Abstract
A widely used method for protein identification couples prefractionation of protein samples by one-dimensional (1D) PAGE with LC/MS/MS. We developed a new label-free quantitative algorithm by combining measurements of spectral counting, ion intensity, and peak area on 1D PAGE-based proteomics. This algorithm has several improvements over other label-free quantitative algorithms: (i) Errors in peak detection are reduced because the retention time is based on each LC/MS/MS run and actual precursor m/z. (ii) Detection sensitivity is increased because protein quantification is based on the combination of peptide count, ion intensity, and peak area. (iii) Peak intensity and peak area are calculated in each LC/MS/MS run for all slices from 1D PAGE for every single identified protein and visualized as a Western blot image. The sensitivity and accuracy of this algorithm were demonstrated by using standard curves (17.4 fmol to 8.7 pmol), complex protein mixtures (30 fmol to 1.16 pmol) of known composition, and spiked protein (34.8 fmol to 17.4 pmol) in complex proteins. We studied the feasibility of this approach using the secretome of angiotensin II (Ang II)-stimulated vascular smooth muscle cells (VSMCs). From the VSMC-conditioned medium, 629 proteins were identified including 212 putative secreted proteins. 26 proteins were differently expressed in control and Ang II-stimulated VSMCs, including 18 proteins not previously reported. Proteins related to cell growth (CYR61, protein NOV, and clusterin) were increased, whereas growth arrest-specific 6 (GAS6) and growth/differentiation factor 6 were decreased by Ang II stimulation. Ang II-stimulated changes of plasminogen activator inhibitor-1, GAS6, cathepsin B, and periostin were validated by Western blot. In conclusion, a novel label-free quantitative analysis of 1D PAGE-LC/MS/MS-based proteomics has been successfully applied to the identification of new potential mediators of Ang II action and may provide an alternative to traditional protein staining methods.
The increased availability of MS-based proteomics is creating new opportunities for routine use as a research tool. Most currently available proteomics studies perform protein identification and quantification as separate, independently optimized processes. Fractionation of complex mixtures prior to LC/MS/MS analysis by one-dimensional (1D)1 PAGE is widely used to improve the detection of low abundance proteins. This sample fractionation technique is popular because it provides protein separation with information regarding molecular weight. The identified proteins are then readily validated using Western blotting. Although Western blot-based methods are also widely used for the quantification of specific proteins, the availability of LC/MS/MS-based proteomics provides new opportunities for the quantitative comparison of all detectable proteins among samples separated by 1D PAGE. However, for a comprehensive 1D PAGE-LC/MS/MS-based approach to be broadly applicable, new methods are needed that enable the large scale comparison of protein abundance among independent samples.
Two main strategies have emerged for quantitative MS-based proteomics, including stable isotopic labeling, which involves the incorporation and monitoring of exogenous isotope labels, and label-free quantitation, which requires spectral counting or measurements of precursor ion intensity. Chemical and metabolic isotopic labeling is a popular method for quantitative proteomics because of its high sensitivity in the detection of differences among sample sets. This approach is well suited for studies among paired or small groups of sample sets that are analyzed at the same time. However, isotope labeling often requires additional sample processing steps, which increase experimental cost and reduce detection sensitivity. Moreover the limited numbers of isotope labels that can be run concurrently create limitations on the comparative analyses among large sample sets (1–3).
Based on the information inherent in chromatographic data, MS spectra, and MS/MS-based peptide assignments, label-free quantitative strategies are attractive alternatives for quantitative LC/MS/MS-based proteomics because of their simplicity, affordability, and flexibility. Although suppressive ionization may affect the quantification, previous studies have demonstrated that spectral counting (peptide count), spectral ion intensity, or peak area of peptide ions correlates well with protein abundance in complex samples (4–12). Pattern-based and identity-based work flows are used in the computation of ion intensity and peak area (1–3, 13). In a pattern-based work flow, ion peaks are isolated and quantified based on a narrow range of m/z tolerance followed by peak matching among the sample set and the assignment of peptide matches. This approach is suitable for MS data with high mass accuracy and high resolution typically generated from TOF or FT detection methods. In an identity-based work flow, the precursor ion m/z and retention time (RT) of an identified peptide are extracted and used as a starting coordinate to extract the peak for this peptide in the remaining measurements. This approach could provide opportunities to routinely quantify spectral data in the absence of ultrahigh resolution instrumentation.
Combining 1D PAGE sample fractionation with LC/MS/MS-based protein identification and label-free quantitation may provide an alternative to Western blotting for comprehensive gel-based protein analyses. In this report we describe a new method for label-free protein quantitation that does not require ultrahigh resolution spectral data. This quantitative approach includes methods to compile protein information among the entire lane and compares results generated from independent samples. We applied this method to the analysis of the effects of angiotensin II (Ang II) on vascular smooth muscle cell (VSMC)-secreted proteins. Our proteomics studies demonstrate the ability of 1D PAGE followed by LC/MS/MS and bioinformatics analysis to reveal new Ang II actions on VSMCs.
EXPERIMENTAL PROCEDURES
Cell Culture and Extraction of the Secreted Proteins—
VSMCs were isolated from Sprague-Dawley rat aorta and cultured in Dulbecco's modified Eagle's medium with 5% fetal bovine serum. VSMCs at passage 6–10 were incubated in 0.1% (w/v) BSA culture medium for overnight followed by rinsing three times with PBS. Cells were then treated with 100 nm Ang II (Sigma-Aldrich) or vehicle in serum-free culture medium for 18 h. Candesartan (Astra Hassle AB, Mölndal, Sweden) and PD123319 (Sigma-Aldrich) were added 30 min in the culture medium before Ang II treatment. The conditioned media were collected, centrifuged, and filtered through a 0.22-μm filter. Proteins in the medium were precipitated by adding 2% sodium deoxycholate (1:100, v/v) for 15 min followed by 100% trichloroacetic acid (1:10, v/v) for 1 h at 4 °C.
Protein Sample Preparation—
To mimic the actual usage of this approach, carbonic anhydrase I (CA-I) ranging from 0.5 to 250 ng, seven-protein mixtures (BSA, cathepsin B, SOD, CA-I, C1 inhibitor, α2-antiplasmin, and angiotensinogen) ranging from 5 to 100 ng, and CA-I ranging from 1 to 500 ng spiked into 1, 5, and 10 mg of protein from VSMC-conditioned medium were separated by 10% SDS-PAGE. Precipitated proteins (200 μg) from conditioned medium were also separated by 10% SDS-PAGE.
