Abstract
Ogre elements are a distinct group of plant Ty3/gypsy-like retrotransposons characterized by several specific features, one of which is a separation of the gag-pol region into two non-overlapping open reading frames: ORF2 coding for Gag-Pro, and ORF3 coding for RT/RH-INT proteins. Previous characterization of Ogre elements from several plant species revealed that part of their transcripts lacks the region between ORF2 and ORF3, carrying one uninterrupted ORF instead. In this work, we investigated a hypothesis that this region represents an intron that is spliced out from part of the Ogre transcripts as a means for preferential production of ORF2-encoded proteins over those encoded by the complete ORF2–ORF3 region. The experiments involved analysis of transcription patterns of well-defined Ogre populations in a model plant Medicago truncatula and examination of transcripts carrying dissected pea Ogre intron expressed within a coding sequence of chimeric reporter gene. Both experimental approaches proved that the region between ORF2 and ORF3 is spliced from Ogre transcripts and showed that this process is only partial, probably due to weak splice signals. This is one of very few known cases of spliced LTR retrotransposons and the only one where splicing does not involve parts of the element’s coding sequences, thus resembling intron splicing found in most cellular genes.
Electronic supplementary material
The online version of this article (doi:10.1007/s00438-008-0376-8) contains supplementary material, which is available to authorized users.
Keywords: Retroelements, Transcription, Splicing, Medicago truncatula, Pisum sativum
Introduction
Long terminal repeat (LTR) retrotransposons represent a group of mobile genetic elements characterized by a replicative (copy-and-paste) mode of transposition involving transcription of the parental element, reverse transcription of the resulting RNA into DNA, and subsequent integration of a new element into the genome. The LTRs, which contain regulatory sequences for transcription, flank the internal region (gag-pol) encoding proteins with structural or enzymatic functions. The gag gene codes for proteins needed for an assembly of virus-like particles and RNA packaging. The pol gene encodes enzymes protease (Pro), reverse transcriptase/RNaseH (RT/RH) and integrase (INT). RT/RH and INT convert the retrotransposon RNA into DNA and integrate it into the genome, respectively. Translation of the gag-pol region is initiated from a single site on full-length RNA and individual functional proteins are released from a precursor polyprotein by the action of protease (Kumar and Bennetzen 1999; Havecker et al. 2004).
While the gag-pol genes are common to all autonomous LTR retrotransposons, there are differences in the structure of their coding regions, which are arranged in single or multiple (overlapping or adjacent) reading frames. Since the structural proteins encoded in the gag region are required in higher numbers than the catalytic proteins encoded in pol, retroelements have developed several mechanisms permitting expression of the Gag protein at higher levels relative to Pol. In the case of translation of the whole polyprotein from a single reading frame, all proteins are produced in equal amounts and their proper ratio can be reached by post-translational degradation of Pol, as observed with yeast retrotransposons Tf1 and Ty5 (Atwood et al. 1996; Irwin and Voytas 2001). On the other hand, division of the gag-pol region into two reading frames suggests the use of translational recoding mechanisms including ribosomal frameshifting and stop codon bypass (Gao et al. 2003; Forbes et al. 2007).
A unique arrangement of the gag-pol region has been described for Ogre elements, a family of LTR retrotransposons occurring in several genera of dicot plants where they often constitute a major fraction of repetitive DNA (Neumann et al. 2003; Neumann et al. 2006; Macas and Neumann 2007). Ogre elements represent a distinct group of Ty3/gypsy-like retrotransposons characterized by the extreme size of the elements (up to 25 kb, with LTRs up to 6 kb), PBS complementary to tRNAarg, the presence of an extra open reading frame (ORF1) coding for an unknown protein upstream of gag-pol, and division of the gag-pol region into two ORFs. The gag-pro domains (ORF2) are separated from rt/rh-int (ORF3) by a region of about 150–350 bp, which includes several stop codons and is surrounded by GT/AG dinucleotides typical of the 5′ and 3′ termini of most introns (Breathnach et al. 1978; Mount 1982; Burset et al. 2000). Although the nucleotide sequences of this region differ between Ogre elements from various plant species, its position within gag-pol and the GT/AG boundaries are conserved. Moreover, removing the region including these boundaries leads to in frame fusion of gag-pro and rt/rh-int, enabling correct translation of the latter domains. Thus, it has been proposed that this region represents an intron that is removed by splicing to reconstitute the full-length gag-pol coding region (Neumann et al. 2003).
Although the splicing has been well documented for some groups of retroelements like retroviruses and LINEs (Rabson and Graves 1997; Belancio et al. 2006; Tamura et al. 2007), it has so far been reported for only a few LTR retrotransposons. It occurs in transcripts of the envelope-class retrotransposon Bagy-2 where it generates a subgenomic RNA lacking almost the entire gag-pol sequence, thus enabling expression of the downstream env gene (Vicient et al. 2001). Alternative splicing of RNA from Drosophila retrotransposon copia was shown to be involved in the regulation of the ratio between Gag and Pol proteins, as the full-length copia RNA containing gag and pol regions is translated to protein at a far lower level than spliced subgenomic RNA encoding gag products only (Brierley and Flavell 1990). In contrast to these cases where splicing always removes part of the coding region, the putatively spliced region within Ogre transcripts does not include any coding sequence.
