Preface
The control and coordination of eukaryotic gene expression rely on transcriptional and post-transcriptional regulatory networks. Although progress has been made in mapping the components and deciphering the function of these networks, the mechanisms by which such intricate circuits originate and evolve remain poorly understood. Here I revisit and expand earlier models proposing that genomic repeats, and in particular transposable elements, have been a rich source of material for the assembly and tinkering of eukaryotic gene regulatory systems.
It has been known for some time that eukaryotic genomes, with rare exceptions, are replete with interspersed repetitive DNA 1. Large-scale DNA sequencing has revealed that most of the repetitive DNA is derived from the activity of transposable elements (TEs), sequences able to move and replicate within the genome. TEs employ different replicative strategies, which involve either RNA (class 1 or retrotransposons) or DNA intermediates (class 2 or DNA transposons) 2. The broad distribution of all major TE classes across the eukaryotic tree of life indicates that they are long-standing residents of eukaryotic genomes 2. Unlike other lasting components of the genome, one needs not bestow TEs with adaptive value to account for their evolutionary persistence. Theoretical considerations and empirical studies show that TEs are best viewed as genomic parasites, which essentially owe their survival to their ability to replicate faster than the host that carries them 3,4. This conjecture, also known as the selfish DNA theory, seems sufficient to explain the maintenance of TEs over long evolutionary time as well as the wide variations in the amount, diversity and chromosomal location of TEs observed between or even sometimes within species 3,5. In spite of -and to some extent because of- this selfish and parasitic nature, the movement and accumulation of TEs have exerted a strong influence on the evolutionary trajectory of their hosts 3–6. Here I review recent discoveries supporting early theories postulating that TEs have played a major role in the evolution of eukaryotic gene regulation. Specifically I explore the properties of TEs that may facilitate their recruitment as building blocks for the assembly of a diversity of systems to regulate and coordinate eukaryotic gene expression.
Life after death: TE exaptation
The co-option of TEs (or exaptation7) to serve cellular function has long been recognized 8–10. But in recent years, the ability to align large amounts of human genomic sequences to their orthologous regions in widely diverged mammals has provided an opportunity to estimate the amplitude of TE exaptation by revealing fixed TE sequences that have been under functional constraint for extended evolutionary time. A pioneering study11 comparing a sample of human-mouse orthologous sequences suggested that a substantial fraction of ancestral repeats (inserted prior to the eutherian radiation) have been subject to strong selective constraint since at least the divergence of human and mouse, implying that mammalian TEs frequently acquire beneficial function for their host.
Recently, this comparative genomics approach was scaled up12, unveiling at least 10,000 TE fragments in the human genome that have evolved under strong purifying selection throughout the eutherian radiation. Furthermore, comparisons of the genome of a marsupial, the opossum, to several eutherian species (human, dog, mouse, rat) revealed that at least 16% of eutherian-specific conserved non-coding elements (CNEs) were derived from various kinds of TEs13. In addition, sensitive sequence similarity searches uncovered thousands of deeply conserved human CNEs (many of them predating the mammalian radiation), including a number of so-called ultraconserved elements, which can be clustered into hundreds of families, suggesting a distant TE origin14–16. To date, only a handful of these CNE families could be unambiguously traced back to TE families15–19. Interestingly, three of these are very ancient families of short interspersed nuclear elements (SINEs) that had previously escaped detection. The apparent enrichment of exapted SINEs may reflect a proclivity of these elements to be recruited for cellular function. Alternatively, it may simply mirror their preponderance in vertebrate genomes and\or the fact that their short size and distinctive sequence signatures make them more readily identifiable as fossil SINE families. Dozens of other CNE families have weak yet significant similarity to known TEs 15, implying that many broadly conserved TE families remain to be characterized.
Individual examples of selectively beneficial TE insertions with apparent regulatory functions have been described in non-mammalian species, especially in Drosophila 20–22. However there has been no attempt to measure the extent of TE exaptation at a genome-wide scale in non-mammalian lineages. Cross-species genome alignments have revealed an abundance of CNEs in species as diverse as dipteran insects, nematodes, yeasts, grasses and crucifers 23,24. Unfortunately, the rapid turnover and decay of TEs in these lineages make it extremely difficult, if at all possible, to recognize ancient elements and assess their contribution to deeply conserved non-coding sequences. But comparative analysis of more closely related species and improved detection and annotation of TEs might be enlightening.
TEs as a supply of regulatory elements
A large body of studies have illustrated the myriad ways by which TEs can directly influence the regulation of nearby gene expression, both at the transcriptional and post-transcriptional levels (Figure 1). Initially these mechanisms were discovered in the laboratory through the analysis of mutations caused by individual TE insertions. But it is now clear that the same changes in gene structure and expression have occurred in the distant past and been preserved by natural selection 9,25,26.
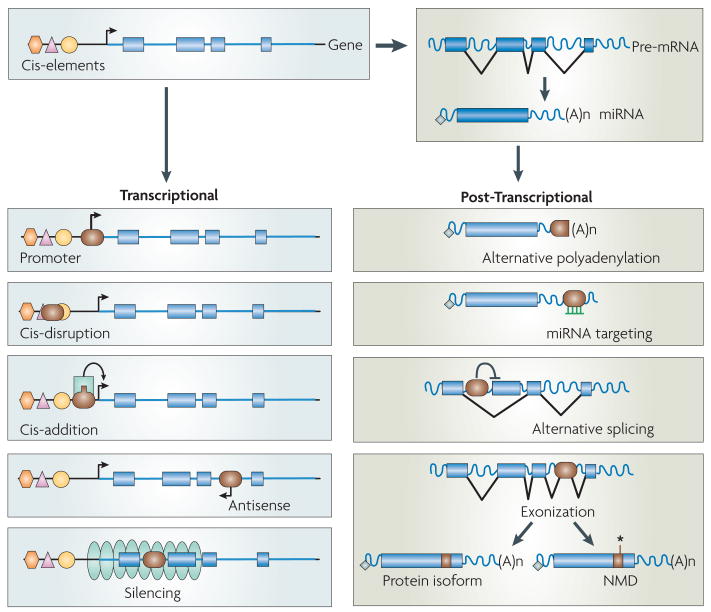
Figure 1. TEs can influence gene expression in many ways.
At the transcriptional level, a TE (shown in brown) inserted upstream of a gene may (i) insert promoter sequences and introduce an alternative transcription start site, (ii) disrupt existing cis-regulatory element(s), or (iii) introduce a new cis element such as a transcription factor binding site. In addition, a TE inserted within an intron may drive antisense transcription and potentially interfere with sense transcription. Finally, a TE may serve as a nucleation center for the formation of heterochromatin potentially silencing the transcription of adjacent gene(s). At the posttranscriptional level, a TE inserted in the 3′ UTR of a gene may introduce an alternative polyadenylation site, a binding site for a microRNA or for RNA-binding protein (not shown). A TE inserted within an intron can interfere with the normal splicing pattern of a pre-mRNA, provoking various forms of alternative splicing (intron retention, exon skipping…etc). A TE inserted within an intron containing cryptic splice sites may be incorporated (‘exonized’) as an alternative exon. This may result in the translation of a new protein isoform, or in the destabilization or degradation of the mRNA via the nonsense mediated decay (NMD) pathway, especially if the exonized TE introduces a premature stop codon (asterisk).
