Abstract
The high selectivity and throughput of tandem mass spectrometry allow for rapid identification and localization of various posttranslational protein modifications from complex mixtures by shotgun approaches. Although sequence database search algorithms provide necessary support to process the potentially enormous quantity of MS/MS spectra generated from large scale tandem mass spectrometry experiments, false positive identifications of peptide modifications may exist even after implementation of stringent identification criteria. In this report, we describe factors that lead to misinterpretation of MS/MS spectra as well as common chemical and experimental artifacts that generate false positives using the proteomics-based identification of tyrosine nitration as an example. In addition to the proposed manual validation criteria, the importance of peptide synthesis and subsequent MS/MS characterization for validation of peptide nitration demonstrated by several examples from earlier publications is also presented.
The advancement in mass spectrometric instrumentation has significantly enhanced proteomics-based research yet presents a new and somewhat difficult challenge in terms of data management and analysis (1–4). Current commercially available mass spectrometers can typically generate thousands of MS/MS spectra in a single HPLC-tandem MS experiment resulting in a large number of high confidence peptide identifications after database searching. Nevertheless numerous spectra remain mis- or unidentified because of a variety of factors such as poor spectral quality, nonspecific protein cleavage, and chemical or posttranslational modifications. For those peptide sequences that are identified from MS/MS spectra based on surpassing statistically significant scores established in various database search algorithms, a number of false positives can be present depending on the stringency of the identification criteria. Recent developments in database search strategies have offered new procedures for the assessment of false positive rates for peptide identification (5, 6); however, to provide a detailed survey of proteins obtained from complex biological samples, a compromise must be established to maximize the number of true positive identifications. Therefore, manual validation of MS/MS-based peptide identifications remains critical in this regard.
The introduction of specific guidelines for reporting peptide and protein identifications (7) has greatly enhanced the quality of publications in the area of proteomics and has provided an essential foundation for the accurate expansion of discovery-based research. Although manual validation still plays a major role in the identification of proteins and their corresponding posttranslational modifications, few uniform guidelines exist to formulate acceptable manual validation criteria. The development of such guidelines is rather complex because a variety of factors can impact the approach used to manually validate MS/MS spectra including mass accuracy and resolving power, energy deposition mechanism for collision-induced dissociation, implementation of alternative forms of dissociation, such as electron capture dissociation or electron transfer dissociation, as well as other issues that may influence spectral quality (i.e. coeluting isobaric peptides, random noise, etc.) and subsequent data interpretation.
The intent of this report is to evaluate criteria that are important in the manual validation of MS/MS-based identification of tyrosine nitration, a low abundance posttranslational modification mediated by the reactive nitrogen species peroxynitrite (ONOO−) and commonly used as a biomarker of cellular oxidative stress under certain pathophysiological conditions (8–10). We have used a previous proteomic data set of the mouse brain endoplasmic reticulum (ER)1 generated in our laboratory (11) to provide a reference for the survey of tyrosine nitration in a complex protein mixture and to evaluate criteria that affect false positive identification. Database searching of the MS/MS spectra was performed at different parent mass tolerances to include mass accuracies obtainable through various commercially available mass spectrometers (quadrupole or linear ion trap, TOF, Orbitrap, and FT mass analyzers). The benefit of high mass resolving power and mass measurement accuracy was apparent from this analysis in terms of eliminating almost all false positive identifications of peptides containing nitrotyrosine, and consequently, these results present new issues regarding isobaric interferences associated with this modification. Additionally we have evaluated several MS/MS spectra selected from previously published research articles to assess the validity of the reported in vivo tyrosine nitration sites and in the process have developed general criteria for the manual validation of peptide modification identification using tandem mass spectrometry.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents—
All chemicals and reagents were purchased from Sigma-Aldrich unless otherwise specified.
Peptide Synthesis—
Y*EEEIK (* denotes tyrosine 3-nitration) and SIHELEK were prepared manually in our laboratory by solid-phase peptide synthesis (SPPS) utilizing standard 9-fluorenylmethloxycarbonyl (Fmoc) chemistry with benzotriazole-1-yloxy-tris-pyrrolidinophosphonium hexafluorophosphate/1-hydroxybenzotriazole/ N,N`-diisopropylethylamine (PyBOP/HOBt/DIPEA) activation using preloaded N-α-Fmoc-N-ɛ-tert-butyloxycarbonyl-l-lysine (0.5 mmol/g) Wang resin (AnaSpec, San Jose, CA) (12). N-α-Fmoc-O-tert-butyl-l-3-nitrotyrosine was purchased from Bachem Bioscience, Inc. (King of Prussia, PA); all other Fmoc-protected amino acids and reagents for SPPS were obtained from AnaSpec. After cleaving from the resin with 95:2.5:2.5 trifluoroacetic acid/water/triisopropylsilane (v/v/v), the crude peptides were purified by semipreparative reversed-phase HPLC using an octadecylsilica column (13, 14). All other peptides were custom synthesized on 2–3-mg scale by Synthetic Biomolecules (San Diego, CA).
Proteomics Analysis of Mouse Brain Microsomes—
ER microsomes were prepared for two-dimensional LC mass spectrometric analysis as described previously (11, 15–18). Briefly 300 μg of the ER tryptic digest was fractionated on a Polysulfoethyl Aspartamide 2.1-mm-inner diameter × 10-cm SCX column (PolyLC, Columbia, MD) using a Surveyor (Thermo, San Jose, CA) HPLC pump operated at a flow rate of 200 μl/min. Eighteen SCX fractions were selected based on the UV absorbance profile at 280 nm and subsequently analyzed via on-line reversed-phase HPLC-tandem mass spectrometric analysis using a hybrid linear ion trap-Fourier transform ion cyclotron resonance (7-Tesla) mass spectrometer (LTQ-FT, Thermo Fisher, San Jose, CA) equipped with an electrospray ionization source and operated with the Xcalibur (version 2.2) data acquisition software.