Our previous data from pea (Pisum sativum) showed that Ogre sequences are transcribed in leaves, roots and flowers and that a significant portion of the transcripts lacks the putative intron sequence (Neumann et al. 2003). However, since there is a small fraction of Ogre copies in the pea genome that also lacks this region, whether the shorter transcripts are produced from these elements instead of the splicing of full-length RNA could not be ruled out. Thus, in this work we investigated transcription and processing of Ogre RNA in more detail employing two different, yet complementary strategies to study this phenomenon: (1) Taking advantage of the available genomic sequence from the model plant Medicago truncatula and of our previous characterization of the Ogre population in this species (Macas and Neumann 2007), we followed transcription patterns of individual Ogre subfamilies using RT-PCR with specific sets of primers. This sequence-specific assay enabled us to exclude the presence of certain spliced Ogre sequences in the genome and thus to detect splicing of the corresponding full-length transcripts. (2) Secondly, the splicing of the putative intron sequence from pea Ogre elements was investigated by its incorporation into the coding sequence of the GFP-GUS reporter gene and expression in transgenic plant tissue. Both these approaches demonstrated that the putative intron sequence can be spliced from the Ogre transcripts but that the splicing is only partial, presumably due to weak splicing signals within the Ogre sequence.
Materials and methods
Plant material and nucleic acid isolation
Seeds of P. sativum L. cv. Carrera were obtained from the Plant Breeding Station at Boršov, Czech Republic. Seeds of M. truncatula cv. Jemalong were provided by the Crop Research Institute at Praha-Ruzyně, Czech Republic. Genomic DNA was extracted from leaves or hairy roots as described by Dellaporta et al. (1983). All DNA concentration measurements were done with PicoGreen dye (Molecular Probes) according to the manufacturer’s instructions. Total RNA was isolated using a ToTALLY RNA Kit (Ambion). RNA isolates were treated with TURBO DNase (TURBO DNA-free kit, Ambion) to remove traces of contaminant DNA.
Sequence analysis
Nucleotide and protein sequence analysis was done using Staden Package software (Staden 1996) and program tools implemented at the Biology workbench server (http://workbench.sdsc.edu). Multiple sequence comparisons were performed with CLUSTALW (Thompson et al. 1994). Splice site analysis was performed at the NetGene2 server (Hebsgaard et al. 1996; http://www.cbs.dtu.dk/services/NetGene2).
Splicing analysis in Medicago truncatula
Transcription profiles of individual Ogre subfamilies were investigated using RT-PCR with subfamily-specific primers designed according to the multiple sequence alignment of previously characterized elements (Macas and Neumann 2007; see Supplementary Fig. S1 for the alignment and primer positions). The following primer pairs were used for individual subfamilies (labeled as MT1–MT4): MT1F (5′ GAC ATT CCY TCA ATC ATG CAT G 3′), MT1R (5′ GAC CAT GCA AAA ATA TCC GG 3′), MT2F (5′ AGA RGA ATG AAG CCT CTA TCT 3′), MT2R (5′ TTT GAG RAG CTC TAT CAC TTG 3′), MT3F (5′ ACT ACA AGA GGT GAA ACA TGC 3′), MT3R (5′ ATT ATC TTT TTC TTG ACA GAC GC 3′), MT4F (5′ TCA TTC CAA GGR TTC TCT GC 3′), MT4R (5′ CTC AGT ACC CAG ATT GAT GAT T 3′). Reverse transcription was carried out using Superscript First-Strand Synthesis System for RT-PCR (Invitrogen) and 4 μg of the template RNA with either reverse (R) or forward (F) primer in order to specifically detect sense or antisense transcripts, respectively. Resulting cDNA was amplified in a subsequent PCR reaction using a 1 μl aliquot of the RT reaction as a template. The PCR was performed in a 50 μl volume of 1× PCR buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.2 μM primers and 2 U of Platinum Taq polymerase (Invitrogen). The PCR profile included initial denaturation (2 min at 94°C), 35 cycles of denaturation (94°C for 1 min), primer annealing (1 min) and extension (72°C for 1 min), followed by final extension at 72°C for 10 min. The annealing temperature was 60°C in the first cycle and was decreased by 1°C at each subsequent cycle down to 50°C, which was used for the rest of the reaction. The RT-PCR products were analyzed by agarose gel electrophoresis, cloned into pCR4-TOPO vector (Invitrogen) and sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). The sequences are available from GenBank under accession numbers FK700536–FK700710.
