Abstract
In contrast to the cell division machineries of bacteria, euryarchaea, and eukaryotes, no division components have been identified in the second main archaeal phylum, Crenarchaeota. Here, we demonstrate that a three-gene operon, cdv, in the crenarchaeon Sulfolobus acidocaldarius, forms part of a unique cell division machinery. The operon is induced at the onset of genome segregation and division, and the Cdv proteins then polymerize between segregating nucleoids and persist throughout cell division, forming a successively smaller structure during constriction. The cdv operon is dramatically down-regulated after UV irradiation, indicating division inhibition in response to DNA damage, reminiscent of eukaryotic checkpoint systems. The cdv genes exhibit a complementary phylogenetic range relative to FtsZ-based archaeal division systems such that, in most archaeal lineages, either one or the other system is present. Two of the Cdv proteins, CdvB and CdvC, display homology to components of the eukaryotic ESCRT-III sorting complex involved in budding of luminal vesicles and HIV-1 virion release, suggesting mechanistic similarities and a common evolutionary origin.
Keywords: cdv, Crenarchaeota, cytokinesis, ftsZ, Sulfolobus
The Archaea constitute a separate domain of life that has evolved in parallel with Bacteria and Eukarya (1). The archaeal domain is currently divided into two main lineages, the Crenarchaeota and the Euryarchaeota, each of which comprises several distinct classes of organisms that thrive in a wide variety of environments. Whereas several aspects of archaeal biology appear to be unique, certain traits resemble those in eukaryotes, including the machineries that govern information storage, maintenance, and processing.
Several features of archaeal cell cycle progression have been elucidated in considerable detail including the overall organization of the cell cycle in certain species, and regulatory and mechanistic aspects of the replication process (2, 3). Conversely, the genome segregation machinery remains essentially uncharacterized in this domain. In archaeal species belonging to the Euryarchaeota phylum, and in bacteria, cell division is mediated by FtsZ protein filaments that form a constricting ring structure (4). In eukaryotes, division occurs with the help of a contractile actin-myosin ring or, in plant cells, by septum formation at a site initially marked by actin and microtubules (5). In contrast to bacteria, euryarchaea, and eukaryotes, no cell division components have been identified in the second main archaeal phylum, Crenarchaeota (2).
Here, we report on the identification of key components of the cell division system in the hyperthermophilic crenarchaeon Sulfolobus acidocaldarius, describe intracellular structures that are formed by the gene products during genome segregation and division, and show that the operon is subject to a checkpoint-like regulation. We also demonstrate that the division machinery is present in all crenarchaeal orders except Thermoproteales, and that it is related to the eukaryotic ESCRT-III sorting complex.
Results
Identification of Genes Involved in Genome Segregation or Cell Division.
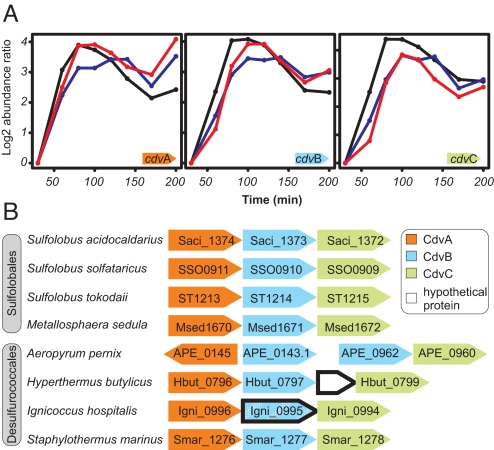
We performed a global mapping of cell-cycle-specifically expressed genes (6), by using synchronized S. acidocaldarius cultures and in-house whole-genome DNA microarrays (7). More than 20 genes were found to be specifically induced around the genome segregation and cell division stages, which, in this organism, occur in close succession (3). These included the three-gene Saci_1374–1372 operon (Fig. 1A), encoding two proteins related to Vps2 (Saci_1373) and Vps4 (Saci_1372), central components of the eukaryotic ESCRT-III protein-sorting machinery (see below; 8, 9), and a gene product of unknown function (Saci_1374).
Fig. 1.
