Abstract
To understand genetic patterns of the morphological and physiological traits in flag leaf of barley, a double haploid (DH) population derived from the parents Yerong and Franklin was used to determine quantitative trait loci (QTL) controlling length, width, length/width, and chlorophyll content of flag leaves. A total of 9 QTLs showing significantly additive effect were detected in 8 intervals on 5 chromosomes. The variation of individual QTL ranged from 1.9% to 20.2%. For chlorophyll content expressed as SPAD value, 4 QTLs were identified on chromosomes 2H, 3H and 6H; for leaf length and width, 2 QTLs located on chromosomes 5H and 7H, and 2 QTLs located on chromosome 5H were detected; and for length/width, 1 QTL was detected on chromosome 7H. The identification of these QTLs associated with the properties of flag leaf is useful for barley improvement in breeding programs.
Keywords: Barley (Hordeum vulgare L.), Double haploid (DH), Flag leaf, Quantitative trait loci (QTL)
INTRODUCTION
Barley (Hordeum vulgare L.) is one of the most important cereal crops in the world, ranking the forth in terms of planting area only after wheat, rice and maize (FAO, 2005). It has wide use, mainly for malting, feed and food. With economic development, more and more barley will be needed for beer processing and animal feed. Hence, improvement of yield potential has been a major objective in barley breeding programs.
Grain yield increase would be effectively rested with the basis of the capabilities of yield components and other closely associated traits (Sharma et al., 2003). In cereal crops, the top three leaves on the stem, especially the top-most leaf, i.e., flag leaf, are the primary source of carbohydrates production (Sicher, 1993). The flag leaf could produce a large proportion of the carbohydrates stored in grains (Li et al., 1998). The morphological traits of flag leaf such as size and shape, and physiological traits of flag leaf such as chlorophyll content and photosynthesis capacity have been considered to be the important determinants of grain yield in cereals (Chen et al., 1995; Hirota et al., 1990). Therefore, flag leaf is one of the greatest components in determining grain yield potential in cereal crops.
In barley, the importance of flag leaf in determining grain yield was also intensively studied (Thorne, 1965; Tungland et al., 1987; Yap and Harvey, 1972; Zheng, 1999). It was found that some flag leaf traits, such as length, width and angle, were inherited quantitatively (Yang and Lu, 1991). Meanwhile, quantitative trait loci (QTL) mapping provides a powerful tool to study complex traits at the molecular level, and considerable progress has been made in the genetic research on yield, yield components, and malting quality traits in barley (Backes et al., 1995; Baum et al., 2003; Bezant et al., 1997; Kjaer and Jensen, 1996; Li J.Z. et al., 2005; 2006; Marquez-Cedillo et al., 2001; Mather et al., 1997; Sameri et al., 2006; Thomas et al., 1995; Yin et al., 1999). However, little research has been done in understanding of genetic mechanisms for flag leaf traits in barley. The objectives in this study were to elucidate the genetics of length, width, length/width and chlorophyll content of flag leaf in barley by QTL mapping using a double haploid (DH) population.
MATERIALS AND METHODS
Plant materials and growing conditions
A DH population was derived from a cross between Yerong (six-rowed) and Franklin (two-rowed) (Li H. et al., 2006; Wenzl et al., 2006). The two parents show the distinct difference in flag leaf traits. The 154 DH lines and their parent lines (Yerong and Franklin lines) were planted in the experimental farm of Huajiachi Campus, Zhejiang University, Hangzhou, China (30°10′ N, 120°12′ E) in the barley growing season. The seeds were sown on early November in 2006. The plot consisted of 156 lines with line length of 2 m and line interval of 0.25 m, and there were 3 replications for each DH line or parent. Fifty seeds were sown in a line. Prior to seeding, 70 kg/ha of N as urea and 180 kg/ha of potassium chloride were applied. Another 70 kg/ha of N was top-dressed at 4-leaf stage.
Measurements
During 10 d after heading, 5 plants were randomly selected from each DH line and parent to measure flag leaf length (FL, from the base of ligula to the tip of leaf, in cm) and width (FW, the widest part of the leaf, in cm), and flag leaf length/width (FLW or FL/FW) was calculated. Meanwhile, chlorophyll content of flag leaf (FC) was determined with a non-destructive method at the mid position of flag leaf using a portable fluorometer (SPAD-502, Minolta, Japan).
Data and QTL analysis
Population distribution and correlation analysis were performed using SPSS 11.0 statistical software for all the traits measured in this study.
