Abstract
Following birth, the breast-fed infant gastrointestinal tract is rapidly colonized by a microbial consortium often dominated by bifidobacteria. Accordingly, the complete genome sequence of Bifidobacterium longum subsp. infantis ATCC15697 reflects a competitive nutrient-utilization strategy targeting milk-borne molecules which lack a nutritive value to the neonate. Several chromosomal loci reflect potential adaptation to the infant host including a 43 kbp cluster encoding catabolic genes, extracellular solute binding proteins and permeases predicted to be active on milk oligosaccharides. An examination of in vivo metabolism has detected the hallmarks of milk oligosaccharide utilization via the central fermentative pathway using metabolomic and proteomic approaches. Finally, conservation of gene clusters in multiple isolates corroborates the genomic mechanism underlying milk utilization for this infant-associated phylotype.
Keywords: carbohydrate metabolism, co-evolution, genomics, human milk oligosaccharides
The establishment of a stable gut microbial consortium is critical to the development of the infant gastrointestinal tract (GIT) (1, 2). Breast-fed infants develop colonic microbiota that is frequently marked by a high concentration of bifidobacteria (3, 4). This numerical advantage confers a substantial health benefit to the neonate by hindering pathogen colonization through competitive exclusion (5). Candidate milk-borne growth factors include oligosaccharides recalcitrant to host digestive enzymes, thus supplied to the infant without a known nutritional purpose (6, 7). Interestingly, these human milk oligosaccharide (HMO) structures possess a vast combinatorial potential, as they incorporate 5 carbohydrate monomers via 13 possible glycosidic linkages to assemble molecules containing 3 to 32 units. Therefore the characterization of only approximately 200 soluble structures within the human milk glycome strongly suggests a functional role for these sugars (8). Accordingly, we have previously demonstrated that select bifidobacterial phylotypes use specific oligosaccharides ubiquitous in human milk and secreted early in the lactation cycle (supporting information (SI) Fig. S1) (9, 10). Of the 200 known compositions, five low molecular weight oligosaccharides abundant in breast milk are the preferred substrates of Bifidobacterium longum subsp. infantis ATCC15697, the archetypical HMO-utilizing bacterium (11). Whereas the physiological capability for particular bifidobacteria to use HMO is indisputable, the molecular mechanisms and networks governing metabolism of these unique carbohydrates remain obscured. Thus, the complete genome of B. longum subsp. infantis was sequenced to enable a systems-level investigation into the unique biology that has emerged from this example of mammal-commensal coevolution.
At present, only two phylotypes (i.e., B. longum subsp. longum and B. adolescentis) have been fully sequenced and deposited into public databases. Recently, a second and near-identical B. longum subsp. longum strain has been sequenced to completion (12). Of the 21 ongoing bifidobacterial genome projects, B. bifidum JCM1255 and B. breve UCC2003 have been reported as fully finished (13).
Results
General Genome Features.
The genome of B. longum subsp. infantis strain ATCC15697T is the largest bifidobacterial genome reported to date with its single circular chromosome consisting of 2,832,748 base pairs (Fig. S2). The protein coding content consists of 2423 sequences annotated as genes, with an additional 91 noncoding RNA genes. General genome features are listed in Table S1. The functional distribution of genes assigned to clusters of orthologous groups of proteins (COGs) is relatively similar for the three fully sequenced bifidobacterial phylotypes (Fig. S3). Although the contribution made by repair, replication, and recombination-related COGs is significantly more than what is observed in ATCC15703 and NCC2705 (10% vs. 7%). This is partially explained by the abundance of mobile genetic elements in ATCC15697 relative to other sequenced bifidobacteria, as predicted for a larger chromosome (14). There is a marked increase in insertion sequence (IS) copy number and family representation as well as several prophage-related genes (Table S2). The appearance of several integrated plasmid remnants is intriguing, as episomal plasmids have not been isolated from B. longum subsp. infantis to date (15).
Proliferation of mobile genetic elements in ATCC15697 may be attributable in part to the presence of rexAB homologs (Blon_0809 and Blon_0810) which have not been described in other bifidobacteria. The exonuclease/helicase RexAB (functionally analogous to RecBCD) participates in double-strand break repair and promotes chromosomal rearrangements and integration of heterologous sequences through recombination (16). Repetitive sequences (e.g., IS elements) prompts chromosomal rearrangements increasing the likelihood of repositioning functionally related genes into clusters and operons.
The larger B. longum subsp. infantis genome possesses 702 unique genes in a comparison of common and taxa-specific genes (Fig. S4). Not surprisingly, ATCC15697 shares a larger fraction of genes with NCC2705 (209 genes) to the exclusion of ATCC15703 than B. adolescentis with NCC2705 (80 genes) or B. adolescentis with ATCC15697 (120 genes). As one would predict, the majority of the genes exclusive to ATCC15697 are annotated as hypothetical with many derived from prophage. A smaller subset includes 171 genes that can be assigned to at least one COG (Fig. S5). Thirty-one percent of these COGs possess an unknown function or a general functional prediction, with an additional 26% functionally grouped as replication/recombination/repair, which reflects the additional transposases and integrases found in the ATCC15697 genome. Most importantly, and related to the HMO+ phenotype, are the additional genes designated carbohydrate transport and metabolism related.