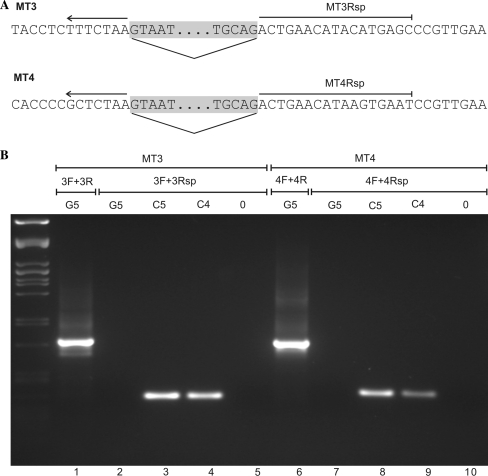
Verification of the absence of intron-less elements in the M. truncatula genome corresponding to spliced Ogre transcripts was performed using PCR with primers specific for subfamilies MT3 and MT4. The reverse primers MT3Rsp (5′ GCT CAT GTA TGT TCA GTT TAG AAA 3′) and MT4Rsp (5′ ATT CAC TTA TGT TCA GTT TAG AGC 3′) spanning the spliced intron position (Fig. 2a) were used in combination with the respective forward primer (MT3F or MT4F). The PCR profile comprised initial denaturation (2 min at 94°C), 35 cycles of 94°C for 1 min, 60°C for 1 min and 72°C for 1 min, followed by final extension at 72°C for 10 min. As templates we used genomic DNA or cloned spliced Ogre cDNA sequences representative of the MT3 (clone 1223, acc. no. FK700657) and MT4 (clone 1277, acc. no. FK700706) subfamilies. The amount of template genomic DNA (48 ng) corresponded to 105 equivalents of haploid M. truncatula genome (1C = 0.48 pg, Arumuganathan and Earle 1991), and the control cDNA templates were used in amounts corresponding to 105 and 104 copies/reaction. The control reactions provided a calibration of PCR sensitivity: successful detection of the expected fragment in the reaction using 105 copies of the control template indicated that the reaction sensitivity was sufficient for detection of a single copy target sequence in 48 ng of M. truncatula genomic DNA, and obtaining the product in the control with 104 template copies per reaction corresponded to about a tenfold higher sensitivity.
Fig. 2.
Investigation of the presence of spliced Ogre copies in the M. truncatula genome. The detection was done using PCR with forward primers specific for either MT3 (primer MT3F) or MT4 (primer MT4F) elements in combination with the respective reverse primer (MT3Rsp or MT4Rsp) shown in panel a The reverse primers spanned regions adjacent to both sides of the intron (gray shading), so it could efficiently anneal only to the spliced sequence variants. b PCR reactions were performed with defined amounts of genomic DNA or control templates in order to confirm that sensitivity was sufficient to detect a single copy of spliced sequence in the genome. Therefore, the amplification using 48 ng of M. truncatula DNA (lanes G5), corresponding to 105 genome equivalents (1C = 0.48 pg), was compared to the reactions including 105 and 104 molecules of control templates (lanes C5 and C4, respectively). Cloned fragments of spliced transcripts were used as the controls (the clone c1223 was used for the MT3 and c1227 for the MT4 subfamily). No amplified fragments were observed in the genomic DNA samples (lanes 2 and 7), whereas the sensitivity of the assay was sufficient to detect even 104 of template molecules (lanes 4 and 9). Lanes 1 and 6 show positive controls (genomic DNA amplified with primers surrounding the intron), lanes 5 and 10 contain reactions with no template (0)
Construction of a reporter gene containing the Ogre intron and its expression in pea hairy root culture
The intron sequence of pea Ogre element was obtained from P. sativum genomic clone Ps-phage20 (Neumann et al. 2003; GenBank accession AY299395, positions 10,863–11,115) by PCR amplification using primers IPS-F (5′ GTA ATT TTC TGT TGT TTT 3′) and IPS-R (5′ CTG CAT GTT TAA TGC 3′). The amplified fragment was cloned into the unique SnaBI restriction site within GUS coding sequence of the plasmid pBGF-D35S, a pBin19-based binary vector containing chimeric NLS-sGFP-GUS gene expressed using a double 35S promoter (Chytilova et al. 1999). The intron PIV2 which was previously employed to disrupt GUS coding sequence (Vancanneyt et al. 1990) was used to prepare a control construct by amplifying the PIV2 sequence using the primers T1 (5′ GTA AGT TTC TGC TTC TAC CTT TGA 3′) and T2 (5′ CTG CAC ATC AAC AAA TTT TG 3′) and cloning it into the same position within the GUS sequence as the Ogre intron. Nucleotide sequences of both constructs were verified by sequencing.
Transgenic hairy root cultures expressing the intron-containing GUS sequences were obtained by transformation of P. sativum plants by Agrobacterium tumefaciens C58C1 carrying hairy root inducing plasmid pRiA4 together with either of the constructs. The transformation was performed by injecting Agrobacterium suspension into stems of 7-day-old seedlings cultivated in vitro on 50% Murashige and Skoog medium (Duchefa). The seedlings were grown at 20°C for 2 weeks (16 h photoperiod) and than transferred to a 25°C growth chamber with identical light conditions. After 2–3 weeks of cultivation, hairy roots emerging from the inoculation sites were excised and placed on solid Gamborg B5 medium (Duchefa) supplemented with ticarcillin (500 mg/l) and cefotaxime (200 mg/l) for elimination of bacteria, and kanamycin (50 mg/l) for selection of lines carrying the GUS constructs. Hairy root cultures were grown in Petri dishes at 24°C in the dark and transferred to fresh B5 medium once a month.