The genetic linkage map was constructed using a total of 408 DArT (diversity arrays technology), SSR (simple sequence repeat) and other markers as described by Wenzl et al.(2006). QTL analysis was performed using software QTLMapper 1.60 that was developed based on the mixed linear model approach (Wang et al., 1999; 2003) to identify QTL with main-effect. QTL was determined with threshold P<0.005. The threshold of LOD (likelihood odds ratio) >2.5 was chosen for claiming a putative QTL. The nomenclature of QTL followed the system of McCouch et al.(1997).
RESULTS
Phenotypic variation and correlation analysis
Phenotypes of the DH lines and the two parents were evaluated for the 4 flag leaf traits, and the grand means, ranges, skewness and kurtosis of measured traits were summarized in Table 1. For FC, FL and FLW of the two-rowed parent Franklin were 50.4, 20.2 cm and 14.4, respectively, being larger than those of the six-rowed parent Yerong (Table 1). The FC, FL, FW and FLW of 154 DH lines averaged 47.4, 15.5 cm, 1.4 cm and 11.1, respectively. The segregation of these traits was continuously distributed among all the DH lines, but showed significant transgressive distribution. The kurtosis and skewness for all the traits were less than 1, indicating their normal distribution. Additionally, transgressive segregation in both directions was observed for all traits, indicating that both parents transmitted favorable alleles for each trait (Table 1).
Table 1.
Phenotypic values of flag leaf traits in DH populations and their parents
| Trait | Yerong | Franklin | DH population |
|||
| Mean±SD | Range | Skewness | Kurtosis | |||
| FC | 46.9 | 50.4 | 47.4±0.27 | 37.8~55.1 | −0.217 | 0.043 |
| FL | 13.4 cm | 20.2 cm | (15.5±0.26) cm | 9.2~26.2 cm | 0.861 | 0.639 |
| FW | 1.5 cm | 1.4 cm | (1.4±0.01) cm | 1.0~2.0 cm | 0.263 | 0.163 |
| FLW | 8.9 | 14.4 | 11.1±0.13 | 7.9~15.7 | 0.494 | 0.120 |
FC: chlorophyll content of flag leaf; FL: flag leaf length; FW: flag leaf width; FLW: length/width of flag leaf
The results of correlation analysis between the flag leaf traits are shown in Table 2. FL was significantly and positively correlated with FW (r=0.679, P<0.01) and FLW (r=0.781, P<0.01), but there was no significant correlation between FW and FLW. In addition, no significant correlation was detected between FC and FL, FW, although there was a positive correlation between FC and FLW (r=0.171, P<0.05), indicating that chlorophyll content in the flag leaves of barley was associated with flag leaf length/width.
Table 2.
The correlation coefficients among four flag leaf traits in the DH population
FC: chlorophyll content of flag leaf; FL: flag leaf length; FW: flag leaf width; FLW: length/width of flag leaf; NS: not significant
significant at 95% probability level
highly significant at 99% probability level
QTL analysis
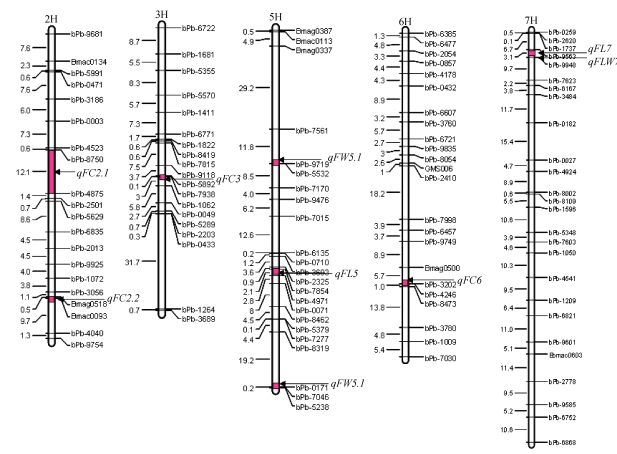
A total of 9 QTLs for the examined flag leaf traits were mapped on barley chromosomes 2H, 3H, 5H, 6H and 7H. The main effects of these QTLs are shown in Table 3 and Fig.1.
Table 3.