Alignments of fully sequenced chromosomes reveal the extent to which genomic rearrangement has influenced their evolution (Fig. S6). Alignment of the ATCC15697 to the NCC2705 genome exhibits a high degree of colinearity interrupted by an inversion around the ori-ter axis. While the alignment to B. adolescentis displays less colinearity, an X-shaped plot diagram is clearly evidenced. This is indicative of multiple large rearrangements around the central axis following divergence from a common ancestor (17). Comparable architectures are observable in alignments of homologous regions across linearlized chromosomes (Fig. S7)
As was observed for the GIT commensal Bacteroides thetaiotaomicron, the preponderance of two-component systems in B. longum subsp. infantis suggests the development of complex regulatory networks to sense dynamic environmental cues and initiate the corresponding physiological response (18). There are 23 sensor histidine kinases and 24 response regulators distributed throughout the ATCC15697 chromosome (Table S3), proving to be more abundant than what would be predicted on genome size alone and two-fold more abundant than other bifidobacterial genomes (19).
Aside from the genetics underlying milk utilization, there is genomic evidence for the molecular interactions between B. longum subsp. infantis and its host. Potential adhesion factors include the 2194 aa residue Blon_1261, which encodes a predicted surface protein containing LPxTG cell wall anchor along with collagen binding motifs (Cna_B, PF05738). Immediately downstream of this gene is a predicted allergen V5/Tpx-1 family protein (Blon_1259) with a myosin tail domain possessing a 26% identity to a protein from the intestinal isolate Collinsella aerofaciens ATCC25986. It is currently unclear whether these proteins contribute to the immunomodulatory effects exhibited by B. longum subsp. infantis, adhesion to the host mucosa, or both (20). It is noteworthy that ATCC15697 does not possess the full complement of cell surface proteins predicted in B. longum subsp. longum NCC2705 (14). A homolog of the glycoprotein binding fimbrae (BL0675) and associated proteins were not found within the B. longum subsp. infantis genome, suggesting an alternate adhesion mechanism adapted specifically for its ecological niche. It is possible that the extensive array of solute binding proteins (SBPs) within the ATCC15697 genome are responsible for binding host cell surface receptors or mucin glycoproteins. Surprisingly, mucus binding proteins and domains were not found in predicted bifidobacterial proteomes including MUB domains from Lactobacillus plantarum and Lactobacillus reuteri, mucBP (PF06458) and several mucus binding proteins characterized in Lactobacillus acidophilus (21–23). In addition, the ATCC15697 genome possesses a serpin homolog originally described in NCC2705 (Blon_0790; 96% nucleotide identity). This secreted protein may facilitate evasion of host proteolytic systems and modulate inflammation in the GIT (24).
Additional interactions of B. longum subsp. infantis with its infant host include the supplementing of specific nutritional requirements. The presence of complete biosynthetic pathways for riboflavin, thiamin, and folate imparts a tangible benefit to its host. Whereas the apparent inability to synthesize biotin, pyridoxine, cobalamin, and panthenate, as was previously described for NCC2705, may define a species characteristic (14).
Furthermore, the presence of a cytochrome P450-like gene (COG2124, Blon_2155) may be indicative of host adaptation, as this gene is commonly detected in intestinal isolates. The gene product is associated with the detoxification of xenobiotics encountered within the colon and may be involved in modulating expression of host hepatic P450 (25).
The complete biosynthetic pathways for purines and pyramidines from glutamine are found intact, as well as at least 19 aa from ammonium with cysteine potentially synthesized from succinylhomoserine and colonic sulfur as was postulated for NCC2705 (14). Furthermore, genes are present for a partial tricarboxylic acid cycle ostensibly to generate anapleurotic metabolites.
It has been previously proposed that B. infantis and B. suis be united with B. longum as a single species and delimited as three biotypes (26), although this taxonomic description remains contentious (27). Accordingly, the ATCC15697 and NCC2705 genomes exhibit a high degree of colinearity and the 96% average nucleotide identity between conserved genes exceeds the threshold for inclusion as a single species (Fig. S7) (28). Nevertheless, the differences in ecological niche (subsp. infantis is typically isolated from the infant GIT) along with physiological dissimilarity merits a distinct taxonomic designation for infantis as a subspecies of B. longum (26, 29). Recently Mattarelli, et al. (15) determined the subspecies designation for suis, longum and infantis to be the most appropriate, albeit without examining the ATCC15697 genome.
