Abstract
The Saccharomyces cerevisiae Mec1–Ddc2 checkpoint kinase complex (the ortholog to human ATR-ATRIP) is an essential regulator of genomic integrity. The S. cerevisiae BRCT repeat protein Dpb11 functions in the initiation of both DNA replication and cell cycle checkpoints. Here, we report a genetic and physical interaction between Dpb11 and Mec1–Ddc2. A C-terminal domain of Dpb11 is sufficient to associate with Mec1–Ddc2 and strongly stimulates the kinase activity of Mec1 in a Ddc2-dependent manner. Furthermore, Mec1 phosphorylates Dpb11 and thereby amplifies the stimulating effect of Dpb11 on Mec1–Ddc2 kinase activity. Thus, Dpb11 is a functional ortholog of human TopBP1, and the Mec1/ATR activation mechanism is conserved from yeast to humans.
Keywords: ATR, checkpoint, DNA damage, TopBP1
Eukaryotic cells have elaborate mechanisms to ensure the faithful maintenance and replication of the genome. Genotoxic stress activates a signal transduction pathway called the DNA damage response (DDR) that coordinates cell cycle transitions, DNA replication, transcription, apoptosis, and DNA repair (1). In Saccharomyces cerevisiae, the PIKK (Phosphatidylinositide 3-kinase-related kinase) kinase Mec1, the mammalian ATR ortholog, senses DNA damage and replication stress and initiates the DNA damage response (2). Mec1 phosphorylates substrates involved in DNA replication and repair, cell cycle checkpoints, RNA metabolism, and transcription (3).
Activation of the DNA damage response requires the colocalization of 2 checkpoint complexes to damaged chromatin. Mec1 is localized to DNA damage through its associated partner, Ddc2, which binds to RPA-coated ssDNA (4–7). Independently of Mec1–Ddc2, the 9-1-1 checkpoint clamp complex composed of Ddc1–Mec3–Rad17 (Rad9–Hus1–Rad1 in mammalian cells) also localizes to sites of DNA damage (8–10). Forced colocalization of the Mec1–Ddc2 complex and the 9-1-1 complex to chromatin can trigger the DNA damage response in the absence of a DNA lesion (11). Furthermore, colocalization of only the Ddc1 subunit of the 9-1-1 complex with Mec1–Ddc2 is sufficient to activate to the DNA damage response (11). This suggests that Ddc1 and/or a Ddc1-interacting protein may function as a direct activator of Mec1. In fact, Ddc1 purified from yeast has been shown to modestly stimulate the kinase activity of Mec1 in vitro under low-salt conditions (12).
Ddc1 interacts with another checkpoint protein Dpb11 (13). Dpb11 and its sequence homologs are essential for the initiation of DNA replication in eukaryotic organisms (14, 15). Specifically, Dpb11 acts as a molecular bridge between the Sld3–Cdc45–MCM helicase complex and the Sld2–DNA polymerase ε complex (16, 17). Dpb11 also has a cell cycle checkpoint function and dpb11 mutants exhibit sensitivity to DNA damaging agents and replication stress (14, 18). In metazoans, the Dpb11 homolog TopBP1 is a general activator of ATR. A region between the sixth and seventh BRCT (BRCA1 C-terminal) domains of TopBP1 called the ATR activation domain (AAD) is sufficient to activate ATR–ATRIP in vitro and in cells (19). However, it is not clear whether Mec1 is regulated in the same manner as ATR because Dpb11 lacks sequence homology to the AAD of TopBP1 and has not been shown to interact with the Mec1–Ddc2 complex.
We recently reported that a ddc2 mutation, termed ddc2-top, causes sensitivity to DNA damage and replication stress and defects in Mec1 checkpoint signaling (20). Here, we took advantage of the ddc2-top mutant to search for a Mec1 activator protein. We found that Ddc2 interacts genetically and physically with Dpb11. Moreover, we discovered a domain of Dpb11 that is sufficient to strongly stimulate the kinase activity of Mec1. Mec1 phosphorylates Dpb11 and this phosphorylation further enhances the ability of Dpb11 to serve as a Mec1 activator. These data demonstrate that Dpb11 is a Mec1 activator linking the Mec1–Ddc2 and 9-1-1 checkpoint complexes.