Putative main effect of the QTL with a LOD score >2.5 for flag leaf traits
| Trait | QTLa | Chr.b | Interval | LODc | Add.d | Var. (%)e |
| FC | qFC2.1 | 2H | bPb-8750~bPb-4875 | 3.12 | 0.88 | 6.4 |
| qFC2.2 | 2H | Bmag0518~Bmac0093 | 8.86 | 1.56 | 20.2 | |
| qFC3 | 3H | bPb-5892~bPb-7938 | 3.88 | 0.87 | 6.3 | |
| qFC6 | 6H | bPb-3202~bPb-4246 | 2.77 | −0.90 | 6.7 | |
| FL | qFL5 | 5H | bPb-7854~bPb-4971 | 3.08 | 0.76 | 5.7 |
| qFL7 | 7H | bPb-9563~bPb-9948 | 2.84 | 0.78 | 5.9 | |
| FW | qFW5.1 | 5H | bPb-9719~bPb-5532 | 15.34 | 0.08 | 18.3 |
| qFW5.2 | 5H | bPb-0171~bPb-7046 | 3.22 | 0.03 | 2.6 | |
| FLW | qFLW7 | 7H | bPb-9563~bPb-9948 | 3.21 | 0.22 | 1.9 |
Individual QTL is designated with the italicized abbreviation of the trait and the chromosome number. When more than one QTL affecting a trait is identified on the same chromosome, they are distinguished by decimal numbers;
Chromosome number;
Maximum-likelihood LOD score for the QTL calculated by QTLMapper 1.60;
Additive effect. The positive or negative value indicates that allele from Yerong or Franklin increases the trait score, respectively;
Variation explained by the putative QTL
Fig. 1.
Chromosomal locations of putative QTLs for flag leaf traits of the DH population and their parents (Yerong and Franklin) in barley
FC: flag leaf chlorophyll content; FL: flag leaf length; FW: flag leaf width; FLW: flag leaf length/width. The distances between markers (cM) are listed to the left of each figure part
Four putative QTLs for chlorophyll content were detected on chromosomes 2H, 3H and 6H. Of these, individual QTL explained the variation from 6.3% to 20.2% of the total phenotypic variation, with QTL qFC2.2 having the largest effect. The alleles in the direction of increasing FC for the QTL qFC6 came from Franklin, while those for the others (qFC2.1, qFC2.2 and qFC3) came from Yerong.
Two putative QTLs (qFL5 and qFL7) for flag leaf length were detected on chromosomes 5H and 7H, respectively. The additive effects of the two QTLs were positive, indicating that the alleles from Yerong increased the FL.
Two QTLs (qFW5.1 and qFW5.2) significantly affecting flag leaf width were detected on chromosome 5H and the additive effect was from Yerong allele. qFW5.1 was a major QTL, which could explain 18.3% of the total phenotypic variation.
One QTL (qFLW7) affecting flag leaf length/width was located on chromosome 7H, and its allele from Yerong could increase FLW. Interestingly, qFLW7 and qFL7 were located on the same region of chromosome 7H.
DISCUSSION
The current results indicate that the 4 traits of flag leaf in the 2 parents and 154 DH lines were quantitatively inherited and could be controlled by multiple genes. The mean values of the population were quite close to the mid-parental values for all traits (Table 1). In addition, significant variation and normal distribution of all traits measured in this study suggest the suitability of the population for QTL analysis.
The improvement in agronomic and physiological traits of flag leaf has been a major objective in rice breeding program. Analysis of QTL controlling morphological and agronomic traits of flag leaf in rice has been intensively investigated (Kang et al., 1999; Kobayashi et al., 2003; Li et al., 2000; Mei et al., 2005; Yue et al., 2006). In barley, there were some reports concerning the relationships between yield and barley plant type, including flag leaf traits (Tungland et al., 1987; Yap and Harvey, 1972). However, genetic control of flag leaf traits has been rarely reported in barley up to date.
Gyenis et al.(2007) detected 30 QTLs controlling 13 morphological traits including flag leaves at 2 to 4 environments using a BC2F8 population from cultivated Harrington and wild accession OUH602 based on 113 SSR markers. They detected 3 QTLs for flag leaf length on chromosomes 3H, 5H and 7H, and 3 QTLs for flag leaf width on chromosomes 2H, 4H and 5H. In the present study, we detected 2 QTLs (qFL5 and qFL7) for flag leaf length on the chromosomes 5H and 7H, and 2 QTLs (qFW5.1 and qFW5.2) for leaf width on chromosome 5H. However, the QTLs controlling length on the chromosome 3H and the QTLs controlling width on chromosomes 2H and 4H were not found. Moreover, fine regions of these QTLs located on the same chromosomes cannot be compared due to the difference in parents, population types, and molecular markers used in the two studies. In addition, the QTL associated with FLW was detected on the same interval with qFL7, and was also supported by significantly positive correlation (r=0.781, P<0.01) between the two traits. The result indicates that close linkage or pleiotropy existed for the two traits, and thus the locus may be useful for marker-assistant selection (MAS) in barley improvement.
