Abstract
Cystathionine β-synthase (CBS), a key enzyme in the metabolism of homocysteine, has previously been shown to require a heme co-factor for maximal activity. However, the biochemical function of the CBS heme is not well defined. Here, we show that expression of human CBS in heme-deficient strains of Saccharomyces cerevisiae and Escherichia coli results in production of an enzyme that is misfolded and degraded. Addition of exogenous heme, porphyrins with non-iron metal, or porphyrin lacking metal entirely produced stable and active CBS enzyme. Purification of recombinant CBS enzyme expressed in the presence of various metalloporphyrins confirmed that Mn(III) and Co(III) had 30–60% of the specific activity of Fe(III)-CBS, and still responded to allosteric activation by S-adenosyl-l-methionine. Treatment of S. cerevisiae with the chemical chaperone trimethylamine-N-oxide resulted in near complete restoration of function to human CBS produced in a heme-deficient strain. Taken together, these results suggest that porphyrin moiety of the heme plays a critical role in proper CBS folding and assembly, but that the metal ion is not essential for this function or for allosteric regulation by S-adenosyl-l-methionine.
Cystathionine β-synthase (CBS)3 is a pyridoxal phosphate (PLP)-dependent enzyme that catalyzes the condensation of serine and homocysteine to cystathionine, the first step of the trans-sulfuration pathway. Deficiency of CBS is the major cause of hyperhomocysteinemia and homocystinuria in humans (1). Mildly elevated levels of homocysteine in plasma represent an independent risk factor for cardiovascular diseases, osteoporosis, age-related dementia, and neural tube defects (2–5). Human CBS is a unique member of the PLP-dependent enzyme family in that it binds an additional cofactor, heme. CBS forms homotetramers comprised of identical subunits of 551 amino acids each with a molecular mass of ∼63 kDa. The CBS monomer has a modular structure in which the N-terminal, the middle, and the C-terminal regions serve as heme binding, catalytic and regulatory domains of the enzyme, respectively (6). The C-terminal regulatory domain consists of a hydrophobic CBS domain tandem named CBS1 and CBS2, which were recently identified as the binding site for the allosteric CBS activator S-adenosyl-l-methionine (AdoMet) (7, 8). It has been shown that the C-terminal region also performs an autoinhibitory function by partial shielding the active site. Binding of AdoMet, deletion of the C-terminal region, increased temperature or specific mutations alleviate this inhibitory effect (9, 10). A truncated 45-kDa form of CBS (1–413 residues), devoid of the C-terminal region forms homodimers, which are constitutively activated and unresponsive to AdoMet (11). This truncated CBS was successfully used to generate crystals used to resolve the CBS tertiary structure (12, 13). Although the functions of the regulatory C-terminal domain and the central catalytic core binding the PLP cofactor have been elucidated, the role of the heme cofactor is still unanswered. The N-terminal heme moiety is coordinated by Cys52 and His65 residues (12). CBS from Saccharomyces cerevisiae, as well as Trypanosoma cruzi, lack the heme cofactor of human CBS (14–16). These enzymes are also not stimulated by AdoMet suggesting that: 1) heme and AdoMet stimulation are new acquisitions in evolution of higher eukaryotes, and 2) that AdoMet activation may be dependent on the presence of heme in CBS. Furthermore, heme-free N-terminal-truncated human enzyme is catalytically active as are human CBS crystals from which the heme was removed (17). The above mentioned facts preclude a direct role of heme in CBS catalysis as does the ∼20-Å distance between the heme moiety and the PLP cofactor (12, 13). Taoka et al. (13, 18) proposed a role for heme as a redox sensor because CBS heme iron reduction has been correlated with a 50% decrease in the enzymatic activity. The potential physiological role for such redox sensing may be to regulate intracellular homocysteine flow by switching between the transmethylation and trans-sulfuration pathways. Our recent results suggest that enzymatic activity of the reduced CBS is indistinguishable from its oxidized form (19). The ferrous CBS is unstable under the assay conditions and undergoes a heme ligand switch inactivating the enzyme (20–22).
