 to 2343. To reduce the number of probes further, we prioritized probes based on their predicted hybridization patterns and selected a total of 1214 probes by eliminating probes that were neither unique to an individual strain nor common to all strains in any of the two Burkholderia species. We conducted hybridization experiments on a total of 1817 probes, which include an additional set of 603 probes. Most of these 603 probes were selected based on sub-optimal specificity thresholds to assess the effect of various specificity criteria on cross hybridization. These 603 probes included 79 probes that were duplicated for verifying consistency of hybridization intensities with individual probes.
Samples from five different strains, B. mallei ATCC 23344, B. mallei NCTC 10229, B. pseudomallei K96243, B. pseudomallei 238, and B. thailandensis E264, were prepared for hybridization by restriction digestion in a cocktail of five restriction endonucleases: Rsa I, Hpa II, Hinp1 I, HpyCH4 IV, and Alu I (New England Biolabs, Ipswich, MA). Fragmented DNA was labeled with a ULYSIS 647 Nucleic Acid Labeling Kit (Invitrogen, Carlsbad, CA), following the manufacturer's instructions. One microgram of each labeled DNA was hybridized onto one segment each of three replicate custom Agilent 8x15K Comparative Genomic Hybridization (CGH) microarray chips (Agilent Technologies, Santa Clara, CA), following the manufacturer's instructions. Each CGH microarray chip consisted of eight identical individual array segments, each with 15000 features. Each individual array segment contained seven replicates each of the 1817 selected Burkholderia probes and seven replicates each of 31 negative control probes. The remaining unspecified features were populated by Agilent control probes, which were not included in the analysis. Hybridized chips were scanned with a GenePix 4000B Axon Scanner, using GenePix Pro 6.1 software (Molecular Devices, Sunnyvale, CA) at 5 μm for individual spot evaluation.
Evaluation of experimental results
We analyzed the hybridization results using microarray data analysis functions available in the MATLAB Bioinformatics Toolbox http://www.mathworks.com. We normalized the data among corresponding arrays in each of the three chips using the quantile normalization method [25] and logarithmically transformed the normalized hybridization intensities. Subsequently, we subtracted the background, which was estimated for each array on each chip using the set of 31 negative control probes. The resulting normalized values were used for evaluating the probes. The complete list of 1817 probes and their normalized hybridization intensities are given in Additional file 1.
Table 4 shows the median (mB) and standard deviation (σ) of the estimated non-logarithmically transformed background intensities. The background intensities were very consistent across the three chips (not shown), as well as among the three Burkholderia strains. Similar to probe design, we used two empirical thresholds RL and RU to classify probes based on these normalized hybridization data. We selected the lower threshold RL to assess cross hybridization with non-targets to be slightly less than 3σ above the background (see Table 4), RL = 0.5. For the upper threshold RU, which is used to assess hybridization with the intended targets, we selected a very conservative value of more than 6σ above the background, RU = 1.0.
Table 4.
Background hybridization intensities averaged over three chips for three Burkholderia genomes
| B. pseudomallei K96243 | B. mallei ATCC 23344 | B. mallei NCTC 10229 | |
| Median background intensity (mB) | 3710 | 3076 | 3890 |
| Standard deviation of background intensity (σ) | 548 | 463 | 569 |
| 0.53 | 0.54 | 0.53 | |
| 0.92 | 0.92 | 0.90 |
To enable consistent comparison between probes designed by TOFI and the experimental results, we re-evaluated the number of in silico fingerprints in a manner that simulates the experimental setup, using the same design thresholds CL and CU indicated in Table 2. Table 5 illustrates the different combinations of the three target strains, B. mallei ATCC 23344, B. mallei NCTC 10229, and B. pseudomallei K96243, that were available for experimental analysis. The first column denotes the five different groups of probes or categories we compared, and the second column identifies the number of re-evaluated in silico (i.e., design) probes. Categories I, II and III correspond to probes unique to individual targets, while treating all other genomes as non-targets. Probes in category IV are common to both B. mallei ATCC 23344 and B. mallei NCTC 10229, considering all other genomes (including B. pseudomallei K96243) as non-targets. Probes in Category V are common to all three target genomes, considering B. thailandensis as the non-target.
Table 5.