Database Searching—
MS/MS data generated by data-dependent acquisition via the LTQ-FT were extracted by BioWorks version 3.3 and searched against a composite International Protein Index (IPI) mouse (version 3.35, 51,490 × 2 entries) protein database containing both forward and randomized sequences using the Mascot (version 2.2.1; Matrix Science, Boston, MA) search algorithm. Separate Mascot searches were performed using a fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of either 5 ppm, 10 ppm, 50 ppm, or 2 Da assuming the digestion enzyme trypsin with the possibility of one missed cleavage. Carbamidomethylation of cysteine was specified as a static modification, whereas oxidation of methionine and nitration of tyrosine were specified as variable modifications in the database search. The software program Scaffold (version Scaffold-01_06_13, Proteome Software Inc., Portland, OR) was then used to compile and validate tandem MS-based peptide and protein identifications. Peptide identifications were accepted at greater than 95.0% probability as determined by the Peptide Prophet algorithm (19). Protein identifications where protein probabilities were assigned by the Protein Prophet algorithm (20) were accepted at greater than 99.0% probability and excluded single peptide-based identifications. These filtering criteria established a false positive identification rate ((number of randomized peptide sequences identified/total number of peptides identified) × 100%) of ∼0% for the mouse brain ER proteome data set based on the reported decoy database search strategy.
Validation of Nitrated Peptide Identification Based on MS/MS Spectra—
MS/MS spectra derived from putative nitrotyrosine-containing tryptic peptides were manually evaluated using theoretical CID MS/MS fragment ion masses obtained from the in silico peptide fragmentation program MS-Product available through ProteinProspector (University of California San Francisco). MS/MS spectra selected from two separate publications reporting in vivo nitration sites (21, 22) were evaluated in the same fashion. In addition, fragment ion masses from these MS/MS spectra were input into the MS-Tag program from ProteinProspector to determine top ranked peptide matches to facilitate de novo sequencing efforts. If high confidence matches corresponding to unmodified (not nitrated) tryptic peptides with alternate sequences were obtained, peptides representing the unmodified as well as the reported nitrated peptides were synthesized and analyzed by tandem mass spectrometry on a quadrupole ion trap instrument (LCQ Classic, Thermo). MS/MS spectra of the nitrated and unmodified synthetic peptides were compared with the reported experimental MS/MS spectra to definitively assess false positive identifications and to develop general criteria for manual validation.
RESULTS AND DISCUSSION
Analysis of the Mouse Brain ER Nitroproteome—
The mouse brain ER sample was digested with trypsin, and the corresponding peptides were fractionated by SCX chromatography followed by LC-MS/MS analysis on an LTQ-FT instrument. After searching the MS/MS data against the IPI mouse version 3.35 database using a precursor ion mass tolerance of 5 ppm (representing FTICR and Orbitrap mass accuracy), 925 proteins containing at least two unique peptides from the mouse brain ER were identified. Nitration of tyrosine was included as a variable modification in the database search to identify potential nitrotyrosine-containing tryptic peptides derived from the ER sample. Of the several hundred proteins identified, only one tryptic peptide sequence corresponding to aldolase A (Aldoa), Y*ASICQQNGIVPIVEPEILPDGDHDLKR where * denotes nitration, was discovered as a putatively nitrated peptide using a precursor ion mass tolerance of 5 ppm.
The mass tolerance was then increased incrementally to a maximum value of 2 Da whereupon 1238 proteins containing at least two unique peptides were identified after database searching the same MS/MS data set. This mass accuracy is typically considered for database searching of tandem MS data acquired on a quadrupole or linear ion trap instrument. The increased number of protein identifications at 2-Da mass tolerance for an instrument (LTQ-FT) that can exhibit such high mass measurement accuracy is a result of two major factors. First, the conversion of asparagine to aspartic/isoaspartic acid through a deamidation reaction is a common protein modification that can occur both enzymatically or as a chemical artifact (23–25). This particular modification imparts a mass shift of +1 Da to a deamidated asparagine-containing peptide compared with its unmodified cognate and consequently would exceed the mass accuracy specification used for FT instrumentation if this modification was not included in the database search. Second, larger molecular weight peptides that exhibit higher mass spectral intensity of their second isotope peak relative to the monoisotopic peak due to increased abundance contribution of the 13C isotope are more likely to be selected for MS/MS in a data-dependent acquisition based on the “13C-based” mass value. This effect would also result in a +1-Da mass shift and exceed the database search mass accuracy specification for FT instrumentation. In addition to the ∼34% increase in protein identifications, the number of putative nitrotyrosine-containing peptides identified significantly increased using a 2-Da precursor ion mass tolerance in the database search. A complete list of the ER-derived putative nitrated tryptic peptides, 22 in total, at this database search mass tolerance is shown in Table I. The ER proteome data set was searched at 50- and then 10-ppm mass tolerance as well, but no novel sites were identified upon comparison with those listed in Table I. In fact, only Y*ASICQQNGIVPIVEPEILPDGDHDLKR, LTLY*DIAHTPGVAADLSHIETR, and GY*LGPEQLPDCLKGCDVVVIPAGVPR remained on the list with 50 ppm, and Y*ASICQQNGIVPIVEPEILPDGDHDLKR was again the sole hit for nitrated peptide with 10-ppm precursor mass tolerance.
Table I.