Identification of Protein by LC/MS/MS—
The gels were stained with Coomassie Blue stain, and the bands containing proteins were cut (for CA-I standard curve, seven-protein mixtures, and CA-I spiking experiments), or each lane was cut into 40 slices (for rat VSMC secretome experiment). Each slice was digested with trypsin and analyzed by nano-LC/MS/MS using a linear ion trap mass spectrometer (Thermo Fisher, San Jose, CA). Assignment of MS/MS data was performed using SEQUEST (Bioworks 3.2, Thermo Fisher) and X!Tandem (version 2006.09.15, The Global Proteome Machine Organization) search against the International Protein Index (IPI) database (Rat 3.22, 41,336 sequences) from the European Bioinformatics Institute and a randomized version of IPI database for calculation of false positive rate (14). The search parameters included potential residue mass modifications of +16.0 Da for oxidized methionine and +71.0 Da for acrylamide alkylated cysteine; peptide tolerance, ±1 Da, fragment ion tolerance, ±0.5 Da.
Compilation of Search Results—
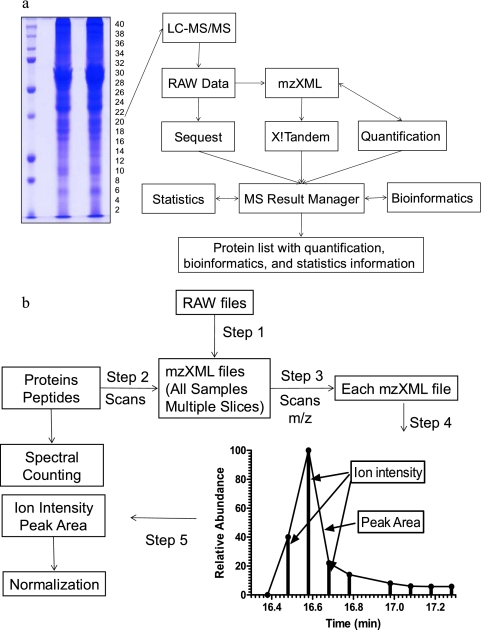
To compile the search results into the MySQL database and perform proteomics computational analyses, we developed software based on the PHP-MySQL-Apache platform named MS Result Manager (Fig. 1a). Data processing occurred in five steps: (i) file parsing in which SEQUEST and X!Tandem search results were parsed into the MySQL database, (ii) peptide-level filtering in which low scoring peptides based on the criteria described previously (15) for the SEQUEST search result and −log(E) ≥ 2.00 for the X!Tandem search result were filtered, (iii) summarizing in which the search results from each sample, generated from 40 gel slices, were combined based on the IPI identifier (ID), (iv) protein-level filtering in which the redundant proteins, contaminates, and proteins that did not have two unique peptides identified from a single slice or adjacent slices were filtered, and (v) reporting in which proteins that were identified in at least two samples were complied into one reporting table. If multiple IDs were assigned for the same peptide match, a uniform ID was selected for the comparison of proteins identified between samples.
Fig. 1.
Schematic diagrams (a and b) for label-free quantitative analysis of 1D PAGE LC/MS/MS proteome.
Bioinformatics Analysis—
Gene Ontology (GO) annotations were extracted from the Gene Ontology Annotation Database (16) and generic GO Slim provided by the European Bioinformatics Institute. Secreted protein identification was carried out in multistep processes involving analysis by GO annotation, TargetP 1.1 (17), SignalP 3.0 (18), TMHMM 2.0 (19), and DAVID (20). (i) The proteins annotated as extracellular by GO or predicted as secreted proteins by TargetP were considered as secreted protein candidates. (ii) Proteins that did not contain a signal peptide predicted by SignalP or those that contained more than one predicted transmembrane segment by TMHMM were discarded. (iii) The proteins that localized in the endoplasmic reticulum lumen, Golgi apparatus, and lysosomes analyzed by DAVID were also discarded. (iv) The information from the European Bioinformatics Institute protein sequence database was taken into account for identification of those suspected secretory proteins.
Relative Protein Quantitation—
Three measurements (spectral counting, ion intensity, and peak area) were used for protein quantitation. Spectral counting for each protein was calculated by the MS Result Manager based on SEQUEST and X!Tandem search results. Quantitation by ion intensity and peak area was performed on the peptide sequences filtered by MS Result Manager and extracted from MS data by the following steps (Fig. 1b). (i) To access MS and MS/MS data, Xcalibur native acquisition files were converted to mzXML (21) files by ReAdW.exe (Institute for Systems Biology). (ii) The scan number of each peptide, which is used to identify precursor peak position from MS data, was extracted by MS Result Manger. (iii) The m/z range of the precursor was calculated from all peptides across all samples. Each MS/MS scan was mapped back to mzXML files, and the m/z of the precursor was extracted. The m/z range was set by the lowest m/z to highest m/z but limited to median ± 0.4 Da (for low to moderate mass resolution LC/MS data) based on an optimization test. (iv) Peak recognition for each peptide was defined by three parameters: retention time (scan number of the precursor), m/z range of the precursor, and the highest intensity of scans that have been identified as the target peptide. The ion intensity in the peak was extracted, normalized to percent total ion current within each scan and added up. The peak area was calculated by the trapezoidal method. To shorten the computation time, the computation process was performed on the first six abundant peptides of each protein based on spectral counting after an optimization test. For some peptides whose multiple charge states of the precursor were identified, this process was repeated for each charge state. If the peptide was dispersed in multiple slices, the ion intensity and peak area were determined by the sum of the value from multiple slices. If a peptide was not successfully identified in every single measurement, the known RT from a paired MS run was used. This software allowed for the batch calculation of entire data sets in an automated process and interacted directly with MySQL database by inserting three measurements into the database. (v) The ion intensity and peak area were normalized by defining the largest value as 100 among the peptides quantified to enable each peptide to contribute to the quantification equally.
Western Blot Analysis—
The extracted proteins were separated by SDS-PAGE and immunoblotted using primary antibodies against plasminogen activator inhibitor-1 (PAI-1) (American Diagnostica, Stamford, CT), cathepsin B, growth arrest-specific 6 (GAS6) (Santa Cruz Biotechnology, Santa Cruz, CA), and periostin (Abcam, Cambridge, MA).
Statistics—
Regression and correlation analyses were performed by GraphPad Prism (GraphPad Software, San Diego, CA). The t test was performed by PHP script incorporated in MS Result Manger based on PHP statistics extension. Results were expressed as -fold change (mean ± S.E.) compared with control.
