Abstract
Articulated jaws are highly conserved structures characteristic of gnathostome evolution. Epithelial-mesenchymal interactions within the first pharyngeal arch (PA1) instruct cephalic neural crest cells (CNCCs) to form the different skeletal elements of the jaws. The endothelin-1 (Edn1)/endothelin receptor type-A (Ednra)→Dlx5/6→Hand2 signaling pathway is necessary for lower jaw formation. Here, we show that the Edn1 signaling is sufficient for the conversion of the maxillary arch to mandibular identity. Constitutive activation of Ednra induced the transformation of upper jaw, maxillary, structures into lower jaw, mandibular, structures with duplicated Meckel's cartilage and dermatocranial jaws constituted by 4 dentary bones. Misexpression of Hand2 in the Ednra domain caused a similar transformation. Skeletal transformations are accompanied by neuromuscular remodeling. Ednra is expressed by most CNCCs, but its constitutive activation affects predominantly PA1. We conclude that after migration CNCCs are not all equivalent, suggesting that their specification occurs in sequential steps. Also, we show that, within PA1, CNCCs are competent to form both mandibular and maxillary structures and that an Edn1 switch is responsible for the choice of either morphogenetic program.
Keywords: craniofacial development, neural crest, pharyngeal arch
The gnathostome skull is characterized by the presence of articulated jaws consisting of the palatoquadrate dorsally and the Meckelian cartilage (MC) ventrally. These prehensile cranial units derive from the maxillary and mandibular processes of the first pharyngeal arch (PA1). Most cartilaginous and dermatocranial derivatives of PA1 are formed by Hox-negative cephalic neural crest cells (CNCCs) emigrating from the mesencephalic and anterior rhombencephalic neural folds (1–6). Before migration, CNCCs lack the topographic information needed to unfold the morphogenetic process of the jaws (7), and receive these instructions from epithelial structures of PA1. In particular, surgical deletion and grafting of different parts of the foregut endoderm has shown that this epithelium harbors instructive signals directing CNCC to form polarized jaw structures such as MC (4, 7, 8). The molecular nature of these instructive signals remains to be determined, but experimental evidence indicates that FGFs, bone morphogenetic proteins (BMPs), endothelin-1 (Edn1), and Sonic hedgehog are involved (9). In particular, loss of Edn1-endothelin receptor type-A (Ednra) signaling (10, 11) results in the homeotic transformation of lower into upper jaw structures, indicating the role of this signaling pathway in the specification of mandibular structures (12, 13). This transformation is accompanied by the down-regulation of Dlx5 and Dlx6 (10, 12–14), and is strongly reminiscent of that observed after targeted inactivation of these 2 genes (15, 16). Dlx homeobox genes, vertebrate orthologues of Drosophila Distal-less, have a fundamental role in the specification of the dorsoventral patterning of PA1 derivatives (17, 18). Edn1 is expressed in the epithelium and mesodermal core of the mandibular part of PA1, whereas Ednra is broadly expressed in CNCC-derived ectomesenchyme of the head [supporting information (SI) Fig. S1] (10, 19, 20), suggesting that all CNCCs might be competent to respond to Edn1 signaling. To test this hypothesis, we generated mice in which Edn1 was knocked-in into the Ednra locus by recombinase-mediated cassette exchange (RMCE) in embryonic stem cells (21), resulting in Ednra constitutive activation throughout the head and pharyngeal mesenchyme. Here, we demonstrate that constitutive activation of Ednra induces the transformation of maxillary structures into mandibular structures with duplicated MCs and dermatocranial jaws constituted by 4 dentary bones. A similar transformation is obtained by forcing the expression of Hand2, a downstream target of the Edn1 pathway, in the Ednra domain. Skeletal transformations are accompanied by neuromuscular remodeling. Thus, within PA1, CNCCs are competent to form both mandibular and maxillary structures and that an Edn1 switch is responsible for the choice of either morphogenetic program.
Results
Constitutive Activation of Ednra Induces the Transformation of Upper Jaw Structures to Lower Jaw Structures.