Genomics of Milk Utilization.
Milk oligosaccharides are defined by an invariable lactose moiety at the reducing end, with structural diversity arising from assorted configurations of glucose, galactose, N-acetylglucosamine, fucose, and sialic acid residues (8). We hypothesized that the complexity of HMO structures must dictate a comparable unique strategy for deconstructing these oligomers, one which has not been described in prior genome sequences (9, 11).
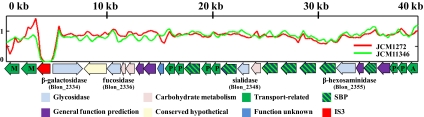
The discovery of a novel 43 kbp gene cluster (Blon_2331-Blon_2361) dedicated to HMO import and processing provides a significant initial validation of this prediction (Fig. 1). Located within this 30-gene cluster are the 4 glycosyl hydrolases predicted to cleave milk oligosaccharides into its constituent monosaccharides. To our knowledge, a sialidase (COG4409, Blon_2348), a fucosidase (COG3669, Blon_2336), an N-acetyl-β-hexosaminidase (COG3525, Blon_2355), and a β-galactosidase (COG3250, Blon_2334) have not been genetically linked within a short contiguous segment in any sequenced bacterial genome. The proximity of these glycosyl hydrolase genes implies potential co-regulation and participation in a common metabolic pathway. Previous studies conducted on ATCC15697 have reported the phenotypic manifestation of these four glycosyl hydrolases (11, 30, 31). Interestingly, fucosidase activity is induced by growth on HMO, whereas sialidase activity appears to be constitutively expressed (11). Thus it is likely that fucose metabolism, and fucosidase expression in particular, is a key regulatory point in hydrolysis of HMO molecules.
Fig. 1.
Glycosyl hydrolases and transport-related genes located in the HMO utilization cluster. The 43 kbp HMO cluster possesses all four glycosyl hydrolases active on HMO linkages. Additionally, family 1 (oligosaccharide-binding) solute binding proteins (SBP) associated with ABC transporters are found at high density in the cluster. HMO cluster sequence depth in JCM1272 and JCM11346 is normalized to ATCC15697 in arbitrary units. With the exception of the IS3 insertion sequence, the entire locus is found to be present in both B. longum subsp. infantis genomes. Transport-related genes are denoted as M: major facilitator superfamily, P: ABC transporter permease component, and A: ABC transporter ATPase subunit.
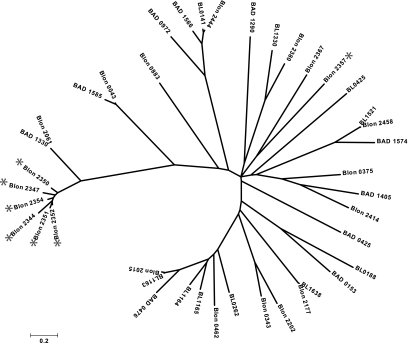
In addition to glycolytic potential, the molecular basis for transmembrane import of milk oligosaccharides into the cytosol is evident and emphasized by redundancy within this gene cluster. There are two ABC transport permeases (COG0395, Blon_2359 and COG1175, Blon_2360) as well as the associated ATPase (COG3839, Blon_2361) in the cluster's 5′ terminus as delineated by the majority of gene orientation. Two additional permease pairs are located in the center of the cluster. Significantly, there are seven extracellular solute binding proteins (SBP; PF01547) predicted to bind oligosaccharides (SBP family 1) and facilitate interactions with proximal permeases in the Gram-positive cell membrane as a component of the ABC transporter complex. Remarkably, six of these HMO cluster lipoproteins exhibit a pronounced evolutionary divergence relative to other SBP family 1 proteins of the three fully sequenced bifidobacteria (Fig. 2). Moreover, the B. longum subsp. infantis genome contains 21 copies of family 1 SBPs, whereas subsp. longum and B. adolescentis possess 10 and 11 respectively. The lineage-specific amplification of this SBP family within the genome coupled with the sequence divergence of HMO cluster proteins underscores their functional importance in the infant GIT. Again, the clustering of these transport-related genes around milk-glycan hydrolases suggests a common operation.
Fig. 2.
Evolutionary relationships of 41 family 1 SBP proteins deduced from the B. longum subsp. infantis ATCC15697 (Blon), B. longum subsp. longum NCC2705 (BL), and B. adolescentis ATCC15703 (BAD) genome sequences. Branch lengths are in the same units (number of amino acid substitutions per site) as those of the evolutionary distances used to construct the tree. SBP proteins with genes located in the HMO utilization cluster are denoted with an asterisk. Interestingly, the B. adolescentis SBP possessing the highest identity to the HMO cluster proteins (BAD_1330) is located in an operon devoted to fructo-oligosaccharide utilization, a substrate that lacks the structural complexity of HMO.