RNA abundance profiles and cdv operon structures. (A) Microarray-determined induction profiles over the cell cycle (6) for cdv genes in synchronized S. acidocaldarius cultures. Each graph represents an independent biological replicate. Initiation of genome segregation in the leading edge of the cell population was estimated to occur at ≈50–60 min, with cell division initiating 10–15 min later. (B) Conserved organization of cdv genes (arrows) across different crenarchaeal species. Only cdvA, cdvB, and cdvC orthologs that occur in operons are displayed. I. hospitalis gene product Igni_0995 displays distant homology to other CdvB proteins, indicated by dashed shading.
The operon, defined through coexpression of the gene products both in terms of kinetics and absolute levels (Fig. 1A), and through the tight clustering of the genes on the genome (intergenic spaces 18 and 11 bp between Saci_1374-Saci_1373 and Saci_1373-Saci_1372, respectively), was designated cdv (cell division; see below). The operon consists of, in direction of transcription, cdvA (Saci_1374), cdvB (Saci_1373), and cdvC (Saci_1372), and is conserved across multiple crenarchaeal species (Fig. 1B).
Colocalization of Cdv Proteins Between Segregating Nucleoids.
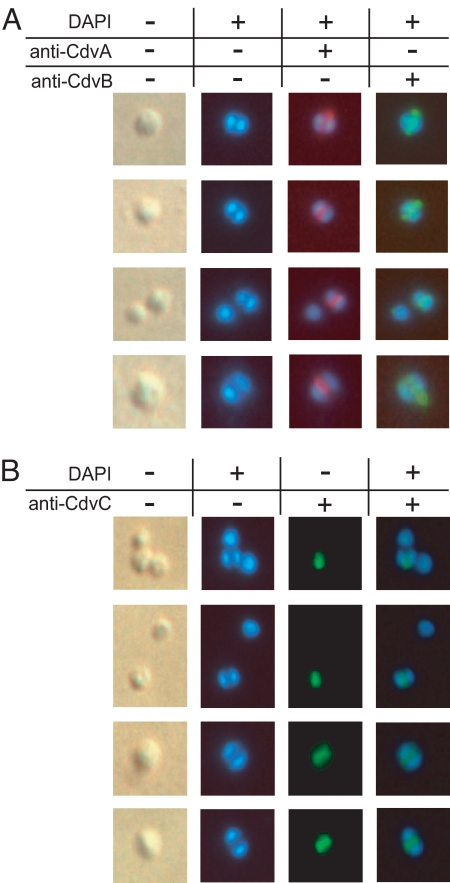
Immunostaining of exponentially growing cell populations revealed that the transcriptional induction was reflected at the protein level, such that all three gene products were strongly correlated to the genome segregation and division stages of the cell cycle. CdvA and CdvB formed colocalized band-like structures between segregating nucleoids, which appeared to span the entire cell width (Fig. 2A). CdvC also formed band-like structures, again restricted to cells with segregated nucleoids (Fig. 2B), although cells with uniform fluorescence across the cell interior also were observed (data not shown).
Fig. 2.
In situ immunofluorescence microscopy of S. acidocaldarius cells. Cultures were sampled in exponential growth phase. The first column depicts phase-contrast illumination of the cells shown in the consecutive columns. Nucleoids were stained with DAPI (4′,6-diamidino-2-phenylindole). Cdv proteins were stained with specific antibodies, followed by fluorescence visualization with Alexa Fluor-labeled secondary antibodies, as described in Materials and Methods. (A) Cells with two segregated nucleoids double-stained with anti-CdvA (red fluorescence) and anti-CdvB (green). (B) Anti-CdvC (green) stained cells. Note the absence of fluorescence signals in single-nucleoid cells (top two rows).