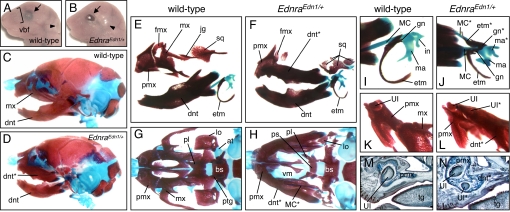
We generated Edn1 knocked-in mice by RMCE in embryonic stem cells (Fig. S2) (21). All heterozygous (EdnraEdn1/+) mice died perinatally with severe craniofacial defects. At E18.5, EdnraEdn1/+ mice were externally characterized by loss of vibrissae, open eyelids, and an anterior shift of the ear position (Fig. 1 A and B). Skeletal preparations revealed that the maxillary components of EdnraEdn1/+ mice were replaced by a second set of mandibular elements (Fig. 1 C–J). Notably, a dentary-like bone carrying an incisor was formed in the maxillary region (Fig. 1 K–N), resulting in jaws constituted by 4 mandibles (Fig. 1 C–H; Fig. S3 and Fig. S4). The tip of this ectopic dentary was fused to the premaxilla (Fig. 1 L and N). The 2 dorsal mandibular-like structures did not form a midline symphysis, possibly due to hindrance by the frontonasal process (Fig. 1 E–H; Fig. S3 and Fig. S4). MC and associated dermal bones (gonial and ectotympanic) were also duplicated in EdnraEdn1/+ mice (Fig. 1 I and J). The incus was fused to the malleus and morphologically changed to a malleus-like structure in mutants (Fig. 1 I and J). Mandibular components were only mildly affected with shortening of coronoid and condylar processes and hyperplasia of the anterior part of the tympanic ring (Fig. S4). The palatine, pterygoid, lamina obturans, and ala temporalis were missing or severely deformed, the squamosal was reduced and divided in 3 to 4 parts as in Dlx1/2 double mutant mice (22), whereas midline structures including the nasal capsule, vomer, and presphenoid were normal (Fig. 1 G and H; Fig. S5 A–C). The premaxilla, the frontal process of maxilla, and the lacrimal bone were maintained in EdnraEdn1/+ mice. The stapes, a derivative of the second PA (PA2), was deformed and fused to the styloid process (Fig. S4).
Fig. 1.
Transformation of maxillary components in EdnraEdn1/+ mice. (A and B) Facial appearance of E18.5 wild-type (A) and EdnraEdn1/+ (B) mice. Vibrissae are absent in mutant mice that have open eyelids (arrows) and an anterior shift of the ear position (arrowheads). (C–L) Bone and cartilage elements of E18.5 wild-type (C, E, G, I, and K) and EdnraEdn1/+ (D, F, H, J, and L) skulls. (C and D) Lateral views of wild-type and EdnraEdn1/+ craniofacial skeletons. (E and F) The EdnraEdn1/+ mutant shows mirror-image duplication of the mandibular elements (dentary, MC, and malleus) at the expense of the maxillary elements (maxilla, jugal, squamosal, and incus). (G and H) Caudal views of wild-type and EdnraEdn1/+ craniofacial skeletons. The dentaries are removed to show the normal and transformed maxillae. Large parts of the palatine, pterygoid, lamina obturans, and ala temporalis are missing or severely deformed in the EdnraEdn1/+ mutant. (I and J) Ectotympanic and gonial bones are also duplicated in association with the ectopic MC in EdnraEdn1/+ mutants. (K and L) Caudal views of the premaxilla-maxilla junction. The tip of the transformed maxilla-dentary in the EdnraEdn1/+ mutant contains an ectopic incisor and fuses to the premaxilla. (M and N) Parasagittal sections of E18.5 wild-type (M) and EdnraEdn1/+ (N) mice stained by Mallory trichromic. The EdnraEdn1/+ mutant shows an ectopic incisor in addition to the orphotopic 1. Fusion between the premaxilla and transformed maxilla-dentary is observed in the mutant. at, ala temporalis; bs, basisphenoid; dnt, dentary; etm, ectotympanic; fmx, frontal process of maxilla; gn, gonial; hy, hyoid; in, incus; jg, jugal; lo, lamina obturans; ma, malleus; mx, maxilla; pl, palatine; pmx, premaxilla; ps, presphenoid; ptg, pterygoid; sq, squamosal; tg, tongue; UI, upper incisor; vbf, vibrissae follicle; vm, vomer; *, ectopic structure.
Maxillomandibular Identity Correlates with Regional Edn1-Ednra Signaling Activity.
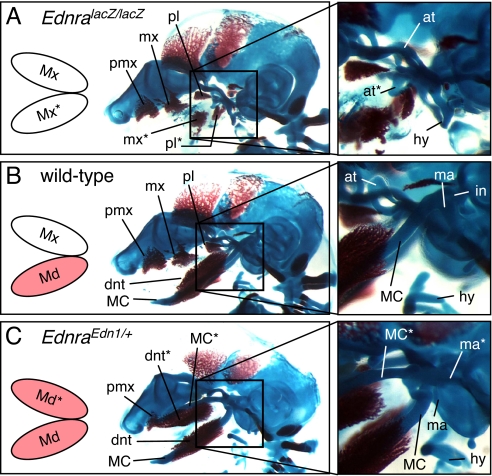
To analyze the effect of Edn1/Ednra signaling on the dorsoventral specification of PA1, we then compared chondrocrania of wild-type, EdnralacZ/lacZ and EdnraEdn1/+ embryos at E15.5. As previously described, EdnralacZ/lacZ embryos showed loss of MC and duplication of the ala temporalis (Fig. 2 A and B; Fig. S5 A and B) (21), indicating a ventral-to-dorsal transformation of PA1. By contrast, EdnraEdn1/+ embryos showed a duplicated MC in place of the ala temporalis (Fig. 2C; Fig. S5 C–I), indicating that an opposite dorsal-to-ventral transformation had taken place. The duplicated MC extends beyond the ectopic dentary bone forming laterally to it, as in case with their orthotopic counterparts (Fig. S5H). The ectopic MC was discontinuous to the transformed incus-malleus in some mutants (Fig. S5H), but was continuous to it in others (Fig. S5I). Thus, Edn1/Ednra signaling is necessary to specify mandibular identity within PA1 and CNCCs colonizing both the maxillary and the mandibular arch are competent to respond to Edn1 signaling.