Bifidobacteria are generally capable of using isomeric lacto-N-tetraose (LNT, Gal(β1–3)GlcNac(β1–3)Gal(β1–4)Glc), which comprise the core HMO structure and is catabolized via a lacto-N-biose (LNB, Gal(β1–3)GlcNAc) intermediate (11). Recently the LNB metabolic pathway has been elucidated in the bifidobacteria which proceeds through the activity of a lacto-N-biose phosphorylase (EC 2.4.1.211, Blon_2174) and associated genes (32). Tellingly, the seven-gene operon responsible for LNB utilization is conserved in B. longum subsp. longum and B. longum subsp. infantis (Blon_2171-Blon_2177) and absent from B. adolescentis ATCC15703 which does not use LNT or other milk oligosaccharide structures (9).
Additional gene clusters predicted to facilitate utilization of milk oligosaccharides and derivatives are a sialidase (Blon_0641-Blon_0651) and two fucosidase clusters (Blon_0243-Blon_0248 and Blon_0423-Blon_0426) illustrated in Fig. S8. Interestingly, these fucosidases and neighboring permeases arose from a recent duplication event as they retain near identity (Fig. S9). One permease and fucosidase pair (Blon_0425 and Blon_0426) seemingly displaced an arabinose isomerase, ribulose 5-phosphate epimerase and a sugar kinase present in B. longum subsp. longum and B. adolescentis. A vestigial fragment of the sugar kinase remains in place of the complete arabinose cluster. This non-orthologous gene displacement suggests a reorientation toward interactions with mammalian glycans containing fucose in lieu of plant-derived arabinose. The apparent dispensability of pentose metabolism is evident throughout the genome, and is consistent with the inability of infant-isolated bifidobacteria to ferment pentose sugars (33). Conversely, B. longum subsp. longum and B. adolescentis have retained the genetic capacity to ferment xylose and arabinose (Table S4); sugars which are not introduced to the infant GIT before weaning.
Glycosyl hydrolases active on HMO linkages, as well as oligosaccharide-binding SBP proteins and permeases, constitute a genomic architecture that was previously unknown in nature. Thus it is difficult to resolve the evolutionary genesis of the entire 43 kbp HMO-utilization cluster, although discrepancies in synonymous codon usage suggest that several genes in the 3′ end (Blon_2331–Blon_2339) may have arisen from an alien source, while the remainder appear to be bifidobacterial in origin (Fig. S10). In contrast, a gross deviation from guanine-cytosine content and dinucleotide genome signature was not detected in the cluster, limiting the evidence for a recent transfer from a heterologous source (Fig. S11). Nonetheless, the linkage of these genes in a common transcriptional orientation evokes the possibility of lateral transfer as functional unit.
Whereas milk carbon is presumably accessed by bifidobacteria as oligosaccharides, the provision of nitrogen to the lower infant GIT is limiting. Accordingly, the protein concentration in human milk is often inadequate to satisfy the infant's increased nitrogen demands during postnatal growth, let alone the metabolic requirements of resident GIT commensals. Interestingly, the urea content of human milk constitutes a substantial fraction (approximately 15%) of total nitrogen and represents a potential source for both the infant and its associated microbiota (34). It is believed that colonic urea is reclaimed through a bacterial-mediated process termed urea recycling or urea nitrogen salvaging (UNS; reviewed in (35)). Adding to milk-delivered urea, enterocytes actively transport urea derived from amino acid catabolism into the intestinal lumen (36). Subsequent bacterial urease activity (EC 5.3.1.5) liberates ammonia from urea, thus availing it for host anabolic processes. While urease activity has been previously reported in bifidobacteria (37), the detection of a complete urease gene cluster (Blon_0104-Blon_0115) connects an observed biochemical process with a specific infant commensal, thus further accentuating the importance of UNS during neonate development. Accordingly, inorganic nitrogen has been demonstrated to be the preferred nitrogen source for most bifidobacteria, as ammonium is predicted to be imported by a dedicated transporter (Blon_0223) conserved within the three completely sequenced bifidobacterial genomes (38). In addition, inorganic nitrogen is likely to be provided to B. longum subsp. infantis through deamination of N-acetylglucosamine residues in HMO.
To understand the phylogenetic distribution of the identified milk utilization gene clusters, five additional HMO-utilizing B. longum isolates were sequenced with sequencing-by-synthesis technology. Phylogenetic analysis of the resulting sequences identified two of these strains as close relatives of the ATCC15697 type strain genome described here (Fig. S12). The three other isolates were determined to be more closely related to B. longum subsp. longum or B. longum subsp. suis and will be described elsewhere. The two additional B. longum subsp. infantis isolates share 231 genes with the 702 unique subsp. infantis genes, further defining the core gene set for this taxon (Table S5). Furthermore, these two isolates possess the 43 kb HMO cluster, thus generalizing the current understanding of oligosaccharide metabolism in this infant-associated subspecies (Fig. 1). It is worth noting that these milk-related gene clusters were also found within an infant gut metagenome (In-M; 39).
in Vivo Metabolism of Human Milk Oligosaccharides.