Here we examine the potential role of the heme moiety in CBS folding and in its interaction with AdoMet and PLP. We have developed two different systems that allowed us to manipulate the heme content of human CBS. For the first system, we have engineered a yeast strain that is deficient in heme biosynthesis and expresses human but not yeast CBS. In this strain, we found that CBS expression and activity were directly dependent on the availability of exogenous heme. Furthermore, various heme analogs were examined to test for the iron requirement in the protoporphyrin. Interestingly, the chemical chaperone trimethylamine N-oxide (TMAO) was able to substitute for a structural function of heme and allowed for the formation of stable and active enzymes. In the second system, we used Escherichia coli deficient in heme synthesis for expression and purification of human CBS containing various heme analogs. Our results suggest that heme plays a key role in proper CBS folding and assembly.
EXPERIMENTAL PROCEDURES
Chemicals—Unless stated otherwise, all chemicals were purchased from Sigma or Fisher Scientific. Protoporhyrins were purchased from Frontier Scientific. l-[U-14C]Serine was obtained from PerkinElmer Life Sciences.
E. coli and S. cerevisae Strains—All bacterial and yeast strains used in this study are listed in Table 1. pHCBS was created as previously described (23). WY254 was derived from W303ΔHEM2 (15) by transformation with a cys4::HIS3 knockout plasmid as previously described (24).
TABLE 1.
Bacterial and yeast strains used in this study
| Strain | Genotype | Ref. |
|---|---|---|
| S. cerevisiae WY254 | α ade2, can1, his3, leu2, trp1, ura3, hem2::LEU2, cys4::HIS3 | See “Experimental Procedures” |
| S. cerevisiae WY79 | α ade2, can1, his3, leu2, trp1, ura3, cys4::LEU2 | This study |
| S. cerevisiae WY61 | α his2, leu2, trp1, ura3, cys4::LEU2 + pHCBS | 23 and 24 |
| E. coli HU227 | hemA41, relA1, spoT1, metB1, rrnB-2, mcrB1, creC510, Hem-p | 45 |
| E. coli RP523 | thr-1, leuB6(Am), fhuA21, lacY1, hemB220, glnV44(AS), e14-, rfbC1, glpR200(glpc), thi-1 | 29 |
CBS Expression and Preparation of Crude Extracts in Yeast—Yeast cells were grown in synthetic complete media supplemented with ergosterol (20 μg/ml) and Tween 80 (5 mg/ml) to an A600 ∼ 1.0 and extracts were prepared by mechanical lysis as previously described (10). To determine the effect of the chemical chaperone, TMAO, it was dissolved in the growth medium in a final concentration of 0.2–0.8 m and filter sterilized. The supernatant was used as a soluble fraction.
CBS Expression and Preparation of Crude Extracts in E. coli—The expression of human CBS in the presence and absence of different porphyrins and preparation of crude extracts from E. coli cells were performed as described previously with some modifications (19). The previously described expression plasmid encoding human CBS as a fusion protein with glutathione S-transferase was used in all E. coli expression experiments (19). Strains were grown in 50 ml of LB medium anaerobically under a layer of sterile mineral oil at 30 °C with gentle mixing on a magnetic stirrer. Bacterial cells used as an inoculum for these cultures were grown twice anaerobically in the absence of any protoporphyrin. The growth medium was supplemented with 100 μg/ml ampicillin, 0.001% thiamine-HCl and the desired protoporphyrin to give a final concentration of 60 μg/ml. The porhyrins were kept sterile in the dark at 4 °C as a 2 mg/ml stock solution dissolved in 0.2 m Na2HPO4, pH 12. Expression was carried overnight (16–20 h) by induction using isopropyl β-d-1-thiogalactopyranoside at a final concentration of 150 μm. Harvested cells were washed in 0.9% NaCl and subsequently lysed in an ice-cold buffer containing 1× TBS, pH 8.6, 1 mm dithiothreitol, 1% Triton X-100, 2 mg/ml lysozyme, 100 μm PLP and Sigma bacterial protease inhibitor mixture. Crude extracts were obtained after shearing the genomic DNA by sonication and removing the insoluble fraction by centrifugation at 20,000 × g for 30 min.