In a seminal study, Jordan et al. reported that nearly 25% of experimentally characterized human promoters contain TE-derived sequences, including empirically defined cis-regulatory elements 27. Further genome-scale analyses showed that many promoters and polyadenylation signals in human and mouse genes are derived from primate-specific and rodent-specific TEs, respectively e.g. 26,28. Another study found that one quarter of DNase I hypersensitive sites identified in human CD4+ T cells overlap with annotated TEs, suggesting that these TEs harbor cis-regulatory sequences. Interestingly, the vast majority of these elements are not deeply conserved, but rather primate-specific. Hence, insertion of these TEs likely contributed to the establishment of lineage-specific patterns of gene expression 29. Further evidence that TEs commonly acquire regulatory function comes from TE fragments that are deeply conserved among mammals. First, these elements tend to cluster around genes involved in development and transcriptional regulation 12. Second, they are over-represented within predicted cis-regulatory modules 30, genomic segments containing dense arrays of transcription factor binding sites (TFBS). Third, about one fifth of eutherian-specific CNEs, thousands of which are derived from ancient TEs, overlap with DNase I hypersensitive sites mapped experimentally in human lymphocytes, which implies that they provide promoter sequences or binding sites for regulatory proteins 13. Finally, there is a growing number of individual cases of highly conserved TEs documented to act as transcriptional enhancers 16,19 or as alternatively spliced exons introducing premature stop codons and triggering non-sense mediated decay, thereby contributing to mRNA homeostasis in the cell 16,31 (Figure 1). Together, these data suggest that TEs have been a profuse source of new regulatory sequences throughout mammalian evolution.
What make TEs such a rich stock of promoter and cis-regulatory elements? One simple explanation is that the pervasive accumulation of TEs creates raw sequence material from which cis-regulatory elements evolve ‘de novo’ by point mutations. Most cis-regulatory elements, such as TFBS, tend to be short and degenerate in sequence 32. Thus it is conceivable that decaying TE sequences provide an abundant material from which cis-regulatory elements emerge ‘de novo’, through the introduction of a single or a few point mutations. Several examples have been reported 9,33–35. Another non-mutually exclusive scenario is that cis-regulatory elements pre-exist within the TEs at the time of its insertion and are co-opted immediately upon insertion or after modifications of the surrounding environment. A wide variety of regulatory elements have been identified in active or reconstructed consensus sequence of active TEs. These include signals normally used by TEs to control their own expression (basal promoters for RNA polymerase II or III, enhancers, insulators, splice sites, polyadenylation signals…) and a plethora of TFBS 36–39. Many empirical studies have demonstrated how such ‘ready-to-use’ cis-elements are incorporated into the ‘natural’ regulatory apparatus of adjacent genes 9,25,35,40–42, including microRNA genes 43.
TE wiring of genetic networks
Regardless of whether the regulatory elements arise ‘de novo’ by a few mutations or are pre-existing within TE sequences, the dispersal of expanding TE families throughout genomes potentially allows the same regulatory motif(s) to be recruited at many chromosomal locations, drawing multiple genes into the same regulatory network (Figure 2). This model, first proposed decades ago by Britten and Davidson 44,45, has recently gained experimental support e.g. 38,46. In a recent study in the human genome, Wang et al.39 found that a set of closely related families of LTR elements affiliated to class I endogenous retroviruses have dispersed more than 1,500 near-perfect binding sites for the ‘master’ regulatory factor p53, encompassing 30% of all p53 binding sites mapped using genome-wide ChIP analysis. In five individual cases further examined, the p53 site within the LTR could be directly associated with p53-dependent transcriptional activation of the closest adjacent gene 39. Interestingly, all the p53-containing LTR families are primate-specific and the p53 sites were apparently present at the time of their insertion. These results strongly suggest that the dispersion of the LTR elements promoted the assembly of a primate-specific transcriptional network of p53-regulated genes (akin to Fig. 2A).
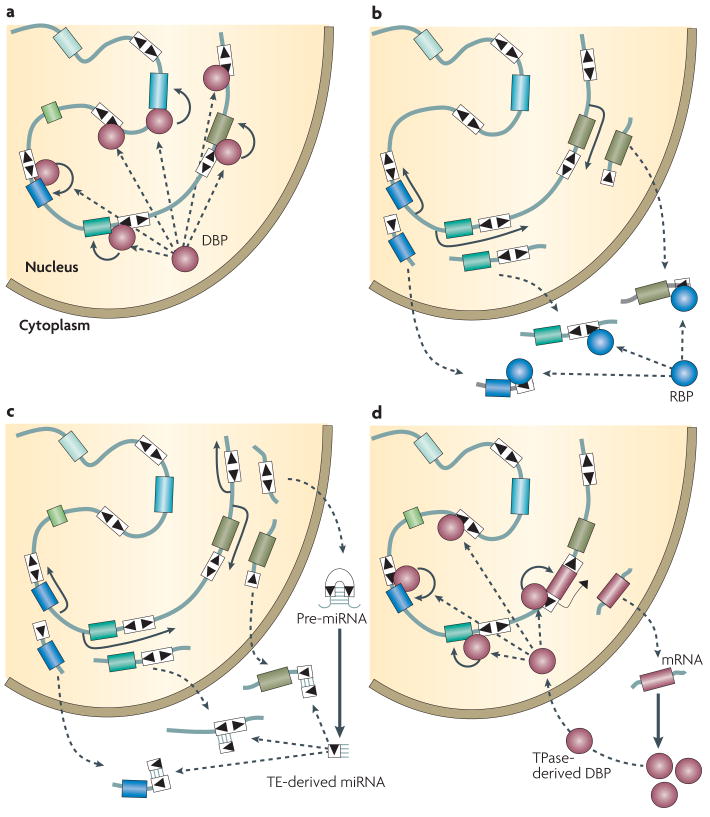
Figure 2. Building regulatory systems with transposable elements.
A family of DNA transposons is shown, with its multiple copies (white boxes) delimited by terminal inverted repeats (black triangles) and interspersed with genes (color boxes) in the genome. For panels A and B, the TE family could be also a retrotransposon family. A: Wiring of a transcriptional regulatory network by TE-derived cis-elements. A binding site for a DNA binding protein (DBP) has been dispersed throughout the genome as part of the TE. If the DBP is a transcription factor, its binding to a TE adjacent to a gene may influence the expression of that gene (arrow pointing at the gene). Multiple genes are brought simultaneously under the control of the transcription factor through their association with different copies of the same TE family. B: Wiring of a post-transcriptional regulatory network by TE-derived cis-elements. Several TE copies are co-transcribed along with their neighboring gene, resulting in the production of different mRNAs containing similar TEs. If the TE contains a binding site for a RNA-binding protein (RBP), it may engage the different mRNAs in the same post-transcriptional pathway of gene regulation. C: De novo assembly of a microRNA network from a TE family. This model combines the idea of TE-host gene co-transcription, as described in B, with the origin of a miRNA precursor containing a TE of the same family. Such precursor may arise by transcription and intramolecular folding of a TE with nearly perfect palindromic structure (e.g. MITEs). The resulting double-stranded RNA may then be processed into a mature miRNA. The resulting TE-derived miRNA can pair with complementary TE sequences embedded within the 3′ UTR of co-transcribed mRNAs. D: De novo assembly of cis and trans components of a transcriptional network from a DNA transposon family. In this model, the DBP is derived from a transposase and thus has the potential to bind to a network of sites previously distributed around the genome by related TEs. If the transposase-derived DBP has transcription factor activity, it may regulate the expression of genes located in proximity to a binding site embedded within a related TE, including its own.