It was reported that leaf chlorophyll content was positively correlated with photosynthetic capacity (Araus et al., 1997) and high chlorophyll content in leaves was considered as a favorable trait in crop production (Teng et al., 2004). QTL for leaf chlorophyll content was rarely reported previously in barley. Chlorophyll content as a drought tolerance criterion was studied, and more than 10 QTLs were identified on chromosomes 2H, 4H, 5H, 6H and 7H using an RIL (recombinant inbred line) population under two water treatments (This et al., 2000). However, the measurement of chlorophyll content was conducted at early growth stage in these studies. Recently, Guo et al.(2008) detected 5 QTLs on chromosomes 2H and 4H associated with chlorophyll content in flag leaves at post-flowering stage under well-watered and drought conditions using an RIL population with 194 lines. In the present study, a total of 4 QTLs controlling FC were mapped on chromosomes 2H, 3H and 6H. However, no similar QTL was found in the current study as that reported previously, indicating that the QTLs detected in the present study were new QTLs for chlorophyll content of flag leaf. In addition, the QTL (qFC2.2) located within interval Bmag0518~Bmac0093 accounted for 20.2% of the phenotypic variation, indicating its importance in determining chlorophyll content of flag leaf and potential use in breeding programs. Moreover, it was found that the additive effects of the detected QTLs, except for qFC6, were positive, indicating that the favorable alleles from Yerong contribute to morphology and chlorophyll content of flag leaf for the DH population used in the current study.
Footnotes
Project supported by the National Natural Science Foundation of China (No. 30630047) and the Project on Absorption of Intellects by Institutions of Higher Education for Academic Disciplinary Innovations (the 111 Project) (No. B06014), China
References
- 1.Araus JI, Bort J, Ceccarelli S, Grando S. Relationship between leaf structure and carbon isotope discrimination in field grown barley. Plant Physiology and Biochemistry. 1997;35(7):533–541. [Google Scholar]
- 2.Backes G, Graner A, Foroughi-Wehr B, Fischbeck G, Wenzel G, Jahoor A. Localization of quantitative trait loci (QTL) for agronomic important characters by the use of an RFLP map in barley (Hordeum vulgare L.) Theoretical and Applied Genetics. 1995;90(2):294–302. doi: 10.1007/BF00222217. [DOI] [PubMed] [Google Scholar]
- 3.Baum M, Grando S, Backes G, Jahoor A, Sabbagh A, Ceccarelli S. QTLs for agronomic traits in the Mediterranean environment identified in recombinant inbred lines of the cross ‘Arta’×H. spontaneum 41-1. Theoretical and Applied Genetics. 2003;107(7):1215–1225. doi: 10.1007/s00122-003-1357-2. [DOI] [PubMed] [Google Scholar]
- 4.Bezant J, Laurie D, Pratchett N, Chojecki J, Kearsey M. Mapping QTLs controlling yield and yield components in a spring barley (Hordeum vulgare L.) cross using marker regression. Molecular Breeding. 1997;3(1):29–38. doi: 10.1023/A:1009648220852. [DOI] [Google Scholar]
- 5.Chen W, Xu Z, Zhang L. Physiological Bases of Super High Yield Breeding in Rice. Shenyang: Liaoning Science and Technology Publishing Company; 1995. pp. 1–2. (in Chinese) [Google Scholar]
- 6.FAO (Food and Agriculture Organization of the United Nations) FAOSTAT. 2005. (Available from: http://faostat.fao.org/)
- 7.Guo P, Baum M, Varshney R, Graner A, Grando S, Ceccarelli S. QTLs for chlorophyll and chlorophyll fluorescence parameters in barley under post-flowering drought. Euphytica. 2008;163(2):203–214. doi: 10.1007/s10681-007-9629-6. [DOI] [Google Scholar]