Fig. 2.
Comparison of craniofacial skeletons among E15.5 Ednra-null (EdnralacZ/lacZ) (A), wild-type (B) and Edn1-misexpressing (EdnraEdn1/+) (C) mice. Activated Edn1/Ednra signaling (colored in pink) correlates to the formation of MC and associated mandibular structures. at, ala temporalis; dnt, dentary; hy, hyoid; in, incus; ma, malleus; mx, maxilla; pl, palatine; pmx, premaxilla; *, ectopic structure.
Edn1-Induced Skeletal Transformations Are Accompanied by Neuromuscular Remodeling.
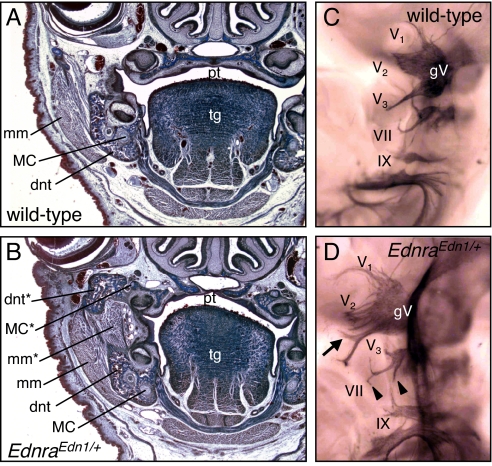
Histological analysis of heads of EdnraEdn1/+ embryos at E18.5 (Fig. 3 A and B) not only confirmed the duplication of the dentary bone and MC, but also revealed the presence of an ectopic profound muscle mass juxtaposed to the normal masseter muscle. This supernumerary muscle, which connected the 2 dentary bones, could be interpreted as a duplicated masseter muscle. To reinforce this notion, neurofilament immunostaining of cranial nerves of wild-type and EdnraEdn1/+ embryos at E10.5 (Fig. 3 C and D) showed that the motor root of the trigeminal nerve, which normally enters the mandibular arch and innervated the masseter, divided in 2 symmetric branches which innervated both the mandibular and the transformed maxillary arch of mutant embryos.
Fig. 3.
Histological and whole-mount neuromuscular analysis of EdnraEdn1/+ mice. (A and B) Frontal sections of E18.5 wild-type (A) and EdnraEdn1/+ (B) mice stained by Mallory trichromic. Constitutive activation of Edn1/Ednra signaling results not only in the transformation of maxillary skeletal elements, but also in the appearance of new muscular components associated to the ectopic dentary, which can be interpreted as a duplicated masseter muscle. (C and D) Cranial nerves of E10.5 wild-type (C) and EdnraEdn1/+ (D) embryos stained by antineurofilament antibody. Ectopic branches of the mandibular (arrow) and facial (arrowheads) nerves are indicated. dnt, dentary; mm, masseter muscle; pl, palatine; tg, tongue; gV, trigeminal ganglion; V1, ophthalmic nerve; V2, maxillary nerve; V1, mandibular nerve; VII, facial nerve; IX, glossopharyngeal nerve; *, ectopic structure.
Mandibular Arch-Specific Genetic Program Is Activated in the Maxillary Arch by Edn1 Signaling.
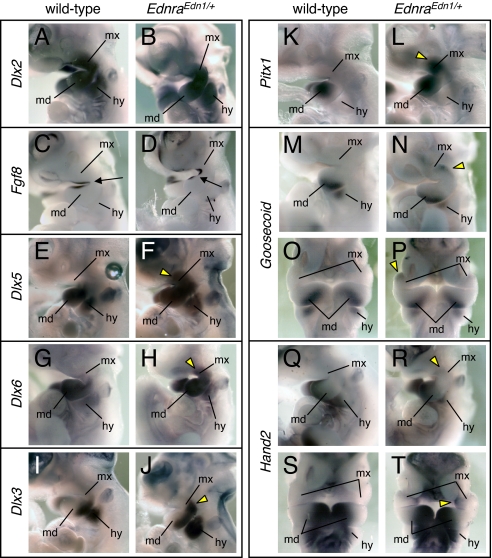
The dorsal-to-ventral transformation of PA1 induced by ectopic Edn1 signaling in the maxillary region suggested that a shift in the genetic program specifying jaw identity might have taken place. To test this hypothesis, we examined the expression of genes involved in PA specification. Dlx2 was expressed in the mesenchyme of PA1 and PA2 in both E10.5 wild-type and EdnraEdn1/+ embryos (Fig. 4 A and B). Fgf8 was similarly expressed in wild-type and EdnraEdn1/+ embryos in the ectoderm around the maxillomandibular junction (Fig. 4 C and D). EdnraEdn1/+ embryos exhibited an extension of the territory of Dlx5/Dlx6 expression to the maxillary (dorsal) region of PA1 (Fig. 4 E–H). Consistently, E10.0 EdnraEdn1/+ embryos carrying the mI56i-lacZ transgene, in which the mI56i enhancer from the intergenic region of the Dlx5/6 cluster drives lacZ expression (23), revealed transgene expression extending to the dorsal region of PA1 and PA2 (Fig. S6), supporting the notion that the mI56i enhancer may mediate the Dlx5/Dlx6 induction by the Edn1/Ednra signaling even in dorsal ectopic locations. The expression of Dlx3 (Fig. 4 I and J), Pitx1 (Fig. 4 K and L), and Goosecoid (Fig. 4 M–P), mandibular markers down-regulated in Dlx5/Dlx6-null and in Edn1/Ednra-null embryos (10, 12, 15, 16), was activated in the maxillary region of EdnraEdn1/+ mutants. Hand2, a downstream target of Dlx6 (24), was also up-regulated in the mutant maxillary arch, although the expression level appeared lower than in the mandibular arch (Fig. 4 Q–T). Ectopic Hand2 expression in the maxillary arch was observed along the nasolacrimal groove with the highest intensity in the medial region abutting the frontonasal process (Fig. S7).