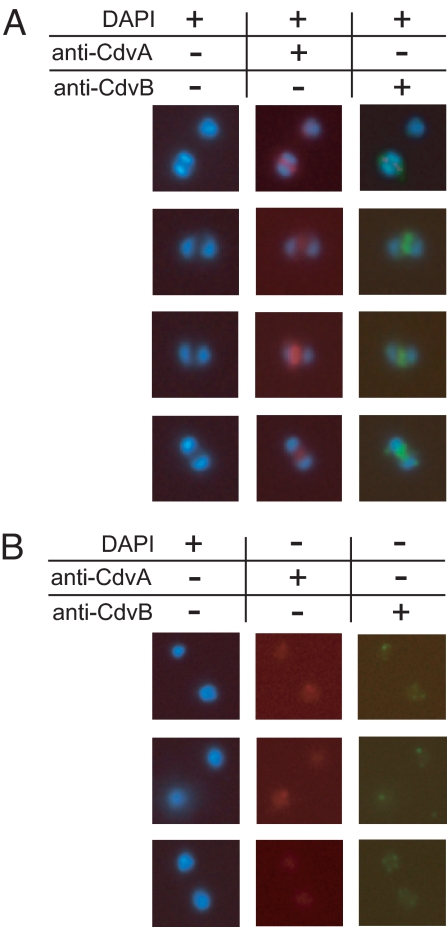
In rare instances, fluorescent bands were observed despite absence of visible nucleoid segregation (Fig. 3A), indicating formation of the structure at the early genome segregation stage. After segregation, CdvA and CdvB persisted in dividing cells, forming a successively smaller fluorescent structure during constriction (Fig. 3B), suggesting that the proteins form an integral part of the division apparatus.
Fig. 3.
In situ immunofluorescence microscopy of S. acidocaldarius cells at different stages of genome segregation and constriction. Growth conditions and staining are specified in Materials and Methods, and in the legend to Fig. 2, respectively. (A) Cells that display a Cdv band together with a single nucleoid. (B) Cdv structures in cells undergoing constriction.
Similarities to the Eukaryotic ESCRT-III Sorting Complex.
Interestingly, the products encoded by cdvB and cdvC display similarity to components involved in type E endosomal protein sorting in eukaryotes. CdvB is homologous to a family of highly charged coiled-coil containing proteins that comprise the ESCRT-III sorting complex [see supporting information (SI) Fig. S1], which includes Snf7, Did2, Vps2, Vps20, Vps24, and Vps60 [yeast nomenclature; (10)]. Furthermore, the genomes of all crenarchaeal species that contain the cdv genes encode at least 2, in some cases up to 4, ESCRT-III homologs, including three additional homologs within the S. acidocaldarius genome, Saci_0451, Saci_1601, and Saci_1416 (Fig. S1). This suggests that additional similarities may exist between the ESCRT-III sorting complex and the Cdv machinery, supported by cyclic induction of the Saci_1601 gene at the same cell cycle stage as the cdv operon (6).
CdvC is the archaeal ortholog of another eukaryotic type E sorting protein, Vps4 (8), an AAA+-type ATPase involved in ATP-mediated disassembly of the ESCRT-III complex (10). Recently, the structural basis for selective recognition of eukaryotic ESCRT-III proteins by Vps4 was elucidated by showing that the N-terminal MIT domain of Vps4 recognizes and binds to a motif located at the C terminus of some of the ESCRT-III proteins (9, 11). The MIT-interacting motif (MIM) was, however, found to be absent from the archaeal ESCRT-III-like proteins. Instead, some of the archaeal CdvB proteins contain a C-terminal predicted helix-turn-helix (HTH) domain, indicating a DNA-binding capability (Fig. S2).
In contrast to the ESCRT-related components, CdvA is a crenarchaea-specific factor (Fig. S3). The protein shares distant homology to the coiled-coil and filament domains of eukaryotic lamins, golgins and cingulin-like proteins (data not shown), which constitute primordial components of the eukaryotic cytoskeleton and nuclear envelope.
Expression of cdv is Subject to Checkpoint-like Control.
All three cdv genes are strongly repressed in both S. acidocaldarius and Sulfolobus solfataricus after UV irradiation of exponentially growing cultures (12, 13), with the S. solfataricus cdvC gene displaying the most dramatic down-regulation in the entire dataset in one study (12). The irradiation was shown to result in a dramatic increase in the number of double-strand chromosome breaks in S. acidocaldarius (13), and may indicate that the high level of DNA damage resulted in induction of a checkpoint-like response to inhibit cell division until completion of DNA repair, replication, and genome segregation. In addition, the operon is down-regulated during transition from exponential growth into stationary phase (our laboratory, unpublished), in accordance with a reduced need for division-related gene products. Further, and in agreement with an essential cellular role, deletion mutants in the S. solfataricus ortholog of the Saci_1372 gene (cdvC) could not be isolated in a previous study (8).