Results
Dpb11 Suppresses the HU Sensitivity of ddc2-top.
We hypothesized that the ddc2-top mutation disrupted an interaction between Ddc2 and a Mec1–Ddc2 activator. If so, overexpression of that protein might be expected to suppress the phenotype of ddc2-top yeast. To search for this protein, we overexpressed the 2 likely candidates, Ddc1 and Dpb11, in ddc2-top yeast. Dpb11 but not Ddc1 partially suppressed the hydroxyurea (HU) sensitivity caused by the ddc2-top mutation. This effect was greater when Dpb11 was expressed on a high-copy (2μ) plasmid compared with a low-copy (cen) plasmid (Fig. 1A). Overexpression of Dpb11 did not restore the viability of Δddc2 yeast in hydroxyurea, demonstrating that the observed suppression was Ddc2-dependent (Fig. 1B). dpb11–1 yeast are sensitive to hydroxyurea and DNA damaging agents and are defective in S-phase checkpoint signaling (14, 18), similar to the ddc2-top phenotype (20). Overexpression of the Dpb11–1 mutant did not suppress the HU sensitivity of ddc2-top yeast indicating allele-specific suppression that is consistent with a direct protein–protein interaction (Fig. 1C).
Fig. 1.
Overexpression of Dpb11 but not Ddc1 suppresses the HU sensitivity of ddc2-top. (A) YDM003, a Δddc2 Δsml1 yeast strain carrying pDM158 (a URA3 CEN plasmid expressing ddc2-top under its endogenous promoter) was transformed with galactose-inducible Dpb11 or Ddc1 high-copy (2μ) or low-copy (cen) expression vectors or an empty vector control. Cells were grown to mid-log phase and serial dilutions were spotted onto galactose plates with the indicated concentration of hydroxyurea (HU) and incubated at 30 °C. (B and C) The HU sensitivity of Δddc2 Δsml1 or ddc2-top yeast strains containing galactose-inducible Dpb11, Dpb11–1, or an empty TRP1 plasmid as indicated were compared on galactose plates as in A.
Dpb11 Associates with Mec1–Ddc2.
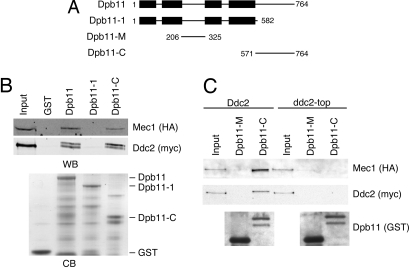
Dpb11 contains four BRCT repeats, which function in tandem as phosphoprotein-interacting domains. The N-terminal pair and C-terminal pair of BRCT domains bind to CDK-phosphorylated residues of Sld3 and Sld2, respectively (16, 17). dpb11–1 encodes a nonsense mutation at residue 583, resulting in a truncation C-terminal to the BRCT domains (21) (Fig. 2A). Given that dpb11–1 is unable to suppress to the HU sensitivity of ddc2-top yeast, we suspected that the C-terminal region of Dpb11 might be responsible for an interaction with Ddc2. To determine whether there is a physical interaction between Dpb11 and Ddc2, we incubated yeast protein lysates with recombinant Dpb11 fragments encoding full-length Dpb11, the Dpb11–1 mutant, or the Dpb11 C-terminal domain, Dpb11-C (amino acid 571–764). Both Dpb11 and Dpb11-C bind to Mec1–Ddc2 but neither GST alone nor the Dpb11–1 protein bind indicating that the C terminus of Dpb11 is both necessary and sufficient to interact with Mec1–Ddc2 (Fig. 2B).
Fig. 2.