Fig. 4.
Gene expression analysis of the pharyngeal arches in EdnraEdn1/+ embryos. (A–T) Whole-mount in situ hybridization for Dlx2 (A and B), Fgf8 (C and D), Dlx5 (E and F), Dlx6 (G and H), Dlx3 (I and J), Pitx1 (K and L), Goosecoid (M–P), and Hand2 (Q–T) in E10.5 wild-type (A, C, E, G, I, K, M, O, Q, and S) and EdnraEdn1/+ (B, D, F, H, J, L, N, P, R, and T) embryos. Arrows indicate the junction of maxillary and mandibular arches. Yellow arrowheads indicate ectopic gene expression in the maxillary arch. hy, hyoid arch; md, mandibular arch; mx, maxillary arch.
These gene expression switches between the maxillary and the mandibular arch are opposite to those observed in Edn1/Ednra-null embryos. Collectively, they indicate that CNCCs colonizing PA1 can respond to Edn1 signaling by activating the genetic program specifying mandibular identity both in dorsal (maxillary) and ventral (mandibular) PA1.
Forced Expression of Hand2 Partially Mimicked Edn1-Induced Skeletal Transformation.
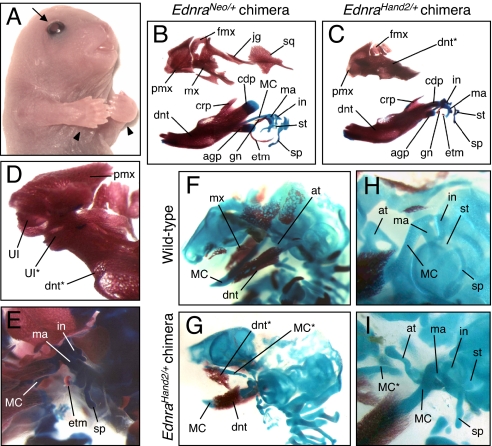
To explore the role of the molecular pathway of jaw specification downstream to the Edn1/Ednra-Dlx5/6 signaling, we used RMCE to replace Ednra with Hand2 (Fig. S2) and examined the phenotype of PA1-derived structures. Because no chimeric EdnraHand2/+ mice survived after birth, we directly analyzed the phenotype of chimeras. Among 93 chimeras, 53 (57%) exhibited mirror-image polydactyly, as previously observed by forced Hand2 expression in the limb bud (25, 26). Chimeras with polydactyly also had craniofacial abnormalities (Fig. 5A; Fig. S8 and Fig. S9). At birth, EdnraHand2/+ chimeras resembled EdnraEdn1/+ mice in lacking vibrissae follicles and having open eyelids (Fig. 5A). Maxillary structures were often transformed into dentary-like bones fused to the premaxilla and carrying a small incisor at the distal end (Fig. 5 B–D; Fig. S8). A MC-like rod-shaped cartilage was present in the maxillary region of 66% (29/44) E15.5 (Fig. 5 F–I) and E16.5 (Fig. S9) EdnraHand2/+ chimeras. However, in EdnraHand2/+ chimeras, the ectopic MC-like cartilage often coexisted with a smaller and deformed ala temporalis and proximal structures of the mandibular and maxillary arches (gonial, ectotympanic, and squamosal) were often malformed or absent rather than duplicated (Fig. 5E; Fig. S9). The premaxilla, the frontal process of maxilla, and the lacrimal bone were maintained in EdnraHand2/+ chimeras (Fig. 5C; Fig. S8). The skeletal abnormalities present in EdnraEdn1/+ and in EdnraHand2/+ mutants were never observed in control EdnraNeo/+ chimeric mice (n = 8) (Fig. 5B; Fig. S8).
Fig. 5.