Britten and Davidson had initially formulated their gene battery model as operating at the RNA level, invoking the co-transcription of cis-elements along with the genes they control 44. A recent study in the trypanosomatid Leishmania major 47 brings support to the idea that the same TE family can be recruited at a genome-wide scale for post-transcriptional regulation (Fig. 2B). In trypanosomatid, most of the protein-coding genes are co-transcribed as large polycistronic transcripts. Individual gene regulation occurs predominantly at the post-transcriptional level and sequences located in 3′ UTRs are known to be important for this process. In L. major, almost all of 1,000 copies of LmSIDER2, an extinct family of retroposons, are located in the 3′ UTRs of predicted mRNAs 47, a strikingly biased distribution suggesting a global function in post-transcriptional regulation. Consistent with this hypothesis, experimental introduction of a LmSIDER2 copy in the 3′ UTR of a reporter gene decreased the stability of the resulting mRNA in vivo. Furthermore, microarray analyses revealed that L. major mRNAs containing LmSIDER2 in their 3′ UTR are generally expressed at lower levels than others 47. Together, these data support the idea that this TE family has been recruited at the whole-genome level to modulate post-transcriptionally the expression of hundreds of genes.
In addition to donating new cis-elements and participating in the de novo assembly of regulatory networks, TEs may also contribute to the rewiring of pre-established networks through their movement and the genomic rearrangements they provoke (Box 1). It is widely believed that the tinkering and reorganization of pre-existing networks is a prominent mode of regulatory evolution 32,48,49.
TEs as microRNAs and their targets
It is now evident that non-coding RNAs are important players in the regulation of eukaryotic gene expression 50. Several classes of small regulatory RNA, including microRNAs (miRNAs), small interfering RNAs (siRNA), repeat-associated small interfering RNAs (rasiRNAs) and piwi-interacting RNAs (piRNAs) — collectively referred hereafter as smRNA — use partially overlapping pathways akin to RNA interference (RNAi) to silence gene expression, via degradation or translation inhibition of mRNAs containing complementary sites. Thus, the logic of post-transcriptional regulation by smRNAs, whereby a single smRNA species can trans-regulate multiple genes through recognition of shared cis-elements, is similar to the logic of transcriptional regulation by transcription factors 53. In addition, there is evidence that some smRNAs are able to mediate homology-dependent transcriptional silencing and participate in the nucleation of heterochromatin 54,55.
Ever since their discovery, the relationship of piRNAs, siRNAs and rasiRNAs to TEs has been apparent. Indeed the ‘natural’ and presumably ancestral function of these smRNA is to silence invasive DNA such as viruses and TEs 52,55. There are now numerous examples illustrating how these genome defense systems and the epigenetic marks deposited to silence TEs have been be co-opted to control adjacent gene expression 35,55.
The evolutionary origins of miRNAs remain more obscure. Although new miRNA genes can arise through duplication of existing miRNA, it appears that the bulk of miRNAs have originated from sequences not previously encoding miRNA 53. One model proposes that new miRNAs arise from pre-existing hairpin structures in the genome that are fortuitously transcribed 51,53. The influence of TEs in the origin, biogenesis and mode of action of miRNA is increasingly being recognized. Several mammalian miRNA precursors have been found to contain or be derived from TE sequences 56. Likewise, a substantial amount of predicted miRNA targets map within members of the same TE families 57,58, again pointing at a model whereby large sets of cis-regulatory sequences have been seeded by transposition (Fig. 2C). The most recent count57 shows that 55 out of 452 (12%) experimentally characterized human miRNA genes originated from TEs. Although this proportion seems lower than what might be expected in relation to the space occupied by TEs in the human genome (~48%), it represents a minimal estimation because many of the miRNAs currently known have deeper evolutionary origins than the most ancient TEs recognizable in the human genome. Also, many uncharacterized miRNAs probably remains to be identified. For example, the same study57 predicted computationally an additional 85 likely miRNAs precursors derived from transcribed TEs.
Several classes and families of TEs show far more overlap with miRNA genes than is expected on the basis of their relative frequency in the genome57. This observation suggests that certain TE families possess characteristics that make them prone to give rise to miRNA. For instance, many MITEs have a palindromic structure, with terminal-inverted repeats joined by little or no spacer DNA 59. Transcription of MITEs is not required for transposition, but because they preferentially integrate in the non-coding portion or immediate vicinity of genes, MITEs are frequently transcribed 59. Following transcription, intramolecular folding of the MITE TIR sequences would produce RNA hairpins that, in principle, could be processed into siRNA 60. Such MITE-derived siRNAs could then act in trans to mediate silencing of multiple genes associated with related MITE sequences, providing a steppingstone toward the emergence of a typical miRNA regulatory circuit (Figure 2C).
In support of this model, it was recently established61 that mir-548, a small family of human miRNAs, is derived from MADE1 TEs spuriously transcribed from adjacent promoters. MADE1 is an anthropoid-specific family of MITEs with an almost perfect palindromic structure. The mRNA targets of mir-548 have not been defined experimentally, but bioinformatic predictions revealed over 3,500 human genes with putative target sites for mir-548 61. Interestingly, a subset of the predicted mir-548 targets are also derived from MADE1 sequences that previously inserted within or close to the 3’ UTRs of genes. Some of these MADE1-containing transcripts are downregulated in colorectal cancer tissues, where mir-548 is upregulated, and they fall within the same functional categories, consistent with the idea that they belong to a network of mir-548-regulated genes assembled through the past propagation of MADE1 61.
Transposases recycled into regulatory proteins
The evolution of complex multicellular organisms in several branches of the tree of life was accompanied, and perhaps facilitated by an expansion and diversification of transcription factors (TFs) 32,48,49,53,62. It is thought that the emergence of new TFs allowed for the elaboration of increasingly complex networks of genes wired by cis-elements recognized by different sets of TFs 48,49,62. Gene duplication and domain shuffling are well-established mechanisms contributing to the emergence of new regulatory proteins 53,62. In the following sections, I argue that DNA transposons and their cognate transposases (TPases) represent another significant, yet largely underappreciated source of the basic components necessary for the co-assembly of new TF and their DNA targets (see Fig. 2D).
Recurrent domestication of transposases
A particular form of TE exaptation, also known as domestication, occurs when TE-encoded proteins or domains become co-opted into functional host proteins 10,63. In principle, any of the activities or domains encoded by TE proteins may be domesticated. However, as the list of TE-derived proteins increases, it is becoming evident that transposases are more prone to domestication than other TE proteins 5,63. The propensity of TPases for domestication was first noticed in the initial analysis of the human genome, which identified 47 genes entirely or mostly derived from TE coding sequences 64, with all but four related to transposases, despite the fact that DNA transposons represent a modest fraction of human TEs (7%) and of the genome (3%)64,65.
Since this influential publication, dozens of additional cases of TE-derived proteins have been identified in animals, fungi and plants, even when stringent criteria were applied to valdiate the functionality of the TE-derived genes, such as syntenic conservation and evidence of strong purifying selection acting in distantly related species e.g. 66,67–69. These studies reveal that several categories of TE coding sequences have been domesticated on multiple independent occasions, such as retroviral gag-like and envelope proteins 63,68, but TPases continue to stand out as a recurrent supply of new proteins in diverse organisms (Figure 3) 5.