The presence of the GUS transgene in kanamycin-resistant lines was verified using PCR with primers GUS-F (5′ AAC TCG ACG GCC TGT GGG C 3′) and GUS-R (5′AGT TCA ACG CTG ACA TCA CC 3′) designed for amplification of the GUS coding sequence surrounding the intron cloning site. Parallel PCR reactions designed to detect eventual Agrobacterium contamination of analyzed hairy root cultures were also run using the primers specific for the vector sequence (BIN-F: 5′ GCA TCA GGC TCT TTC ACT CC 3′; BIN-R: 5′ TCA AAC GTC CGA TTC ATT CA 3′). Selected hairy root lines were subjected to RNA extraction and reverse transcription reactions (using oligo-dT primer) as described above. The PCR reaction employed for intron splicing detection using primers GUS-F and GUS-R included initial denaturation (2 min at 94°C), 35 cycles of 1 min at 94°C, 1 min at 55°C and 1 min at 72°C, and a final extension step (10 min at 72°C) using a 1 μl aliquot of the RT reaction as a template. Proportions of amplified fragments corresponding to unspliced and spliced transcripts were estimated by quantification of band intensities on ethidium bromide-stained agarose gels using BioCapt program (ver. 11, Vilber Lourmat). The proportions were calculated as percentages of unspliced and spliced molecules within the samples, taking into account band intensities (DNA amounts within the bands) and length of the amplified transcripts.
Polyribosome isolation and analysis
Polyribosome isolation was performed according to Davies et al. (1972) and Jackson and Larkins (1976) with certain modifications. 1 g of hairy roots was frozen in liquid nitrogen and ground to a fine powder with a mortar and pestle. The powder was thawed in polysome extraction buffer (0.2 M sucrose, 0.2 M Tris–HCl pH 8.5, 0.4 M KCl, 35 mM MgCl2, 25 mM EGTA, 5 mM DTT) and the mixture was gently homogenized. The homogenate was strained through sterile cheesecloth and the filtrate was centrifuged at 2,000 g for 5 min at 4°C. The supernatant was adjusted to 1% (v/v) for Triton X-100 and centrifuged at 30,000g for 10 min at 4°C. The supernatant was then layered on a discontinuous gradient consisting of one volume of 2 M sucrose and one volume of 1.5 M sucrose. Polyribosomes were pelleted by centrifugation for 5 h at 200,000g at 4°C. RNA extraction from pellet and supernatant was performed using a ToTALLY RNA Kit (Ambion). Reverse transcription using oligo-dT primer and PCR detection of transgene transcripts using GUS-F and GUS-R primers were carried out as described above. Pea actin transcripts were detected using primers actin-F (5′ CCC TAA GGC TAA TCG TGA GA 3′) and actin-R (5′ ATA TTC TGC CTT TGC AAT CC 3′) with the same PCR profile.
Results
Transcription patterns and splicing of Ogre elements in Medicago truncatula
The population of Ogre elements in the M. truncatula genome can be classified into four basic subfamilies (MT1–MT4) based on the divergence of their non-coding sequences. A set of 85 previously identified full-length elements representing these subfamilies (Macas and Neumann 2007) was used to design subfamily-specific pairs of primers targeted to amplification of regions surrounding a putative intron (Supplementary Fig. S1). RT-PCR reactions using these primers and carried out with RNA extracted from different organs (roots, leaves, flowers) revealed variations in transcription patterns between individual subfamilies. While the majority of MT1 and MT2 transcripts corresponded to unspliced sequences, there were similar proportions of unspliced and spliced transcripts detected in MT3 and MT4 subfamilies, especially in the root RNA samples (Fig. 1). Cloning and sequencing of RT-PCR products from this tissue confirmed the specificity of primers used for each respective subfamily and revealed that shorter products indeed corresponded to transcripts spliced at the predicted donor and acceptor sites. This was the case for all fragments corresponding to the band labeled as “S” on Fig. 1, sequenced from the MT1 (23 clones), MT2 (4 clones), MT3 (16 clones) and MT4 (28 clones) subfamilies. There was an additional shorter band (labeled as “Sx” on Fig. 1) detected in subfamily MT3 that was predominantly (13 out of 14 sequenced clones) composed of Ogre-like transcripts with relatively divergent sequence compared to known elements (similarity of 78% or lower). Due to this sequence divergence it was not possible to determine whether this sequence was spliced or merely transcribed from an element containing an internal deletion. The remaining clone sequenced from this RT-PCR band corresponded to an aberrantly spliced MT3 element. We also tested for the presence of antisense transcripts in the MT3 and MT4 subfamilies (Fig. 1) by performing reverse transcription using the corresponding forward instead of reverse primers employed in standard RT-PCR reactions. Although the antisense transcripts were detected, they corresponded to unspliced sequences only, thus providing additional evidence that shorter fragments are generated by splicing, which acts on the sense transcripts containing the intron sequence in proper orientation.
Fig. 1.