The C terminus of Dpb11 interacts with Mec1–Ddc2. (A) Schematic of wild-type Dpb11, the Dpb11–1, Dpb11-M, and Dpb11-C proteins used in this study. Boxes indicate BRCT domains. (B) Dpb11 and Dpb11-C interact with Mec1–Ddc2. Recombinant GST, Dpb11, Dpb11–1, or Dpb11-C proteins bound to glutathione beads were incubated with yeast protein extracts. After extensive washing, the proteins bound to the beads were eluted, separated by SDS/PAGE and immunoblotted with antibodies to detect Mec1 and Ddc2 (WB). The GST-tagged recombinant proteins used in this experiment were separated by SDS/PAGE and visualized with Coomassie blue staining (CB). (C) Yeast protein extracts from cells expressing Ddc2 or Ddc2-top were incubated with recombinant GST-tagged Dpb11 fragments bound to glutathione beads. Proteins bound to the beads were eluted, separated by SDS/PAGE, and immunoblotted with antibodies to detect Mec1, Ddc2, and the GST-tagged Dpb11 fragments. Input represents 10% of the lysate loaded in the binding reactions.
We then examined whether the Dpb11 interaction with Mec1–Ddc2 is disrupted by the ddc2-top mutation as suggested by the genetic suppression results. Yeast protein lysates from Ddc2 or ddc2-top cells were incubated with Dpb11-C or the other non-BRCT region of Dpb11, Dpb11-M (amino acids 206–325) as a control. Unlike wild-type Ddc2, Ddc2-top does not associate with Dpb11-C (Fig. 2C). Furthermore, in the presence of Ddc2-top, Mec1 is no longer able to associate with Dpb11, indicating that the observed association of Mec1 with Dpb11 is Ddc2-dependent. We conclude that Ddc2 contains a binding site for Dpb11 that is necessary for the interaction of the Mec1–Ddc2 complex with the C terminus of Dpb11.
Dpb11 Stimulates the Kinase Activity of Mec1–Ddc2.
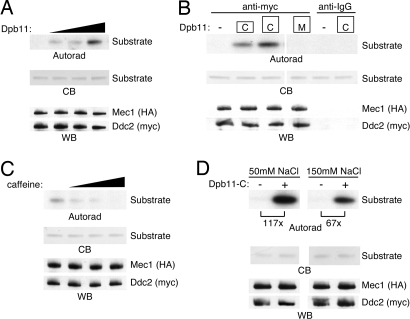
We next tested whether Dpb11 could function as a Mec1 activator. We immunopurified Mec1–Ddc2 from yeast lysates and incubated the complexes with recombinant Dpb11 and an established substrate, MCM2 (22, 23). Addition of Dpb11 strongly stimulated the kinase activity of Mec1 in a dose-dependent manner (Fig. 3A). Dpb11-C also stimulated Mec1 kinase activity, whereas Dpb11-M did not (Fig. 3B). Thus, the Dpb11 C-terminal domain is sufficient to activate Mec1. Dpb11-C did not produce an increase in substrate phosphorylation in control immunoprecipitation reactions lacking Mec1 (Fig. 3B). To confirm that the observed kinase activity was due to Mec1, we carried out the Mec1 kinase assay in the presence of Dpb11-C and increasing amounts of caffeine, an inhibitor of PIK kinases (24). Caffeine effectively inhibited the kinase activity of the activated Mec1 (Fig. 3C). These Mec1 kinase assays were performed in low-salt (50 mM NaCl) conditions. Under these conditions, Dpb11 stimulated the kinase activity of Mec1 >100-fold. We wanted to determine whether Dpb11 could stimulate Mec1 in the presence of a physiological salt concentration, so the Mec1 kinase assay was performed in the presence of 150 mM NaCl. In this case, the basal Mec1 kinase activity and the stimulated Mec1 kinase activity were both slightly lower, yet the degree of Dpb11-dependent activation was still very large (67-fold) (Fig. 3D). Thus, Dpb11 is a potent Mec1 activator.
Fig. 3.
Dpb11 activates Mec1–Ddc2. (A) HA-Mec1-myc-Ddc2 complexes were immunopurified from yeast lysates and were incubated with recombinant Dpb11, substrate, and γ-32P ATP. Kinase reactions were separated by SDS/PAGE, stained with Coomassie blue (CB) to visualize the amount of substrate, and exposed to film (autorad). A duplicate gel was immunoblotted with anti-HA and anti-myc antibodies to detect Mec1 and Ddc2, respectively (WB). (B) Mec1–Ddc2 complex kinase reactions were performed in the absence (−) or presence of recombinant Dpb11-M or Dpb11-C fragments. Anti-IgG antibody was used a control for the immunopurifications. (C) Mec1–Ddc2 complex kinase reactions containing Dpb11-C were performed in the presence of 0, 0.2, 1, and 5 mM caffeine. (D) Mec1–Ddc2 complex kinase reactions were performed with buffer containing 50 mM or 150 mM NaCl. Substrate phosphorylation was quantitated by using a PhosphorImager.