Expression and Purification of CBS Substituted with Heme Analogs—The starter cultures were grown as described above. Overnight expressions were carried out in tightly sealed 2-liter bottles (Nalgene) degassed for 15 min with sterile nitrogen. For purification of the fusion proteins, the crude extracts were incubated for 10 min at room temperature with 2 mm ATP and 10 mm MgSO4 to prevent nonspecific interaction of E. coli DNA K protein with the affinity resin. The fusion proteins were purified on a glutathione-Sepharose column as described previously (19). The isolated GST-hCBS fusion proteins were cleaved with Prescission protease (GE Healthcare) in 1× cleavage buffer (50 mm Tris-HCl pH 7.0, 150 mm NaCl, 1 mm EDTA, and 1 mm dithiothreitol) at 4 °C for 12 h at a final concentration of 0.5 units/mg of protein. The cleaved fusion protein was subsequently concentrated on Amicon YM-30 (Millipore) with three buffer exchanges for 20 mm HEPES, pH 7.4, 1 mm Tris-(2-carboxyethyl)phosphine, and 0.01% Tween 20.
Spectroscopic Measurements—The UV-visible spectra of purified CBS proteins were measured at room temperature in a HP diode array model 8453 UV-visible spectrophotometer in 20 mm HEPES, pH 7.4, 1 mm Tris-(2-carboxyethyl)phosphine, 0.01% Tween 20.
CBS Activity Assay—The CBS activity was determined by a previously described radioisotope assay using l-[U-14C]serine as the labeled substrate (25). The reaction was started by the addition of 150 μg of the crude extract or 25 μg of concentrated cleaved fusion protein, respectively. Protein concentrations were determined by the Lowry procedure using bovine serum albumin as a standard (26).
Protein Gel Electrophoresis and Western Blot—Denatured proteins were separated by SDS-PAGE (27) using a 9% separating gel with a 4% stacking gel. The commercially available apparatus (Criterion, Bio-Rad) was used for separation of either denatured proteins by SDS-PAGE using 10% Tris-HCl precast gels or native samples using 4–12% polyacrylamide gradient Tris-HCl precast gels. For visualization, gels were stained with Simple Blue (Invitrogen).
Western blot analysis of crude cell lysates under denaturing conditions was performed as described previously (17). After electrophoresis, the separated proteins were transferred onto a polyvinylidene difluoride membrane (Millipore) and probed with affinity-purified polyclonal or commercial monoclonal anti-CBS antibody (Abnova), respectively. The Precision Plus Standard Kaleidoscope (Bio-Rad) was used as molecular weight marker for SDS-PAGE gels.
RESULTS
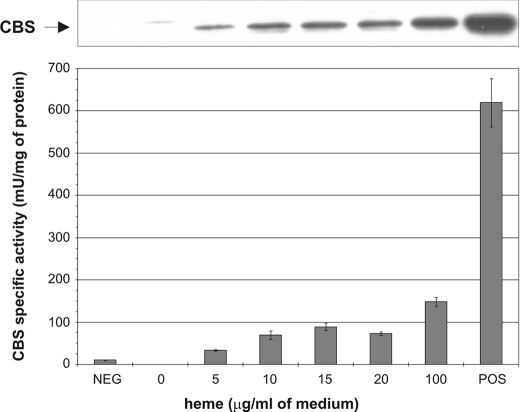
CBS Expression and Activity Is Dependent on Heme Availability—We developed an expression system for human CBS in a yeast strain deficient in both heme and yeast CBS biosynthesis to study the role of heme in human CBS expression, folding, and activity. Strain WY254 (Table 1) contains a deletion of the yeast CBS gene (Δcys4) and contains a mutation in the HEM2 enzyme, which catalyzes the conversion of δ-ALA to porphobilinogen, the second step in the heme biosynthetic pathway. This strain can grow in media supplemented with ergosterol and Tween 80 as a source of unsaturated fatty acids and sterols (15). We transformed WY254 with an expression vector carrying human WT CBS and created WY254pHCBS (23). To study the effect of heme availability on CBS we grew this strain in the minimal medium supplemented with 2% ergosterol and 0.5% Tween 80, and 30 μg/ml glutathione in the presence of exogenous hemin in concentrations ranging from 0 to 100 μg/ml of medium. Fig. 1 shows that expression of CBS as well as its specific activity increase with increasing hemin concentration in the growth medium. Without hemin supplementation there was no signal for CBS on a Western blot probed with monoclonal anti-CBS antibody. As the concentration of hemin gradually increased, the CBS band intensity increased as well (Fig. 1, top). Similarly, there was no CBS activity without added hemin and activity gradually grew in response to higher hemin concentrations (Fig. 1, bottom). The maximum activity achieved was ∼25% of the positive control (WY61 WT yeast strain with deleted endogenous yeast CBS and transformed with plasmid expressing human WT CBS). At the highest concentration of hemin in the medium (100 μg/ml) we still did not attain the intensity of the control CBS band on Western blot. The signal saturation was not achieved even if the concentration of hemin was increased to 300 μg/ml (data not shown). At concentrations higher than 100 μg/ml, the yeast growth was substantially slower and precipitation of hemin in the growth medium was observed. Western blot analysis did not detect any CBS in insoluble fractions (data not shown). This result suggests that CBS expressed in yeast in the absence of heme is likely misfolded and degraded.