Putative nitrated peptides from two-dimensional LC-MS/MS data of mouse brain ER microsomes
The search was performed against a composite IPI mouse database (version 3.35 containing both forward and randomized sequences) using Mascot with a 2-Da precursor ion mass tolerance. Listed sequences had greater than 95% probability as determined by the Peptide Prophet algorithm. All values in the table were taken directly from Scaffold output. All cysteine residues shown were modified via carbamidomethylation (+57.02 Da). Asterisks indicate nitration (+44.98 Da). See text for discussion about the validity of these identifications using more stringent criteria (parent ion tolerance of 5, 10, and 50 ppm) and about the combination of factors that may collectively generate isobaric interferences.
| IPI accession number(s) | Gene symbol | Putative nitrated peptide sequence | Mascot ion score | Mascot identity score (p < 0.05) | Observed m/z | Charge state | Mass error |
|---|---|---|---|---|---|---|---|
| ppm | |||||||
| IPI00757353 | spna2 | ALSSEGKPY*VTKEELYQNLTR | 49.89 | 41.62 | 824.094 | 3 | −395 |
| IPI00122048 | atp1a3 | KY*NTDCVQGLTHSK | 47.99 | 42.89 | 847.4022 | 2 | −1169 |
| IPI00338039 | tubb2a | LTTPTY*GDLNHLVSATMSGVTTCLR | 41.39 | 41.12 | 1377.617 | 2 | 327 |
| GHY*TEGAELVDSVLDVVRK | 52.87 | 42.24 | 1065.560 | 2 | −915 | ||
| IPI00473320 | actb | KDLY*ANTVLSGGTTMYPGIADR | 41.91 | 41.81 | 1194.098 | 2 | −404 |
| IPI00110753 | tuba1 | GHY*TIGKEIIDLVLDR | 54.92 | 42.62 | 629.350 | 3 | −510 |
| IPI00221402 | aldoa | Y*ASICQQNGIVPIVEPEILPDGDHDLKR | 57.57 | 40.33 | 1074.538 | 3 | 1.97 |
| IPI00554928 | nefl | Y*LKEYQDLLNVK | 46.98 | 43.06 | 784.920 | 2 | −1262 |
| IPI00331590 | mdh2 | HGVY*NPNKIFGVTTLDIVR | 41.32 | 42.12 | 1094.096 | 2 | −443 |
| LTLY*DIAHTPGVAADLSHIETR | 69.80 | 41.68 | 813.426 | 3 | 13.0 | ||
| GY*LGPEQLPDCLKGCDVVVIPAGVPR | 78.00 | 40.91 | 1427.742 | 2 | 18.3 | ||
| IPI00223377 | mbp | Y*LATASTMDHAR | 48.38 | 43.26 | 690.322 | 2 | −1435 |
| IPI00263013 | plp1 | MY*GVLPWNAFPGKVCGSNLLSICK | 45.9 | 41.42 | 1378.194 | 2 | −347 |
| IPI00663736 | syngap1 | Y*LKDAIGEFIR | 51.55 | 43.24 | 684.515 | 2 | −1235 |
| IPI00230192 | gnao1 | QY*KPVVYSNTIQSLAAIVR | 76.23 | 42.04 | 1097.138 | 2 | −872 |
| IPI00121550 | atp1b1 | Y*NPNVLPVQCTGKR | 43.81 | 42.84 | 844.930 | 2 | −1174 |
| IPI00465810 | sv2a | GGQY*FNDKFIGLR | 43.82 | 43.03 | 779.396 | 2 | −1269 |
| IPI00116498 | ywhaz | Y*LAEVAAGDDKK | 65.53 | 43.29 | 661.830 | 2 | −1502 |
| IPI00110658 | hba-a1 | TY*FPHFDVSHGSAQVK | 43.14 | 42.70 | 931.949 | 2 | −1059 |
| IPI00117978 | cox4i1 | DY*PLPDVAHVTMLSASQK | 74.73 | 42.41 | 1008.000 | 2 | −981 |
| IPI00132390 | ndufb4 | Y*KPAPLATLPSTLDPAEYDVSPETR | 40.68 | 41.12 | 926.476 | 3 | 376 |
| IPI00132874 | mgll | Y*DELAHMLKGLDMLVFAHDHVGHGQSEGER | 38.93 | 39.92 | 1145.760 | 3 | −383 |
Factors that Contribute to MS-based Nitrotyrosine Misidentification—
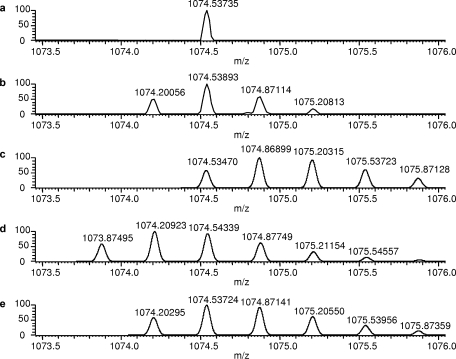
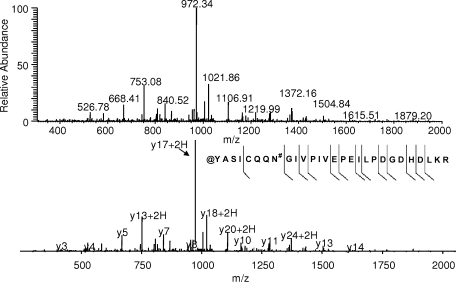
A surprising finding from the database search results using a lower mass accuracy specification was the considerable increase in nitrated peptide identification. Before evaluation of the mass spectrometric data to determine the cause of the larger mass errors for these peptides and the corresponding validity of the nitrotyrosine identifications, the Aldoa tryptic peptide was examined first because the mass error obtained (2 ppm) was within the mass measurement accuracy of the LTQ-FT (<5 ppm). It is interesting to note that this particular nitrated tryptic peptide was also found in a recent proteomics survey of endogenously nitrated proteins from mouse brain (26) using an LTQ instrument. Fig. 1a shows an expanded region of the full scan mass spectrum that was utilized for the data-dependent MS/MS acquisition of a peptide at m/z 1074.54 representing potentially the Aldoa-derived nitrated tryptic peptide Y*ASICQQNGIVPIVEPEILPDGDHDLKR. Upon averaging the full-scan mass spectra over the retention time window of the peptide, however, the peak selected for MS/MS was actually the 13C-based isotope rather than the monoisotopic peak as shown in Fig. 1b. Therefore, we decided to perform a theoretical isotope simulation of the nitrated peptide sequence and found a 1-Da error associated with this mass measurement (Fig. 1c). After assessment of modifications similar in mass to nitration that could occur under the experimental conditions used, carbamoylation was considered because of the presence of urea in the sample. Because the putative nitrotyrosine residue comprises the N terminus of the peptide, a similar modification could occur in which the urea decomposition product, isocyanic acid, reacts with the N terminus of the peptide after trypsin digestion to impart a +43-Da mass shift. The isotopic simulation of the N-terminal carbamoylated Aldoa tryptic peptide, shown in Fig. 1d, indicated a −1-Da difference in mass compared with the experimentally derived isotope cluster. After close inspection of the Aldoa tryptic peptide sequence, we also noted an asparagine residue on the N-terminal side of the peptide that could also be potentially modified by deamidation. Because this particular asparagine precedes a glycine residue, deamidation has been shown to occur readily at this sequence motif (27). Together with carbamoylation and deamidation, the theoretical isotope cluster was then in good agreement with the experimentally derived isotope cluster as shown in Fig. 1e. To further support this finding, the annotated MS/MS spectrum (Fig. 2) showed y ion coverage that included the asparagine residue within the peptide sequence. The shift in mass of +1 Da for the y242+ ion indicates deamidation of the asparagine residue.