Dpb11-Dependent Mec1 Activation Requires a Ddc2-Dpb11 Interaction.
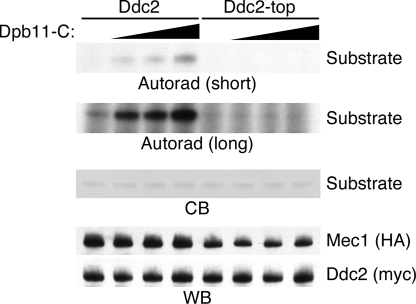
We previously reported that ddc2-top yeast have a defect in checkpoint signaling after replication stress and DNA damage (20). To determine whether this defect is due to an inability of Mec1–Ddc2-top complexes to be activated by Dpb11, Mec1–Ddc2 and Mec1–Ddc2-top complexes were isolated and incubated with increasing amounts of the Dpb11-C fragment. In the presence of wild-type Ddc2, we observed increased Mec1 kinase activity with increasing amounts of Dpb11-C. However, even high concentrations of Dpb11-C did not stimulate Mec1–Ddc2-top complexes (Fig. 4). Therefore, an interaction between Ddc2 and Dpb11 is required for Dpb11 to activate Mec1. After the high-stringency washes used for the kinase assays, we did note that the Ddc2-top mutant samples contained slightly less associated Mec1 suggesting the mutation may slightly alter the affinity of Mec1 for Ddc2 (Fig. 4).
Fig. 4.
Ddc2-top does not support Mec1 activation by Dpb11. Mec1–Ddc2 or Mec1–Ddc2-top complexes were purified and incubated with increasing amounts of Dpb11-C, substrate, and γ-32P ATP. Kinase reactions were separated by SDS/PAGE, stained with Coomassie blue (CB) to visualize the substrate, and exposed to film (autorad). Both long and short exposures of the autoradiogram are shown. A duplicate gel was blotted and probed with anti-HA and anti-myc antibodies to detect Mec1 and Ddc2, respectively (WB).
Dpb11 Phosphorylation Potentiates Mec1 Activation.
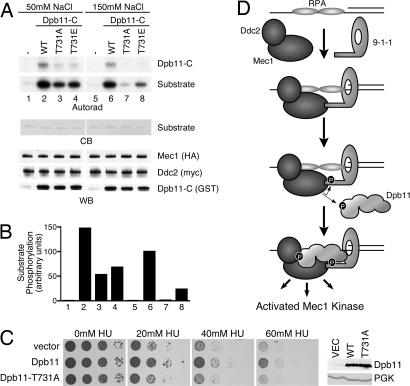
Mec1 targets its substrates at SQ/TQ motifs (3, 25). In the course of our kinase assays, we noticed that Dpb11-C itself served as a Mec1 substrate. This region of Dpb11 contains a single Mec1 consensus site, threonine 731. Mutation of T731 to alanine largely abolished the phosphorylation of Dpb11 by Mec1 (Fig. 5A).
Fig. 5.
Dpb11 T731 phosphorylation potentiates Mec1 activation. (A) Mec1–Ddc2 complexes were purified and incubated with wild-type or mutant forms of Dpb11-C, substrate, and γ-32P ATP. Kinase reactions were separated by SDS/PAGE, stained with Coomassie blue (CB) to visualize the substrate, and exposed to film (autorad). A duplicate gel was blotted and probed with anti-HA and anti-myc antibodies to detect Mec1 and Ddc2, respectively. An anti-GST antibody was used to detect Dpb11-C proteins (WB). (B) Quantification of substrate phosphorylation corresponding to the numbered lanes in A. (C) YDM003, a Δddc2 Δsml1 yeast strain carrying pDM158 (a URA3 CEN plasmid expressing ddc2-top under its endogenous promoter) was transformed with galactose-inducible Dpb11 or Dpb11 T731A (2μ) expression vectors or an empty vector control. Cells were grown to mid-log phase and serial dilutions were spotted onto galactose plates with the indicated concentration of hydroxyurea (HU) and incubated at 30 °C. The expression level of the wild-type and T731A Dpb11 proteins was confirmed to be equivalent by Western blotting. Anti-PGK antibodies were used as a loading control. (D) Simplified model of Mec1–Ddc2 activation described in Discussion. In this model, we propose that Dpb11 functions downstream of 9-1-1. Our data do not exclude the possibility that Dpb11 could function upstream of 9-1-1 in some circumstances. In this alternative scenario, an initial weak Dpb11-Ddc2 interaction could promote partial Mec1 activation, leading to Ddc1 phosphorylation. Phosphorylated Ddc1 could then stabilize the Dpb11 protein at the damaged site and allow further amplification of Mec1 signaling.