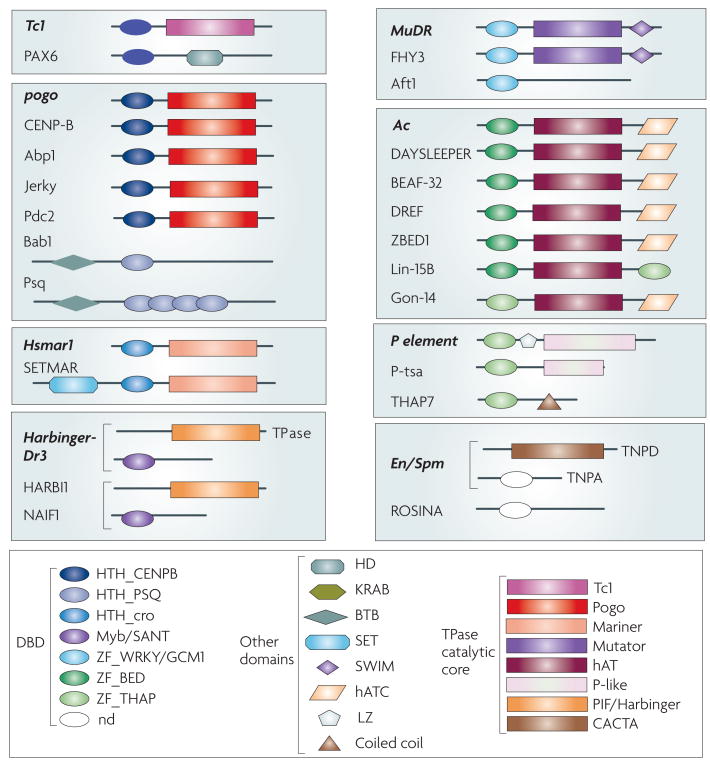
Figure 3. DNA-binding proteins and transcription factors derived from transposases.
The domain structure of several well-documented cases of host DBPs and TF derived from TPases (names in bold) and their closest TPase relatives. The different proteins are grouped according to the corresponding TPase superfamily. Tc1, pogo and Hsmar1 belong to three different subgroups of Tc1/mariner; Harbinger-Dr3, PIF/Harbinger; MuDR, Mutator; Ac, hAT (hobo/Activator/Tam3); P element, P; En/Spm, CACTA 2. Gene names (the species where they were originally described and a brief description of their function, unless listed in Table 1): Abp1, ARS-binding protein 1 (Saccharomyces cerevisiae, centromeric heterochromatin formation, chromosome segregation, retrotransposon repression); Jerky (Mus musculus, probable neuronal translational and transcriptional regulator); Pdc2, pyruvate decarboxylase 2 (S. cerevisiae, transcription factor involved in pyruvate and thiamin metabolism); Bab1, Bric-à-brac 1 (D. melanogaster, transcriptional regulator); Psq, pipsqueak (D. melanogaster. transcriptional repressor, embryonic and adult development); SETMAR (Homo sapiens, histone modification, DNA repair); NAIF1, nuclear apoptosis-inducing factor 1 (H. sapiens, directly interacts with and mediates nuclear translocation of HARBI1); FHY3, far-red elongated hypocotyls (Arabidopsis thaliana) 3; Aft1, Activator of Ferrous Transport 1 (S. cerevisiae, transcription factor involved in iron utilization and homeostasis); DAYSLEEPER (Arabidopsis thaliana, probable developmental regulator) BEAF-32, boundary element-associated factor of 32 kDa (D. melanogaster, insulator function, gene regulation and chromosome organization); DREF, DNA replication-related element-binding factor (D. melanogaster); ZBED1, Zinc finger BED domain containing protein 1 (H. sapiens, transcriptional activator of cell proliferation and ribosomal proteins); Lin-15B, abnormal cell LINeage 15B (Caenorhabditis elegans, developmental regulator through cell cycle control); Gon-14, gonadogenesis deficient 14 (C. elegans, required for gonadogenesis and other developmental processes); P-tsa, P-neogene (Drosophila tsacasi, unknown function); THAP7, Thanatos-Associated Protein 7 (H. sapiens, transcriptional repressor); ROSINA (Anthirinium majus, floral organ development) Domains: HTH, helix-turn-helix; ZF, zinc-finger; SANT, Swi3-Ada2-NCoR-TFIIIB; GCM1, Glial Cell Missing 1; BED, BEAF and DREF; HD, homeodomain; KRAB, Kruppel-associated box; BTB, Broad-Complex, Tramtrack, and Bric-à-brac; SET, Su(var)3–9, E(z) and Trithorax; SWIM, SWI2/SNF2 and MuDR; hATC, hAT C-terminal dimerization; LZ, leucine zipper.
Transposases as a source of DNA-binding domains
Like TFs, TPases must translocate to the nucleus to recognize specific DNA sites on the chromosomes. To achieve this, most TPases utilize a nuclear localization signal and an N-terminal DNA-binding domain (DBD) that interacts specifically with a short DNA motif located near each of the transposon ends, often within the TIRs 70. Also, like other DNA-binding proteins (DBPs), TPases must either promote their own access to open chromatin, for example by recruiting host chromatin remodeling complexes see ref. 71 or take advantage of transient relaxing of chromatin at certain regions of the genome 72.
TPase DBDs are structurally diverse and many can be allied to those found in established families of TFs and DBPs, 67,73–77 (Table 1) e.g. 78,79, but it was often unclear originally whether the host’s DBD derived from the TPase or vice-versa. With the accumulation of genomic sequence and the mining of diverse transposons across the eukaryotic tree, it is becoming increasingly clear that these DBDs first originated in TPases 73,77,80–83. Typically, the TPases have a broader taxonomic distribution than the related host DBPs and phylogenetic reconstructions point to the association of the DBD with the TPase catalytic core as the ancestral state, while the host proteins are derived from a subclade of TPases. The origination of DBDs from TPases has involved all major TPase superfamilies (Table 1 and Figure 3) and has occurred repeatedly in the evolution of plants, fungi and animals 5,77,81,82,84,85, including some ‘master’ developmental regulators (e.g. Pax proteins, see Table 1 and Figure 3).
Table 1.