Evaluation of in silico (design) probes against hybridization results with RU = 1.0 and RL = 0.5 for five categories of probes
| Category (targets) | In silico | Experimental | ||
| Class A | Class B | Class C | ||
| I (B. mallei ATCC 23344) | 0 | 0 (0%) | 0 (0%) | 0 (0%) |
| II (B. mallei NCTC 10229) | 2 | 0 (0%) | 2 (100%) | 0 (0%) |
| III (B. pseudomallei K96243) | 523 | 420 (80%) | 53 (10%) | 50 (10%) |
| IV (B. mallei ATCC 23344 and B. mallei NCTC 10229) | 21 | 17 (81%) | 2 (10%) | 2 (10%) |
| V (B. mallei ATCC 23344, B. mallei NCTC 10229 and B. pseudomallei K96243) | 431 | 184 (43%) | 12 (3%) | 236 (55%) |
Probes that behave as expected are categorized as Class A; i.e., these probes have normalized hybridization intensity greater than RU with intended targets and less than RL with non-targets. Class B probes have normalized hybridization intensity greater than RL with non-targets, and Class C probes have normalized hybridization intensity less than RU with the intended targets.
Based on the experimental results, we classify the probes in each category into three classes (Table 5). Class A corresponds to probes that have normalized hybridization intensity greater than RU with the intended targets and normalized hybridization intensity less than or equal to RL with non-targets. Class B designates probes that have normalized hybridization intensity greater than RL with non-targets, whereas Class C designates probes that have normalized hybridization intensity less than or equal to RU with the intended targets. Therefore, Class A probes are the probes that behave as expected. Note that some probes can be in both Class B and Class C, and that the purpose of the design criteria CU and CL is to maximize the number of probes in Class A and minimize the number of probes in Class B and Class C.
Due to the high similarity between the two B. mallei genomes, the probes in Categories I and II (both in silico and experimental) are too few for analysis. According to the design criteria, 523 probes out of the 1817 are unique to B. pseudomallei (Category III). According to the experimental thresholds, 420 (80%) of these are in Class A, meaning that they hybridize with B. pseudomallei and do not hybridize with any of the other genomes. Relatively few probes in this category are in Class B or Class C. The probes unique to both strains of B. mallei (Category IV) perform similarly. A large fraction (81%) of these probes is in Class A. The probes in Category V, which are expected to hybridize with all three target genomes, behaved differently. Less than half of these probes (43%) are in Class A, whereas the majority (55%) of these probes are in Class C, meaning that they are failing to hybridize with some or all of the intended targets.
Performance against an unsequenced target
The robustness of common probes designed to identify a group of targets can be evaluated by testing their hybridization against another member of the group that was not included in the design process. Accordingly, we obtained the hybridization results of the 1817 probes with B. pseudomallei 238, for which the genome sequence is not available from NCBI. Hybridization results with this strain might provide insights into how common probes designed based on a limited set of strains of B. pseudomallei and B. mallei would perform on unsequenced strains of these pathogens. Table 6 shows the performance of these group-specific probes as the result of hybridization experiments with B. pseudomallei 238. The 302 in silico probes in Category VI represent the subset of the 1817 probes that are expected to identify all eight target genomes listed in Table 1. Similarly, the 92 probes in Category VII are expected to identify all four B. pseudomallei genomes in Table 1.
Table 6.
Experimental hybridization results of group-specific in silico probes tested against B. pseudomallei 238
| Category (targets) | In silico | Probes hybridizing with B. pseudomallei 238 |
| VI (B. mallei and B. pseudomallei) | 302 | 236 (78%) |
| VII (B. pseudomallei specific) | 92 | 60 (65%) |
Out of the 302 probes in Category VI, 236 (78%) have normalized hybridization intensity greater than RU = 1.0 with B. pseudomallei 238 and less than RL = 0.5 with B. thailandensis E264. Similarly, out of the 92 B. pseudomallei-specific probes in Category VII, 60 (65%) probes have normalized hybridization intensity greater than RU with B. pseudomallei 238, and less than RL with the two strains of B. mallei and the strain of B. thailandensis tested. These results indicate that these group-specific probes can be used to identify B. pseudomallei 238, and possibly other unsequenced strains of B. mallei and B. pseudomallei.