To assess the functional significance of this Dpb11 phosphorylation, we tested whether T731 phosphorylation regulates the ability of Dpb11 to activate Mec1. In low-salt (50 mM) conditions, Dpb11-C T731A stimulated Mec1 kinase activity ≈3-fold less efficiently that wild-type Dpb11-C (Fig. 5 A and B). However, at more physiologically relevant salt concentrations, Dpb11-C T731A is 100-fold less efficient at activating Mec1 compared with wild-type Dpb11-C (Fig. 5 A and B). These data suggest that Mec1-catayzed Dpb11 T731 phosphorylation increases the ability of Dpb11 to activate the Mec1–Ddc2 complex. We also created a Dpb11-C phosphorylation-mimetic mutant at this site. The Dpb11-C T731E mutant partially restored Mec1 activation compared with the T731A mutant at the physiological salt concentration (Fig. 5 A and B), supporting the idea that T731 phosphorylation promotes the ability of Dpb11 to activate Mec1.
Finally, to determine the effect of the T731 mutation on Mec1–Ddc2 function in vivo we asked whether the T731A protein could function like wild-type Dpb11 to suppress the HU hypersensitivity of the ddc2-top yeast. In contrast to wild-type Dpb11, overexpression of Dpb11 T731A does not reduce the HU sensitivity of the ddc2-top yeast (Fig. 5C). Thus, phosphorylation of Dpb11 is important to activate Mec1–Ddc2 complexes in vitro and regulate Mec1–Ddc2 activity in vivo.
Discussion
The Mec1/ATR kinase is an essential regulator of genome integrity (2). In this study, we examined how the kinase activity of Mec1 is regulated and discovered that an essential replication and checkpoint protein, Dpb11, functions as a direct Mec1 activator. Just as Dpb11 bridges the helicase and polymerase complexes during the initiation of replication, our data suggests that Dpb11 also serves as a link between the Mec1–Ddc2 kinase complex and 9-1-1 checkpoint clamp complex in the DNA damage response. Significantly, phosphorylation of Dpb11 by Mec1 increases its ability to serve as an activator providing a means of signal amplification.
The Mec1 activation domain of Dpb11 is located C-terminal to its BRCT domains. This Dpb11-C domain appears to be functionally equivalent to the TopBP1 ATR activation domain (AAD) (19) despite the lack of sequence conservation. The TopBP1 AAD binds the ATR–ATRIP complex through interactions with ATRIP and the ATR PIKK regulatory domain (PRD) (20). An ATRIP allele (-top) that eliminates TopBP1 binding to ATR–ATRIP cannot be activated. Similarly, we found an equivalent mutation in Ddc2 (-top) also cannot be activated by Dpb11. Given that very little sequence homology exists between Ddc2 and ATRIP, it is not surprising that the sequence of the TopBP1 AAD is not similar to the Dpb11-C domain. Although, given the similar mechanisms of ATR and Mec1 activation, we expect that Dpb11-C and the TopBP1 AAD proteins adopt a similar tertiary structure.
Both Dpb11 and TopBP1 contain a phosphorylated SQ/TQ site within their Mec1/ATR activation domains that enhance their ability to activate Mec1–Ddc2 or ATR–ATRIP complexes, respectively (19, 26). For Dpb11, our data indicate that this site serves as a positive-feedback amplification loop for Mec1 activation. For TopBP1, it is still unclear whether ATR autoamplifies by using this mechanism; however, ATM phosphorylation of this site potentiates the ability of TopBP1 to activate ATR (27).