Fig. 1.
a, expanded region of an FTICR survey scan at a single chromatographic time point obtained from data-dependent LC-MS/MS analysis of a fraction derived from SCX HPLC separation of the mouse brain ER tryptic digest. The isotope peak at m/z 1074.54 was selected for subsequent MS/MS analysis. b, after averaging the full-scan mass spectra over the peptide elution window, a detailed isotope profile was obtained demonstrating that the monoisotopic peak was not selected for MS/MS analysis. Mass spectra of several peptide sequences containing different isobaric modifications were simulated to determine the correct sequence of the peptide based on previous database search results: Y*ASICQQNGIVPIVEPEILPDGDHDLKR (c), @YASICQQNGIVPIVEPEILPDGDHDLKR (d), and @YASICQQN#GIVPIVEPEILPDGDHDLKR (e) where *, @, and # indicate nitration, carbamoylation of the N terminus, and deamidation, respectively.
Fig. 2.
MS/MS spectra obtained from data-dependent LC-MS/MS analysis of the peptide ion shown in Fig. 1. The exact modification type is difficult to determine because there is minimal b and y ion coverage for the N-terminal portion of the peptide that is modified; however, the +1-Da shift for y242+ provides further evidence for the sequence @YASICQQN#GIVPIVEPEILPDGDHDLKR (where @ and # indicate carbamoylation of the N terminus and deamidation, respectively) determined by the accurate mass measurement presented in Fig. 1.
The isobaric interferences associated with the analysis of tyrosine nitration complicate the definitive identification of this particular modification. Most sites identified from the database search at 2-Da precursor ion mass tolerance were due most likely to a combination of factors including carbamoylation of the N terminus or basic residues in the peptide sequence, deamidation, and 13C-based data-dependent isotope selection. This result is evident from the high frequency of putative nitrotyrosine residues in close proximity to the N terminus or to a basic residue in the peptide sequence as shown in Table I. Definitive assignment of tyrosine nitration may be complicated both by the data-dependent mass measurement accuracy of lower resolution instruments as well as limited b or y ion coverage that would provide sequence information regarding the N-terminal portion of the peptide if carbamoylation occurs at this particular location.
Evaluation of in Vivo Nitration Sites Reported in Selected Previous Publications—
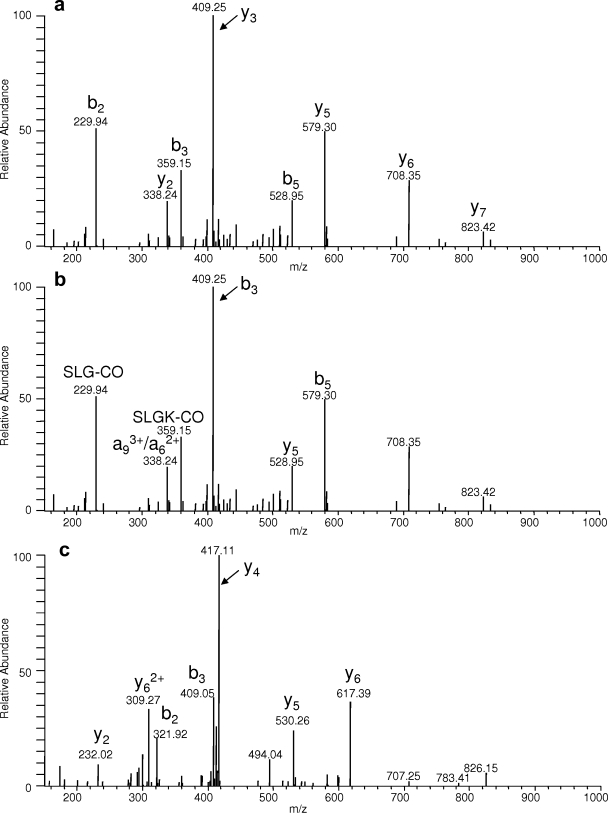
Two publications, Hong et al. (21) and Gokulrangan et al. (22), that reported in vivo protein nitration sites using mass spectrometry were evaluated to assess needs for adherence to criteria concerning guidelines about manual validation of nitration based on MS/MS data and to propose revision of such criteria and introduction of new ones for the application to modification identification in general if required. Twelve tyrosine nitration sites were reported in Hong et al. (21) of which five representative annotated MS/MS product ion spectra were presented. Fig. 4 from Hong et al. (21) displays an MS/MS spectrum corresponding to the nitrated tryptic peptide Y*EEEIK derived from tropomyosin. Upon close inspection of the MS/MS spectrum, two issues were apparent from the published annotation. First, a peak corresponding to >55% relative intensity was not identifiable (which consequently did not meet the manual validation criterion established in the study), and second, a fragment ion corresponding to three neutral losses of water was observed at considerable relative intensity (>20%). To facilitate de novo sequencing of this particular peptide, MS-Tag (University of California San Francisco) was used to search the MS/MS spectral information against the Swiss-Prot database in which the enzyme specificity, taxonomy, and precursor and fragment ion masses were known. After the database search, a tryptic peptide of SIHELEK from myosin was identified as a top match. The theoretical fragment