Experimental inconsistencies
Based on the two design thresholds, CU and CL, all 431 probes in Category V are expected to hybridize to all three targets. However, as shown in Table 5, only 184 of these are in Class A, whereas many (236) are in Class C. To understand why this might be happening, we looked at the number of probes out of these 431 that hybridize to each of the three individual targets. We found that 343 probes (80%) hybridized with B. mallei ATCC 23344, 408 probes (95%) hybridized with B. mallei NCTC 10229, and only 196 probes (45%) hybridized with B. pseudomallei K96243. This clearly indicates that most of the probes in category V failed to hybridize with B. pseudomallei K96243.
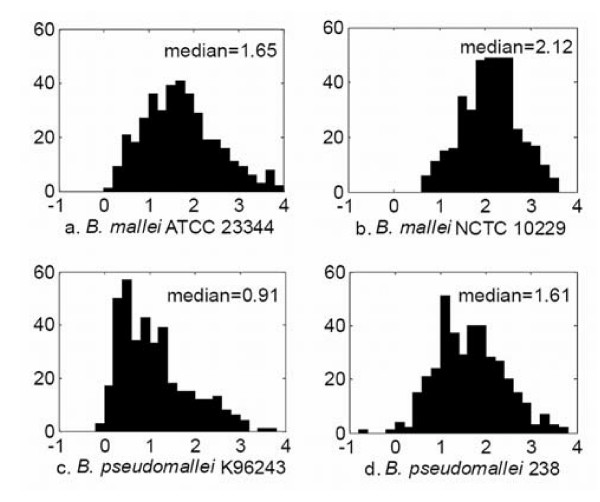
Further insights can be gained by looking at the probes that have 100% identity with each of the three targets. There are 382 such probes, and all of them are expected to hybridize to all three targets. Figure 2 shows the histograms of the normalized hybridization intensities for these 382 probes. Figures 2a, 2b, and 2c indicate that hybridization intensities with the B. mallei strains are significantly higher than those with B. pseudomallei K96243. The median hybridization intensities for both strains of B. mallei are substantially above the experimental threshold RU = 1.0, which is not the case for B. pseudomallei K96243. Hybridization with the unsequenced strain B. pseudomallei 238 in Figure 2d is also comparable to that of the two B. mallei strains, with a median value (1.61) higher than the threshold (1.0), indicating that a large fraction of these 382 probes can be used to detect this strain. Hybridization with B. thailandensis E264 (not shown) is at the background level.
Figure 2.
Histograms of normalized hybridization intensities for the 382 probes that have 100% identity with the three target genomes. The X-axis shows the normalized hybridization intensities and the Y-axis shows the number of probes that have a given normalized hybridization intensity. Many of the 382 probes fail to hybridize with B. pseudomallei K96243 even though all these probes have 100% identity with this genome, whereas hybridization intensities for the other three genomes are as expected.
Based on these histograms, one might be tempted to conclude that the hybridization intensities with B. pseudomallei K96243 are lower than those for the remaining strains due to some experimental anomaly. However, this does not seem to be the case because most of the 523 probes in category III (Table 5) that are expected to hybridize only to B. pseudomallei K96243 perform as expected. The median hybridization for these 523 probes is 2.44, which is well above the threshold 1.0. Currently, we do not have an explanation for why the strain-specific probes are hybridizing as expected with B. pseudomallei K96243, whereas group-specific probes are failing. We are investigating the causes for the observed discrepancies.
Conclusion
The enhanced TOFI pipeline can efficiently design microarray fingerprints for multiple, related bacterial and viral genomes. We designed probes for eight pathogenic Burkholderia genomes, covering probes unique to single targets as well as probes common to groups of targets. Probe design results show that the presented method is effective in taking advantage of the commonalities among the genomes to considerably reduce the overall computation time (about a four-fold reduction in this case, with larger gains for larger number of input targets). This indicates that the pipeline can be used to design fingerprints for a large number of related microbial genomes in a single run. In addition, the computational efficiency of the pipeline allows quick reevaluation of the probes as new target/non-target sequences become available.
This study also allowed us to assess and experimentally validate new specificity criteria recently introduced by Vijaya Satya et al. [12]. Preliminary hybridization results, with three targets, one unsequenced target, and one non-target, demonstrate that only a small percentage of the designed probes (≤10%) cross hybridize with non-targets (last three rows of Class B in Table 5). However, additional tests with a larger panel of non-target genomes are necessary to qualify the selected probes for diagnostic assays. More than 65% of the group-specific probes identify the unsequenced B. pseudomallei 238 strain, which suggests that these probes might be useful in identifying new strains of B. mallei or B. pseudomallei.