RESULTS
Validating Label-free Protein Quantification with Protein Standard Curve and Seven-protein Mixtures—
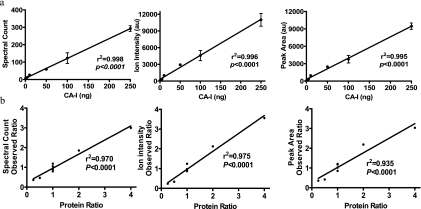
Prior to application of the quantification algorithm to complex protein mixtures, we first tested the linearity of spectral counting, ion intensity, and peak area with CA-I as standard ranging from 0.5 to 250 ng (17.4 fmol to 8.7 pmol). The CA-I was run on 1D PAGE, and the band containing CA-I was digested with trypsin. The digests were analyzed by LC/MS/MS. Spectral counting was calculated by summing the number of identified peptides. Spectral counting was linearly correlated with protein ranging from 0.5 to 250 ng (r2 = 0.998, p < 0.0001, n = 3). Ion intensity and peak area of the MS/MS precursor were extracted and calculated for each identified peptide (an extracted peptide from four different samples is depicted in supplemental Fig. S1). Using this algorithm, we generated a standard curve showing the correlation between ion intensity/peak area and protein abundance for CA-I. Over the range of 0.5–250 ng of protein, this standard curve has an r2 = 0.996 for ion intensity (p < 0.0001, n = 3) and r2 = 0.995 for peak area (p < 0.0001, n = 3) (Fig. 2a). The coefficients of variation (CVs) of spectral counting, ion intensity, and peak area were 27, 26.3, and 25.4%, respectively.
Fig. 2.
Validation of label-free protein quantification with a single protein standard curve and seven-protein mixtures. a, standard curves for spectral counting, ion intensity, and peak area versus the amount of CA-I (mean ± S.E. from three independent experiments). Error bars represent standard error of the mean (S. E.). b, linear correlation in the seven-protein mixtures between the measured ratios by spectral counting, ion intensity, and peak area and known ratio of protein mixtures. au, arbitrary units.
Two groups of seven-protein mixtures were used to test the feasibility of our algorithm in the quantification of relative changes of proteins in a mixture. Seven-protein mixtures were prepared so that the concentrations of three components were kept constant (BSA, cathepsin B, and SOD), whereas the remaining four varied at ratios of 1:2 (CA-1), 2:1 (C1 inhibitor), 4:1 (α2-antiplasmin), and 1:4 (angiotensinogen) (Table I). 100 ng of BSA was added as an interfering protein. Other protein concentrations ranged from 5 to 50 ng (90–760 fmol). These mixtures were run on 1D PAGE and stopped before they were separated. The bands containing the seven proteins were digested with trypsin. To measure the analytical variability of LC/MS/MS, the digests were divided into three equal amounts (30–1160 fmol of protein) and analyzed by LC/MS/MS. The spectral counting, ion intensity, and peak area were calculated as described under “Experimental Procedures.” The spectral counting, ion intensity, and peak area for each protein correlated with the expected relative amounts of protein (r2 = 0.970, r2 = 0.975, and r2 = 0.935, respectively; p < 0.0001) (Table I and Fig. 2b). Spectral counting showed a greater percent error of ratio (16.94%) than ion intensity (14.14%) and peak area (13.56%) but a lesser CV (10.76%) than ion intensity (23.31%) and peak area (23.13%).
Table I.
Observed ratios by spectral counting, ion intensity, and peak area of protein mixtures
| Protein | Mixture
|
Ratio | Observed ratio
|
|||
|---|---|---|---|---|---|---|
| I | II | Spectral counting | Ion intensity | Peak area | ||
| ng | ||||||
| Albumin | 100 | 100 | 1 | 1.18 ± 0.09 | 1.25 ± 0.08 | 1.13 ± 0.11 |
| α2-Plasmin inhibitor | 20 | 5 | 4 | 3.00 ± 0.20a | 3.56 ± 1.78b | 2.85 ± 1.29c |
| Angiotensinogen | 10 | 40 | 0.25 | 0.36 ± 0.07b | 0.23 ± 0.08b | 0.27 ± 0.09b |
| Carbonic anhydrase I | 10 | 20 | 0.5 | 0.47 ± 0.04a | 0.33 ± 0.04a | 0.38 ± 0.04b |
| Cathepsin B | 50 | 50 | 1 | 1.05 ± 0.13 | 0.87 ± 0.14 | 0.99 ± 0.17 |
| C1 inhibitor | 40 | 20 | 2 | 1.85 ± 0.16b | 2.13 ± 0.15a | 1.97 ± 0.22b |
| SOD | 20 | 20 | 1 | 0.80 ± 0.06 | 0.87 ± 0.06 | 0.88 ± 0.10 |
| Trypsin | 1 | 0.90 ± 0.05 | 0.97 ± 0.08 | 0.80 ± 0.10 | ||
p < 0.001, comparison between mixtures I and II; n = 3.
p < 0.01, comparison between mixtures I and II; n = 3.
p < 0.05, comparison between mixtures I and II; n = 3.
Validating Label-free Protein Quantification with Protein Spiked in Biological Complexes at Three Concentrations—
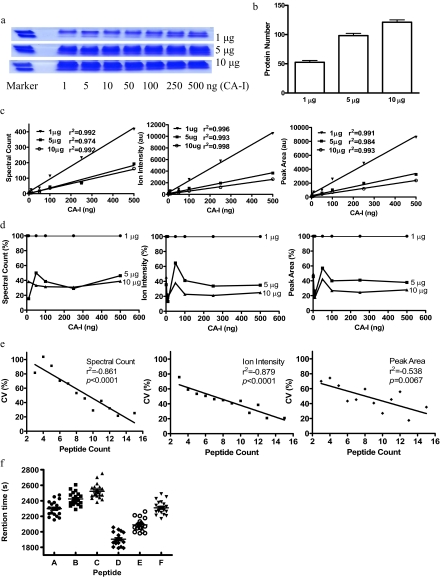
The utility and performance were further tested by spiking varying amounts of standard proteins into varying amounts of complex proteins. A wide range of CA-I from 1 ng (34.8 fmol) to 500 ng (17.4 pmol) was spiked into 1, 5, and 10 μg of precipitated proteins from VSMC-conditioned medium, run on 10% 1D PAGE, and stopped before they were separated (Fig. 3a). The ratios of CA-I to mixture protein were from 1:2 (500 ng in 1 μg of complex proteins) to 1:10,000 (1 ng in 10 μg of complex proteins). The amounts of complex proteins (1, 5, and 10 μg) were arbitrarily selected to be approximately equal to the protein amount of each slice of the gel from the actual experiment that contains 40 slices with total protein of 200 μg of proteins/lane. After LC/MS/MS analysis and database searching, 54.14 ± 1.8 (false discovery rate (FDR), 1.68 ± 0.48%), 98.14 ± 3.94 (FDR, 0.85 ± 0.26%), and 121.1 ± 4.05 (FDR, 0.82 ± 0.24%) unique proteins were detected in 1, 5, and 10 μg of complex proteins, respectively (Fig. 3b). The detection sensitivities for spiked CA-I protein were 1 ng in 1 μg of complex proteins and 10 ng in 5 and 10 μg of complex proteins, suggesting that the detection limit in our experiment is ∼1:1000. The linearity between spectral counting, ion intensity, and peak area versus amount of added CA-I was observed in three different complex proteins with r2 between 0.974 and 0.998 and p < 0.0001 (Fig. 3c). This result demonstrated that the observed amounts of spiked protein were correlated with expected amounts in complex samples.