ions from this sequence using MS-Product were then compared with the MS/MS spectrum in Hong et al. (21) to assure the validity of the identification. To further validate this finding, peptides representing Y*EEEIK and SIHELEK were synthesized and analyzed via MS/MS on an LCQ instrument. As shown in Fig. 3, the MS/MS spectrum of the synthetic peptide SIHELEK (Fig. 3a) is in good agreement with the reported MS/MS spectrum that was annotated as Y*EEEIK by Hong et al. (21) (the MS/MS spectrum of SIHELEK is annotated as Y*EEEIK for reference in Fig. 3b). To determine whether Y*EEEIK would exhibit an MS/MS spectrum similar to that of SIHELEK, the synthetic nitrated peptide was analyzed by MS/MS, and the corresponding spectrum is shown in Fig. 3c. Upon comparison of the MS/MS spectra, very few similarities were found based on the observed fragment ion masses and corresponding intensity distribution. Of five published MS/MS spectra in Hong et al. (21), four were determined to be misidentified using the described validation methodology. These results are summarized in Table II, and the related MS/MS spectra of the synthetic peptides used in the validation analysis are presented as supplemental data (supplemental Figs. 1–3).
Fig. 4.
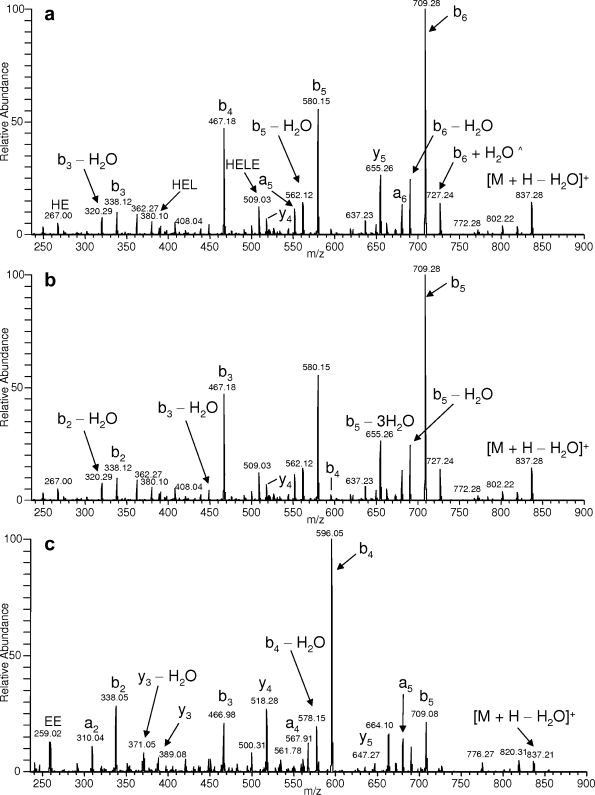
a, ESI-MS/MS spectrum of the synthetic peptide NDEGIAYR. b, the same MS/MS spectrum annotated assuming the peptide sequence LY*SLGKGR reported in Gokulrangan et al. (22) where * indicates nitration. c, the synthetic peptide LY*SLGKGR was also synthesized and showed little similarity to the MS/MS spectrum of NDEGIAYR.
Fig. 3.
a, ESI-MS/MS spectrum of the synthetic peptide SIHELEK (^ indicates a peeling sequence ion that can be generated from basic residue-containing peptides (bn − 1 or bn − 2 + H2O allowed where n equals the total number of residues)). b, the same MS/MS spectrum annotated assuming the peptide sequence Y*EEEIK reported in Hong et al. (21) where * indicates nitration. c, the synthetic peptide Y*EEEIK was also synthesized and showed little similarity to the MS/MS spectrum of SIHELEK.
Table II.
Summary of misidentified nitrotyrosine peptides from Hong et al. (21) as verified by de novo sequencing and ESI-MS/MS analysis of synthetic peptides
| Reported nitrated peptide sequencea | Charge state | Actual peptide sequence | Charge state | Theoretical m/z difference | Approximate theoretical resolution (M/ΔM) required to resolve ESI-generated ions of reported nitrated peptide from those of actual peptide |
| EY*RKDLEESIR | 2 | KVQHELDEAEER | 2 | 0.00 | N/Ab |
| ERYAAWoxMIY*TY*SGLFCVTVNPYK | 3 | DQEGQDVLLFIDNIFR | 2 | −0.38 | 2,500 |
| Y*EEEIK | 1 | SIHELEK | 1 | −0.084 | 10,000 |
| SYKY*LLLSMVK | 2 | NLVHIITHGEEK | 2 | −0.0019 | 350,000 |
oxM, oxidized methionine; *, nitration.
Not applicable.
The remaining MS/MS spectrum in this study could not be definitively assigned because of the presence of differential cysteine alkylation in the peptide sequence TGTCAY*CGLQFK as indicated by the fragment ion masses reported in Hong et al. (21). Because we were unable to selectively alkylate one of the two cysteine residues of the synthetic nitrated peptide, the proposed validation methodology could not be implemented; however, after searching for homologous sequences via NCBI BLAST (Basic Local Alignment Search Tool), the sequence TGTCGYCGLQFK from another NADH dehydrogenase (ubiquinone) Fe-S protein 6 isoform containing a single amino acid variation (Ala → Gly) was identified. Assuming quantitative cysteine alkylation (+57 Da), no tyrosine nitration (−45 Da), and replacement of alanine with a glycine residue (−14 Da), the nominal peptide mass is the same within typical parent mass tolerances (2–4 Da) enabled for database searching of quadrupole ion trap tandem MS data. Furthermore b and y type ions subsequently could be assigned for peaks that were labeled as less probable internal fragment ions; e.g. TCAY(NO2)CGLQ−28 annotation for m/z 916.5 could be replaced with y7.