Authors' contributions
RVS conceived and implemented the algorithmic enhancements and KK upgraded the user interface for processing multiple genomes. LW designed and guided the experiments. EB and SP conducted the hybridization experiments. JG provided overall guidance for the hybridization experiments. NZ analyzed the hybridization data. JR provided overall project guidance for the computational efforts. RVS, NZ, LW and JR were the main writers of the manuscript. All authors reviewed the manuscript.
Availability and requirements
• Project name: TOFI
• Project home page: http://www.bhsai.org/downloads/tofi_multi.tar.gz
• Operating systems: Linux
• Programming Language: Perl
• Other Requirements: mpiBLAST 1.4.0 or higher, MUMmer 3.19 or higher, and OMP developer edition
Supplementary Material
List of probes and their normalized hybridization intensities. The file contains the list of 1817 probes that were tested experimentally. The first three columns give the probe name, probe sequence, and probe length, respectively. Columns 4 through 8 provide the average normalized hybridization intensity values for B. mallei ATCC 23344, B. mallei NCTC 10229, B. pseudomallei K96243, B. pseudomallei 238, and B. thailandensis E264, respectively. The averages were computed across three chips, where each probe was replicated seven times on each chip.
Acknowledgments
Acknowledgements
This work was sponsored by the U.S. Department of Defense High Performance Computing Modernization Program, under the High Performance Computing Software Applications Institutes initiative, and the U.S. Defense Threat Reduction Agency project 8.10006 07 RD B.
The opinions and assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the U. S. Army or of the U. S. Department of Defense. This paper has been approved for public release with unlimited distribution.
Contributor Information
Ravi Vijaya Satya, Email: rvijaya@bioanalysis.org.
Nela Zavaljevski, Email: nelaz@bioanalysis.org.
Kamal Kumar, Email: kamal@bioanalysis.org.
Elizabeth Bode, Email: Elizabeth.Ardis.Bode@us.army.mil.
Susana Padilla, Email: Susana.Lopez2@amedd.army.mil.
Leonard Wasieloski, Email: leonard.wasieloski@us.army.mil.
Jeanne Geyer, Email: jeanne.geyer@us.army.mil.
Jaques Reifman, Email: jaques.reifman@us.army.mil.
References
- Wilson WJ, Strout CL, DeSantis TZ, Stilwell JL, Carrano AV, Andersen GL. Sequence-specific identification of 18 pathogenic microorganisms using microarray technology. Mol Cell Probes. 2002;16:119–127. doi: 10.1006/mcpr.2001.0397. [DOI] [PubMed] [Google Scholar]
- Bryant PA, Venter D, Robins-Browne R, Curtis N. Chips with everything: DNA microarrays in infectious diseases. Lancet Infect Dis. 2004;4:100–111. doi: 10.1016/S1473-3099(04)00930-2. [DOI] [PubMed] [Google Scholar]
- Fitch JP, Gardner SN, Kuczmarski TA, Kurtz S, Myers R, Ott LL, Slezak TR, Vitalis EA, Zemla AT, McCready PM. Rapid Development of Nucleic Acid Diagnostics. Proceedings of the IEEE. 2002;90:1708–1720. doi: 10.1109/JPROC.2002.804680. [DOI] [Google Scholar]
- Phillippy AM, Mason JA, Ayanbule K, Sommer DD, Taviani E, Huq A, Colwell RR, Knight IT, Salzberg SL. Comprehensive DNA signature discovery and validation. PLoS Comput Biol. 2007;3:e98. doi: 10.1371/journal.pcbi.0030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slezak T, Kuczmarski T, Ott L, Torres C, Medeiros D, Smith J, Truitt B, Mulakken N, Lam M, Vitalis E, et al. Comparative genomics tools applied to bioterrorism defence. Brief Bioinform. 2003;4:133–149. doi: 10.1093/bib/4.2.133. [DOI] [PubMed] [Google Scholar]
- Kaderali L, Schliep A. Selecting signature oligonucleotides to identify organisms using DNA arrays. Bioinformatics. 2002;18:1340–1349. doi: 10.1093/bioinformatics/18.10.1340. [DOI] [PubMed] [Google Scholar]
- Rimour S, Hill D, Militon C, Peyret P. GoArrays: highly dynamic and efficient microarray probe design. Bioinformatics. 2005;21:1094–1103. doi: 10.1093/bioinformatics/bti112. [DOI] [PubMed] [Google Scholar]