Effects of Antibiotic Treatments and Conditional-lethal Mutants.
Treatment of S. acidocaldarius cells with the nucleoside antibiotic tunicamycin inhibits cell division, presumably by blocking protein glycosylation required for proper invagination (14). A threefold increase in the proportion of cells displaying Cdv bands was observed after treatment (Fig. 4A), in accordance with cell cycle arrest at the division stage. The anthracycline daunomycin causes postreplicative cell cycle arrest in S. acidocaldarius, accompanied by a dispersed nucleoid conformation (14). No Cdv bands could be detected in daunomycin-treated cells (Fig. 4B), in agreement with daunomycin-dependent interference with transition from G2 into genome segregation and division, and consequent lack of cdv induction. S. acidocaldarius cultures were also treated with the macrolactone radicicol, which inhibits DNA topoisomerase VI (15), a possible chromosome decatenation enzyme in archaea. Cdv bands were, however, still observed (data not shown), indicating continued genome segregation and cell division, in accordance with the constitutive expression of the top6A and top6B genes during the cell cycle (6), which conflicts with models in which the enzyme mainly would act in preparation for genome segregation.
Fig. 4.
In situ immunofluorescence microscopy of exponentially growing S. acidocaldarius cells 6–8 h after antibiotic addition. Growth conditions, antibiotic concentrations, and staining are specified in the Material and Methods, and in the legend to Fig. 2, respectively. (A) Double-staining of tunicamycin-treated cells. (B) Double-staining of daunomycin-treated cells.
We also investigated Cdv structures in the S. acidocaldarius conditional-lethal mutants DG132 and DG134, in which genome segregation and cell division, respectively, are blocked when the temperature is increased from 70°C to 81°C (16). Although Cdv bands were detected in both mutants (data not shown), often with aberrant localization and morphology, loss of cell integrity and viability at nonpermissive temperature resulted in a high frequency of dead and irregularly shaped cells that displayed abnormal nucleoid morphology, precluding clear-cut interpretations. No mutations were identified by DNA sequencing of PCR-amplified complete cdv coding regions from either mutant (data not shown).
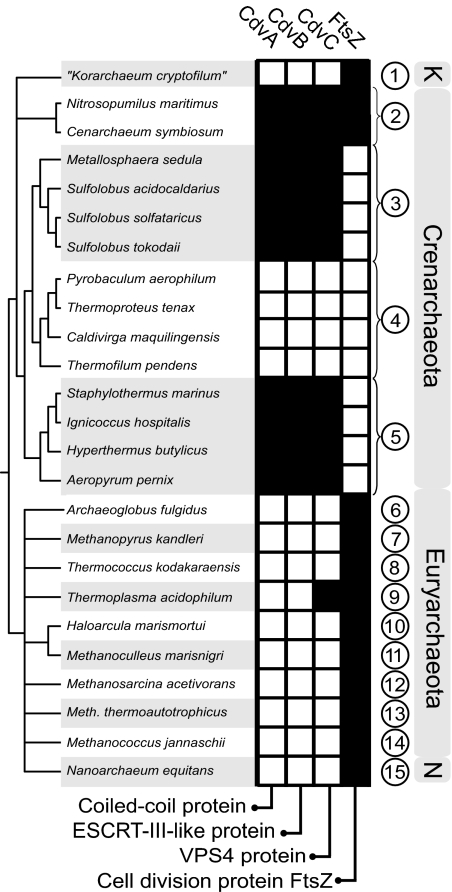
The Phylogenetic Distribution of cdv Genes Complements that of ftsZ.