Transcription analysis of Ogre subfamilies MT1-MT4 in M. truncatula. RT-PCR reactions were performed with total RNA isolated from flowers (F), leaves (L) or roots (R) and primers specific for individual subfamilies (+ and − indicate the presence or absence of reverse transcriptase in otherwise identical RT reactions). The primers specific for individual subfamilies were designed to amplify a region between ORF2 and ORF3 including the potential intron (Supplementary Fig. S1). Positions of amplified full-length (unspliced) fragments on the gels are indicated with U, and their spliced variants are marked with S or Sx. The lanes marked G and 0 include control PCR reactions with genomic DNA and no template, respectively, and lane M is the DNA size marker (lambda DNA digested with PstI). The lanesAS in panels MT3 and MT4 show detection of antisense transcripts from roots (using forward primers for RT reaction), whereas all other RT reactions were performed using reverse primers, thus detecting the sense transcripts. Sequences of RT-PCR products are available from GenBank under accession numbers FK700536–FK700710
PCR reactions using genomic DNA as a template (Fig. 1) as well as bioinformatic analysis of M. truncatula genomic data revealed that a small fraction of MT1 and MT2 elements present in the genome lacks the intron sequence. Thus, whether the spliced fragments detected by RT-PCR actually represented transcripts of these elements instead of the products of full-length RNA splicing could not be ruled out. Although the intron-lacking elements were not found for subfamilies MT3 and MT4 by either of the two approaches, this may be due to incomplete genomic sequence available for analysis and insufficient sensitivity of the PCR assay. Therefore, we designed primers spanning the intron site that should selectively amplify only the spliced targets (Fig. 2a) and we calibrated the PCR sensitivity using known amounts of the template copies in order to ensure that it was sufficient to detect even a single copy of the corresponding spliced element in the M. truncatula genome. This assay confirmed that there are no spliced copies of these elements present in the genome (Fig. 2b) and thus that the intron-less transcripts detected for MT3 and MT4 subfamilies originated by splicing their full-length RNA.
Splicing analysis of pea Ogre intron expressed within GUS coding sequence
An alternative approach for functional analysis of the predicted intron sequence was employed for Ogre elements from pea (P. sativum). The intron region with the highest prediction score as calculated at the NetGene2 server (confidence values of 0.96 for both donor and acceptor splice sites, branch point score of −0.95) was selected from the available pea Ogre clones (Neumann et al. 2003) and subcloned into a coding region of the GUS reporter gene. This chimeric sequence was expressed in transgenic pea hairy root cultures and its splicing was studied using RT-PCR with primers surrounding the cloning site. A modified intron 2 of potato ST-LS1 gene (PIV2 intron, Vancanneyt et al. 1990) cloned into the same position within the GUS sequence and expressed in the same way as the Ogre construct was used as a control (Fig. 3a).
Fig. 3.
Splicing of intron-containing GUS constructs expressed in transgenic pea hairy root lines. a Schematic representation of GUS coding sequence (gray boxes) containing intron from pea Ogre element or a control intron PIV2 (white boxes). Authentic donor and acceptor sites are connected with solid lines, and the alternative splicing involving the cryptic donor site within the GUS sequence is marked with dashed lines. b Splicing patterns detected in transgenic lines using RT-PCR with primers GUS-F and GUS-R surrounding the intron cloning site within the GUS sequence. Fragment sizes corresponding to spliced (S) and unspliced (U) transcripts are indicated. RT-PCR reactions performed with RNA from five independent lines expressing pea Ogre intron are shown (marked as Ogre) along with the line expressing the control intron (PIV2). The lanes marked with + and – indicate the presence or absence of reverse transcriptase in RT reactions. The lane GUS + int shows the control PCR amplification of the GUS construct containing the Ogre intron. The marker is lambda DNA digested with PstI. Proportions of unspliced and spliced transcripts (in %) within the RT + samples are given below the gel
Splicing of the Ogre intron from the chimeric GUS transcripts was evident in four out of five lines of transgenic hairy roots tested (Fig. 3b). The only line where almost no spliced transcripts were detected (PO25b) showed reduced growth rate and was viable for only a few months. In the other four lines the expression patterns varied considerably and the splicing was always only partial, although in two of them (PO10d and PO25d) the spliced transcripts clearly prevailed. On the other hand, the splicing in the line expressing GUS with the control intron PIV2 was complete and no unspliced transcripts were detected (Fig. 3b, line PIV2).
Splicing accuracy was checked by cloning and sequencing RT-PCR-amplified spliced transcripts from lines PO25d (47 clones), PO23d (19 clones), PO10d (9 clones) and PO5a (7 clones). Surprisingly, from a total of 82 analyzed sequences only two clones from the line PO23d contained transcripts spliced at the expected donor splice sites of the Ogre intron, whereas other clones were spliced using a cryptic donor site occurring within the GUS sequence four nucleotides upstream of the Ogre intron (Fig. 3a). On the contrary, all 36 sequenced fragments from the control PIV2 line were spliced at the PIV2 intron acceptor and donor sites.