- 8.Gyenis L, Yun SJ, Smith KP, Steffenson BJ, Bossolini E, Sanguineti MC, Muehlbauer GJ. Genetic architecture of quantitative trait loci associated with morphological and agronomic trait differences in a wild by cultivated barley cross. Genome. 2007;50(8):714–723. doi: 10.1139/G07-054. [DOI] [PubMed] [Google Scholar]
- 9.Hirota O, Oka M, Takeda T. Sink activity estimation by sink size and dry matter increase during the ripening stage of barley (Hordeum vulgare) and rice (Oryza sativa) Annals of Botany. 1990;65(4):349–354. [Google Scholar]
- 10.Kang HJ, Cho YG, Lee YT, et al. Mapping QTLs for Flag Leaf Length and Width, Panicle Exertion Length, and Awn Length Using Recombinant Inbred Population of Rice (Oryza sativa L.). Plant & Animal Genome VII Conference; Town & Country Hotel, San Diego. 1999. p. 309. [Google Scholar]
- 11.Kjaer B, Jensen J. Quantitative trait loci for grain yield and yield components in a cross between a six-rowed and a two-rowed barley. Euphytica. 1996;90(1):39–48. [Google Scholar]
- 12.Kobayashi S, Fukuta Y, Morita S, Sato T, Osaki M, Khush GS. Quantitative trait loci affecting flag leaf development in rice (Oryza sativa L.) Breeding Science. 2003;53(3):255–262. doi: 10.1270/jsbbs.53.255. [DOI] [Google Scholar]
- 13.Li H, Vaillancourt RE, Zhou M, et al. Large-scale Segregation Distortion in Barley Revealed by a Genetic Map Based on DArT, AFLP and SSR Markers; Proceedings of the 13th Australasian Plant Breeding Conference; 18-21 April; Christchurch, New Zealand. 2006. pp. 660–665. [Google Scholar]
- 14.Li JZ, Huang XQ, Heinrich F, Ganal MW, Röder MS. Analysis of quantitative trait loci for yield, yield components and malting quality in a BC3-DH population of spring barley. Theoretical and Applied Genetics. 2005;110(2):356–363. doi: 10.1007/s00122-004-1847-x. [DOI] [PubMed] [Google Scholar]
- 15.Li JZ, Huang XQ, Heinrichs F, Ganal MW, Röder MS. Analysis of QTLs for yield components, agronomic traits, and disease resistance in an advanced backcross population of spring barley. Genome. 2006;49(5):454–466. doi: 10.1139/G05-128. [DOI] [PubMed] [Google Scholar]
- 16.Li SG, He P, Wang YP, Li HY, Chen Y, Zhou KD, Zhu LH. Genetic analysis and gene mapping of the leaf traits in rice (Oryza sativa L.) Acta Agronomica Sinica. 2000;26(3):261–265. (in Chinese) [Google Scholar]
- 17.Li Z, Pinson SRM, Stansel JW, Paterson AH. Genetic dissection of the source-sink relationship affecting fecundity and yield in rice (Oryza sativa L.) Molecular Breeding. 1998;4(5):419–426. doi: 10.1023/A:1009608128785. [DOI] [Google Scholar]
- 18.Marquez-Cedillo LA, Hayes PM, Kleinhofs A, Legge WG, Rossnagel BG, Sato K, Ullrich SE, Wesenberg DM. QTL analysis of agronomic traits in barley based on the doubled-haploid progeny of two elite North American varieties representing different germplasm groups. Theoretical and Applied Genetics. 2001;103(4):625–637. doi: 10.1007/PL00002919. [DOI] [Google Scholar]
- 19.Mather D, Tinker NA, LaBerge DE, Edney M, Jones BL, Rossnagel BG, Legge WG, Briggs KG, Irvine RB, Falk DE, et al. Regions of the genome that affect grain and malt quality in a North American two-row barley cross. Crop Science. 1997;37(2):544–554. [Google Scholar]
- 20.McCouch SR, Cho YG, Yano M, Paul E, Blinstrub M. Report on QTL nomenclature. Rice Genetics Newsletter. 1997;14:11–13. [Google Scholar]
- 21.Mei HW, Luo LJ, Ying CS, Wang YP, Yu XQ, Guo LB, Paterson AH, Li ZK. Gene actions of QTLs affecting several agronomic traits resolved in a recombinant inbred rice population and two backcross populations. Theoretical and Applied Genetics. 2005;110(4):649–659. doi: 10.1007/s00122-004-1890-7. [DOI] [PubMed] [Google Scholar]