Morphological transformation in EdnraHand2/+ ES cell-derived chimeric mice. (A) Representative surface appearance of E18.5 EdnraHand2/+ chimeric mice. Mutant mice demonstrate loss of vibrissae follicles, open eyelids (arrows), and polydactyly (arrowheads). (B–E) Craniofacial bone and cartilage elements of E18.5 EdnraNeo/+ ES cell-derived (B) and EdnraHand2/+ ES cell-derived (C–E) chimeric mice. Whereas EdnraNeo/+-chimeric mice show normal skeletal morphology (B), EdnraHand2/+-chimeric mice demonstrate transformation of the maxilla into dentary-like bone (C), which fuses to the premaxilla and contains an ectopic incisor at the tip (D). The condylar and angular processes of the dentary, gonial, ectotympanic, and squamosal bones were often malformed or absent (E). (F–I) Craniofacial skeletons of E15.5 wild-type (F and H) and EdnraHand2/+-chimeric (G and I) mice. Ectopic formation of MC-like, rod-shaped cartilage is present often with small and deformed ala temporalis in EdnraHand2/+-chimeric mice. The malleus, incus, stapes, and styloid process are deformed. agp, angular process; at, ala temporalis; cdp, condylar process; crp, coronoid process; dnt, dentary; etm, ectotympanic; fmx, frontal process of maxilla; gn, gonial; hy, hyoid; in, incus; jg, jugal; ma, malleus; mx, maxilla; pmx, premaxilla; sp, styloid process; sq, squamosal; st, stapes; UI, upper incisor; *, ectopic structure.
Discussion
In the gnathostome craniofacial development, environmental signals have central roles in the fate determination of CNCCs and regional patterning. CNCCs constituting PAs and adjacent facial prominences are highly plastic at early stages and their identities can be separately specified by signaling cues (27–29). Here, we show that a single Edn1 signal can convert the maxillary arch into a mandibular fate, indicating a dorsal-to-ventral transformation within PA1. Previously, we and other groups have reported a ventral-to-dorsal transformation by Edn1/Ednra-null mutations in mice (12, 13) and zebrafish (30, 31). These findings indicate that an Edn1 switch is necessary and sufficient for the determination of mandibular identity.
The significance of endoderm- and ectoderm-derived epithelia as sources of instructive signals for facial skeletal patterning has also been demonstrated (7, 32, 33). Edn1 is produced in the ventral aspect of the pharyngeal endoderm (20), a region which can induce the formation of MC and associated membranous bones (7), suggesting that Edn1 could be the endodermal signal that specifies ventral PA identity. Notably, distal-to-proximal polarity within PA1 was maintained in EdnraEdn1/+ mutants. Determination of intra-PA polarity is essential for modular development of upper and lower jaws (9, 34). The nested expression of Dlx genes seems to have a central role in determining the dorsoventral polarity of PA1, whereas the proximodistal polarity of the maxillary and mandibular regions appears to depend on the integration of signals from the proximal “hinge” and distal “caps” territories of PA1 (9). Thus, dorsoventral identity and intraregional polarity may be separately regulated within PA1.
The fact that constitutive activation of the Edn1/Ednra signaling pathway confers mandibular identity to maxillary CNCCs indicates that CNCCs within PA1 are competent to respond to Edn1. By contrast, the effects of Ednra constitutive activation, such as Dlx5/Dlx6 induction, were mostly restricted to PA1, although Edn1expression took place throughout the head mesenchyme and in other regions of the embryo including the limb bud (data not shown). This fact suggests that the competence to respond to Edn1/Ednra signaling may be limited to a subpopulation of Hox-negative CNCCs, although the possibility that Edn1 peptide ligands may not be efficiently produced outside PA1 cannot be completely excluded. The limited, but not negligible, response of PA2 CNCCs to the constitutive activation of Endra suggests that the absence of Hox expression is not necessary for Edn1 competence. It is worth remembering that targeted disruption of Hoxa2 results in the transformation of PA2 into a mandibular PA1 (35), suggesting that the expression of Hox genes might have an active role in determining the morphogenetic consequence of Edn1 signaling to CNCCs.
The transformation of the maxillary skeleton was accompanied by changes in the associated musculature and corresponding innervation. Previous reports have suggested a tight communication between CNCCs and cranial mesodermal cells involved in patterning the craniofacial neuromuscular system (3, 5, 36). Thus, in EdnraEdn1/+ mutants, changes in CNCCs may cause the muscle duplication and concurrent changes in regional axon guidance signals. Indeed, the changes in innervation are likely to be due to changes in such guidance signals in the transformed region rather than changes intrinsic to motor neurons, because Ednra is not expressed in neurons in the E9.5–10.5 neural tube (21); therefore, Edn1 does not directly act on them. These neuromuscular changes may affect muscle attachment to the bone and subsequently the formation of coronoid processes. Analysis of differential gene expression in EdnraEdn1/+ mutants and RMCE-induced knock-in experiments used in this study may give a clue to the mechanism underlying the region-specific muscle patterning and axon guidance.