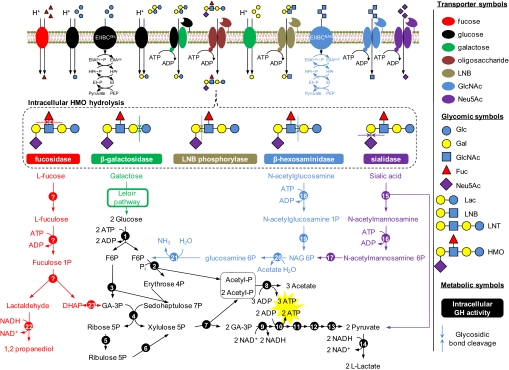
The bifid shunt is a catabolic pathway that uses fructose-6-phosphate phosphoketolase (EC 4.1.2.22, Blon_1722) to catalyze a thiamin-diphosphate-dependent conversion of D-fructose-6-phosphate (F-6P) into D-erythrose-4-phosphate and acetyl phosphate. This pathway, of which all genes are identifiable in the ATCC15697 genome, produces 1.5 moles acetate and 1 mole lactate for every mole hexose that enters. Similarly, the pathways that feed hydrolyzed HMO residues into central metabolism were reconstructed from the genome sequence (Fig. 3). The acetate derived from sugars via the bifid shunt is both secreted and incorporated de novo fatty into acids where it is a hallmark of metabolic flux through this pathway. Fatty acid synthesis is presumably accomplished with the multidomain fatty acid synthase I (Blon_2284) which is common to Actinobacteria and eukaryotes (40). Proteomics has confirmed that most predicted HMO catabolic proteins are detected during HMO fermentation, in addition to the expression of several 43 kb cluster proteins (Fig. S13 and Table S6).
Fig. 3.
B. longum subsp. infantis metabolism of human milk oligosaccharides and derivatives. Carbohydrates transverse the cell membrane by ABC transporters (HMO, lactose, LNB, N-acetylglucosamine, and sialic acid), major facilitator superfamily permeases (fucose, glucose, galactose, and lactose), and PTS (glucose and N-acetylglucosamine). Intracellular glycosyl hydrolases (Table S7) process sugar polymers into constituent carbohydrates which are further degraded before entering the catabolic pathways. The central fermentative pathway (Bifid shunt) is denoted in black with stoichiometric coefficients representing utilization of two hexoses. Genes involved in intracellular metabolism are listed in Table S8. Note that galactose is predicted to be catabolized by a modified Leloir pathway determined experimentally in B. longum subsp. longum (32). PTS, phosphotransferase system; Pi, phosphate; G6P, glucose 6-phosphate; F6P, fructose 6-phosphate; GA-3P, glyceraldehyde 3-phosphate; DHAP, dihydroxyacetone phosphate; GH, glycosyl hydrolase; Glc, glucose; Gal, galactose; GlcNAc, N-acetylglucosamine; Fuc, fucose; Neu5Ac, sialic acid; Lac, lactose; LNB, lacto-N-biose; LNT, lacto-N-tetraose; HMO, human milk oligosaccharide.
Isotopic labeling experiments with 13C-fructose and 13C-acetate confirm that the bifid shunt is the predominant pathway for carbohydrate fermentation by B. longum subsp. infantis (A.A., T.R.W. and N.P.P., unpublished work). The fractional distribution of 13C into carbohydrate and fatty acid metabolites were analyzed by GC-EI-MS. Isotopic labeling with universally-labeled [U-13C]fructose resulted in time dependent incorporation of 13C into cellular carbohydrates (galactose, glucose, ribose, and rhamnose) and fatty acids (myristate, palmitate, stearate, and oleate), examples of which are shown in Fig. S14 A–C.
Fully labeled ATCC15697 cells were also chased by switching to growth on unlabeled precursors, specifically human milk oligosaccharides or N-acetylneuraminic acid (NeuNAc). The depletion of 13C/12C ratios, and the concomitant reincorporation of 12C from the unlabeled chase, were monitored by GC-MS over a 24 h growth period (Fig. S10D). After 1–4 h of growth, HMO catabolism was apparent by the chase of 13C from the cellular metabolites of [U-13C]fructose-grown ATCC15697. A similar chase of 13C from intracellular galactose and myristate (and other small metabolites) was also observed with unlabeled NeuNAc (data not shown). To confirm these data, ATCC15697 cells were grown on N-acetyl-[2-13C]neuraminic acid as a sole carbon source. 13C-Labeled fatty acids were produced during growth (Fig. S10C) demonstrating that this bacterium is able to ferment sialic acid as a sole carbon source. Stable isotope from [2-13]NeuNAc was not incorporated into cellular sugars (data not shown). These results are consistent with the utilization pathway reconstructed from the genome sequence, as the 9-carbon carbohydrate backbone of N-acetyl-[2-13C]neuraminic acid was metabolized into 3-carbon [2-13]pyruvate, plus unlabeled N-acetylmannosamine (ManNAc). As predicted, ManNAc is likely phosphorylated by NanK (E.C. 2.7.1.60; Blon_0644), epimerized by NanE (E.C. 5.-; Blon_0645), deacetylated by NagA (E.C. 3.5.1.25; Blon_0882), and subsequently deaminated into fructose 6-phosphate by NagB (E.C. 3.5.99.6; Blon_0881), at which point it enters the catabolic bifid shunt.