A recent publication demonstrated that the loading of the 9-1-1 clamp onto DNA can stimulate the phosphorylation of Mec1 substrates in vitro (12). Because Mec1–Ddc2 also associates with DNA (28), the loaded clamp may serve as a scaffold for the recruitment of other Mec1 substrates. Purified Ddc1 can also modestly stimulate Mec1 kinase activity, but only when the kinase reactions are performed in low-salt conditions (12). Ddc1 associates with Mec1–Ddc2; however, the sites of interaction on Ddc1 and Mec1–Ddc2 have not been identified. We have found that recombinant Dpb11 greatly stimulates Mec1–Ddc2 kinase activity even in physiological salt concentrations. Furthermore, we identified mutations in Dpb11 and in Mec1–Ddc2 that disrupt their interactions. Taken together with our previous characterization of the ddc2-top mutant (20) and the work of others on the dpb11–1 mutant (14, 18), these data indicate that the interaction between Dpb11 and Mec1–Ddc2 is critical for Mec1 checkpoint signaling and cellular resistance to DNA damage and replication stress. It will be interesting to examine the simultaneous effect of Dbp11 and Ddc1 in Mec1 kinase assays. If these proteins have distinct modes of interaction with Mec1–Ddc2, then there might be an additive effect on Mec1 activation. On the other hand, if they use similar binding surfaces on Mec1–Ddc2, then competition between these proteins might be expected.
Our data are consistent with the following dual sensor and dual amplification model for Mec1 activation (Fig. 5D). RPA-coated ssDNA, generated as a consequence of replication stress or DNA damage, independently recruits the Mec1–Ddc2 and 9-1-1 complexes (7–10). The colocalization of these 2 complexes allows Ddc1 to initially stimulate Mec1. Mec1 then phosphorylates residues on Ddc1 necessary for the recruitment of Dpb11 (13, 29), which allows Mec1 to phosphorylate Dpb11. The phosphorylated form of Dpb11 serves as a potent Mec1 activator to further amplify Mec1 kinase activity toward its substrates. Mec1 phosphorylation of adaptor proteins, such as Rad9 and Mrc1, facilitates the recruitment of additional Mec1 substrates.
An alternative model is that Dpb11 starts the activation process in a parallel pathway to 9-1-1. This model would require Dpb11 recruitment to the stalled forks before 9-1-1 recruitment, perhaps through interactions with other replication proteins, such as Sld3 and Sld2. Once Dpb11 is recruited, a weak interaction between Dpb11 and Mec1–Ddc2 could activate Mec1, resulting in the phosphorylation of Ddc1. The phosphorylation of Ddc1 and Dpb11 could then stabilize the interaction between Dpb11 and Mec1–Ddc2, leading to full Mec1 activation. Further research on how Dpb11 is recruited to sites of DNA damage is required to test this alternative hypothesis.
Activation of the Mec1 signaling pathway does not require Dpb11 in all cases. Rad53 phosphorylation after UV radiation occurs in dpb11–1 yeast, consistent with the presence of redundant pathways of Mec1 activation (30). Further experiments will be necessary to determine why there are 2 Mec1 activators. The type of DNA damage and cell cycle position of the damaged cells affect which pathways are used to activate Mec1 (31). The Dpb11 pathway may be more important in the response to stalled replication forks or when DNA damage occurs during S-phase.
In conclusion, Dpb11 is a Mec1 activator. Activation requires Ddc2 due to a direct protein–protein interaction between Dpb11 and Ddc2. Moreover, Mec1 activation results from a positive-feedback loop whereby Mec1 phosphorylation of Dpb11 stimulates its ability to activate Mec1. Thus, Dpb11 is a functional ortholog of human TopBP1 and the Mec1/ATR activation mechanism is conserved from yeast to humans.
Methods
Kinase Assays.