FIGURE 1.
Dependence of CBS expression and activity on heme concentration. Western blot of yeast strain WY254 bearing plasmid expressing human CBS. Cell lysates (20 μg/lane) were electrophoresed on a 9% SDS-gel. Following Western transfer, CBS was detected using anti-CBS monoclonal antibody (upper panel). The cell lysates were assayed for CBS activity and the results were plotted (bottom panel). The strains WY61 and WY79 served as positive and negative controls, respectively.
The enzyme activity was fully responsive to AdoMet (not shown), as can be expected for human enzyme but not for the yeast enzyme (15). These results indicate that availability of heme is a limiting factor for CBS expression, thus one of the functions of heme may be to nucleate proper folding and/or assembly of the enzyme.
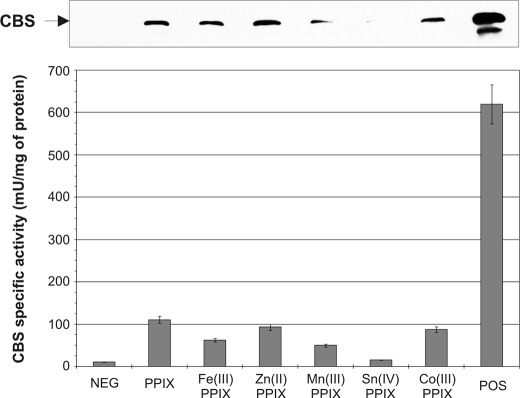
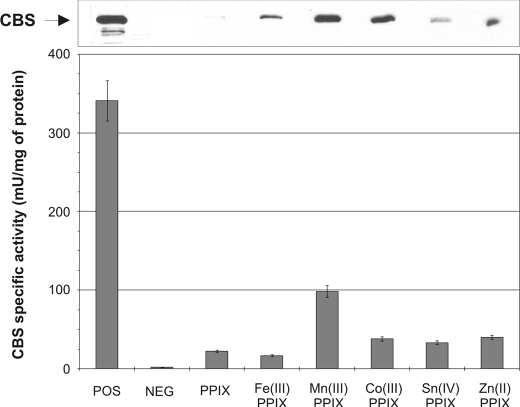
The Iron in Heme May Be Replaceable by Other Metals—To further support one of the possible heme roles as a structural scaffold during CBS synthesis and/or assembly we decided to express CBS from the heme-deficient yeast strain in the presence of various porphyrins coordinated to different metals at a final concentration of 100 μg/ml. The selected porphyrins share the same structure of the porhyrin ring but they differ in the metal in the central position. We also included protoporphyrin IX, which does not possess any metal within its structure. We detected CBS in each yeast extract on a Western blot probed with monoclonal anti-CBS antibody (Fig. 2, top). The amounts of detected CBS on Western blot roughly correlate with CBS activities for the same samples (Fig. 2, bottom). Protoporhyrin IX (PPIX), Zn(II)-PPIX, and Co(III)-PPIX gave the highest activities roughly equivalent to 17% of a positive control. We also examined human CBS expressed in heme-deficient E. coli. Again, we found that all of the porphyrins tested resulted in increased CBS protein and activity (Fig. 3).
FIGURE 2.