DNA-binding domain families likely originated from transposases
| DBD family | Example of DBP and its function | Distribution Host Proteins | TE origin (superfamily) | Distribution TE Superfamily | References |
|---|---|---|---|---|---|
| Paired box (HTH) | PAX6 (development of sensory organs and brain) | M | Tc1 | M,F,E | 80 |
| CENPB (HTH) | CENP-B (centromere function) | Vertebrates*, F* | Pogo | M,F,E,A,P | 82 |
| PSQ (HTH) | Bric-a-Brac1 (development of ovaries, appendages and abdomen) | Insects | Pogo | M,F | 74 |
| Myb/SANT (HTH) | NAIF1 (apoptosis/cell cycle regulation?) | M*, A* | PIF/Harbinge r | M,F,A,D,T | Sinzelle L.& Ivics Z.,pers. com. |
| WRKY/GCM1 (ZF) | FHY3 (far-red light signaling) | A*, Yeasts*, Insects* | Mutator | M,F,E,A,P,T | 77 |
| BED (ZF) | DREF (regulation of cell proliferation and differentiation) | M*, A* | hAT | M,F,E,A,C,P,T | 73 |
| THAP (ZF) | THAP1 (apoptosis and cell cycle regulation) | M* | P-element | M,C | 75 |
M= Metazoans (Opistokonts)
F= Fungi (Opistokonts)
E= Entamoeba (Lobosa)
A= Angiosperms (Plantae)
C= Chlamydomonas reinhardti (Green algae)
D= Diatom (Stramenopiles)
P= Phytopthora infestans (Oomycetes)
T= Trichomonas vaginalis (Trichomonads)
= indicates multiple independent domestication events within that clade
Transposases possess intrinsic regulatory activities
In many instances, the sequence similarity of TPase-derived proteins with their ancestral TPase is not limited to the DBD but spans their entire sequence, including the catalytic core of the TPase (see Figure 3). However the acidic residues essential for catalysis often have been altered, compromising cleavage and/or strand transfer activities e.g. 86,87,88. Nonetheless, the overall conservation of full-length TPase sequence and architecture suggests that biochemical activities other than DNA-binding have been coopted. These may include oligomerization activity, which allows the pairing of DNA sites bound simultaneously by different TPase molecules and ‘looping’ of the intervening DNA 70. The inherent ability of TPases to ‘loop’ and ‘bundle’ DNA to which they are attached may predispose them to be recruited as proteins that package and organize the genome into functionally independent chromatin domains, akin to ‘insulator’ proteins. Indeed, BEAF-32 is a Drosophila insulator protein entirely derived from a hAT transposase (see ref. 73 and Figure 3) that binds the scs chromatin boundary element and connects chromatin to the nuclear matrix 89. In fission yeast, Abp1 and its two paralogs, collectively known as CENP-B homologs, are centromere-binding proteins involved in chromosome segregation that originated from a fungi lineage of pogo-like transposases 82. Recently it was shown that the fission yeast CENP-Bs can also bind noncentromeric interspersed DNA repeats and promote the bundling of these repeats at the nuclear periphery 90. Furthermore, Abp1 interacts directly with histone deacetylases and directs them to its associated DNA, thereby triggering a local nucleation of heterochromatin and repressing transcription of adjacent genes at several chromosomal locations 90 (see Figure 2A). It is unknown whether the ability of CENP-Bs to interact with histone deacetylases was drawn from the progenitor TPase. However it is evident that the DNA-binding and self-dimerization activities, which are necessary for bundling and tethering of DNA to the nuclear periphery, directly descend from the domesticated transposases.
In Arabidopsis thaliana, FHY3 and FAR1 are two closely related proteins entirely derived from Mutator-like TPases 85,91. FHY3 and FAR1 are bonafide TFs that bind directly to promoter regions and activate several genes involved in far-red light and circadian signaling 85. The transcriptional activation domains of FHY3 and FAR1 are physically separable from their DBD and this activity requires two residues highly conserved in Mutator-like transposases 85. Thus, it is conceivable that Mutator-like TPases possess intrinsic TF activity, which may explain repeated domestication events of this superfamily of TPases in plants, fungi and animals 77,84. Interestingly, the TPase encoded by the maize MuDR element, and also TnpA, one of the two proteins encoded by the maize Spm transposon, can function as transcriptional regulators of their own expression 92,93. Such transcriptional self-regulation might offer an opportunity for a newborn TPase-derived TF to instantly acquire a regulatory feedback loop, a characteristic of most regulatory circuits 32,48,49 (see Figure 2D).
Birth of a genetic network
In vitro and in vivo studies have shown that TPases often cross-interact with distantly related transposons with similar termini 94–97. Thus, not only the amplification of an active transposon, but also the past accumulation of evolutionarily related elements, result in a buildup of TPase binding sites throughout the genome. Domestication may occur when one or several TPase-DNA interactions become selectively advantageous for the host (Figure 2D). This selective benefit may arise as the result of a transposon insertion in the proximity of a host gene, bringing the latter under the regulatory influence of the TPase. Alternatively, domestication may be initiated by mutational events at a TPase-encoding locus, leading to the emergence of a modified transposase with new trans-regulatory activities. This can occur by mutations in the transposase sequence, fusion of flanking exons to the transposase65 and/or a change in the pattern of transposase expression. Natural selection may further shape and expand this primordial regulatory network by acting on newly arisen or polymorphic transposon insertions to retain those bringing beneficial interactions with the domesticated TPase, while removing those with deleterious consequences5.
Testing this model is hampered by the fact that most DBPs known to derive from TPases are of relatively ancient origin, making it impractical to trace the origin of their binding sites to ancestral transposons because sequences surrounding the TPase binding site would likely be erased by extended period of neutral evolution. In addition, TPase binding sites are short and fast-evolving motifs and consequently they tend to be poorly conserved, even among related transposon families 70,96,97. Therefore, to validate the model, it is necessary to study recently emerged TPase-derived proteins that can be tied to transposon families still recognizable in the same genome.
One promising candidate is SETMAR, a primate-specific protein that results from the fusion of a pre-existing SET histone methyltransferase gene to the transposase gene of a Hsmar1 transposon 86. The amplification of the Hsmar1 family and its allies (MADE1 MITEs) occurred approximately 45 Myr ago65 and was concomitant to the emergence of SETMAR. Evolutionary sequence analyses of SETMAR across anthropoid primate species have revealed that the DBD, but not the catalytic region, has been subject to continuous purifying selection since its domestication 86. Consistent with these observations, in vitro experiments demonstrated that the transposase region of SETMAR has retained robust DNA-binding activity, but lost some of its catalytic abilities 86–88. The 19-bp binding site recognized by SETMAR is reiterated in about 1,500 perfect or nearly perfect copies in the human genome and almost all of these sites map within the TIRs of Hsmar1 and MADE1 86. Thus, the TPase region of SETMAR might be used to target the SET domain to multiple genomic sites, where it might modify the surrounding chromatin and modulate the transcription of adjacent genes.
Outlook
The recent discoveries and models presented herein echo the visionary predictions of McClintock and of a few pioneers on the significance of TEs for eukaryotic gene regulation. Meanwhile, it is being realized that the most influential contributions of TEs to macroevolution may arise and persist long after transposition activity has ceased and typically emerged as a side-product of their selfish and parasitic lifestyle. It is these characteristics that ensure the long-term survival of TEs and entails their intimate co-evolutionary relationship with the host genome. Ironically, the breadth and versatility of TE exaptation has become most apparent in mammalian genomes, where repetitive DNA has traditionally been perceived as a hurdle to geneticists rather than as a valuable source of genomic information. While the wide diversity, large-scale amplification and relatively slow mutational decay of TEs in mammals have likely promoted co-option, these characteristics have also allowed the process to be observable [Au:OK? YES]. Evaluating quantitatively the functional heritage of TEs in organisms with faster sequence turnover is more difficult, but there is increasing evidence that TEs have been co-opted repeatedly for cellular and regulatory function in various eukaryotic lineages. Together these findings are likely to recapitulate a prominent evolutionary process that has been shaping eukaryotic genomes ever since their origins. Our appreciation of the impact of TEs in regulatory evolution will certainly gain from a broadening of the diversity of organisms under genomic scrutiny. This will necessitate the development of new tools to accelerate the discovery and automate the genomic annotation of TEs. A more rigorous and quantitative examination of the dynamics and mode of molecular evolution of TEs is also needed. For example, it remains unclear how transposition mechanisms, chromosomal distribution and epigenetic markings of TEs affect their rate of evolution and influence their propensity toward exaptation. Finally, there is a need to continue to develop experimental approaches to further probe the models developed here and elsewhere, which position TEs and their derived proteins as central players in the evolution of eukaryotic gene regulation.
