Enveloped viruses acquire a lipid membrane when they bud across a cellular membrane during virus assembly. In cell entry, the viral membrane must be fused to the host-cell membrane to deliver the viral genome into the cytoplasm for replication. Membrane fusion is therefore an essential step in the life cycle of enveloped viruses, and a great deal of research in recent years has been directed at identifying inhibitors of viral membrane fusion. One notable success in this area is the HIV fusion inhibitor enfurvirtide (T-20, Fuzeon) (1), which has become part of the standard treatment for patients who have detectable viral loads after treatment with protease and reverse transcriptase inhibitors. Although influenza remains a primary global health problem, there are no clinically useful fusion inhibitors availableagainst influenza virus. In a recent issue of PNAS, Russell et al. (2) report the crystal structure of the influenza virus hemagglutinin (HA) envelope protein bound to a compound that was reported to inhibit membrane fusion and infectivity of certain strains of influenza. The structure provides an excellent starting framework for the rational design of more effective membrane fusion inhibitors for use as therapeutics against influenza.
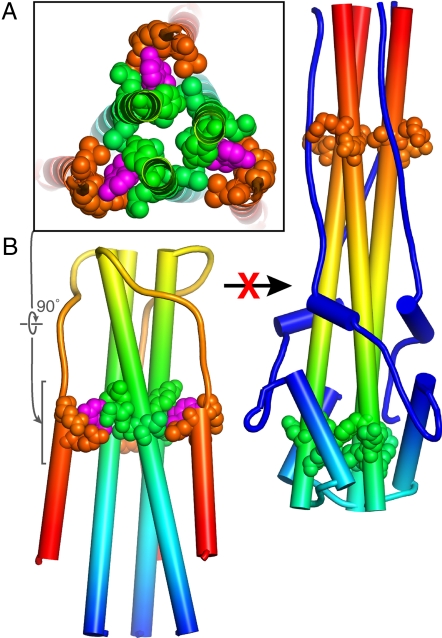
For 2 membranes to fuse, they must be bent toward each other until they are separated by only a fraction of a nanometer. Bending membranes requires energy, which in viral membrane fusion is provided by envelope proteins anchored in the viral membrane as they undergo a large, spontaneous, “fusogenic” conformational change (3). The fusogenic conformational change of HA is well understood from numerous biophysical and biochemical studies, making HA the prototype of viral fusion proteins (4). Moreover, various small hydrophobic molecules such as tert-butyl hydroquinone (TBHQ) have been found to block influenza virus infectivity in cell culture by inhibiting (or prematurely inducing) the fusogenic conformational change in HA (5–7). The development of more effective fusion inhibitors has, however, been limited by the lack of crystal structures of relevant HA complexes and the failure of known fusion inhibitors to neutralize all influenza subtypes. Russell et al. (2) report the crystal structures of HA from 2 subtypes of influenza, H14 and H3, in complex with TBHQ. TBHQ specifically inhibits the fusogenic conformational change of HA and reduces viral infectivity of group 2 influenza subtypes such as H3 and H14 (6). The structures show that TBHQ does not bind near the fusion peptide of HA as predicted by in silico docking analyses (6). Instead, TBHQ binds in a hydrophobic pocket at the interface between monomers in the prefusion HA trimer (Fig. 1). This binding pocket is present only in group 2 HAs, which explains the failure of TBHQ to inhibit membrane fusion and the infectivity of group 1 viruses. The apparent mechanism for fusion inhibition is that TBHQ stabilizes the prefusion conformation of HA, thus increasing the energy barrier for the fusogenic conformational change to the point that it no longer responds to the acidic environment of the endosomal lumen (2). Specifically, TBHQ locks HA in its prefusion conformation by extending the hydrophobic core between the “outer layer” and “inner layer” α-helices (Fig. 1). In other words, TBHQ acts as a sort of molecular glue. To exclude water and other solvent molecules, hydrophobic compounds must fit tightly into their binding pockets, with a high degree of shape complementarity. The snug fit of TBHQ into its binding pocket leaves little room for additional hydrophobic substitutions. However, the structure shows that space for additional polar substituents on TBHQ exists in the highly polar membrane distal region of the binding site (2). Because drug-like molecules typically require a balance between polar and hydrophobic properties, structure-based design of TBHQ derivatives with polar substituents holds promise as a strategy for developing new treatments against influenza.
Fig. 1.
The structural basis of the inactivation of the influenza virus HA envelope protein by TBHQ. (A) TBHQ (shown schematically in magenta) binds in a hydrophobic pocket near the C terminus of the short α-helix, at the interface between monomers in the prefusion HA trimer. For clarity, only the core regions of HA2 are shown. In group 1 influenza subtypes, the TBHQ binding site is absent, explaining the failure of TBHQ to inhibit membrane fusion of group 1 viruses. (B) TBHQ stabilizes the prefusion trimer, thereby inhibiting the conformational rearrangements that are required for membrane fusion and viral entry.