Expression and activity of human CBS in the presence of various heme analogs in yeast system. Cell lysates (20 μg/lane) were electrophoresed on a 9% SDS gel. Following Western transfer, CBS was detected using anti-CBS monoclonal antibody (upper panel). Graph of corresponding CBS activities (bottom panel). Strains WY61 and WY79 served as positive and negative controls, respectively.
FIGURE 3.
Expression and activity of human CBS in the presence of various heme analogs in E. coli system. Western blot of E. coli RP523 bearing plasmid expressing human CBS. Cell lysates (40 μg/lane) were electrophoresed on a 9% SDS gel. Following Western transfer, CBS was detected using anti-CBS monoclonal antibody (upper panel). Graph of corresponding CBS activities (bottom panel).
Taken together, these results demonstrate that the presence and availability of heme or a porphyrin analog is a limiting factor in CBS expression. CBS enzymes with different porphyrins retained their AdoMet responsiveness (results not shown). Our results further suggest that heme in CBS can be replaced with a similar structure either possessing no metal or a different metal ion in the central location.
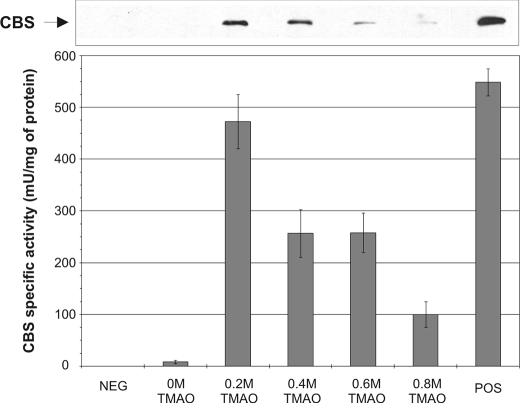
The Function of Heme in CBS Folding Can Be Substituted by a Chemical Chaperone—Recently, it was shown that chemical chaperones can facilitate proper folding and increase specific activity of misfolded or unassembled mutant CBS enzymes (28). We examined if a chemical chaperone could be beneficial for folding and/or assembly of wild-type human CBS expressed in the absence of heme. We tested the effect of different concentrations of the osmolyte chemical chaperone TMAO on expression and activity of CBS using our yeast system in the absence of exogenous heme (Fig. 4). We observed that 0.2 m TMAO is capable in rescuing expression as well as activity of CBS to 82% of WT in the absence of exogenous hemin. However, higher TMAO concentrations led to a decreased expression (Fig. 4, top) as well as lower specific activity of CBS (Fig. 4, bottom). This result further supports the chaperone-like function of heme in proper folding and/or assembly of CBS.
FIGURE 4.
Impact of chemical chaperone TMAO on human CBS expression and activity in the absence of heme. Western blot of yeast bearing plasmid expressing human CBS. Crude extracts (35 μg/lane) were electrophoresed on a 9% SDS gel. Following Western transfer, CBS was detected using anti-CBS monoclonal antibody (upper panel). Graph of corresponding CBS activities (bottom panel). Strains WY61 and WY79 served as positive and negative controls, respectively.
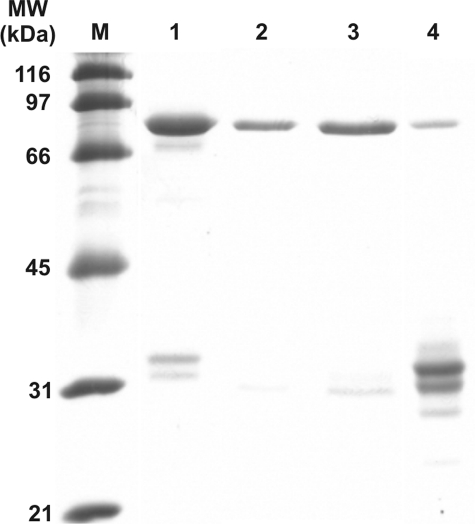
Expression and Purification of CBS Containing Heme Analogs—The finding that the heme cofactor in CBS can be replaced by a different porphyrin or even be completely missing led us to purification and characterization of such modified enzymes. For these experiments we chose to use the heme-deficient E. coli expression system, as it utilized a fusion tag that facilitated large scale purification of CBS. Therefore, we employed the hemA mutant E. coli HU227 and hemB mutant E. coli RP523 containing an uncharacterized permeability alteration that render the bacteria hemin-permeable (29). These auxotrophic mutants are able to grow without exogenous heme supplementation under anaerobic conditions. Preliminary experiments suggested that the CBS expression levels were somewhat better in the RP523 strain than in the HU227, thus all subsequent expressions were done in the former strain. We transformed it with a plasmid expressing a glutathione S-transferase (GST)-CBS fusion protein (19). We used this bacterial system for expression of human CBS in the presence of porphyrins containing Fe(III), Co(III), and Mn(III) metal ion at a final concentration of 10 μg/ml as well as in the absence of any porphyrin. SDS-PAGE shows that the isolated proteins are highly purified, whereas some degradation is visible in the last lane containing the heme-free CBS (Fig. 5).