Fig. 3.
Validation of label-free protein quantification with protein spiked in biological complexes. a, CA-I from 1 to 500 ng was spiked into 1, 5, and 10 μg of precipitated proteins from VSMC-conditioned medium and run on 10% 1D PAGE. b, the protein number detected in 1, 5, and 10 μg of complexes. Error bars represent standard error of the mean (S. E.). c, linear correlation between spectral counting, ion intensity, and peak area versus amount of added CA-I. d, the suppressive ionization of protein spiked in different mixtures. The spectral counting, ion intensity, and peak area of CA-I in 1 μg were expressed as 100%. e, linear correlation between average coefficient of variation of identified proteins versus unique peptide number. f, peak retention time distribution of six peptides. au, arbitrary units.
The suppressive ionization effect was estimated by comparing the same amount of CA-I spiked in different amounts of complex samples. The spectral counting, ion intensity, and peak area of CA-I were about 65 and 70% lower in 5 and 10 μg of complex samples than in 1 μg of complex samples (Fig. 3d). The suppressive ionization was significant if the difference between two samples was higher than 5-fold, but it is acceptable if the difference between two samples was less than 2-fold. It suggested that a limitation of this method is that suppressive ion effects could be an important factor if the protein composition of sample sets were markedly different.
The reproducibility and variability of this approach to analyze complex samples were analyzed from the identified proteins in an experiment using 5 μg of complex sample. The linearity between CV versus unique peptide number was observed in three measurements (Fig. 3e). The CV generated from spectral counting was higher than from ion intensity and peak area when there are a few unique peptides identified in each protein. The average CVs of spectral counting, ion intensity, and peak area were 55.21, 43.60, and 48.20%, respectively. This result suggested that the more abundant the proteins are the more confident the results generated will be and that the measurement by ion intensity is the best among the three measurements.
The drift of RT was also analyzed from this data set. Six abundant peptides were randomly selected, and the peak retention times were extracted from 21 LC/MS/MS data. The average drift of RT (maximum peak RT − minimum peak RT) was 388 ± 67 s (n = 6) (Fig. 3f), indicating that peak detection based on the RT from each MS run is a better approach than using the reference MS run.
Bioinformatics Analysis of Rat VSMC Secretome—
VSMCs were incubated with 100 nm Ang II for 18 h, and the proteins from the conditioned medium from three independent experiments were fractionated by 10% SDS-PAGE. Each lane was cut into 40 slices followed by tryptic digestion and nano-LC/MS/MS analysis. The LC/MS/MS data were analyzed by SEQUEST and X!Tandem. 30% more peptides and 20% more proteins were found by the combination of SEQUEST and X!Tandem search results compared with SEQUEST or X!Tandem alone. The analysis of 240 gel slices from six samples led to the identification of 629 proteins with 3% FDR (supplemental Table 1). These proteins were analyzed by GO annotation, TargetP (17), SignalP (18), TMHMM (19), and Visualization and Integrated Discovery (DAVID) (20). 82 proteins were annotated as extracellular proteins by GO annotation, and 290 proteins were predicted to be secreted by TargetP. After a multistep bioinformatics process, 210 proteins were determined to be secreted proteins (supplemental Table 2). Among the secreted proteins, only nine proteins have been identified previously. The discovery of these additionally secreted proteins significantly enriched our knowledge of the secretory function of VSMCs. GO annotations were analyzed using the GO Slims list provided by Gene Ontology. 184 of 210 proteins were associated with at least one GO ID. The major functions of these proteins are related to metabolism, development, regulation of biological process, catalytic activity, and signal transducer activity (supplemental Fig. S2, A and B). We used spectral counting as a measurement of relative protein abundances within the VSMC secretome. Absolute protein amount was estimated by spectral number of one protein/total spectral × 200 μg. 49% of proteins identified had less than 100 total peptides in six samples presumably because of their relatively low abundances (less than 50 ng). The detection sensitivity was 0.89 ng. 20 proteins were identified with peptides >5000 (supplemental Fig. S2C) including extracellular matrix, cell adhesion proteins, and metabolic enzymes.
The Effect of Ang II on Rat VSMC Secretome—
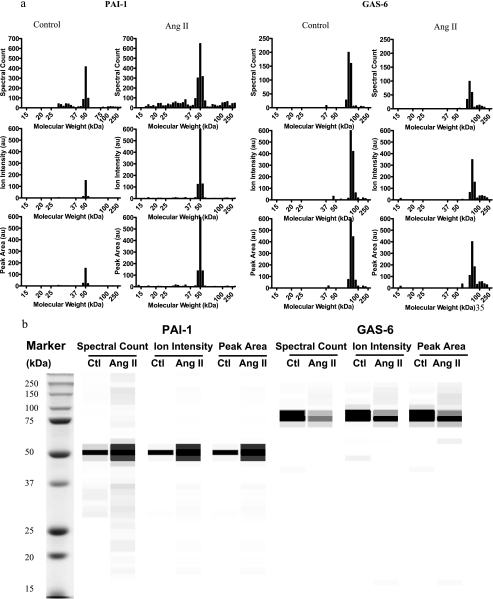
The protein change between the control and Ang II-stimulated group was quantified by the summary of spectral counting, ion intensity, and peak area from multiple slices (supplemental Table 2). The proteins were usually identified in multiple slices, which can be visualized as a bar graph showing quantitative indices versus slice number (Fig. 4a) or grayscale digital image (Fig. 4b). The protein molecular weight, intensity, and distribution displayed on the grayscale digital image can be visualized as displayed in a manner similar to that of a Western blot image. Levels of 14, 12, and 15 secreted proteins from control and Ang II-stimulated conditioned media were different significantly as measured by the spectral counting, ion intensity, and peak area, respectively. Taken together, 26 proteins were differentially expressed in control and Ang II-stimulated VSMC medium. The changes of eight proteins were consistent with other reports at the mRNA level and four proteins at the protein level. Proteins with similar functions were classified into the following groups: cell growth, cell adhesion, calcium ion homeostasis, endopeptidase inhibitor activity, endopeptidase activity, and others (Table II).