BOX1: TE-mediated tinkering of cis-regulatory networks
In addition to donating cis-elements and creating new regulatory networks, the movement and accumulation of TEs are likely to participate in the rewiring of pre-established regulatory networks. First, TE insertions may disrupt and effectively eliminate cis-regulatory elements, thereby removing some genes from an existing network. Examples of TE insertions altering gene regulation have been described in the context of deleterious mutations causing disease e.g. 98 or mutant alleles recovered in the laboratory 99. The depletion of TEs in certain regions of the human genome enriched in regulatory sequences, such as Hox gene clusters and other transposon-free regions 100, attests to the deleterious nature of TE insertions into genomic environments containing a high density of cis-regulatory elements. However, like any mutational events, it is conceivable that these disruptive insertions might be beneficial under some circumstances. Potentially adaptive TE insertions disrupting promoter function have been identified at the Hsp70 locus in natural populations of Drosophila melanogaster 20,101. A screen for naturally occurring P elements within Hsp promoters recovered over 200 independent insertions, suggesting they are hotspots for P insertions 102. Many of these insertions are present at high frequency in populations and some are associated with decrease in Hsp expression and reduced thermotolerance, suggesting that they are phenotypically consequential 102.
A less destructive route for TEs to modify existing gene network is through TE-mediated shuffling and duplication of cis-regulatory elements 49. It is well established that TEs can promote the mobilization, rearrangement and duplication of host sequences through various mechanisms, including recombination between TE copies, aberrant transposition events, and transduction 5,6. Thus, TEs have the potential to shuffle regulatory sequence information into new genomic contexts. A concrete example is the acquisition of a functional binding site for the NFAT transcription factor that is required for transcriptional activation of the Interferon-gamma gene in human lymphocytes 103. The intact NFAT binding site was brought in the promoter as part of a short DNA segment co-mobilized by an Alu element inserted 22–34 Myr ago. Furthermore, subsequent nucleotide substitutions next to the NFAT site created another TFBS (this time for NF-kappa B) that remains polymorphic in human populations 103.
The preferential insertion and accumulation of some TEs, notably SINEs and DNA transposons, into the vicinity of genes may further enhance the scrambling of cis-regulatory elements. In addition, the excision of cut-and-paste DNA transposons is often imprecise, leaving behind small stretches of sequences (footprints) or rearranging flanking host sequences, which may offer yet another mechanism for generating subtle alterations of adjacent regulatory sequences. This is well illustrated by allelic series of regulatory mutations, with a range of pigmentation phenotypes, recovered in the laboratory following aberrant transposition and imprecise excision events of Tam3 in the nivea promoter of snapdragon 104 and of Tol2 in the promoter of the tyrosinase gene of medaka fish 105.
Acknowledgments
I owe many thanks to C. Casola, J-M. Deragon, J. Fondon, I.K. Jordan, A. Pires da Silva, E. Pritham and D. Voytas for stimulating discussions and insightful comments during the preparation of this article. I also thank the two anonymous reviewers for their constructive comments and useful suggestions. The author apologizes to many colleagues whose relevant work and original articles could not be cited due to space limitations. Research in the C.F. laboratory is supported by grants from the U.S. National Institutes of Health.
Footnotes
About the author
Cédric Feschotte received his B.S degree in 1996 from the University of Toulouse and his Ph.D. in 2001 from the University of Paris 6 (Université Pierre and Marie Curie). He carried out his postdoctoral research on plant transposable elements in the group of Susan Wessler at the University of Georgia, Athens, USA. Since 2004, he has been a tenure-track assistant professor in the Department of Biology at the University of Texas, Arlington, USA. His research interests focus on the evolution of transposable elements, their impact on genome structure and function and their role in the emergence of genetic innovations.
References
- 1.Britten RJ, Kohne DE. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968;161:529–40. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- 2.Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–82. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 3.Brookfield JF. The ecology of the genome - mobile DNA elements and their hosts. Nat Rev Genet. 2005;6:128–36. doi: 10.1038/nrg1524. [DOI] [PubMed] [Google Scholar]
- 4.Kidwell MG, Lisch DR. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution Int J Org Evolution. 2001;55:1–24. doi: 10.1111/j.0014-3820.2001.tb01268.x. [DOI] [PubMed] [Google Scholar]
- 5.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annual Review of Genetics. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deininger PL, Moran JV, Batzer MA, Kazazian HH., Jr Mobile elements and mammalian genome evolution. Curr Opin Genet Dev. 2003;13:651–8. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 7.Gould SJ, Vrba ES. Exaptation-a missing term in the science of form. Paleobiology. 1983;8:4–15. [Google Scholar]
- 8.Brosius J. Retroposons - seeds of evolution. Science. 1991;251:753. doi: 10.1126/science.1990437. [DOI] [PubMed] [Google Scholar]
- 9.Britten RJ. Cases of ancient mobile element DNA insertions that now affect gene regulation. Mol Phylogenet Evol. 1996;5:13–7. doi: 10.1006/mpev.1996.0003. [DOI] [PubMed] [Google Scholar]
- 10.Miller WJ, McDonald JF, Nouaud D, Anxolabehere D. Molecular domestication--more than a sporadic episode in evolution. Genetica. 1999;107:197–207. [PubMed] [Google Scholar]
- 11.Silva JC, Shabalina SA, Harris DG, Spouge JL, Kondrashovi AS. Conserved fragments of transposable elements in intergenic regions: evidence for widespread recruitment of MIR- and L2-derived sequences within the mouse and human genomes. Genet Res. 2003;82:1–18. doi: 10.1017/s0016672303006268. [DOI] [PubMed] [Google Scholar]
- 12.Lowe CB, Bejerano G, Haussler D. Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc Natl Acad Sci U S A. 2007;104:8005–10. doi: 10.1073/pnas.0611223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mikkelsen TS, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–77. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- 14.Bejerano G, Haussler D, Blanchette M. Into the heart of darkness: large-scale clustering of human non-coding DNA. Bioinformatics. 2004;20(Suppl 1):i40–8. doi: 10.1093/bioinformatics/bth946. [DOI] [PubMed] [Google Scholar]
- 15.Xie X, Kamal M, Lander ES. A family of conserved noncoding elements derived from an ancient transposable element. Proc Natl Acad Sci U S A. 2006;103:11659–64. doi: 10.1073/pnas.0604768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bejerano G, et al. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 17.Kamal M, Xie X, Lander ES. A large family of ancient repeat elements in the human genome is under strong selection. Proc Natl Acad Sci U S A. 2006;103:2740–5. doi: 10.1073/pnas.0511238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishihara H, Smit AF, Okada N. Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 2006;16:864–74. doi: 10.1101/gr.5255506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santangelo AM, et al. Ancient exaptation of a CORE-SINE retroposon into a highly conserved mammalian neuronal enhancer of the proopiomelanocortin gene. PLoS Genet. 2007;3:1813–26. doi: 10.1371/journal.pgen.0030166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maside X, Bartolome C, Charlesworth B. S-element insertions are associated with the evolution of the Hsp70 genes in Drosophila melanogaster. Curr Biol. 2002;12:1686–91. doi: 10.1016/s0960-9822(02)01181-8. [DOI] [PubMed] [Google Scholar]