The cdv genes exhibit complementary phylogenetic ranges compared with FtsZ-based archaeal division systems (Fig. 5). In genomes representing the Euryarchaeota phylum, and in Nanoarchaeum equitans, which divide using FtsZ, no Cdv homologs could be identified, except for distant cdvC homologs in Thermoplasma spp. The genes were also found to be absent from the genomes of the Thermoproteales order of the Crenarchaeota, in agreement with the unusual cell division mechanisms reported, for example, for Pyrobaculum and Thermoproteus species, which appear to carry out rapid “snapping” division rather than gradual invagination (17, 18), suggesting that these organisms rely on yet another division machinery. In contrast, all genome-sequenced members of the Sulfolobales and Desulfurococcales orders contain all three cdv genes, and are thus likely to carry out cell division in a similar fashion. The phylogenetic position of low-temperature members of Crenarchaeota is currently under debate, including the suggestion that they represent a distinct archaeal phylum (19). The organisms display a mixture of features considered unique either to the Eury- or Crenarchaeota and it is, therefore, noteworthy that a combination of ftsZ and cdv genes are present in both Nitrosopumilus maritimus and Cenarchaeum symbiosum, perhaps indicating joint participation in a division process that combines features from the two other phyla. The genome of the only sequenced member of the proposed Korarchaeota phylum, “Candidatus Korarchaeum cryptofilum”, contains seven FtsZ homologues but no cdv genes.
Fig. 5.
Phylogenetic distribution of division genes. Orthology-based representation of the phylogenetic distribution of cell division genes across sequenced archaeal genomes, based on the ArCOG database (31). Shaded and empty boxes represent the presence and absence of an ortholog in a given species, respectively. Abbreviations: 1, “Korarchaeota”; 2, Cenarchaeales; 3, Sulfolobales; 4, Thermoproteales; 5, Desulfurococcales; 6, Archaeoglobales; 7, Methanopyrales; 8, Thermococcales; 9, Thermoplasmatales; 10, Halobacteriales; 11, Methanomicrobiales; 12, Methanosarcinales; 13, Methanobacteriales; 14, Methanococcales; 15, Nanoarchaeales. N, Nanoarchaeota; K, Korarchaeota.
Discussion
We report the identification of key components of the archaeal cell division machinery, Cdv. The constituents do not resemble the bacterial and eukaryotic machineries that have been characterized in considerable detail. Instead, two of the Cdv proteins, CdvB and CdvC, display homology to proteins of the eukaryotic ESCRT-III complex, part of the endosomal protein sorting machinery required for budding events in several important biological processes, including the formation of intraluminal vesicles. The observations on this machinery have been brought together in the concentric circle model of multivesicular body cargo sorting (20), which suggests that ESCRT-III proteins form a ring-like configuration (21) on the endosomal membrane, and mediate membrane scission and/or detachment of intraluminal vesicles. Analgously, we propose that the archaeal ESCRT-III homologs, together with CdvA, form a complex at the division site, in agreement with the colocalization of the immunofluorescence signals. S. solfataricus CdvB and CdvC have recently been shown to interact in fluorescence anisotropy assays (9), which further supports the analogy between the respective machineries. The observation that CdvB contains a predicted C-terminal HTH domain rather than a MIM motif, as in eukaryotic ESCRT-III proteins that bind Vps4, suggests that, if such an interaction exists, it will be different in nature. Furthermore, it is possible that the HTH domain could be involved in positioning of the division machinery relative to the nucleoids, thus providing an interesting topic for further investigation.
Parallel roles for the ESCRT machinery in the abscission stage of cytokinesis and retroviral budding have been demonstrated in human cells (22), lending further support to a role for ESCRT-related gene products in cell division. Another interesting aspect is provided by eukaryotic CHMP (chromatin modifying, or charged multivesicular) proteins. A dual role in cell cycle progression and chromatin structure has been proposed for CHMP1 (23), which belongs to the same orthologous group as CdvB (Fig. S1). Further, budding of HIV type 1 (HIV-1) is mediated by the ESCRT machinery in a process topologically similar to the budding of luminal vesicles (24), and Vps4 plays an essential role also in this process (25). Intriguingly, the cdv operon is strongly induced during infection of S. solfataricus with Sulfolobus-turreted icosahedral virus [STIV; (26)], suggesting that Cdv proteins could be recruited also during archaeal virus budding.