Fig. 4.
Extracted spectral counting, ion intensity, and peak area of PAI-1 and GAS6 from control and Ang II-stimulated conditioned media displayed as a bar graph (a) or grayscale digital image (b). The result was from one representative experiment. The spectral counting, ion intensity, and peak area from each slice were converted to 256 intensities (i.e. shades of gray). Black represents the strongest intensity, and white represents the weakest intensity. Ctl, control; au, arbitrary units.
Table II.
Proteins were differentially expressed by Ang II stimulation
| Gene symbol | IPI ID | Protein name | -Fold change
|
||
|---|---|---|---|---|---|
| Spectral counting | Ion intensity | Peak area | |||
| Cell growth | |||||
| CYR61 | IPI00551794 | Protein CYR61 | +a | 2.89b | 2.07c |
| INHBA | IPI00190775 | Inhibin β A chain (34) | 10.3c | 2.31c | 1.98b |
| TGFB1 | IPI00214538 | Transforming growth factor β-1 (35, 36) | 4c | 2.05b | 1.86 |
| Clu | IPI00198667 | Clusterin precursor | 1.64c | 1.71 | 1.80 |
| NOV | IPI00215256 | Protein NOV homolog | 1.77 | 1.55c | 1.5b |
| IGFBP7 | IPI00361798 | Insulin-like growth factor-binding protein 7 | 0.59 | 0.84c | 0.81c |
| Ogn | IPI00362931 | Mimecan (osteoglycin) isoform 1 (37) | 0.64c | 0.73 | 0.76 |
| GDF6 | IPI00454588 | Growth/differentiation factor 6 | 0.53c | 0.69 | 0.72 |
| GAS6 | IPI00201622 | Growth arrest-specific protein 6 | 0.32c | 0.51c | 0.64 |
| TWSG1 | IPI00197515 | Twisted gastrulation homolog 1 | 0.57 | 0.5 | 0.52b |
| Cell adhesion | |||||
| POSTN | IPI00190088 | Periostin (38) | 3.46 | 4.56c | 3.78c |
| SPP1 | IPI00327895 | Osteopontin (39, 40) | 1.84c | 1.86b | 1.78b |
| COL6A1 | IPI00371853 | Collagen α-1(VI) (41) | 3.88c | 1.77 | 1.55 |
| LOXL2 | IPI00767994 | Lysyl oxidase-like 2 | + | 1.69 | 1.52 |
| PRELP | IPI00190287 | Prolargin | 2.7 | 1.61 | 1.61c |
| Calcium ion homeostasis | |||||
| S100A11 | IPI00365443 | S100 calcium-binding protein A11 | 3.34c | 2.21 | 1.83 |
| NUCB1 | IPI00205022 | Nucleobindin-1 | 1.93c | 1.85c | 1.91c |
| NUCB2 | IPI00200070 | Nucleobindin-2 | 1.61 | 1.69d | 1.65d |
| Endopeptidase activity | |||||
| CTSF | IPI00201100 | Cathepsin F | 2.03c | 1.86 | 1.93 |
| CTSK | IPI00206378 | Cathepsin K | 6.59c | 1.71 | 1.71c |
| CTSB | IPI00212811 | Cathepsin B | 1.49c | 1.35 | 1.24 |
| Endopeptidase inhibitor activity | |||||
| SERPINE1 | IPI00204483 | Plasminogen activator inhibitor 1 (40, 42) | 4.42b | 5.2b | 4.44b |
| TIMP1 | IPI00205805 | Metalloproteinase inhibitor 1 (40, 43) | 1.97c | 2.02 | 1.67 |
| SERPINB6A | IPI00189148 | Serine (or cysteine) peptidase inhibitor, clade B, member 6a | 1.95c | 1.30 | 1.27 |
| Others | |||||
| LOXL3 | IPI00365102 | Lysyl oxidase-like 3 | 2.31 | 1.48 | 1.4c |
| Man2b2 | IPI00382210 | Ab2-450 | 1.50 | 1.48 | 1.73c |
+, only found in Ang II group; n = 3.
p < 0.01 compared with a control; n = 3.
p < 0.05 compared with a control; n = 3.
p < 0.001 compared with a control; n = 3.
Western Blot Analysis—
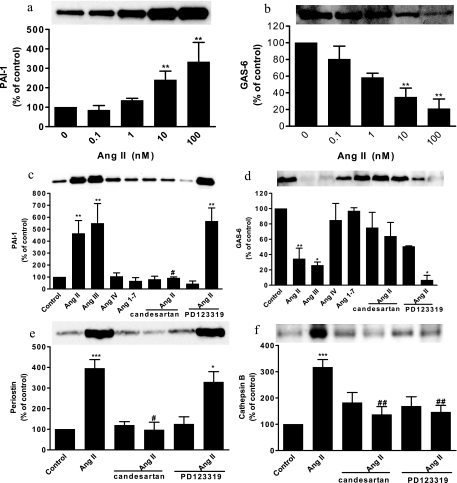
The effects of Ang II on PAI-1 and GAS6, cathepsin B, and periostin released from VSMCs by Ang II were confirmed by Western blot analyses. Ang II increased PAI-1 and decreased GAS6 secretion in a concentration-dependent manner (Fig. 5, a and b). Ang II (100 nm) increased PAI-1, periostin, and cathepsin B secretion by 4.65-, 3.95-, and 3.17-fold, respectively, and decreased GAS6 secretion by 34% (Fig. 5, c–f). The contribution of AT1 or AT2 receptors to these actions was investigated with pretreated AT1 (1 μm candesartan) or AT2 receptor blockers (10 μm PD123319). Pretreatment with candesartan reduced Ang II-stimulated PAI-1 and periostin secretion to basal levels (Fig. 5e) and reduced Ang II-mediated inhibition of GAS6 secretion by 85% (Fig. 5d). Pretreatment with candesartan or PD123319 decreased cathepsin B secretion by 47 and 53%, respectively (Fig. 5f). These results suggest that the AT1 receptor contributes to Ang II-mediated PAI-1 and periostin stimulation and GAS6 secretion inhibition, whereas both the AT1 and AT2 receptors contribute to Ang II-stimulated cathepsin B secretion.