The use of hydrophobic compounds to inhibit pH-dependent conformational changes in viral entry is not unprecedented. Hydrophobic compounds such as disoxaril (WIN 51711) and pleconaril (WIN 63843) bind in a hydrophobic pocket within the core of the picornavirus capsid protein VP1. Binding of these compounds rigidifies the protein shell, thereby inhibiting the acid-induced disassembly of the capsid in the endosome (8, 9). Structural studies have gone hand in hand with the development of more efficacious antiviral agents against rhinoviruses and enteroviruses (8). After disoxaril failed phase I clinical trials, the crystallographic determination of the structure of disoxaril bound to human rhinovirus (9) guided the design of the improved compound pleconaril (WIN 63843), which reached phase III clinical trials.
Similarly, a hydrophobic ligand-binding pocket was discovered in the envelope (E) protein of dengue type 2 virus. In the crystal structure of dengue E, a molecule of β-octyl glucoside (β-OG) is bound in a hydrophobic pocket (10). It has been suggested (but not verified) that molecules with similar hydrophobic moieties to β-OG, but with added polar substituents to increase binding affinity to E, might block the fusogenic conformational rearrangement in dengue and other flaviviruses by an analogous mechanism as for TBHQ and HA, that is, by stabilizing the prefusion conformation of E and preventing the fusogenic transition to the postfusion conformation, which lacks the β-OG binding pocket (10, 11).
TBHQ specifically inhibits the fusogenic conformational change of HA.
The 3 types of antiviral compounds discussed above each have in common that they bind in hydrophobic pockets in the core of viral envelope (or capsid) proteins, thus locking the proteins in their prefusion (or preentry) conformations. In contrast, the HIV membrane fusion inhibitor enfurvirtide is a 36-mer peptide with a sequence of largely polar amino acids corresponding to the C-terminal helical heptad repeat (HR-2) of the HIV envelope protein gp41. Enfuvirtide does not stabilize the prefusion conformation of the envelope protein, but rather prevents it from reaching its final postfusion conformation by competing with the intramolecular foldback of HR-2 onto the inner core of the envelope protein, formed by the trimeric N-terminal helical heptad repeat, HR-1 (1). The mechanism of action of enfvirtide is therefore different from that of TBHQ. The latter is potentially superior in that (i) it binds to the prefusion form of the envelope protein rather than to a short-lived intermediate in the fusogenic transition, (ii) it is much easier to synthesize, and (iii) it has more drug-like chemical properties.
Looking to the future, the work by Russell et al. (2) provides a structural framework for the development of small-molecule compounds to prevent influenza virus from entering the host cell. The development of more effective compounds that inhibit membrane fusion by influenza has been limited by not knowing where and how known fusion inhibitors bind to HA and inhibit its fusogenic conformational change. The structure of HA bound to TBHQ reveals a potential binding site for larger or additional substituents on TBHQ. This finding opens the door to a structure-based drug design program for identifying novel small-molecule membrane fusion inhibitors. Proof of principle for this strategy is provided by the encouraging results of related approaches discussed above to develop antiviral compounds against picornaviruses and HIV. New therapeutics resulting from these efforts would make possible drug combination therapies that avoid the selection of resistant viruses. Indeed, combination chemotherapy against influenza with amantadine and oseltamivir has already been shown to reduce the emergence of drug resistance (12).
Acknowledgments.
My work is supported by the Burroughs Wellcome Fund, the Lupus Research Institute, and the Roche Organ Transplant Research Foundation.
Footnotes
The author declares no conflict of interest.
See companion article on page 17736 in issue 46 of volume 105.
References
- 1.Kilby JM, et al. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 2.Russell RJ, et al. Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc Natl Acad Sci USA. 2008;105:17736–17741. doi: 10.1073/pnas.0807142105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 5.Yu KL, et al. Structure–activity relationships for a series of thiobenzamide influenza fusion inhibitors derived from 1,3,3-trimethyl-5-hydroxy-cyclo-hexylmethylamine. Bioorg Med Chem Lett. 2002;12:3379–3382. doi: 10.1016/s0960-894x(02)00761-8. [DOI] [PubMed] [Google Scholar]
- 6.Bodian DL, et al. Inhibition of the fusion-inducing conformational change of influenza hemagglutinin by benzoquinones and hydroquinones. Biochemistry. 1993;32:2967–2978. doi: 10.1021/bi00063a007. [DOI] [PubMed] [Google Scholar]
- 7.Hoffman LR, Kuntz ID, White JM. Structure-based identification of an inducer of the low-pH conformational change in the influenza virus hemagglutinin: Irreversible inhibition of infectivity. J Virol. 1997;71:8808–8820. doi: 10.1128/jvi.71.11.8808-8820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hadfield AT, Diana GD, Rossmann MG. Analysis of three structurally related antiviral compounds in complex with human rhinovirus 16. Proc Natl Acad Sci USA. 1999;96:14730–14735. doi: 10.1073/pnas.96.26.14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith TJ, et al. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science. 1986;233:1286–1293. doi: 10.1126/science.3018924. [DOI] [PubMed] [Google Scholar]
- 10.Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci USA. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 12.Ilyushina NA, Bovin NV, Webster RG, Govorkova EA. Combination chemotherapy, a potential strategy for reducing the emergence of drug-resistant influenza A variants. Antiviral Res. 2006;70:121–131. doi: 10.1016/j.antiviral.2006.01.012. [DOI] [PubMed] [Google Scholar]