In contrast to the ESCRT-related components, CdvA, which appears to play a central role in the division process, is a crenarchaea-specific factor, that shares weak homology with proteins that constitute basal components of the eukaryotic cytoskeleton and nuclear envelope. Regardless of the relevance of these similarities, the Cdv proteins appear to form part of the actual constriction complex, and may thus be comparable to bacterial and archaeal FtsZ in terms of mechanistic and structural roles during cytokinesis. The phylogenetic distribution of the cdv and ftsZ division systems parallels and confirms the suggested main archaeal evolutionary lineages. Thus, although the overall unity of the Archaea domain remains undisputed, species belonging to different archaeal phyla differ fundamentally regarding central cellular and physiological characteristics, including cell division mechanisms.
The observed analogy between the cdv system and the eukaryotic ESCRT-III complex is compelling and suggests a shared origin for these systems. After the ancient divergence of eukaryotes and archaea, the primordial division machinery may have been retained in archaea, while evolving into its present-day function in protein sorting in the eukaryotic lineage, although an ancient horizontal gene transfer event cannot be ruled out. In addition to these evolutionary considerations, the current study represents an onset in terms of regulatory and mechanistic characterization of Cdv-based cell division systems. The cdv operon is preceded by a potential transcription regulatory site, the CCR-1 box (6), which is likely to define a regulon under the control of a common transcription factor. Thus, the immunostainings provide support that the other CCR-1 regulated genes, consisting of three transcription factors and two hypothetical proteins, either control or directly participate in genome segregation and/or division, or coordinate these processes with subsequent initiation of chromosome replication. These factors, and other genes that are up-regulated upon onset of genome segregation and division (6), provide excellent targets for identification of additional components and clarification of the molecular details of archaeal genome segregation and Cdv-based cell division.
Materials and Methods
Strains and Growth Conditions.
Growth of S. acidocaldarius strain DSM639 cultures, and treatment with daunomycin (300 μg/ml) or tunicamycin (3 μg/ml), was performed as in ref. 14 and 27. The temperature-sensitive S. acidocaldarius mutants DG132 and DG134, and the parental DG64 strain, were cultivated and phenotypically induced as described in ref. 16.
PCR Amplification, Cloning, and Transformation.
S. acidocaldarius genomic DNA was extracted as described in ref. 27. Target genes were amplified by PCR with primers CdvA for (5′-CACCATGGGCATTCCGGTTGAGG-3′), CdvA rev (5′-TCATAAAGTATTTATTACCCTTACTG-3′), CdvB for (5′-TATGGATCCCATGTTTGATAAGTTATCGATAA-3′), CdvB rev (5′-TATAAGCTTAACCCTCAAGAACAATTAG-3′), CdvC for (5′-CACCATGTCTGCCCAAGTAATGC-3′), and CdvC rev (5′-TTATAGAGCCTTATACTTCTCGTG-3′). The cdvA and cdvC PCR products were cloned into expression vector pET100/D-Topo (Invitrogen). The cdvB product was digested with restriction endonucleases and cloned into pET-45b(+) (Novagen). In both vectors, the protein is expressed with an N-terminally fused histidine tag of 6 aa. All constructs were verified with DNA sequencing (Big Dye Terminator v3.1 Cycle Sequencing kit, Applied Biosystems), and transformed into Escherichia coli strain Rosetta DE3 by electroporation.
Protein Expression and Purification.
E. coli Rosetta DE3 cultures were grown at 37°C in LB medium containing 50 μg/ml ampicillin and 34 μg/ml chloramphenicol to an OD600 of 0.6–0.7. After incubation on ice for 30 min, gene expression was induced by addition of IPTG (isopropyl β-d-1-thiogalactopyranoside) to a final concentration of 0.4 mM. The cultures were incubated at 37°C for approximately 20 h, after which the cells were harvested by centrifugation, resuspended in binding buffer (20 mM sodium phosphate pH 7.4, 0.5 M NaCl, and 20 mM imidazole) and lysed by sonication. Heat-labile proteins were denatured by incubation at 80°C for 10–30 min, and cell debris was removed by centrifugation at 23 000 × g for 45 min. His-tagged proteins were purified from the supernatant by immobilized metal ion affinity chromatography (IMAC), by using the His GraviTrap system (GE Healthcare). The affinity columns were equilibrated with binding buffer, loaded with protein sample, washed with binding buffer, and eluted with 3 × 1 ml of elution buffer (20 mM sodium phosphate pH 7.4, 0.5 M NaCl, and 500 mM imidazole). The protein-containing fraction was dialysed overnight against 20 mM phosphate buffer, pH 7.4, containing 0.5 M NaCl at a ratio of 1:1,000 (vol/vol).