Fig. 5.
Western blot analysis of PAI-1, GAS6, periostin, and cathepsin B. Effects of Ang II on expression of PAI-1 (a) and GAS6 (b) are shown. The role of AT1 receptors on Ang II-induced PAI-1 (c), GAS6 (d), periostin (e), and cathepsin B (f) expression is shown. (*, p < 0.05; **, p < 0.01; and ***, p < 0.001 compared with a control. #, p < 0.05; and ##, p < 0.01 compared with Ang II-treated groups.) Values are expressed as the percent change compared with controls. Error bars represent standard error of the mean (S. E.).
DISCUSSION
Quantifying changes in protein abundance based on label-free quantitative proteomics approaches requires calculation of spectral counting, ion intensity, or peak area. Because the quantitative proteomics approach based on spectral counting is convenient, fast, and easy to implement, it is widely applied on a large scale and in highly complex samples (15, 22–24). However, the major limitations of the spectral counting method are its dependence on the quality of MS/MS peptide identifications and the unreliability for the proteins with a few peptides identified (3). To improve spectral counting performance, the combination of two search engines, SEQUEST and X!Tandem, was used in our study, increasing the identified peptide number and protein reliability.
Various software solutions can perform label-free quantification from the information inherent at the MS1 level (3). However, these software solutions were in most cases applied to data acquired on high mass accuracy and high resolution mass spectrometers, such as the Rosetta Elucidator system (Rosetta Biosoftware, Seattle, WA), SuperHirn (25), PEPPeR (platform for experimental proteomic pattern recognition) (13), MsInspect (26), and MSight (27). To our knowledge, these software solutions cannot be applied to the 1D PAGE-LC/MS/MS proteome approach that is a commonly used method for proteomics. Errors in peak detection and drifts in RT can influence the label-free quantification result. To minimize these effects, our algorithm introduced three strategies. (i) The m/z range of the MS/MS precursor was statistically calculated from all of the MS run; this is a valuable strategy for the analysis of low to moderate mass resolution LC/MS data. (ii) The peak detection was based on the RT from each MS run instead of the reference MS run so that fluctuations in the LC systems do not affect peak detection. (iii) The intensity of the identified precursor was used to determine the peak position. Spectral counting, ion intensity, and peak area represent the information from different MS levels. The combination of spectral counting, ion intensity, and peak area data increases the detection sensitivity for protein changes.
The sensitivity and accuracy of our algorithm were demonstrated using standard protein, complex protein mixtures of known composition, and spiked protein in complex protein mixtures. This study revealed that spectral counting, ion intensity, and peak area of the protein increased linearly with increasing amount from 17.4 fmol to 8.7 pmol with no apparent saturation effect. The analysis of standards and mixtures showed larger measurement error by spectral counting than by ion intensity and peak area. The measurements by ion intensity and peak area showed similar correlation, percent error of ratio, and coefficient variation. The analysis of protein spiked in a biological complex showed that observed protein amounts by three measurements were correlated with the expected amount of spiked protein. The suppressive ionization was acceptable if the difference between two samples was less than 2-fold. These results suggested that spectral counting, ion intensity, and peak area generated during LC/MS/MS are viable for relative quantification of proteins. From three validation analyses, the measurement by ion intensity seems to be the best among the three measurements, but spectral counting and peak area also provide useful information.
Validation of the protein quantification algorithm was performed by studying the effect of Ang II on the VSMC secretome. Proteins secreted by VSMCs can act locally on VSMCs and endothelial cells or can be released into the bloodstream (28). The identity of secreted proteins isolated from cell supernatants is often complicated by proteins from dead cells occurring during cell culture and originally present in the medium. The proteins that are not secreted by the investigated cells can be identified by bioinformatics analysis (29). The contamination of proteins from serum-derived cultures can be minimized by stringently rinsing cells (30). By bioinformatics analysis, 210 proteins were identified as secreted proteins; this is the largest secretome data set to our knowledge with the majority of them not yet identified by proteomics (31) or other studies. Thus, the discovery of additional novel secreted proteins in the present study significantly enriched our understanding of the secretory function of VSMCs. The putative detection limit is 1 ng using spectral counting as the relative measurement method. Considering that the loading protein (200–500 μg) on 1D PAGE for LC/MS/MS is 10-fold greater than that used in Western blot (20–50 μg), the quantification information provided by 1D PAGE-LC/MS/MS is comparable to Western blot.
The changes in secreted protein abundance between control and stimulated VSMCs were quantified by spectral counting, ion intensity, and peak area. The combination of three measurements resulted in 26 differently expressed proteins, yielding almost double the number of proteins detected by a single measurement. Of the 26 proteins, 22 proteins have not been reported, 21 proteins showed enhanced secretion by Ang II stimulation, and five proteins showed inhibited secretion. After annotation analysis, seven novel identified proteins (protein CYR61, clusterin (32), protein NOV, insulin-like growth factor-binding protein 7, growth/differentiation factor 6, GAS6 (33), and twisted gastrulation homolog 1) related to cell proliferation were regulated by Ang II. Ang II also induces extracellular matrix formation and participates in vascular fibrosis. Using GO analysis, 64 extracellular matrix- and cell adhesion-related proteins were secreted by VSMCs. Of them, periostin, collagen α-1(VI), prolargin, osteopontin, CYR61, transforming growth factor β, and lysyl oxidase-like 2 were up-regulated by Ang II.
The application of our algorithm on the effect of Ang II on the VSMC secretome demonstrates several significant improvements over other label-free quantitative algorithms. First, errors in peak detection are reduced because the retention time is based on each LC/MS/MS run and statistically calculated m/z range. Second, detection sensitivity is increased because protein quantification is based on the combination of spectral counting, ion intensity, and peak area. Last, ion intensity and peak area are calculated in each LC/MS/MS run for all slices from 1D PAGE for every identified peptide. These improvements make the software solution an efficient approach to identification, quantification, and annotation in parallel. In conclusion, a label-free proteomics approach by a combination of spectral counting, ion intensity, and peak area was applied to globally identify and quantify proteins in large scale complex biological samples fractionated by 1D PAGE. This report describes the largest composition of the VSMC secretome and identifies novel potential mediators of Ang II action.
Supplementary Material
Acknowledgments
We thank Xiaohong Chen for technical assistance and Drs. Stephen Kolwicz, Jr. and Jia Liu for critical comments regarding this manuscript.