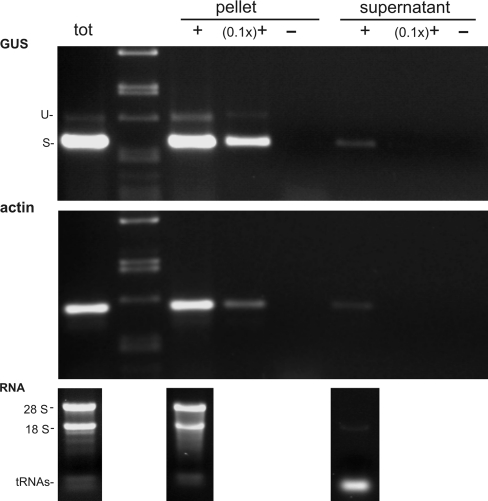
Intron-containing RNAs can be retained in the nucleus to await splicing or degradation or they can represent a type of alternative splicing—intron retention. Thus, we investigated whether the unspliced transcripts occur outside the nucleus by testing for their presence in the mRNA fraction associated with polyribosomes. In all three tested transgenic lines (PO5a, PO10d, PO23d), the same ratios of spliced to unspliced transcripts were observed when RT-PCR reactions with total cellular or polyribosome-associated RNA samples were compared (an example is shown in Fig. 4). These findings suggest that transcripts retaining the intron are exported from the nucleus at a similar rate as the spliced ones and are also associated with polyribosomes.
Fig. 4.
Detection of polyribosome-associated RNA. Cytoplasmatic extract from hairy root line PO10d was loaded onto a sucrose cushion gradient and RNA was extracted from the resulting polysomal pellet and corresponding supernatant. The RNA panel shows that large and small ribosomal RNAs (28S and 18S) were effectively concentrated in the pellet, indicating the presence of polyribosomes. RNA was used for the reverse transcription reaction and the subsequent PCR was performed with undiluted (+) or 10× diluted [(0.1x)+] cDNA with primers GUS-F and GUS-R (unspliced and spliced transcripts are indicated as U and S, respectively) or act-F and act-R designed for amplification of part of the actin transcript (used as a control). Eventual false-positive results caused by contamination with genomic DNA were excluded in the RT− reaction (–). The result of RT-PCR with total RNA is also presented (tot). The marker is lambda DNA digested with PstI
Discussion
The results obtained using two different experimental approaches concurrently demonstrated that Ogre LTR retrotransposons possess functional introns, non-coding regions that are spliced out from the element transcripts. It has been shown that in spite of the presence of intron-less Ogre copies in the M. truncatula genome, at least some of the transcripts present in the cells originate by splicing of the RNA expressed from intron-containing elements. The ratio of spliced to unspliced transcripts varied between Ogre subfamilies, being the highest in MT3 and the lowest in MT2. As all subfamilies include large proportions of recently transposed elements (Macas and Neumann 2007) carrying intact donor and acceptor splice sites (Supplementary Fig. S1), this variation was probably not caused by differences in sequence degeneration of the elements. On the other hand, there are differences in sequence composition of introns and their adjacent regions between the subfamilies (Supplementary Fig. S1) which could provide more likely explanation for their different splicing efficiencies (Buratti and Baralle 2004). However, it should be noted that in some cases the splicing patterns of the same subfamily differed between the organs (for example, MT4 in roots vs. leaves/flowers, Fig. 1), suggesting that some tissue-specific factors could also influence the proportion of spliced transcripts.
Investigation of the dissected Ogre intron sequence expressed within the GUS gene also confirmed its splicing, although this process was incomplete in all transgenic lines tested. Moreover, it was found to predominantly involve a cryptic donor site within the surrounding sequence instead of the one in the investigated intron. These results can be directly compared to those reported by Ibrahim et al. (2001) who used the same experimental strategy for investigation of potato ST-LS1 and pea legumin introns. They showed that the splicing accuracy of investigated introns differed when expressed in transgenic plants or protoplasts and that it depended to a lesser extent on the intron sequence. Although the activation of the same GUS cryptic splice site as in the Ogre intron was also observed, the authentic splice sites were always utilized preferentially. The difference in splicing accuracy was even more evident in comparison with the intron PIV2 (a derivative of ST-LS1 intron, Vancanneyt et al. 1990) used as a control in our experiments, which was spliced correctly in all analyzed transcripts. Based on these observations we conclude that the splice signals within the Ogre intron are less efficient compared to the other introns investigated. It is also possible that efficiency of the authentic donor site is in part determined by Ogre sequences surrounding its intron, which were missing in the GUS construct. This is in agreement with our observation that the splicing of Ogre intron is correct when present within the element sequence. Nevertheless, the weak splice signals probably contribute to only partial splicing of the corresponding transcripts.
Transcript splicing is known to occur in several groups of retroelements, including retroviruses, LINEs, and Penelope-like elements (Rabson and Graves 1997; Tamura et al. 2007; Arkhipova et al. 2003), but it has been reported for only a few LTR retrotransposons. A retrovirus-like element Bagy-2 in barley employs splicing to generate subgenomic transcripts lacking most of its coding region in order to express the env gene located at its 3′ end (Vicient et al. 2001). A similar strategy, but involving removal of the pol coding region from part of the transcripts, is used by the copia element in Drosophila in order to regulate the ratio of Gag and Pol proteins produced (Brierley and Flavell 1990). Alternative splicing as a mechanism of Gag to Pol ratio regulation was also proposed for CRR, a Ty3/gypsy-like centromeric retrotransposon of rice. In spliced transcripts of these elements the removal of the RT-coding domain together with alternative usage of several different donor splice sites suppresses translation of the pol region while gag-pro domains remain unaffected (Neumann et al. 2007). Splicing as a mechanism for modulation of the proportion of element-encoded proteins can also be envisioned for Ogre retrotransposons investigated in this work. Removal of the sequence containing several stop codons between protease and reverse transcriptase domains allows translation of the rt/rh-int region in frame with the upstream gag-pro sequence, while the unspliced transcripts would be translated into gag-pro proteins only. It should be noted that in this case the splicing does not involve removal of any coding sequence, contrary to the cases of other spliced LTR retrotransposons described above. Thus, the intron in Ogre elements closely resembles those occurring in most genes.