Antibody Generation.
Antibodies against purified proteins were generated by Innovagen. Antibodies against CdvB and CdvC were produced in rabbit and IgG purified. CdvA antibodies were produced in chicken and IgY purified according to the standard protocol of the manufacturer.
Immunostaining.
Samples from S. acidocaldarius cell cultures were diluted in ethanol to a final concentration of 70%. Aliquots of 0.5–1 ml were then centrifuged and washed with PBST (PBS containing 0.05% Tween 20), incubated with primary antibodies, diluted in PBST, and incubated for 2 h at 37°C and then overnight at 4°C. The cells were washed three times with PBST, incubated with secondary antibodies (Alexa Fluor 568 goat anti-chicken IgG, Alexa Fluor 488 goat anti-rabbit IgG, Alexa Fluor 568 goat anti-rabbit IgG, or Alexa Fluor 488 goat anti-chicken IgG; Invitrogen), diluted in PBST, incubated for 2 h, washed three times with PBST, and finally resuspended in 30 μl of PBS. The immunostained cells were placed on a thin layer of 10% agarose containing 0.2 μg/ml DAPI (4′,6-diamidino-2-phenylindole) on a glass slide. A coverslip was then added and sealed with nail polish.
Microscopy.
Mounted cells were visualized with an 100× PL Fluotar objective (NA 1.30) in a DMRXE epifluorescence microscope (Leica). For each examined area four images were recorded: phase contrast, UV filter A for DAPI illumination, and filters I3 or L5 and filter N2.I for Alexa Fluor 488 and Alexa Fluor 568 emission respectively. Images were recorded with an Orca IIIm CCD camera (Hamamatsu).
Protein Sequence Analysis of Cdv Proteins.
Protein sequences were retrieved from the National Center for Biotechnology Information (NCBI) by using PSI-BLAST (28) searches (BLOSUM62 matrix; inclusion threshold 1E-04; low complexity filter enabled) seeded with the respective S. acidocaldarius sequence, that is, Saci_1373 for CdvB and Saci_1374 for CdvA. The homology of the C-terminal domain of CdvB proteins with the HTH domains of DNA-binding transcriptional regulators was further explored by similarity searches with the C-terminal region of the S. acidocaldarius CdvB protein (Saci_1373, residues 191–264) by using PSI-BLAST (28) (settings as above) and the PFAM database (29). Retrieved proteins were aligned by using Kalign (30) and alignments were manually edited as required.
Microarray Datasets.
The microarray datasets covering cdv expression in synchronized S. acidocaldarius cultures (6), and in S. acidocaldarius and S. solfataricus cultures exposed to UV irradiation (13), are available in the ArrayExpress repository (http://www.ebi.ac.uk/microarray-as/ae/), accession numbers E-MEXP-936 and E-MEXP-1252, respectively.
Supplementary Material
Acknowledgments.
This work was supported by the Swedish Research Council, the Carl Trygger Foundation, and by Marie Curie Intra-European (European Union) and Rubicon (NWO) fellowships (T.J.G.E).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 18653.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809467105/DCSupplemental.