Discussion
Various bifidobacterial strains (some infant-associated) are incapable of fermenting HMO to high cell density (9, 11), contrasting with the broad metabolic capacity exhibited by B. longum subsp. infantis. The clustering of requisite glycosidases with oligosaccharide transport proteins within the ATCC15697 genome presents a molecular rationale for the consumption of a diverse assortment of milk glycans. These features hint tantalizingly at a competitive strategy predicated on the efficient capture of milk carbon by this member of the infant microbial consortium.
Several ecological surveys confirm that bifidobacteria are a major constituent of the infant GIT (particularly breast-fed infants), although species representation and relative concentration among other phylogenetic types exhibit inter-study heterogeneity (41–43). If a discernible population structure exists, the complexities in resolving the core infant microbiome will mirror those experienced in defining the adult by population dynamics or functional redundancy. Thus the elucidation of a milk-active gene suite provides a salient target to test their relative contribution to a core infant microbiome, their impact on colonization and succession, and their phylogenetic distribution. It remains unknown if these genomic features constitute a unique competitive strategy evolved in subsp. infantis and perhaps other bifidobacteria. Likewise, these encoded operations may characterize a segment of the infant microbiota's aggregate physiology; thus dictating the distal GIT metabolome in the first few months of life irrespective of phylogenetic composition.
Initially, milk-related genes and pathways were identified by their functional predictions, as well as their proximity within the genome (e.g., 43 kb HMO cluster). Therefore, it is conceivable that some of these encoded activities are also active on host or milk glycoconjugates, although B. longum subsp. infantis cannot ferment mucoglycoproteins in vitro (44). Predicted HMO metabolism was functionally confirmed by isotopic studies which indicate that milk carbon is shunted through the central fermentative pathway for energy generation and integrated into cellular biomass as fatty acids. This is only possible if the oligosaccharide molecule is first digested into constituent monosaccharides by glycosidases and further degraded before entry into the bifid shunt. Although our previous studies have shown HMOs to be degraded concomitant with bacterial growth, intracellular metabolism has not been described in vivo to this point (9, 11). Moreover, the proteomic profile obtained during growth on HMO is consistent with the metabolic reconstruction extrapolated from the ATCC15697 genome sequence.
In addition, and of a practical matter, the discovery of a rexAB helicase/exonuclease might impact future genetic research. Previous efforts to adapt standard genetic tools to the bifidobacteria have been severely hindered by low recombination frequencies, which have been attributed to the conspicuous absence of recBCD (or rexAB) genes in previously sequenced bididobacteria (14). It remains to be determined whether the rec+ phenotype exists in B. longum subsp. infantis and is linked to the putative rexAB, thus enabling the study of bifidobacterial genetics.
As an infant commensal, the evolutionary heritage of B. longum subsp. infantis is necessarily linked to milk and thus its genome dramatically recapitulates the pressures encountered in the mammalian GIT. Clearly, the refinement of milk through the millennia is reflected in benefits to the infant which offset the significant energetic costs exacted from the mother during lactation. Whereas the Darwinian argument for milk's role in enhancing mammalian fitness is inexorably linked to its nutritive value, the potential for milk to deliver exogenous materials to resident GIT microbiota cannot be ignored. Thus, the tripartite relationship of B. longum subsp. infantis, its infant host, and milk provides a fascinating example of coevolution to be explored in subsequent studies of the human microbiome. Moreover, it is through this evolutionary perspective that the present composition of milk must be considered, so as to fully appreciate the extent to which purifying selection has culled superfluous components from this fluid secretion and sustained beneficial properties including the enrichment of select commensals.
Materials and Methods
Whole genome shotgun sequencing and assembly were carried out at the Department of Energy Joint Genome Institute. The ATCC15697 genome was sequenced to approximately 8X depth and assembled with PHRAP, PGA, and Arachne (45) assemblers. Finishing was carried out using CONSED (46). Automated gene modeling was performed using multiple databases and modeling packages as detailed elsewhere (47).
Additional sequencing, bioinformatic, metabolomic, and proteomic methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments.