Inactivation of Edn1-dependent Hand2 expression in the ventrolateral PA regions results in malformations of MC and associated bones (37). Our results confirm the importance of the Edn1/Ednra→Dlx→Hand2 regulatory cascade for mandibular specification as forced expression of Hand2 in the Ednra domain induces the partial transformation of upper jaw structures into lower jaw elements. Apparently, incomplete penetrance of the phenotype is probably due to different contribution of ES cells to each chimeric mouse. The coexistence of the MC-like cartilage and the ala temporalis in Hand2-knock-in chimeras may reflect such variation, and suggest that Hand2-positive and negative cells might behave differently within the same maxillary region. It is also noteworthy that the gonial and ectotympanic bones were malformed in EdnraHand2/+ chimeras. This fact indicates that Hand2 may antagonize the genetic program for the formation of the proximal mandibular structures.
Together with the grafting experiments of Couly et al. (7), in which small aliquots of premigratory crest could rescue the deletion of all CNCCs, our data would suggest a multistep specification of CNCCs. Before migration, CNCCs constitute an equivalence group. The first step in their specification would be the definition of large craniofacial territories, such as the Edn1-responsive PA1 and the Edn1-unresponsive frontonasal prominence. Successive signaling from restricted epithelial regions would then specify specific subregions such as the maxillary and mandibular territories of PA1.
Materials and Methods
Mice.
Mice carrying the EdnralacZ allele (21) and mI56i-lacZ transgenic mice (23) have been previously described. To obtain mice carrying the EdnraEdn1 (Edn1-knock-in) or EdnraHand2 (Hand2-knock-in) allele, we performed RMCE on the Ednraneo/+ ES cells, in which an exchangeable floxed site was introduced into the Ednra locus as previously described (21). Briefly, PCR-amplified fragments encoding the ORF of mouse Edn1 and Hand2 cDNA were introduced into the knock-in vector p66–2272 containing multiple cloning sites between lox66 and lox2272 (38). The resultant plasmids were transfected into Ednraneo/+ ES cells with Cre-expressing adenovirus (39). Targeted ES clones were injected into ICR blastocysts to generate chimeras. To obtain mice carrying the EdnraEdn1 allele, 2 independent male germ-line chimeras were crossed with ICR females. EdnraEdn1/lacZ mice, derived by crossing the germ-line chimeras and EdnralacZ/+ females heterozygous for Ednra-null mutation, exhibited the typical Ednra-null phenotype, confirming that Ednra was responsible for the effect of ectopic Edn1 signaling. All of the chimeras from EdnraHand2 ES cells were died before or at the time of birth; therefore, chimeric embryos from EdnraHand2 ES cells and wild-type ICR blastocysts were prepared at different stages for phenotype analysis. Genotypes were determined by PCR by using primers specific for RMCE-mediated recombination. All of the animal experiments were performed in accordance with the guidelines of the University of Tokyo Animal Care and Use Committee.
Skeletal Staining.
Alizarin red/alcian blue staining was performed, as previously described (40).
Histological Analysis.
Embryos were fixed in Bouin's solution and embedded in paraffin. Sections were then subjected to Mallory trichromic staining.
Whole-Mount Immunostaining.
Whole-mount neurofilament staining was performed and visualized with 3–3′-diaminobenzidine tetrahydrochloride/NiCl2, as previously described (41).
In Situ Hybridization.
Whole-mount in situ hybridization was performed as described previously (42). Probes for Hand2 (43) and Goosecoid (44) were generously provided by D. Srivastava (University of California, San Francisco, CA) and G. Yamada (Kumamoto University, Kumamoto, Japan), respectively. Other probes were prepared by RT-PCR.
β-Galactosidase Staining.
LacZ expression was detected by staining with X-gal for β-galactosidase activity. Whole-mount staining was performed as previously described (41) with minor modifications.
Supplementary Material
Acknowledgments.
We thank K. Yamamura (Kumamoto University, Kumamoto, Japan) and K. Araki (Kumamoto University, Kumamoto, Japan) for lox-containing plasmids; I. Saito (University of Tokyo, Tokyo) for Cre-expressing adenovirus; D. Srivastava (University of California, San Francisco) and G. Yamada (Kumamoto University, Kumamoto, Japan) for probes; N. Hoya, Y. Fujisawa, S. Kushiyama, and T. Tsuchida for technical assistance; and F. Ikemoto, M. Nishikibe, and colleagues in Banyu Pharmaceutical Co., Ltd. for supporting this work. This work was supported in part by Global Centers of Excellence (COE) Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms), Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan; and grants-in-aid for scientific research from MEXT and the Ministry of Health, Labour, and Welfare, Japan. G.L. is partially supported by the European Union Consortium CRESCENDO (LSHM-CT-2005-018652) and the National Agency for Research project “GENDACTYL.”
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807345105/DCSupplemental.