References
- 1.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lundgren M, Bernander R. Archaeal cell cycle progress. Curr Opin Microbiol. 2005;8:662–668. doi: 10.1016/j.mib.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Bernander R. The cell cycle of Sulfolobus. Mol Microbiol. 2007;66:557–562. doi: 10.1111/j.1365-2958.2007.05917.x. [DOI] [PubMed] [Google Scholar]
- 4.Margolin W. FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol. 2005;6:862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan DO. In: The Cell Cycle: Principles of Control. Lawrence E, editor. Sunderland: New Science Press Ltd.; 2007. pp. 157–173. [Google Scholar]
- 6.Lundgren M, Bernander R. Genome-wide transcription map of an archaeal cell cycle. Proc Natl Acad Sci USA. 2007;104:2939–2944. doi: 10.1073/pnas.0611333104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersson A, Bernander R, Nilsson P. Dual-genome primer design for construction of DNA microarrays. Bioinformatics. 2005;21:325–332. doi: 10.1093/bioinformatics/bti001. [DOI] [PubMed] [Google Scholar]
- 8.Hobel CFV, Albers SV, Driessen AJM, Lupas AN. The Sulfolobus solfataricus AAA protein Sso0909, a homologue of the eukaryotic ESCRT Vps4 ATPase. Biochem Soc Trans. 2008;36:94–98. doi: 10.1042/BST0360094. [DOI] [PubMed] [Google Scholar]
- 9.Obita T, et al. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 10.Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stuchell-Brereton MD, et al. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 12.Fröls S, et al. Response of the hyperthermophilic archaeon Sulfolobus solfataricus to UV damage. J Bacteriol. 2007;189:8708–8718. doi: 10.1128/JB.01016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Götz D, et al. Responses of hyperthermophilic crenarchaea to UV irradiation. Genome Biol. 2007;8:R220. doi: 10.1186/gb-2007-8-10-r220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hjort K, Bernander R. Cell cycle regulation in the hyperthermophilic crenarchaeon Sulfolobus acidocaldarius. Mol Microbiol. 2001;40:225–234. doi: 10.1046/j.1365-2958.2001.02377.x. [DOI] [PubMed] [Google Scholar]
- 15.Gadelle D, Bocs C, Graille M, Forterre P. Inhibition of archaeal growth and DNA topoisomerase VI activities by the Hsp90 inhibitor radicicol. Nucleic Acids Res. 2005;33:2310–2317. doi: 10.1093/nar/gki526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernander R, Poplawski A, Grogan DW. Altered patterns of cellular growth, morphology, replication and division in conditional-lethal mutants of the thermophilic archaeon Sulfolobus acidocaldarius. Microbiology. 2000;146:749–757. doi: 10.1099/00221287-146-3-749. [DOI] [PubMed] [Google Scholar]
- 17.Horn C, Paulmann B, Kerlen G, Junker N, Huber H. In vivo observation of cell division of anaerobic hyperthermophiles by using a high-intensity dark-field microscope. J Bacteriol. 1999;181:5114–5118. doi: 10.1128/jb.181.16.5114-5118.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lundgren M, Malandrin L, Eriksson S, Huber H, Bernander R. Cell cycle characteristics of Crenarchaeota: Unity among diversity. J Bacteriol. 2008;190:5362–5367. doi: 10.1128/JB.00330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: Proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 20.Nickerson DP, Russell MRG, Odorizzi G. A concentric circle model of multivesicular body cargo sorting. EMBO Rep. 2007;8:644–650. doi: 10.1038/sj.embor.7401004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: A role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 23.Stauffer DR, Howard TL, Nyun T, Hollenberg SM. CHMP1 is a novel nuclear matrix protein affecting chromatin structure and cell-cycle progression. J Cell Sci. 2001;114:2383–2393. doi: 10.1242/jcs.114.13.2383. [DOI] [PubMed] [Google Scholar]
- 24.Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 25.von Schwedler UK, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 26.Ortmann AC, et al. Transcriptome analysis of infection of the archaeon Sulfolobus solfataricus with Sulfolobus turreted icosahedral virus. J Virol. 2008;82:4874–4883. doi: 10.1128/JVI.02583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundgren M, Andersson A, Chen L, Nilsson P, Bernander R. Three replication origins in Sulfolobus species: Synchronous initiation of chromosome replication and asynchronous termination. Proc Natl Acad Sci USA. 2004;101:7046–7051. doi: 10.1073/pnas.0400656101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lassmann T, Sonnhammer ELL. Kalign, Kalignvu and Mumsa: Web servers for multiple sequence alignment. Nucleic Acids Res. 2006;34:W596–599. doi: 10.1093/nar/gkl191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makarova KS, Sorokin AV, Novichkov PS, Wolf YI, Koonin EV. Clusters of orthologous genes for 41 archaeal genomes and implications for evolutionary genomics of archaea. Biol Direct. 2007;2:33. doi: 10.1186/1745-6150-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.