Yeast expressing HA-Mec1 and myc-Ddc2 were spheroplasted by using Quantazyme lyticase (Qbiogene). A complete list of the yeast strains used in this work is in Table 1. Spheroplasts were lysed in 50 mM Hepes (pH 7.4), 100 mM KCl, 0.1 mM EDTA, 0.2% Tween 20, 1 mM diothiothreitol, 5 μg/ml aprotinin, 5 μg/ml leupeptin, 1 mM NaF, 50 mM β-glycerolphosphate, 1 mM sodium vanadate, 1 mM phenylmethylsulfonyl fluoride. Mec1–Ddc2 complexes were immunoprecipitated from cleared lysate by using anti-myc 9E10 antibody (Covance) and protein G agarose beads (Invitrogen). Immunoprecipitates were processed and used for kinase reactions as described (20). Where indicated, kinase reactions contained 150 mM NaCl instead of 50 mM NaCl. Quantification of kinase assays was performed by using a FLA-5100 PhosphorImager (Fuji Film).
Table 1.
Yeast strains used in this study
| Strain | Description | Source |
|---|---|---|
| DMP2995/1B | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 sml1Δ::KanMX4 ddc2Δ::KanMX4 | (4) |
| yDM003 | DMP2995/1B [pDM158:myc-ddc2-top-URA3-CEN] | This study |
| yDM011 | yDM003 [pDM172:pGAL-Dpb11-TRP1-CEN] | This study |
| yDM012 | yDM003 [pDM173:pGAL-Ddc1-TRP1-CEN] | This study |
| yDM013 | yDM003 [pDM174:pGAL-Dpb11-TRP1-2 μ] | This study |
| yDM014 | yDM003 [pDM175:pGAL-Ddc1-TRP1-2 μ] | This study |
| yDM016 | yDM003 [p1216:TRP1-CEN] | This study |
| yDM017 | yDM003 [p1221:TRP1-2 μ] | This study |
| yDM020 | DMP2995/1B [pDM174:pGAL-Dpb11-TRP1-2 μ] [p1220:URA3-CEN] | This study |
| yDM022 | DMP2995/1B [p1221:TRP1-2 μ] [p1220:URA3-CEN] | This study |
| yDM029 | yDM003 [pDM183:pGAL-dpb11-1-TRP1-2 μ] | This study |
| yDM063 | yDM003 [pDM209:pGAL-dpb11T731A-TRP1-2 μ] | This study |
| HSY1597 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 MEC1-HA::sml1Δ HIS3 ddc2Δ::HGR | (32) |
| yDM007 | HSY1597 [pDM158:myc-ddc2-top-URA3-CEN] | This study |
| yDM010 | HSY1597 [pNML1:myc-DDC2-URA3-CEN] | This study |
Protein Interactions.
GST-tagged Dpb11 fragments were purified from Escherichia coli with glutathione Sepharose 4B beads according to the manufacturer's instructions (GE Healthcare). Yeast were harvested and lysed in low-salt buffer [20 mM Hepes-KOH (pH 7.5), 0.1% Tween 20, 20% glycerol, 1 mM diothiothreitol, 5 μg/ml aprotinin, 5 μg/ml leupeptin, 1 mM NaF, 50 mM β-glycerolphosphate, 1 mM sodium vanadate, 1 mM phenylmethylsulfonyl fluoride] by using glass beads. Lysates was cleared by centrifugation and an equal volume of high-salt buffer [20 mM Hepes-KOH (pH 7.5), 350 mM NaCl, 25% glycerol, 1 mM diothiothreitol] was added. Then, lysates were incubated with the GST-tagged protein bound to glutathione beads overnight at 4 °C. Beads were washed 4 times in wash buffer [25 mM Hepes-KOH (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.1% Tween 20, 10% glycerol, 1 mM diothiothreitol, 5 μg/ml aprotinin, 5 μg/ml leupeptin, 1 mM NaF, 50 mM β-glycerolphosphate, 1 mM sodium vanadate, 1 mM phenylmethylsulfonyl fluoride]. Bound proteins were eluted in 2× SDS sample buffer, and processed for SDS/PAGE.
Acknowledgments.