References
- 1.Couly GF, Coltey PM, Le Douarin NM. The triple origin of skull in higher vertebrates: A study in quail-chick chimeras. Development. 1993;117:409–429. doi: 10.1242/dev.117.2.409. [DOI] [PubMed] [Google Scholar]
- 2.Creuzet S, Couly G, Vincent C, Le Douarin NM. Negative effect of Hox gene expression on the development of the neural crest-derived facial skeleton. Development. 2002;129:4301–4313. doi: 10.1242/dev.129.18.4301. [DOI] [PubMed] [Google Scholar]
- 3.Köntges G, Lumsden A. Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Development. 1996;122:3229–3242. doi: 10.1242/dev.122.10.3229. [DOI] [PubMed] [Google Scholar]
- 4.Noden DM, Trainor PA. Relations and interactions between cranial mesoderm and neural crest populations. J Anat. 2005;207:575–601. doi: 10.1111/j.1469-7580.2005.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trainor PA, Tam PP. Cranial paraxial mesoderm and neural crest cells of the mouse embryo: Co-distribution in the craniofacial mesenchyme but distinct segregation in branchial arches. Development. 1995;121:2569–2582. doi: 10.1242/dev.121.8.2569. [DOI] [PubMed] [Google Scholar]
- 6.Le Douarin NM, Kalcheim C. The Neural Crest. 2nd Ed. Cambridge, UK: Cambridge Univ Press; 1999. [Google Scholar]
- 7.Couly G, Creuzet S, Bennaceur S, Vincent C, Le Douarin NM. Interactions between Hox-negative cephalic neural crest cells and the foregut endoderm in patterning the facial skeleton in the vertebrate head. Development. 2002;129:1061–1073. doi: 10.1242/dev.129.4.1061. [DOI] [PubMed] [Google Scholar]
- 8.Ruhin B, et al. Patterning of the hyoid cartilage depends upon signals arising from the ventral foregut endoderm. Dev Dyn. 2003;228:239–246. doi: 10.1002/dvdy.10380. [DOI] [PubMed] [Google Scholar]
- 9.Depew MJ, Simpson CA. 21st century neontology and the comparative development of the vertebrate skull. Dev Dyn. 2006;235:1256–1291. doi: 10.1002/dvdy.20796. [DOI] [PubMed] [Google Scholar]
- 10.Clouthier DE, et al. Cranial and cardiac neural crest defects in endothelin-A receptor-deficient mice. Development. 1998;125:813–824. doi: 10.1242/dev.125.5.813. [DOI] [PubMed] [Google Scholar]
- 11.Kurihara Y, et al. Elevated blood pressure and craniofacial abnormalities in mice deficient in endothelin-1. Nature. 1994;368:703–710. doi: 10.1038/368703a0. [DOI] [PubMed] [Google Scholar]
- 12.Ozeki H, Kurihara Y, Tonami K, Watatani S, Kurihara H. Endothelin-1 regulates the dorsoventral branchial arch patterning in mice. Mech Dev. 2004;121:387–395. doi: 10.1016/j.mod.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Ruest LB, Xiang X, Lim KC, Levi G, Clouthier DE. Endothelin-A receptor-dependent and -independent signaling pathways in establishing mandibular identity. Development. 2004;131:4413–4423. doi: 10.1242/dev.01291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukuhara S, Kurihara Y, Arima Y, Yamada N, Kurihara H. Temporal requirement of signaling cascade involving endothelin-1/endothelin receptor type A in branchial arch development. Mech Dev. 2004;121:1223–1233. doi: 10.1016/j.mod.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 15.Beverdam A, et al. Jaw transformation with gain of symmetry after Dlx5/Dlx6 inactivation: Mirror of the past? Genesis. 2002;34:221–227. doi: 10.1002/gene.10156. [DOI] [PubMed] [Google Scholar]
- 16.Depew MJ, Lufkin T, Rubenstein JL. Specification of jaw subdivisions by Dlx genes. Science. 2002;298:381–385. doi: 10.1126/science.1075703. [DOI] [PubMed] [Google Scholar]
- 17.Depew MJ, Simpson CA, Morasso M, Rubenstein JL. Reassessing the Dlx code: The genetic regulation of branchial arch skeletal pattern and development. J Anat. 2005;207:501–561. doi: 10.1111/j.1469-7580.2005.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merlo GR, et al. Multiple functions of Dlx genes. Int J Dev Biol. 2000;44:619–626. [PubMed] [Google Scholar]
- 19.Kurihara Y, et al. Aortic arch malformations and ventricular septal defect in mice deficient in endothelin-1. J Clin Invest. 1995;96:293–300. doi: 10.1172/JCI118033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maemura K, et al. Sequence analysis, chromosomal location, and developmental expression of the mouse preproendothelin-1 gene. Genomics. 1996;31:177–184. doi: 10.1006/geno.1996.0029. [DOI] [PubMed] [Google Scholar]
- 21.Sato T, et al. Recombinase-mediated cassette exchange reveals the selective use of Gq/G11-dependent and -independent endothelin 1/endothelin type A receptor signaling in pharyngeal arch development. Development. 2008;135:755–765. doi: 10.1242/dev.012708. [DOI] [PubMed] [Google Scholar]
- 22.Qiu M, et al. Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: Mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev Biol. 1997;185:165–184. doi: 10.1006/dbio.1997.8556. [DOI] [PubMed] [Google Scholar]