Footnotes
Published, MCP Papers in Press, August 2, 2008, DOI 10.1074/mcp.M800104-MCP200
The abbreviations used are: 1D, one-dimensional; RT, retention time; CV, coefficient of variation; FDR, false discovery rate; GO, Gene Ontology; Ang II, angiotensin II; VSMC, vascular smooth muscle cell; GAS6, growth arrest-specific 6; CA-I, carbonic anhydrase I; SOD, superoxide dismutase; IPI, International Protein Index; ID, identifier; DAVID, Database for Annotation, Visualization, and Integrated Discovery; PAI-1, plasminogen activator inhibitor-1.
This work was supported, in whole or in part, by National Institutes of Health Grants DK60165 and DK36836. This work was also supported by the Adler Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Domon, B., and Aebersold, R. ( 2006) Mass spectrometry and protein analysis. Science 312, 212–217 [DOI] [PubMed] [Google Scholar]
- 2.Nesvizhskii, A. I., Vitek, O., and Aebersold, R. ( 2007) Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat. Methods 4, 787–797 [DOI] [PubMed] [Google Scholar]
- 3.Mueller, L. N., Brusniak, M. Y., Mani, D. R., and Aebersold, R. ( 2008) An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J. Proteome Res. 7, 51–61 [DOI] [PubMed] [Google Scholar]
- 4.Bondarenko, P. V., Chelius, D., and Shaler, T. A. ( 2002) Identification and relative quantitation of protein mixtures by enzymatic digestion followed by capillary reversed-phase liquid chromatography-tandem mass spectrometry. Anal. Chem. 74, 4741–4749 [DOI] [PubMed] [Google Scholar]
- 5.Chelius, D., and Bondarenko, P. V. ( 2002) Quantitative profiling of proteins in complex mixtures using liquid chromatography and mass spectrometry. J. Proteome Res. 1, 317–323 [DOI] [PubMed] [Google Scholar]
- 6.Wang, W., Zhou, H., Lin, H., Roy, S., Shaler, T. A., Hill, L. R., Norton, S., Kumar, P., Anderle, M., and Becker, C. H. ( 2003) Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal. Chem. 75, 4818–4826 [DOI] [PubMed] [Google Scholar]
- 7.Wiener, M. C., Sachs, J. R., Deyanova, E. G., and Yates, N. A. ( 2004) Differential mass spectrometry: a label-free LC-MS method for finding significant differences in complex peptide and protein mixtures. Anal. Chem. 76, 6085–6096 [DOI] [PubMed] [Google Scholar]
- 8.Liu, H., Sadygov, R. G., and Yates, J. R., III ( 2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 9.Venable, J. D., Dong, M. Q., Wohlschlegel, J., Dillin, A., and Yates, J. R. ( 2004) Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods 1, 39–45 [DOI] [PubMed] [Google Scholar]
- 10.Old, W. M., Meyer-Arendt, K., Aveline-Wolf, L., Pierce, K. G., Mendoza, A., Sevinsky, J. R., Resing, K. A., and Ahn, N. G. ( 2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4, 1487–1502 [DOI] [PubMed] [Google Scholar]
- 11.Meng, F., Wiener, M. C., Sachs, J. R., Burns, C., Verma, P., Paweletz, C. P., Mazur, M. T., Deyanova, E. G., Yates, N. A., and Hendrickson, R. C. ( 2007) Quantitative analysis of complex peptide mixtures using FTMS and differential mass spectrometry. J. Am. Soc. Mass Spectrom. 18, 226–233 [DOI] [PubMed] [Google Scholar]
- 12.Lu, P., Vogel, C., Wang, R., Yao, X., and Marcotte, E. M. ( 2007) Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25, 117–124 [DOI] [PubMed] [Google Scholar]
- 13.Jaffe, J. D., Mani, D. R., Leptos, K. C., Church, G. M., Gillette, M. A., and Carr, S. A. ( 2006) PEPPeR, a platform for experimental proteomic pattern recognition. Mol. Cell. Proteomics 5, 1927–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elias, J. E., and Gygi, S. P. ( 2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 15.Gao, B. B., Clermont, A., Rook, S., Fonda, S. J., Srinivasan, V. J., Wojtkowski, M., Fujimoto, J. G., Avery, R. L., Arrigg, P. G., Bursell, S. E., Aiello, L. P., and Feener, E. P. ( 2007) Extracellular carbonic anhydrase mediates hemorrhagic retinal and cerebral vascular permeability through prekallikrein activation. Nat. Med. 13, 181–188 [DOI] [PubMed] [Google Scholar]
- 16.Camon, E., Barrell, D., Lee, V., Dimmer, E., and Apweiler, R. ( 2004) The Gene Ontology Annotation (GOA) Database—an integrated resource of GO annotations to the UniProt Knowledgebase. In Silico Biol. 4, 5–6 [PubMed] [Google Scholar]
- 17.Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. ( 2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 18.Bendtsen, J. D., Nielsen, H., von Heijne, G., and Brunak, S. ( 2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 19.Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. ( 2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 20.Dennis, G., Jr., Sherman, B. T., Hosack, D. A., Yang, J., Gao, W., Lane, H. C., and Lempicki, R. A. ( 2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 21.Pedrioli, P. G., Eng, J. K., Hubley, R., Vogelzang, M., Deutsch, E. W., Raught, B., Pratt, B., Nilsson, E., Angeletti, R. H., Apweiler, R., Cheung, K., Costello, C. E., Hermjakob, H., Huang, S., Julian, R. K., Kapp, E., McComb, M. E., Oliver, S. G., Omenn, G., Paton, N. W., Simpson, R., Smith, R., Taylor, C. F., Zhu, W., and Aebersold, R. ( 2004) A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 22, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 22.Andersen, J. S., Wilkinson, C. J., Mayor, T., Mortensen, P., Nigg, E. A., and Mann, M. ( 2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570–574 [DOI] [PubMed] [Google Scholar]
- 23.Blondeau, F., Ritter, B., Allaire, P. D., Wasiak, S., Girard, M., Hussain, N. K., Angers, A., Legendre-Guillemin, V., Roy, L., Boismenu, D., Kearney, R. E., Bell, A. W., Bergeron, J. J., and McPherson, P. S. ( 2004) Tandem MS analysis of brain clathrin-coated vesicles reveals their critical involvement in synaptic vesicle recycling. Proc. Natl. Acad. Sci. U. S. A. 101, 3833–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilchrist, A., Au, C. E., Hiding, J., Bell, A. W., Fernandez-Rodriguez, J., Lesimple, S., Nagaya, H., Roy, L., Gosline, S. J., Hallett, M., Paiement, J., Kearney, R. E., Nilsson, T., and Bergeron, J. J. ( 2006) Quantitative proteomics analysis of the secretory pathway. Cell 127, 1265–1281 [DOI] [PubMed] [Google Scholar]