We thank Stephen Elledge for providing reagents. This work was supported by National Cancer Institute Grant R01CA102729 and the Ingram Charitable Fund. D.A.M. was supported by Public Health Service award T32GM07347 from the National Institute of General Medical Studies for the Vanderbilt Medical-Scientist Training Program and a Department of Defense Breast Cancer Research Program Predoctoral Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Harper JW, Elledge SJ. The DNA damage response: Ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 2.Cimprich KA, Cortez D. ATR: An essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci USA. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paciotti V, Clerici M, Lucchini G, Longhese MP. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 2000;14:2046–2059. [PMC free article] [PubMed] [Google Scholar]
- 5.Rouse J, Jackson SP. LCD1: An essential gene involved in checkpoint control and regulation of the MEC1 signalling pathway in Saccharomyces cerevisiae. EMBO J. 2000;19:5801–5812. doi: 10.1093/emboj/19.21.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wakayama T, Kondo T, Ando S, Matsumoto K, Sugimoto K. Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:755–764. doi: 10.1128/MCB.21.3.755-764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 8.Melo JA, Cohen J, Toczyski DP. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondo T, Wakayama T, Naiki T, Matsumoto K, Sugimoto K. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science. 2001;294:867–870. doi: 10.1126/science.1063827. [DOI] [PubMed] [Google Scholar]
- 10.Majka J, Burgers PM. Yeast Rad17/Mec3/Ddc1: A sliding clamp for the DNA damage checkpoint. Proc Natl Acad Sci USA. 2003;100:2249–2254. doi: 10.1073/pnas.0437148100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonilla CY, Melo JA, Toczyski DP. Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Mol Cell. 2008;30:267–276. doi: 10.1016/j.molcel.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majka J, Niedziela-Majka A, Burgers PM. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell. 2006;24:891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Elledge SJ. Genetic and physical interactions between DPB11 and DDC1 in the yeast DNA damage response pathway. Genetics. 2002;160:1295–1304. doi: 10.1093/genetics/160.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Araki H, Leem SH, Phongdara A, Sugino A. Dpb11, which interacts with DNA polymerase II(epsilon) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc Natl Acad Sci USA. 1995;92:11791–11795. doi: 10.1073/pnas.92.25.11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia V, Furuya K, Carr AM. Identification and functional analysis of TopBP1 and its homologs. DNA Repair. 2005;4:1227–1239. doi: 10.1016/j.dnarep.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka S, et al. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 17.Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Elledge SJ. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1999;96:3824–3829. doi: 10.1073/pnas.96.7.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR–ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 20.Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamimura Y, Masumoto H, Sugino A, Araki H. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol Cell Biol. 1998;18:6102–6109. doi: 10.1128/mcb.18.10.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cortez D, Glick G, Elledge SJ. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc Natl Acad Sci USA. 2004;101:10078–10083. doi: 10.1073/pnas.0403410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoo HY, Shevchenko A, Dunphy WG. Mcm2 is a direct substrate of ATM and ATR during DNA damage and DNA replication checkpoint responses. J Biol Chem. 2004;279:53353–53364. doi: 10.1074/jbc.M408026200. [DOI] [PubMed] [Google Scholar]
- 24.Sarkaria JN, et al. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- 25.Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 26.Yamane K, Wu X, Chen J. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol Cell Biol. 2002;22:555–566. doi: 10.1128/MCB.22.2.555-566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoo HY, Kumagai A, Shevchenko A, Dunphy WG. Ataxia-telangiectasia mutated (ATM)-dependent activation of ATR occurs through phosphorylation of TopBP1 by ATM. J Biol Chem. 2007;282:17501–17506. doi: 10.1074/jbc.M701770200. [DOI] [PubMed] [Google Scholar]
- 28.Rouse J, Jackson SP. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol Cell. 2002;9:857–869. doi: 10.1016/s1097-2765(02)00507-5. [DOI] [PubMed] [Google Scholar]
- 29.Puddu F, et al. Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol Cell Biol. 2008;28:4782–4793. doi: 10.1128/MCB.00330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puddu F, et al. Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol Cell Biol. 2008;28:4782–4793. doi: 10.1128/MCB.00330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barlow JH, Lisby M, Rothstein R. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell. 2008;30:73–85. doi: 10.1016/j.molcel.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim HS, Brill SJ. MEC1-dependent phosphorylation of yeast RPA1 in vitro. DNA Repair. 2003;2:1321–1335. doi: 10.1016/j.dnarep.2003.07.004. [DOI] [PubMed] [Google Scholar]