- 21.Schlenke TA, Begun DJ. Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc Natl Acad Sci U S A. 2004;101:1626–31. doi: 10.1073/pnas.0303793101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung H, et al. Cis-regulatory elements in the Accord retrotransposon result in tissue-specific expression of the Drosophila melanogaster insecticide resistance gene Cyp6g1. Genetics. 2007;175:1071–7. doi: 10.1534/genetics.106.066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inada DC, et al. Conserved noncoding sequences in the grasses. Genome Res. 2003;13:2030–41. doi: 10.1101/gr.1280703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brosius J. The contribution of RNAs and retroposition to evolutionary novelties. Genetica. 2003;118:99–116. [PubMed] [Google Scholar]
- 26.Marino-Ramirez L, Lewis KC, Landsman D, Jordan IK. Transposable elements donate lineage-specific regulatory sequences to host genomes. Cytogenet Genome Res. 2005;110:333–41. doi: 10.1159/000084965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jordan IK, Rogozin IB, Glazko GV, Koonin EV. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68–72. doi: 10.1016/s0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 28.van de Lagemaat LN, Landry JR, Mager DL, Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003;19:530–6. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Marino-Ramirez L, Jordan IK. Transposable element derived DNaseI-hypersensitive sites in the human genome. Biol Direct. 2006;1:20. doi: 10.1186/1745-6150-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gentles AJ, et al. Evolutionary dynamics of transposable elements in the short-tailed opossum Monodelphis domestica. Genome Res. 2007;17:992–1004. doi: 10.1101/gr.6070707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ni JZ, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–18. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wray GA, et al. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 33.Hambor JE, Mennone J, Coon ME, Hanke JH, Kavathas P. Identification and characterization of an Alu-□containing, T-cell-specific enhancer located in the last intron of the human CD8 alpha gene. Mol Cell Biol. 1993;13:7056–7070. doi: 10.1128/mcb.13.11.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou YH, Zheng JB, Gu X, Saunders GF, Yung WK. Novel PAX6 binding sites in the human genome and the role of repetitive elements in the evolution of gene regulation. Genome Res. 2002;12:1716–22. doi: 10.1101/gr.188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medstrand P, et al. Impact of transposable elements on the evolution of mammalian gene regulation. Cytogenet Genome Res. 2005;110:342–52. doi: 10.1159/000084966. [DOI] [PubMed] [Google Scholar]
- 36.Thornburg BG, Gotea V, Makalowski W. Transposable elements as a significant source of transcription regulating signals. Gene. 2006;365:104–10. doi: 10.1016/j.gene.2005.09.036. [DOI] [PubMed] [Google Scholar]
- 37.Polak P, Domany E. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics. 2006;7:133. doi: 10.1186/1471-2164-7-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson R, et al. Identification of the REST regulon reveals extensive transposable element-mediated binding site duplication. Nucleic Acids Res. 2006;34:3862–77. doi: 10.1093/nar/gkl525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang T, et al. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci U S A. 2007;104:18613–8. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wessler SR, Bureau TE, White SE. LTR-retrotransposons and MITEs: important players in the evolution of plant genomes. Curr Opin Genet Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- 41.Ferrigno O, et al. Transposable B2 SINE elements can provide mobile RNA polymerase II promoters. Nat Genet. 2001;28:77–81. doi: 10.1038/ng0501-77. [DOI] [PubMed] [Google Scholar]
- 42.Romanish MT, Lock WM, van de Lagemaat LN, Dunn CA, Mager DL. Repeated recruitment of LTR retrotransposons as promoters by the anti-apoptotic locus NAIP during mammalian evolution. PLoS Genet. 2007;3:e10. doi: 10.1371/journal.pgen.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 44.Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165:349–57. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- 45.Britten RJ, Davidson EH. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol. 1971;46:111–38. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- 46.Peaston AE, et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 47.Bringaud F, et al. Members of a large retroposon family are determinants of post-transcriptional gene expression in Leishmania. PLoS Pathog. 2007;3:1291–307. doi: 10.1371/journal.ppat.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilkins AS. The evolution of developmental pathways. Sinauer; Sunderland, MA: 2002. [Google Scholar]
- 49.Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. Academic; New York: 2006. [Google Scholar]
- 50.Mattick JS. A new paradigm for developmental biology. J Exp Biol. 2007;210:1526–47. doi: 10.1242/jeb.005017. [DOI] [PubMed] [Google Scholar]
- 51.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 52.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–4. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 53.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 54.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 55.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–85. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 56.Smalheiser NR, Torvik VI. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005;21:322–6. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 57.Piriyapongsa J, Marino-Ramirez L, Jordan IK. Origin and evolution of human microRNAs from transposable elements. Genetics. 2007;176:1323–37. doi: 10.1534/genetics.107.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smalheiser NR, Torvik VI. Alu elements within human mRNAs are probable microRNA targets. Trends Genet. 2006;22:532–6. doi: 10.1016/j.tig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 59.Feschotte C, Jiang N, Wessler SR. Plant transposable elements: where genetics meets genomics. Nat Rev Genet. 2002;3:329–41. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- 60.Sijen T, Plasterk RH. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426:310–4. doi: 10.1038/nature02107. [DOI] [PubMed] [Google Scholar]
- 61.Piriyapongsa J, Jordan IK. A Family of Human MicroRNA Genes from Miniature Inverted-Repeat Transposable Elements. PLoS ONE. 2007;2:e203. doi: 10.1371/journal.pone.0000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–51. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 63.Volff JN. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays. 2006;28:913–22. doi: 10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- 64.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 65.Pace JK, 2nd, Feschotte C. The evolutionary history of human DNA transposons: Evidence for intense activity in the primate lineage. Genome Res. 2007;17:422–432. doi: 10.1101/gr.5826307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zdobnov EM, Campillos M, Harrington ED, Torrents D, Bork P. Protein coding potential of retroviruses and other transposable elements in vertebrate genomes. Nucleic Acids Res. 2005;33:946–54. doi: 10.1093/nar/gki236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Casola C, Lawing AM, Betran E, Feschotte C. PIF-like transposons are common in drosophila and have been repeatedly domesticated to generate new host genes. Mol Biol Evol. 2007;24:1872–88. doi: 10.1093/molbev/msm116. [DOI] [PubMed] [Google Scholar]
- 68.Campillos M, Doerks T, Shah PK, Bork P. Computational characterization of multiple Gag-like human proteins. Trends Genet. 2006;22:585–9. doi: 10.1016/j.tig.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 69.Muehlbauer GJ, et al. A hAT superfamily transposase recruited by the cereal grass genome. Mol Genet Genomics. 2006;275:553–63. doi: 10.1007/s00438-006-0098-8. [DOI] [PubMed] [Google Scholar]