- 25.Mueller, L. N., Rinner, O., Schmidt, A., Letarte, S., Bodenmiller, B., Brusniak, M. Y., Vitek, O., Aebersold, R., and Muller, M. ( 2007) SuperHirn—a novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics 7, 3470–3480 [DOI] [PubMed] [Google Scholar]
- 26.Bellew, M., Coram, M., Fitzgibbon, M., Igra, M., Randolph, T., Wang, P., May, D., Eng, J., Fang, R., Lin, C., Chen, J., Goodlett, D., Whiteaker, J., Paulovich, A., and McIntosh, M. ( 2006) A suite of algorithms for the comprehensive analysis of complex protein mixtures using high-resolution LC-MS. Bioinformatics 22, 1902–1909 [DOI] [PubMed] [Google Scholar]
- 27.Palagi, P. M., Walther, D., Quadroni, M., Catherinet, S., Burgess, J., Zimmermann-Ivol, C. G., Sanchez, J. C., Binz, P. A., Hochstrasser, D. F., and Appel, R. D. ( 2005) MSight: an image analysis software for liquid chromatography-mass spectrometry. Proteomics 5, 2381–2384 [DOI] [PubMed] [Google Scholar]
- 28.Berk, B. C. ( 2001) Vascular smooth muscle growth: autocrine growth mechanisms. Physiol. Rev. 81, 999–1030 [DOI] [PubMed] [Google Scholar]
- 29.Bortoluzzi, S., Scannapieco, P., Cestaro, A., Danieli, G. A., and Schiaffino, S. ( 2006) Computational reconstruction of the human skeletal muscle secretome. Proteins 62, 776–792 [DOI] [PubMed] [Google Scholar]
- 30.Pellitteri-Hahn, M. C., Warren, M. C., Didier, D. N., Winkler, E. L., Mirza, S. P., Greene, A. S., and Olivier, M. ( 2006) Improved mass spectrometric proteomic profiling of the secretome of rat vascular endothelial cells. J. Proteome Res. 5, 2861–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dupont, A., Corseaux, D., Dekeyzer, O., Drobecq, H., Guihot, A. L., Susen, S., Vincentelli, A., Amouyel, P., Jude, B., and Pinet, F. ( 2005) The proteome and secretome of human arterial smooth muscle cells. Proteomics 5, 585–596 [DOI] [PubMed] [Google Scholar]
- 32.Sivamurthy, N., Stone, D. H., Logerfo, F. W., and Quist, W. C. ( 2001) Apolipoprotein J inhibits the migration, adhesion, and proliferation of vascular smooth muscle cells. J. Vasc. Surg. 34, 716–723 [DOI] [PubMed] [Google Scholar]
- 33.Melaragno, M. G., Cavet, M. E., Yan, C., Tai, L. K., Jin, Z. G., Haendeler, J., and Berk, B. C. ( 2004) Gas6 inhibits apoptosis in vascular smooth muscle: role of Axl kinase and Akt. J. Mol. Cell. Cardiol. 37, 881–887 [DOI] [PubMed] [Google Scholar]
- 34.Pawlowski, J. E., Taylor, D. S., Valentine, M., Hail, M. E., Ferrer, P., Kowala, M. C., and Molloy, C. J. ( 1997) Stimulation of activin A expression in rat aortic smooth muscle cells by thrombin and angiotensin II correlates with neointimal formation in vivo. J. Clin. Investig. 100, 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibbons, G. H., Pratt, R. E., and Dzau, V. J. ( 1992) Vascular smooth muscle cell hypertrophy vs. hyperplasia. Autocrine transforming growth factor-β1 expression determines growth response to angiotensin II. J. Clin. Investig. 90, 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber, H., Taylor, D. S., and Molloy, C. J. ( 1994) Angiotensin II induces delayed mitogenesis and cellular proliferation in rat aortic smooth muscle cells. Correlation with the expression of specific endogenous growth factors and reversal by suramin. J. Clin. Investig. 93, 788–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shanahan, C. M., Cary, N. R., Osbourn, J. K., and Weissberg, P. L. ( 1997) Identification of osteoglycin as a component of the vascular matrix. Differential expression by vascular smooth muscle cells during neointima formation and in atherosclerotic plaques. Arterioscler. Thromb. Vasc. Biol. 17, 2437–2447 [DOI] [PubMed] [Google Scholar]
- 38.Li, G., Oparil, S., Sanders, J. M., Zhang, L., Dai, M., Chen, L. B., Conway, S. J., McNamara, C. A., and Sarembock, I. J. ( 2006) Phosphatidylinositol-3-kinase signaling mediates vascular smooth muscle cell expression of periostin in vivo and in vitro. Atherosclerosis 188, 292–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.deBlois, D., Lombardi, D. M., Su, E. J., Clowes, A. W., Schwartz, S. M., and Giachelli, C. M. ( 1996) Angiotensin II induction of osteopontin expression and DNA replication in rat arteries. Hypertension 28, 1055–1063 [DOI] [PubMed] [Google Scholar]
- 40.Campos, A. H., Zhao, Y., Pollman, M. J., and Gibbons, G. H. ( 2003) DNA microarray profiling to identify angiotensin-responsive genes in vascular smooth muscle cells: potential mediators of vascular disease. Circ. Res. 92, 111–118 [DOI] [PubMed] [Google Scholar]
- 41.Ford, C. M., Li, S., and Pickering, J. G. ( 1999) Angiotensin II stimulates collagen synthesis in human vascular smooth muscle cells. Involvement of the AT(1) receptor, transforming growth factor-β, and tyrosine phosphorylation. Arterioscler. Thromb. Vasc. Biol. 19, 1843–1851 [DOI] [PubMed] [Google Scholar]
- 42.Gao, B. B., Hansen, H., Chen, H. C., and Feener, E. P. ( 2006) Angiotensin II stimulates phosphorylation of an ectodomain-truncated platelet-derived growth factor receptor-β and its binding to class IA PI3K in vascular smooth muscle cells. Biochem. J. 397, 337–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castoldi, G., Di Gioia, C. R., Pieruzzi, F., D’Orlando, C., Van De Greef, W. M., Busca, G., Sperti, G., and Stella, A. ( 2003) ANG II increases TIMP-1 expression in rat aortic smooth muscle cells in vivo. Am. J. Physiol. 284, H635–H643 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.