The discovery of the functional intron within the Ogre sequence raises several interesting questions regarding the replication strategy of this element. In a similar way to other retroelements, Ogre is supposed to use its transcripts as a replication intermediate, serving as a template for reverse transcription and reintegration of new elements into the genome. The results of our experiments suggested that both full-length and spliced transcripts are present in the cytoplasm and thus could potentially serve as a replication template. However, replication of the spliced transcripts should lead to gradual replacement of intron-containing elements with their spliced variants during genome evolution, as there is no reverse mechanism available. Although the spliced Ogre copies have been detected in genomes of several investigated species, they represented only a minority of the Ogre populations. In pea, there are only about 1.5% of spliced elements (Neumann et al. 2003) and similar or smaller proportions of spliced copies were estimated for Vicia pannonica (our unpublished data). Using bioinformatics analysis we estimated that there were 3.2% of spliced elements in the M. truncatula genome (data not shown). Thus, there is probably some mechanism favoring intron-containing transcripts as a replication template or preventing reintegration of spliced copies into the genome. The mechanism of recognition of full-length RNA for encapsidation has been well studied in retroviruses. In general the RNA sequences necessary for RNA encapsidation, which are usually present only in the unspliced genomic RNA, are recognized by the viral unprocessed Gag polyprotein (Jewell and Mansky 2000). Thus we can speculate of the presence of some signal within the Ogre intron sequence that would allow the integration of unspliced copies only into the genome. On the other hand, the presence of intron-less Ogre copies in the genome suggests that this selection is not complete. This is in agreement with observations reported from retrovirus HIV-1, where spliced RNAs were found to be at a very low frequency packaged into the virus particles, in spite of the lack of corresponding signals (Luban and Goff 1994; Houzet et al. 2007b), and can also be reverse transcribed into cDNAs (Liang et al. 2004; Houzet et al. 2007a).
Splicing of the intron sequence separating the ORF2 and ORF3 reported in this work is only one of the features that distinguishes Ogre elements from other groups of LTR retrotransposons. The other one is an additional open reading frame (ORF1) upstream of the gag-pol ORFs, coding for a protein with unknown function that is present in all Ogre copies identified so far (Macas and Neumann 2007). Moreover, the complex structure of the Ogre sequence, including multiple ORFs, raises questions about the mechanisms facilitating translation of the ORFs downstream from ORF1. As the Ogre populations in several investigated species include intact, recently transposed elements that are also transcriptionally active (Neumann et al. 2003, 2006; Macas and Neumann 2007), there is the potential to study these aspects of Ogre structure by experimental approaches similar to those described in this paper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Ms. J. Látalová and Ms. H. Štěpančíková for excellent technical assistance. This work was supported by grants IAA500960702 and AVOZ50510513 from the Academy of Sciences of the Czech Republic, and LC06004 from the Ministry of Education, Youth and Sport of the Czech Republic.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s00438-008-0376-8) contains supplementary material, which is available to authorized users.
References
- Arkhipova IR, Pyatkov KI, Meselson M, Evgen’ev MB (2003) Retroelements containing introns in diverse invertebrate taxa. Nat Genet 33:123–124 [DOI] [PubMed]
- Arumuganathan K, Earle E (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9:208–218 [DOI]
- Atwood A, Lin JH, Levin HL (1996) The retrotransposon Tf1 assembles virus-like particles that contain excess Gag relative to integrase because of a regulated degradation process. Mol Cell Biol 16:338–346 [DOI] [PMC free article] [PubMed]
- Belancio VP, Hedges DJ, Deininger P (2006) LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res 34:1512–1521 [DOI] [PMC free article] [PubMed]
- Breathnach R, Benoist C, O’Hare K, Gannon F, Chambon P (1978) Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc Natl Acad Sci USA 75:4853–4857 [DOI] [PMC free article] [PubMed]
- Brierley C, Flavell AJ (1990) The retrotransposon copia controls the relative levels of its gene products post-transcriptionally by differential expression from its two major mRNAs. Nucleic Acids Res 18:2947–2951 [DOI] [PMC free article] [PubMed]
- Buratti E, Baralle FE (2004) Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol 24:10505–10514 [DOI] [PMC free article] [PubMed]
- Burset M, Seledtsov IA, Solovyev VV (2000) Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res 28:4364–4375 [DOI] [PMC free article] [PubMed]