- 70.Craig NL, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. American Society for Microbiology Press; Washington, D.C: 2002. [Google Scholar]
- 71.Makarova KS, Aravind L, Koonin EV. SWIM, a novel Zn-chelating domain present in bacteria, archaea and eukaryotes. Trends Biochem Sci. 2002;27:384–6. doi: 10.1016/s0968-0004(02)02140-0. [DOI] [PubMed] [Google Scholar]
- 72.Ros F, Kunze R. Regulation of activator/dissociation transposition by replication and DNA methylation. Genetics. 2001;157:1723–33. doi: 10.1093/genetics/157.4.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aravind L. The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends Biochem Sci. 2000;25:421–3. doi: 10.1016/s0968-0004(00)01620-0. [DOI] [PubMed] [Google Scholar]
- 74.Siegmund T, Lehmann M. The Drosophila Pipsqueak protein defines a new family of helix-turn-helix DNA-binding proteins. Dev Genes Evol. 2002;212:152–7. doi: 10.1007/s00427-002-0219-2. [DOI] [PubMed] [Google Scholar]
- 75.Roussigne M, et al. The THAP domain: a novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem Sci. 2003;28:66–9. doi: 10.1016/S0968-0004(02)00013-0. [DOI] [PubMed] [Google Scholar]
- 76.Kapitonov VV, Jurka J. Harbinger transposons and an ancient HARBI1 gene derived from a transposase. DNA Cell Biol. 2004;23:311–24. doi: 10.1089/104454904323090949. [DOI] [PubMed] [Google Scholar]
- 77.Babu MM, Iyer LM, Balaji S, Aravind L. The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res. 2006;34:6505–20. doi: 10.1093/nar/gkl888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tudor M, Lobocka M, Goodwell M, Pettitt J, O’Hare K. The pogo transposable element family of Drosophila melanogaster. Mol Gen Genet. 1992;232:126–134. doi: 10.1007/BF00299145. [DOI] [PubMed] [Google Scholar]
- 79.Franz G, Loukeris TG, Dialektaki G, Thompson CR, Savakis C. Mobile Minos elements from Drosophila hydei encode a two-exon transposase with similarity to the paired DNA-binding domain. Proc Natl Acad Sci USA. 1994;91:4746–4750. doi: 10.1073/pnas.91.11.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Breitling R, Gerber JK. Origin of the paired domain. Dev Genes Evol. 2000;210:644–650. doi: 10.1007/s004270000106. [DOI] [PubMed] [Google Scholar]
- 81.Quesneville H, Nouaud D, Anxolabehere D. Recurrent recruitment of the THAP DNA-binding domain and molecular domestication of the P-transposable element. Mol Biol Evol. 2005;22:741–6. doi: 10.1093/molbev/msi064. [DOI] [PubMed] [Google Scholar]
- 82.Casola C, Hucks D, Feschotte C. Convergent Domestication of pogo-Like Transposases Into Centromere-Binding Proteins in Fission Yeast and Mammals. Mol Biol Evol. 2008;25:29–41. doi: 10.1093/molbev/msm221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Piriyapongsa J, Rutledge MT, Patel S, Borodovsky M, Jordan IK. Evaluating the protein coding potential of exonized transposable element sequences. Biol Direct. 2007;2:31. doi: 10.1186/1745-6150-2-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cowan RK, Hoen DR, Schoen DJ, Bureau TE. MUSTANG is a novel family of domesticated transposase genes found in diverse angiosperms. Mol Biol Evol. 2005;22:2084–9. doi: 10.1093/molbev/msi202. [DOI] [PubMed] [Google Scholar]
- 85.Lin R, et al. Transposase-derived transcription factors regulate light signalling in Arabidopsis. Science. 2007;318:1302–1305. doi: 10.1126/science.1146281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cordaux R, Udit S, Batzer MA, Feschotte C. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc Natl Acad Sci USA. 2006;103:8101–8106. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu D, et al. The human SETMAR protein preserves most of the activities of the ancestral Hsmar1 transposase. Mol Cell Biol. 2007;27:1125–32. doi: 10.1128/MCB.01899-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miskey C, et al. The Ancient Mariner Sails Again: Transposition of the Human Hsmar1 Element by a Reconstructed Transposase and Activities of the SETMAR Protein on Transposon Ends. Mol Cell Biol. 2007;27:4589–600. doi: 10.1128/MCB.02027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pathak RU, Rangaraj N, Kallappagoudar S, Mishra K, Mishra RK. Boundary element-associated factor 32B connects chromatin domains to the nuclear matrix. Mol Cell Biol. 2007;27:4796–4806. doi: 10.1128/MCB.00305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cam HP, Noma KI, Ebina H, Levin HL, Grewal SI. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature. 2008;451:431–436. doi: 10.1038/nature06499. [DOI] [PubMed] [Google Scholar]
- 91.Hudson ME, Lisch DR, Quail PH. The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J. 2003;34:453–471. doi: 10.1046/j.1365-313x.2003.01741.x. [DOI] [PubMed] [Google Scholar]
- 92.Raizada MN, Brewer KV, Walbot V. A maize MuDR transposon promoter shows limited autoregulation. Mol Genet Genomics. 2001;265:82–94. doi: 10.1007/s004380000393. [DOI] [PubMed] [Google Scholar]
- 93.Cui H, Fedoroff NV. Inducible DNA demethylation mediated by the maize Suppressor-mutator transposon-encoded TnpA protein. Plant Cell. 2002;14:2883–99. doi: 10.1105/tpc.006163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Atkinson PW, Warren WD, O’Brochta DA. The hobo transposable element of Drosophila can be cross-mobilized in houseflies and excises like the Ac element of maize. Proc Natl Acad Sci U S A. 1993;90:9693–7. doi: 10.1073/pnas.90.20.9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rezsohazy R, van Luenen HGAM, Durbin RM, Plasterk RHA. Tc7, a Tc1-hitch hiking transposon in Caenorhabditis elegans. Nucleic Acids Res. 1997;25:4048–4054. doi: 10.1093/nar/25.20.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lampe DJ, Walden KK, Robertson HM. Loss of transposase-DNA interaction may underlie the divergence of mariner family transposable elements and the ability of more than one mariner to occupy the same genome. Mol Biol Evol. 2001;18:954–61. doi: 10.1093/oxfordjournals.molbev.a003896. [DOI] [PubMed] [Google Scholar]
- 97.Feschotte C, Osterlund MT, Peeler R, Wessler SR. DNA-binding specificity of rice mariner-like transposases and interactions with Stowaway MITEs. Nucleic Acids Res. 2005;33:2153–65. doi: 10.1093/nar/gki509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wallace MR, et al. A de novo Alu insertion results in neurofibromatosis type 1. Nature. 1991;353:864–6. doi: 10.1038/353864a0. [DOI] [PubMed] [Google Scholar]
- 99.Girard L, Freeling M. Regulatory changes as a consequence of transposon insertion. Dev Genet. 1999;25:291–6. doi: 10.1002/(SICI)1520-6408(1999)25:4<291::AID-DVG2>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 100.Simons C, Pheasant M, Makunin IV, Mattick JS. Transposon-free regions in mammalian genomes. Genome Res. 2006;16:164–72. doi: 10.1101/gr.4624306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lerman DN, Feder ME. Naturally occurring transposable elements disrupt hsp70 promoter function in Drosophila melanogaster. Mol Biol Evol. 2005;22:776–83. doi: 10.1093/molbev/msi063. [DOI] [PubMed] [Google Scholar]
- 102.Walser JC, Chen B, Feder ME. Heat-shock promoters: targets for evolution by P transposable elements in Drosophila. PLoS Genet. 2006;2:e165. doi: 10.1371/journal.pgen.0020165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ackerman H, Udalova I, Hull J, Kwiatkowski D. Evolution of a polymorphic regulatory element in interferon-gamma through transposition and mutation. Mol Biol Evol. 2002;19:884–90. doi: 10.1093/oxfordjournals.molbev.a004145. [DOI] [PubMed] [Google Scholar]
- 104.Martin C, Lister C. Genome juggling by transposons: Tam3-induced rearrangements in Antirrhinum majus. Dev Genet. 1989;10:438–51. doi: 10.1002/dvg.1020100605. [DOI] [PubMed] [Google Scholar]
- 105.Koga A, Iida A, Hori H, Shimada A, Shima A. Vertebrate DNA transposon as a natural mutator: the medaka fish Tol2 element contributes to genetic variation without recognizable traces. Mol Biol Evol. 2006;23:1414–9. doi: 10.1093/molbev/msl003. [DOI] [PubMed] [Google Scholar]