- Rouillard JM, Herbert CJ, Zuker M. OligoArray: genome-scale oligonucleotide design for microarrays. Bioinformatics. 2002;18:486–487. doi: 10.1093/bioinformatics/18.3.486. [DOI] [PubMed] [Google Scholar]
- Tembe W, Zavaljevski N, Bode E, Chase C, Geyer J, Wasieloski L, Benson G, Reifman J. Oligonucleotide fingerprint identification for microarray-based pathogen diagnostic assays. Bioinformatics. 2007;23:5–13. doi: 10.1093/bioinformatics/btl549. [DOI] [PubMed] [Google Scholar]
- Wang D, Urisman A, Liu YT, Springer M, Ksiazek TG, Erdman DD, Mardis ER, Hickenbotham M, Magrini V, Eldred J, et al. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003;1:E2. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Tillier ER. A fast and flexible approach to oligonucleotide probe design for genomes and gene families. Bioinformatics. 2007;23:1195–1202. doi: 10.1093/bioinformatics/btm114. [DOI] [PubMed] [Google Scholar]
- Vijaya Satya R, Zavaljevski N, Kumar K, Reifman J. A high-throughput pipeline for designing microarray-based pathogen diagnostic assays. BMC Bioinformatics. 2008;9:185. doi: 10.1186/1471-2105-9-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putonti C, Chumakov S, Mitra R, Fox GE, Willson RC, Fofanov Y. Human-blind probes and primers for dengue virus identification. Febs J. 2006;273:398–408. doi: 10.1111/j.1742-4658.2005.05074.x. [DOI] [PubMed] [Google Scholar]
- He Z, Wu L, Li X, Fields MW, Zhou J. Empirical establishment of oligonucleotide probe design criteria. Appl Environ Microbiol. 2005;71:3753–3760. doi: 10.1128/AEM.71.7.3753-3760.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SantaLucia J, Jr, Hicks D. The thermodynamics of DNA structural motifs. Annu Rev Biophys Biomol Struct. 2004;33:415–440. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Darling A, Carey L, Feng W. 4th International Conference on Linux Clusters: The HPC Revolution 2003 in conjunction with the ClusterWorld Conference & Expo. San Jose, CA; 2003. The Design, Implementation, and Evaluation of mpiBLAST. [Google Scholar]
- Kane MD, Jatkoe TA, Stumpf CR, Lu J, Thomas JD, Madore SJ. Assessment of the sensitivity and specificity of oligonucleotide (50mer) microarrays. Nucleic Acids Res. 2000;28:4552–4557. doi: 10.1093/nar/28.22.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, He Z, Zhou J. Selection of optimal oligonucleotide probes for microarrays using multiple criteria, global alignment and parameter estimation. Nucleic Acids Res. 2005;33:6114–6123. doi: 10.1093/nar/gki914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klau GW, Rahmann S, Schliep A, Vingron M, Reinert K. Optimal robust non-unique probe selection using Integer Linear Programming. Bioinformatics. 2004;20:I186–I193. doi: 10.1093/bioinformatics/bth936. [DOI] [PubMed] [Google Scholar]
- Schliep A, Rahmann S. Decoding non-unique oligonucleotide hybridization experiments of targets related by a phylogenetic tree. Bioinformatics. 2006;22:e424–430. doi: 10.1093/bioinformatics/btl254. [DOI] [PubMed] [Google Scholar]
- Schliep A, Torney DC, Rahmann S. Group testing with DNA chips: generating designs and decoding experiments. Proc IEEE Comput Soc Bioinform Conf. 2003;2:84–91. [PubMed] [Google Scholar]
- Liebich J, Schadt CW, Chong SC, He Z, Rhee SK, Zhou J. Improvement of oligonucleotide probe design criteria for functional gene microarrays in environmental applications. Appl Environ Microbiol. 2006;72:1688–1691. doi: 10.1128/AEM.72.2.1688-1691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of probes and their normalized hybridization intensities. The file contains the list of 1817 probes that were tested experimentally. The first three columns give the probe name, probe sequence, and probe length, respectively. Columns 4 through 8 provide the average normalized hybridization intensity values for B. mallei ATCC 23344, B. mallei NCTC 10229, B. pseudomallei K96243, B. pseudomallei 238, and B. thailandensis E264, respectively. The averages were computed across three chips, where each probe was replicated seven times on each chip.