- Chytilova E, Macas J, Galbraith DW (1999) Green fluorescent protein targeted to the nucleus, a transgenic phenotype useful for studies in plant biology. Ann Bot 83:645–654 [DOI]
- Davies E, Larkins BA, Knight RH (1972) Polyribosomes from peas: an improved method for their isolation in the absence of ribonuclease inhibitors. Plant Physiol 50:581–584 [DOI] [PMC free article] [PubMed]
- Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 1:19–21 [DOI]
- Forbes EM, Nieduszynska SR, Brunton FK, Gibson J, Glover LA, Stansfield I (2007) Control of gag-pol gene expression in the Candida albicans retrotransposon Tca2. BMC Mol Biol 8:94. doi:10.1186/1471-2199-8-94 [DOI] [PMC free article] [PubMed]
- Gao X, Havecker ER, Baranov PV, Atkins JF, Voytas DF (2003) Translational recoding signals between gag and pol in diverse LTR retrotransposons. RNA 9:1422–1430 [DOI] [PMC free article] [PubMed]
- Havecker ER, Gao X, Voytas DF (2004) The diversity of LTR retrotransposons. Genome Biol 5:225. doi:10.1186/gb-2004-5-6-225 [DOI] [PMC free article] [PubMed]
- Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouze P, Brunak S (1996) Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res 24:3439–3452 [DOI] [PMC free article] [PubMed]
- Houzet L, Morichaud Z, Mougel M (2007a) Fully-spliced HIV–1 RNAs are reverse transcribed with similar efficiencies as the genomic RNA in virions and cells, but more efficiently in AZT-treated cells. Retrovirology 4:30. doi:10.1186/1742-4690-4-30 [DOI] [PMC free article] [PubMed]
- Houzet L, Paillart JC, Smagulova F, Maurel S, Morichaud Z, Marquet R, Mougel M (2007b) HIV controls the selective packaging of genomic, spliced viral and cellular RNAs into virions through different mechanisms. Nucleic Acids Res 35:2695–2704 [DOI] [PMC free article] [PubMed]
- Ibrahim AF, Watters JA, Clark GP, Thomas CJ, Brown JW, Simpson CG (2001) Expression of intron-containing GUS constructs is reduced due to activation of a cryptic 5′ splice site. Mol Genet Genomics 265:455–460 [DOI] [PubMed]
- Irwin PA, Voytas DF (2001) Expression and processing of proteins encoded by the Saccharomyces retrotransposon Ty5. J Virol 75:1790–1797 [DOI] [PMC free article] [PubMed]
- Jackson AO, Larkins BA (1976) Influence of ionic strength, pH, and chelation of divalent metals on isolation of polyribosomes from tobacco leaves. Plant Physiol 57:5–10 [DOI] [PMC free article] [PubMed]
- Jewell NA, Mansky LM (2000) In the beginning: genome recognition, RNA encapsidation and the initiation of complex retrovirus assembly. J Gen Virol 81:1889–1899 [DOI] [PubMed]
- Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33:479–532 [DOI] [PubMed]
- Liang C, Hu J, Russell RS, Kameoka M, Wainberg MA (2004) Spliced human immunodeficiency virus type 1 RNA is reverse transcribed into cDNA within infected cells. AIDS Res Hum Retroviruses 20:203–211 [DOI] [PubMed]
- Luban J, Goff SP (1994) Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J Virol 68:3784–3793 [DOI] [PMC free article] [PubMed]
- Macas J, Neumann P (2007) Ogre elements—a distinct group of plant Ty3/gypsy-like retrotransposons. Gene 390:108–116 [DOI] [PubMed]
- Mount SM (1982) A catalogue of splice junction sequences. Nucleic Acids Res 10:459–472 [DOI] [PMC free article] [PubMed]
- Neumann P, Pozarkova D, Macas J (2003) Highly abundant pea LTR retrotransposon Ogre is constitutively transcribed and partially spliced. Plant Mol Biol 53:399–410 [DOI] [PubMed]
- Neumann P, Koblizkova A, Navratilova A, Macas J (2006) Significant expansion of Vicia pannonica genome size mediated by amplification of a single type of giant retroelement. Genetics 173:1047–1056 [DOI] [PMC free article] [PubMed]
- Neumann P, Yan H, Jiang J (2007) The centromeric retrotransposons of rice are transcribed and differentially processed by RNA interference. Genetics 176:749–761 [DOI] [PMC free article] [PubMed]
- Rabson AB, Graves BJ (1997) Synthesis and processing of viral RNA. In: Coffin JM, Hughes SH, Varmus HE (eds) Retroviruses. Cold Spring Harbor Laboratory Press, New York, pp 205–262 [PubMed]
- Staden R (1996) The Staden sequence analysis package. Mol Biotechnol 5(3):233–241 [DOI] [PubMed]
- Tamura M, Kajikawa M, Okada N (2007) Functional splice sites in a zebrafish LINE and their influence on zebrafish gene expression. Gene 390:221–231 [DOI] [PubMed]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680 [DOI] [PMC free article] [PubMed]
- Vancanneyt G, Schmidt R, O’Connor-Sanchez A, Willmitzer L, Rocha-Sosa M (1990) Construction of an intron-containing marker gene: splicing of the intron in transgenic plants and its use in monitoring early events in Agrobacterium-mediated plant transformation. Mol Gen Genet 220:245–250 [DOI] [PubMed]
- Vicient CM, Kalendar R, Schulman AH (2001) Envelope-class retrovirus-like elements are widespread, transcribed and spliced, and insertionally polymorphic in plants. Genome Res 11:2041–2049 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.