In a separate publication by Gokulrangan et al. (22), four nitrotyrosine sites were reported along with their corresponding MS/MS spectra. Fig. 8 in Gokulrangan et al. (22) shows an MS/MS spectrum annotated as the fragmentation pattern of the nitrated tryptic peptide LY*SLGKGR of 2,3-cyclic-nucleotide phosphodiesterase. After inspection of this spectrum, a relatively high intensity peak (>20%) was not identified, and fragment ion signals exhibiting considerable intensity and representing internal peptide fragmentation were present. Internal fragment ions can be produced under CID conditions; however, this fragmentation pathway is more typical with higher energy deposition-based fragmentation or peptide ions containing an internal proline (28, 29) and probably would not represent a major mechanism of CID fragmentation for multiply charged ions in an ion trap instrument. To assess the validity of the annotated MS/MS spectrum for LY*SLGKGR, the fragment ion information was searched against the Swiss-Prot database, and the tryptic peptide NDEGIAYR derived from Peroxiredoxin 2 was identified as the top match. The peptides NDEGIAYR and LY*SLGKGR were then synthesized, analyzed by tandem mass spectrometry, and compared with the reference spectrum in Gokulrangan et al. (22). The annotated MS/MS spectrum of NDEGIAYR and LY*SLGKGR including the MS/MS spectrum of NDEGIAYR annotated as LY*SLGKGR (reference for Gokulrangan et al. (22)) are shown in Fig. 4. The CID fragmentation pattern of NDEGIAYR is in good agreement with the MS/MS spectrum reported, and the fragment ions represent more convincing mechanisms of dissociation (i.e. b and y type ions rather than internal fragment ions or multiply charged a type ions). In addition, the MS/MS spectrum of the synthetic peptide LY*SLGKGR showed no major similarities with NDEGIAYR and, taken together, confirm the misidentification listed in Gokulrangan et al. (22).
Of the remaining three MS/MS spectra, two spectra could not be definitively assigned to peptide sequences due to the lack of mass spectral information one can obtain from the published figures, whereas one MS/MS spectrum indicated the presence of selective cysteine alkylation that could not be produced with a routine SPPS method for synthetic nitrated peptides. Figs. 5 and 7 from Gokulrangan et al. (22) represent two examples in which MS/MS spectra derived from the synthetic version of the nitrated peptide could be compared with those spectra reported in the present study. The reported sequences QRGHY*VGVPT(oxM)RDDPK and LPKNY(oxM)(oxM)SNGY*K both contain oxidized methionine residues (denoted as (oxM)) and, upon CID-based MS/MS analysis of the synthetic nitrated peptides, demonstrated strong neutral loss of SOCH3 (supplemental Fig. 4), which is a common fragmentation pathway observed for this particular modification in an ion trap instrument. Moreover the fragmentation profiles obtained for the synthetic nitrated peptides showed no similarities upon comparison with the reported MS/MS spectra in Figs. 5 and 7 of Gokulrangan et al. (22).
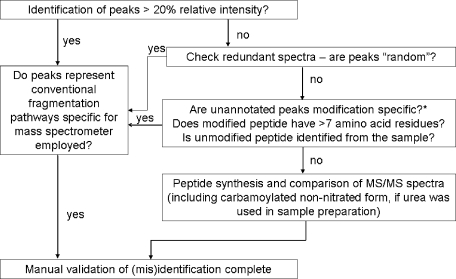
Fig. 5.
General protocol for the manual validation of MS/MS-based identification of nitrated peptides derived from the tyrosine nitration analyses presented in this study. Carbamoylation and deamidation should be explored as potential isobaric artifacts given that these particular modifications can occur in vivo in addition to the possibility of in vitro modification if urea was used for sample processing. Other important factors that should be considered include the size of the peptide (>7 amino acid residues) and identification of the unmodified form of the nitrated peptide given the substoichiometric nature of nitration. *, based on MS/MS analyses of nitrated peptide standards from this study, no modification-specific fragmentation was observed for nitration.
To further evaluate in vivo nitration site identifications from the literature, the report by Sacksteder et al. (26) was assessed because of the high quality annotation reported for a representative MS/MS spectrum corresponding to the nitrated peptide sequence DSY*VAIANACCAPR. This particular nitrated peptide was synthesized, analyzed by MS/MS on an LTQ ion trap instrument, and compared with the reference spectrum in Sacksteder et al. (26). The CID fragmentation pattern of DSY*VAIANACCAPR (supplemental Fig. 5A) was in good agreement with the MS/MS spectrum reported in terms of matching fragment ions as well as their corresponding intensity distribution, and consequently, this particular example would represent a true positive nitropeptide identification based on our validation protocol. Although no urea was used in the reported sample preparation procedures and consequently would preclude carbamoylation as a potential N-terminal modification, we decided to test whether the contribution of chemical artifacts such as N-terminal carbamoylation and deamidation could mimic the MS/MS profile obtained for the nitrated peptide identified. The peptide DSYVAIADACCAPR (where N was replaced with D) was synthesized and incubated in 8 m urea in 25 mm ammonium bicarbonate at 37 °C for 2 h. A small amount of carbamoylated peptide was detected and subsequently analyzed by MS/MS (supplemental Fig. 5B). Not surprisingly, the fragmentation pattern contained similarities to that of the nitrated peptide, although the spectrum was complicated by a heterogeneous distribution of N- and C-terminal (arginine-) modified peptides at the precursor ion mass selected for MS/MS analysis. Nonetheless it is apparent from this observation that careful consideration should be taken in the choice of buffer components for proteomics studies involving the characterization of protein nitration.