The authors thank C. Bevins and J. Eisen for their critical review of the manuscript, K. Ahrens for technical assistance, R. Ward for providing HMO, D. O'Sullivan for his gift of DJO10A, and B. Phinney for proteomics assistance. The project described was supported by National Institutes of Health NIGMS T32-GM08799 (DAS), University of California Discovery Grant Program and California Dairy Research Foundation (DAM) and by USDA NRI-CSREES Award 2008–35200-18776 (DAS and DAM).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession no. CP001095 (B. longum subsp. infantis ATCC15697)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0809584105/DCSupplemental.
References
- 1.Stappenbeck T, Hooper L, Gordon J. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Favier C, de Vos W, Akkermans A. Development of bacterial and bifidobacterial communities in feces of newborn babies. Anaerobe. 2003;9:219–229. doi: 10.1016/j.anaerobe.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Kleessen B, Bunke H, Tovar K, Noack J, Sawatzki G. Influence of two infant formulas and human milk on the development of the faecal flora in newborn infants. Acta Paediatr. 1995;84:1347–1356. doi: 10.1111/j.1651-2227.1995.tb13567.x. [DOI] [PubMed] [Google Scholar]
- 5.Lievin V, et al. Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut. 2000;47:646–652. doi: 10.1136/gut.47.5.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liepke C, et al. Human milk provides peptides highly stimulating the growth of bifidobacteria. Eur J Biochem. 2002;269:712–718. doi: 10.1046/j.0014-2956.2001.02712.x. [DOI] [PubMed] [Google Scholar]
- 7.Gyorgy P, Kuhn R, Rose C, Zilliken F. Bifidus factor II. Its occurrence in milk from different species and in other natural products. Arch Biochem Biophys. 1954;48:202–208. doi: 10.1016/0003-9861(54)90324-0. [DOI] [PubMed] [Google Scholar]
- 8.Ninonuevo M, et al. A strategy for annotating the human milk glycome. J Agric Food Chem. 2006;54:7471–7480. doi: 10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- 9.Ward R, Ninonuevo M, Mills D, Lebrilla C, German J. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol. 2006;72:4497–4499. doi: 10.1128/AEM.02515-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ninonuevo M, et al. Daily variations in oligosaccharides of human milk determined by microfluidic chips and mass spectrometry. J Agric Food Chem. 2008;56:618–626. doi: 10.1021/jf071972u. [DOI] [PubMed] [Google Scholar]
- 11.Locascio R, et al. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrate strain specific, preferential consumption of small chain glycans secreted in early human lactation. Journal of Agriculture and Food Chem. 2007;55:8914–8915. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- 12.Lee J, et al. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics. 2008;9:247. doi: 10.1186/1471-2164-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides N. The genomes on line database (GOLD) in 2007: Status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2008;36:D475–D479. doi: 10.1093/nar/gkm884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schell M, et al. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci USA. 2002;99:14422–14427. doi: 10.1073/pnas.212527599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattarelli P, Bonaparte C, Pot B, Biavati B. Proposal to reclassify the three biotypes of Bifidobacterium longum as three subspecies: Bifidobacterium longum subsp. longum subsp. nov., Bifidobacterium longum subsp. infantis comb. nov. and Bifidobacterium longum subsp. suis comb. nov. Int J Syst Evol Microbiol. 2008;58:767–772. doi: 10.1099/ijs.0.65319-0. [DOI] [PubMed] [Google Scholar]
- 16.Prudhomme M, Libante V, Claverys J-P. Homologous recombination at the border: Insertion-deletions and the trapping of foreign DNA in Streptococcus pneumoniae. PNAS. 2002;99:2100–2105. doi: 10.1073/pnas.032262999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisen J, Heidelberg J, White O, Salzberg S. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol. 2000;1(6):RESEARCH0011.1–0011.9. doi: 10.1186/gb-2000-1-6-research0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 19.Ulrich L, Koonin E, Zhulin I. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 2005;13:52–56. doi: 10.1016/j.tim.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Mahony L, et al. Lactobacillus and bifidobacterium in irritable bowel syndrome: Symptom responses and relationship to cytokine profiles. Gastroenterology. 2005;128:541–551. doi: 10.1053/j.gastro.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 21.Boekhorst J, Helmer Q, Kleerebezem M, Siezen R. Comparative analysis of proteins with a mucus-binding domain found exclusively in lactic acid bacteria. Microbiology. 2006;152:273–280. doi: 10.1099/mic.0.28415-0. [DOI] [PubMed] [Google Scholar]
- 22.Buck B, Altermann E, Svingerud T, Klaenhammer T. Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl Environ Microbiol. 2005;71:8344–8351. doi: 10.1128/AEM.71.12.8344-8351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roos S, Jonsson H. A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology. 2002;148:433–442. doi: 10.1099/00221287-148-2-433. [DOI] [PubMed] [Google Scholar]