- 22.Sameri M, Takeda K, Komatsuda T. Quantitative trait loci controlling agronomic traits in recombinant inbred lines from a cross of oriental- and occidental-type barley cultivars. Breeding Science. 2006;56(3):243–252. doi: 10.1270/jsbbs.56.243. [DOI] [Google Scholar]
- 23.Sharma SN, Sain RS, Sharma RK. The genetic control of the flag leaf length in normal and late sown durum wheat. The Journal of Agricultural Science. 2003;141(3-4):323–331. doi: 10.1017/S0021859603003642. [DOI] [Google Scholar]
- 24.Sicher RC. Assimilate PARTITIONIng within Leaves of Small Grain Cereals. In: Yash PA, Prasanna M, Govindjee D, editors. Photosynthesis Photoreactions to Plant Productivity. Dordrecht, the Netherlands: Kluwer Academic Publishers; 1993. pp. 351–360. [Google Scholar]
- 25.Teng S, Qian Q, Zeng D, Kunihiro Y, Fujimoto K, Huang D, Zhu L. QTL analysis of leaf photosynthetic rate and related physiological traits in rice (Oryza sativa L.) Euphytica. 2004;135(1):1–7. doi: 10.1023/B:EUPH.0000009487.89270.e9. [DOI] [Google Scholar]
- 26.This D, Borries C, Souyris I, Teulat B. QTL study of chlorophyll content as a genetic parameter of drought tolerance in barley. Barley Genetics Newsletter. 2000;30:20–23. [Google Scholar]
- 27.Thomas WTB, Powell W, Waugh R, Chalmers KJ, Barua UM, Jack P, Lea V, Forster BP, Swanston JS, Ellis RP, et al. Detection of quantitative trait loci for agronomic, yield, grain, and disease characters in spring barley (Hordeum vulgare L) Theoretical and Applied Genetics. 1995;91(6-7):1037–1047. doi: 10.1007/BF00223917. [DOI] [PubMed] [Google Scholar]
- 28.Thorne GN. Photosynthesis of ear and flag leaves of wheat and barley. Annals of Botany. 1965;29(3):317–329. [Google Scholar]
- 29.Tungland L, Chapko LB, Wiersma JV, Rasmusson DC. Effect of erect leaf angle on grain yield in barley. Crop Science. 1987;27(1):37–40. [Google Scholar]
- 30.Wang DL, Zhu J, Li ZK, Paterson AH. Mapping QTLs with epistatic effects and QTL×environment interactions by mixed linear model approaches. Theoretical and Applied Genetics. 1999;99(7-8):1255–1264. doi: 10.1007/s001220051331. [DOI] [Google Scholar]
- 31.Wang DL, Zhu J, Li ZK, et al. QTLMapper Version 1.6: A Computer Software for Mapping Quantitative Trait Loci (QTLs) with Additive Effects, Epistatic Effects and QTL×Environment Interactions. 2003. (Available from: http://ibi.zju.edu.cn/software/qtlmapper/index.htm)
- 32.Wenzl P, Li H, Carling J, Zhou M, Raman H, Paul E, Hearnden P, Maier C, Xia L, Caig V, et al. A high-density consensus map of barley linking DArT markers to SSR, RFLP and STS loci and agricultural traits. BMC Genomics. 2006;7(1):206. doi: 10.1186/1471-2164-7-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang YF, Lu DZ. Genetic analysis on morpho-physiological traits of barley flag leaf. Scientia Agricultura Sinica. 1991;24(1):20–26. (in Chinese) [Google Scholar]
- 34.Yap TC, Harvey RL. Relations between grain yield and photosynthetic parts above the flag leaf node in barley. Canadian Journal of Plant Science. 1972;52:241–246. [Google Scholar]
- 35.Yin X, Stam P, Johan Dourleijn C, Kropff MJ. AFLP mapping of quantitative trait loci for yield-determining physiological characters in spring barley. Theoretical and Applied Genetics. 1999;99(1-2):244–253. doi: 10.1007/s001220051230. [DOI] [Google Scholar]
- 36.Yue B, Xue WY, Luo LJ, Xing YZ. QTL analysis for flag leaf characteristics and their relationship with yield and yield traits in rice. Acta Genetica Sinica. 2006;33(9):824–832. doi: 10.1016/S0379-4172(06)60116-9. [DOI] [PubMed] [Google Scholar]
- 37.Zheng T. Effects of some photosynthetic organs on milking and grain yield of barley. Barley Science. 1999;1:21–22. (in Chinese) [Google Scholar]