Summary of Guidelines for MS/MS Validation of Nitration—
Upon review of the nitrated peptides identified from the mouse brain ER proteomics analysis (Table I) at 2-Da parent ion mass tolerance, most (but not all) could be eliminated as true positives based on the mass measurement accuracy and resolving power of the LTQ-FT instrument (Table I). When utilizing low mass accuracy-based methods such as standard data-dependent acquisition on a quadrupole or linear ion trap, the definitive assignment of tyrosine nitration may be difficult because of a combination of factors that collectively generate an isobaric interference that exists within the 2–3-Da precursor ion mass tolerance generally enabled in the subsequent database search. Manual evaluation of MS/MS spectra is therefore an important step upon identification of peptide nitration or other modifications in general, and we have developed a simple strategy for this purpose as summarized in Fig. 5.
An important criterion to consider is the identification of all fragment ion peaks above a considerable threshold value, which we have determined from manual analysis of misidentified MS/MS spectra from this study to be ∼20–25% relative intensity. If the fragment ion peaks represent reasonable fragmentation pathways based on the primary sequence of the peptide, mode of energy deposition, and modification type, then manual validation is generally achieved. However, certain factors may complicate the MS/MS spectra including random noise spikes, modification-specific fragmentation, or selection of coeluting isobaric peptide ions. Typically multiple MS/MS spectra are acquired for a given peptide ion in a data-dependent acquisition for most commercial mass spectrometers, and issues regarding random noise or isobaric coeluting peptides can be clarified. In terms of modification-specific fragmentation, we have observed retention of the nitration modification on the tyrosine residue during CID fragmentation with no further fragmentation of the actual NO2 group for nitrated peptide standards analyzed on a linear ion trap instrument from this study. If the identification of the peptide modification is still unresolved, the nitrated peptide should be synthesized and analyzed by MS/MS using experimental parameters similar to those used upon proteomics analysis. Although peptide synthesis is an important step in this stage of the validation process, it is nevertheless beneficial to provide additional supporting evidence for the identification of modified peptides even when the corresponding MS/MS spectra can be definitively assigned. Other factors that are important to consider in the validation process include the substoichiometric nature of nitration (10−6 mol of nitrotyrosine/mol of protein derived tyrosine (30)) in which the unmodified form of the nitrated peptide should be detected in the proteome data set. Additionally short peptides of <7 amino acid residues should not be used for nitration site identification because the limited structural information would provide insufficient evidence for sequence and modification identification.
It is important to note that certain situations will arise in which the experimentally derived nitrated peptide fragmentation spectra will not meet all of the validation requirements and still represent true positive database identifications. The criteria described here for the validation of MS/MS-based identification of peptide nitration are therefore general guidelines to address the need for a uniform validation protocol for the identification of posttranslational modifications by MS/MS, and consequently, deviations from this protocol should be systematically evaluated and addressed.
Supplementary Material
Acknowledgments
We thank Professor Peter Koulen for providing the mouse brain microsomes for the study.
Footnotes
Published, MCP Papers in Press, August 16, 2008, DOI 10.1074/mcp.M800065-MCP200
The abbreviations used are: ER, endoplasmic reticulum; SPPS, solid-phase peptide synthesis; SCX, strong cation exchange; Fmoc, 9-fluorenylmethloxycarbonyl; IPI, International Protein Index; Aldoa, aldolase A.
This work was supported, in whole or in part, by National Institutes of Health Grant AG025384. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Topaloglou, T. ( 2006) Informatics solutions for high-throughput proteomics. Drug Discov. Today 11, 509–516 [DOI] [PubMed] [Google Scholar]
- 2.Mead, J. A., Shadforth, I. P., and Bessant, C. ( 2007) Public proteomic MS repositories and pipelines: available tools and biological applications. Proteomics 7, 2769–2786 [DOI] [PubMed] [Google Scholar]
- 3.Smith, J. C., Lambert, J. P., Elisma, F., and Figeys, D. ( 2007) Proteomics in 2005/2006: developments, applications and challenges. Anal. Chem. 79, 4325–4344 [DOI] [PubMed] [Google Scholar]
- 4.Matthiesen, R. ( 2007) Methods, algorithms and tools in computational proteomics: a practical point of view. Proteomics 7, 2815–2832 [DOI] [PubMed] [Google Scholar]
- 5.Peng, J., Elias, J. E., Thoreen, C. C., Licklider, L. J., and Gygi, S. P. ( 2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2, 43–50 [DOI] [PubMed] [Google Scholar]
- 6.Huttlin, E. L., Hegeman, A. D., Harms, A. C., and Sussman, M. R. ( 2007) Prediction of error associated with false-positive rate determination for peptide identification in large-scale proteomics experiments using a combined reverse and forward peptide sequence database strategy. J. Proteome Res. 6, 392–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradshaw, R. A., Burlingame, A. L., Carr, S., and Aebersold, R. ( 2006) Reporting protein identification data. The next generation of guidelines. Mol. Cell. Proteomics 5, 787–788 [DOI] [PubMed] [Google Scholar]