- 24.Ivanov D, et al. A serpin from the gut bacterium Bifidobacterium longum inhibits eukaryotic elastase-like serine proteases. J Biol Chem. 2006;281:17246–17252. doi: 10.1074/jbc.M601678200. [DOI] [PubMed] [Google Scholar]
- 25.John G, et al. The presence of a cytochrome P450-like protein in the human intestinal microflora Eubacterium aerofaciens. Microbial Ecology in Health and Disease. 2001;13:3–8. [Google Scholar]
- 26.Sakata S, et al. Unification of Bifidobacterium infantis and Bifidobacterium suis as Bifidobacterium longum. Int J Syst Evol Microbiol. 2002;52:1945–1951. doi: 10.1099/00207713-52-6-1945. [DOI] [PubMed] [Google Scholar]
- 27.Klein G. International committee on systematics of prokaryotes; subcommittee on the taxonomy of Bifidobacterium, Lactobacillus and related organisms:. Int J Syst Evol Microbiol; Minutes of the meetings; 30 August and 1 September 2006; Bologna, Italy. 2007. pp. 1367–1369. [Google Scholar]
- 28.Konstantinidis K, Tiedje J. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reuter G. The Lactobacillus and Bifidobacterium microflora of the human intestine: Composition and succession. Curr Issues Intest Microbiol. 2001;2:43–53. [PubMed] [Google Scholar]
- 30.Moller P, Jorgensen F, Hansen O, Madsen S, Stougaard P. Intra- and extracellular {beta}-galactosidases from Bifidobacterium bifidum and B. infantis: Molecular cloning, heterologous expression, and comparative characterization. Appl Environ Microbiol. 2001;67:2276–2283. doi: 10.1128/AEM.67.5.2276-2283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noriega L, Gueimonde M, Sanchez B, Margolles A, de los Reyes-Gavilan CG. Effect of the adaptation to high bile salts concentrations on glycosidic activity, survival at low PH, and cross-resistance to bile salts in Bifidobacterium. Int J Food Microbiol. 2004;94:79–86. doi: 10.1016/j.ijfoodmicro.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 32.Nishimoto M, Kitaoka M. The complete lacto-N-biose I/galacto-N-biose metabolic pathway in Bifidobacterium longum: identification of N-acetylhexosamine 1-kinase. Appl Environ Microbiol. 2007;73:6444–6449. doi: 10.1128/AEM.01425-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scardovi V. Genus Bifidobacterium, Orla-Jensen 1924, 472AL. In: Sneath PHA, Mair NS, Sharpe MS, Holt JG, editors. Bergey's Manual of Systematic Biology. Vol 2. Baltimore: The Williams and Wilkins Co.; 1986. pp. 1418–1434. [Google Scholar]
- 34.Donovan S, Lonnerdal B. Non-protein nitrogen and true protein in infant formulas. Acta Paediatr Scand. 1989;78:497–504. doi: 10.1111/j.1651-2227.1989.tb17927.x. [DOI] [PubMed] [Google Scholar]
- 35.Fuller M, Reeds PJ. Nitrogen cycling in the gut. Annu Rev Nutr. 1998;18:385–411. doi: 10.1146/annurev.nutr.18.1.385. [DOI] [PubMed] [Google Scholar]
- 36.You G, et al. Cloning and characterization of the vasopressin-regulated urea transporter. Nature. 1993;365:844–847. doi: 10.1038/365844a0. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki K, et al. Urease-producing species of intestinal anaerobes and their activities. Appl Environ Microbiol. 1979;37:379–382. doi: 10.1128/aem.37.3.379-382.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka R, Mutai M. Improved medium for selective isolation and enumeration of Bifidobacterium. Appl Environ Microbiol. 1980;40:866–869. doi: 10.1128/aem.40.5.866-869.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurokawa K, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schweizer E, Hofmann J. Microbial type I fatty acid synthases (FAS): Major players in a network of cellular FAS systems. Microbiol Mol Biol Rev. 2004;68:501–517. doi: 10.1128/MMBR.68.3.501-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Favier C, Vaughan E, De Vos W, Akkermans AD. Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol. 2002;68:219–226. doi: 10.1128/AEM.68.1.219-226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harmsen H, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. 2000;30:61–67. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 43.Palmer C, Bik E, DiGiulio D, Relman D, Brown P. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crociani F, Alessandrini A, Mucci M, Biavati B. Degradation of complex carbohydrates by Bifidobacterium spp. Int J Food Microbiol. 1994;24:199–210. doi: 10.1016/0168-1605(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 45.Jaffe D, et al. Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res. 2003;13:91–96. doi: 10.1101/gr.828403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gordon D, Desmarais C, Green P. Automated finishing with autofinish. Genome Res. 2001;11:614–625. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chain P, et al. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc Natl Acad Sci USA. 2006;103:15280–15287. doi: 10.1073/pnas.0606924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.