- 23.Zerucha T, et al. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J Neurosci. 2000;20:709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charite J, et al. Role of Dlx6 in regulation of an endothelin-1-dependent, dHAND branchial arch enhancer. Genes Dev. 2001;15:3039–3049. doi: 10.1101/gad.931701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charite J, McFadden DG, Olson EN. The bHLH transcription factor dHAND controls Sonic hedgehog expression and establishment of the zone of polarizing activity during limb development. Development. 2000;127:2461–2470. doi: 10.1242/dev.127.11.2461. [DOI] [PubMed] [Google Scholar]
- 26.Fernandez-Teran M, et al. Role of dHAND in the anterior-posterior polarization of the limb bud: Implications for the Sonic hedgehog pathway. Development. 2000;127:2133–2142. doi: 10.1242/dev.127.10.2133. [DOI] [PubMed] [Google Scholar]
- 27.Hu D, Helms JA. The role of sonic hedgehog in normal and abnormal craniofacial morphogenesis. Development. 1999;126:4873–4884. doi: 10.1242/dev.126.21.4873. [DOI] [PubMed] [Google Scholar]
- 28.Lee SH, Fu KK, Hui JN, Richman JM. Noggin and retinoic acid transform the identity of avian facial prominences. Nature. 2001;414:909–912. doi: 10.1038/414909a. [DOI] [PubMed] [Google Scholar]
- 29.Vieux-Rochas M, et al. Molecular dynamics of retinoic acid-induced craniofacial malformations: Implications for the origin of gnathostome jaws. PLoS ONE. 2007;2:e510. doi: 10.1371/journal.pone.0000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller CT, Schilling TF, Lee K, Parker J, Kimmel CB. sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development. 2000;127:3815–3828. doi: 10.1242/dev.127.17.3815. [DOI] [PubMed] [Google Scholar]
- 31.Kimmel CB, Ullmann B, Walker M, Miller CT, Crump JG. Endothelin 1-mediated regulation of pharyngeal bone development in zebrafish. Development. 2003;130:1339–1351. doi: 10.1242/dev.00338. [DOI] [PubMed] [Google Scholar]
- 32.Hu D, Marcucio RS, Helms JA. A zone of frontonasal ectoderm regulates patterning and growth in the face. Development. 2003;130:1749–1758. doi: 10.1242/dev.00397. [DOI] [PubMed] [Google Scholar]
- 33.Benouaiche L, Gitton Y, Vincent C, Couly G, Levi G. Sonic hedgehog signalling from foregut endoderm patterns the avian nasal capsule. Development. 2008;135:2221–2225. doi: 10.1242/dev.020123. [DOI] [PubMed] [Google Scholar]
- 34.Kuratani S. Evolution of the vertebrate jaw: Comparative embryology and molecular developmental biology reveal the factors behind evolutionary novelty. J Anat. 2004;205:335–347. doi: 10.1111/j.0021-8782.2004.00345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santagati F, Minoux M, Ren SY, Rijli FM. Temporal requirement of Hoxa2 in cranial neural crest skeletal morphogenesis. Development. 2005;132:4927–4936. doi: 10.1242/dev.02078. [DOI] [PubMed] [Google Scholar]
- 36.Rinon A, et al. Cranial neural crest cells regulate head muscle patterning and differentiation during vertebrate embryogenesis. Development. 2007;134:3065–3075. doi: 10.1242/dev.002501. [DOI] [PubMed] [Google Scholar]
- 37.Yanagisawa H, Clouthier DE, Richardson JA, Charite J, Olson EN. Targeted deletion of a branchial arch-specific enhancer reveals a role of dHAND in craniofacial development. Development. 2003;130:1069–1078. doi: 10.1242/dev.00337. [DOI] [PubMed] [Google Scholar]
- 38.Araki K, Araki M, Yamamura K. Site-directed integration of the cre gene mediated by Cre recombinase using a combination of mutant lox sites. Nucleic Acids Res. 2002;30:e103. doi: 10.1093/nar/gnf102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanegae Y, et al. Efficient gene activation in mammalian cells by using recombinant adenovirus expressing site-specific Cre recombinase. Nucleic Acids Res. 1995;23:3816–3821. doi: 10.1093/nar/23.19.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McLeod MJ. Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology. 1980;22:299–301. doi: 10.1002/tera.1420220306. [DOI] [PubMed] [Google Scholar]
- 41.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo: A Laboratory Manual. 3rd Ed. Plainview, NY: Cold Spring Harbor Lab Press; 2003. [Google Scholar]
- 42.Wilkinson D. In Situ Hybridization: A Practical Approach. Oxford, UK: IRL Press; 1992. [Google Scholar]
- 43.Srivastava D, Cserjesi P, Olson EN. A subclass of bHLH proteins required for cardiac morphogenesis. Science. 1995;270:1995–1999. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- 44.Yamada G, et al. Targeted mutation of the murine goosecoid gene results in craniofacial defects and neonatal death. Development. 1995;121:2917–2922. doi: 10.1242/dev.121.9.2917. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.