- 8.Pacher, P., Beckman, J. S., and Liaudet, L. ( 2007) Nitric oxide and peroxynitrite in health and disease. Physiol. Rev. 87, 315–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radi, R., Cassina, A., Hodara, R., Quijano, C., and Castro, L. ( 2002) Peroxynitrite reactions and formation in mitochondria. Free Radic. Biol. Med. 33, 1451–1464 [DOI] [PubMed] [Google Scholar]
- 10.Radi, R. ( 2004) Nitric oxide, oxidants, and protein tyrosine nitration. Proc. Natl. Acad. Sci. U. S. A. 101, 4003–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stevens, S. M., Jr., Duncan, R. S., Koulen, P., and Prokai, L. ( 2008) Proteomic analysis of mouse brain microsomes: identification and bioinformatic characterization of endoplasmic reticulum proteins in the mammalian central nervous system. J. Proteome Res. 7, 1046–1054 [DOI] [PubMed] [Google Scholar]
- 12.Wang, S.-S. ( 1973) p-Alkoxybenzyl alcohol resin and p-alkoxybenzyloxycarbonylhydrazide resin for solid phase synthesis of protected peptide fragments. J. Am. Chem. Soc. 95, 1328–1333 [DOI] [PubMed] [Google Scholar]
- 13.Prokai, L., Prokai-Tatrai, K., Zharikova, A. D., Nguyen, V., Perjesi, P., and Stevens, S. M., Jr. ( 2004) Centrally acting and metabolically stable thyrotropin-releasing hormone analogues by replacement of histidine with substituted pyridinium. J. Med. Chem. 47, 6025–6033 [DOI] [PubMed] [Google Scholar]
- 14.Prokai-Tatrai, K., Nguyen, V., Zharikova, A. D., Braddy, A. C., Stevens, S. M., and Prokai, L. ( 2003) Prodrugs to enhance central nervous system effects of the TRH-like peptide pGlu-Glu-Pro-NH2. Bioorg. Med. Chem. Lett. 13, 1011–1014 [DOI] [PubMed] [Google Scholar]
- 15.Hwang, S.-Y., Wei, J., Westhoff, J. H., Duncan, R. S., Ozawa, F., Volpe, P., Inokuchi, K., and Koulen, P. ( 2003) Differential functional interaction of two Vesl/Homer protein isoforms with ryanodine receptor type 1: a novel mechanism for control of intracellular calcium signaling. Cell Calcium 34, 177–184 [DOI] [PubMed] [Google Scholar]
- 16.Westhoff, J. H., Hwang, S.-Y., Duncan, R. S., Ozawa, F., Volpe, P., Inokuchi, K., and Koulen, P. ( 2003) Vesl/Homer proteins regulate ryanodine receptor type 2 function and intracellular calcium signaling. Cell Calcium 34, 261–269 [DOI] [PubMed] [Google Scholar]
- 17.Koulen, P., and Ehrlich, B. E. ( 2000) Reversible block of the calcium release channel/ryanodine receptor by protamine, a heparin antidote. Mol. Biol. Cell 11, 2213–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leite, M. F., Thrower, E. C., Echevarria, W., Koulen, P., Hirata, K., Bennett, A. M., Ehrlich, B. E., and Nathanson, M. H. ( 2003) Nuclear and cytosolic calcium are regulated independently. Proc. Natl. Acad. Sci. U. S. A. 100, 2975–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keller, A., Nesvizhskii, A. I., Kolker, E., and Aebersold, R. ( 2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 20.Nesvizhskii, A. I., Keller, A., Kolker, E., and Aebersold, R. ( 2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 21.Hong, S. J., Gokulrangan, G., and Schöneich, C. ( 2007) Proteomic analysis of age dependent nitration of rat cardiac proteins by solution isoelectric focusing coupled to nanoHPLC tandem mass spectrometry. Exp. Gerontol. 42, 639–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gokulrangan, G., Zaidi, A., Michaelis, M. L., and Schöneich, C. ( 2007) Proteomic analysis of protein nitration in rat cerebellum: effect of biological aging. J. Neurochem. 100, 1494–1504 [DOI] [PubMed] [Google Scholar]
- 23.Mycek, M. J., and Waelsch, H. ( 1960) Enzymatic deamidation of proteins. J. Biol. Chem. 235, 3513–3517 [PubMed] [Google Scholar]
- 24.Stephenson, R. C., and Clarke, S. ( 1989) Succinimide formation from aspartyl and asparaginyl peptides as a model for the spontaneous degradation of proteins. J. Biol. Chem. 264, 6164–6170 [PubMed] [Google Scholar]
- 25.Krokhin, O. V., Antonovici, M., Ens, W., Wilkins, J. A., and Standing, K. G. ( 2006) Deamidation of -Asn-Gly-sequences during sample preparation for proteomics: consequences for MALDI and HPLC-MALDI analysis. Anal. Chem. 78, 6645–6650 [DOI] [PubMed] [Google Scholar]
- 26.Sacksteder, C. A., Qian, W. J., Knyushko, T. V., Wang, H., Chin, M. H., Lacan, G., Melega, W. P., Camp, D. G., II, Smith, R. D., Smith, D. J., Squier, T. C., and Bigelow, D. J. ( 2006) Endogenously nitrated proteins in mouse brain: Links to neurodegenerative disease. Biochemistry 45, 8009–8022 [DOI] [PubMed] [Google Scholar]
- 27.Tyler-Cross, R., and Schirch, V. ( 1991) Role of serine residues in the nonenzymatic mechanism of deamidation of asparaginyl residues and peptide bond cleavage reactions. J. Biol. Chem. 266, 22549–22556 [PubMed] [Google Scholar]
- 28.Leymarie, N., Berg, E. A., McComb, M. E., O'Connor, P. B., Grogan, J., Oppenheim, F. G., and Costello, C. E. ( 2002) Tandem mass spectrometry for structural characterization of proline-rich proteins: application to salivary PRP-3. Anal. Chem. 74, 4124–4132 [DOI] [PubMed] [Google Scholar]
- 29.Breci, L. A., Tabb, D. L., Yates, J. R., III, and Wysocki, V. H. ( 2003) Cleavage N-terminal to proline: analysis of a database of peptide tandem mass spectra. Anal. Chem. 75, 1963–1971 [DOI] [PubMed] [Google Scholar]
- 30.Duncan, M. W. ( 2003) A review of approaches to the analysis of 3-nitrotyrosine. Amino Acids 25, 351–361 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.