FIGURE 5.
SDS-PAGE analysis of purified recombinant GST human CBS fusion proteins. 10 μg of each protein was separated on 9% SDS-PAGE gel and stained with SimplyBlue SafeStain (Invitrogen). Lanes: M, broad range SDS-PAGE marker (Bio-Rad); 1, Fe(III)-PPIX hCBS; 2, Mn(III)-PPIX hCBS; 3, Co(III)-PPIX hCBS; 4, GST-hemeless hCBS fusion proteins. The small bands visible in lane 4 when examined on a Western blot are not detectable with anti-CBS or anti-GST antibody.
Characterization of the Purified Enzymes—For spectral analysis and activity determinations some of the purified GST-CBS fusion proteins were cleaved with Prescission protease to remove the GST tag (all the heme analog containing CBS), whereas others were only concentrated as fusion proteins (heme-free CBS).
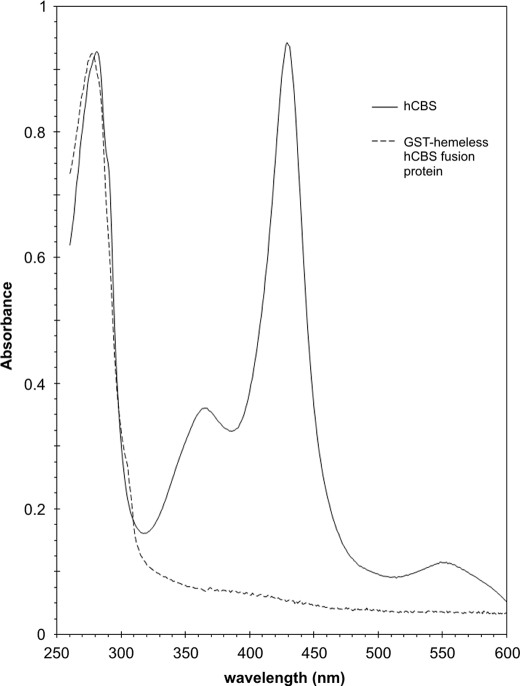
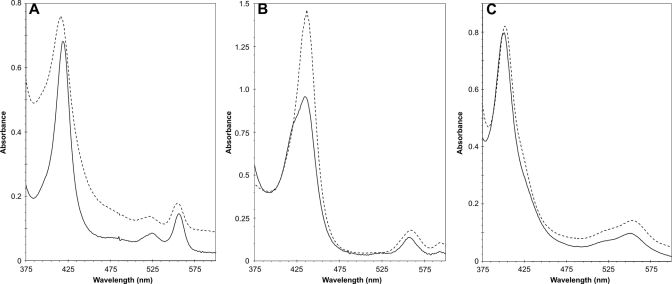
The absence of heme in the heme-free CBS was ascertained by recording absorption spectra in the visible region. Fig. 6 shows spectra of WT and heme-free CBS. In the ferric CBS there was the characteristic 428-nm Soret absorption peak for ferric CBS with a broad α/β adsorption centered at ∼550 nm. The A428/A280 ratio was equal to 1. On the other hand, the heme-free GST-CBS fusion protein spectrum did not have any significant adsorption peak in the visible spectrum. This result also means that PLP binding to the heme-free CBS was impaired consistent with our earlier observation that PLP binding depends on the presence of heme moiety in CBS (30). Fig. 7 shows the spectra of pyridine hemochrome of the reduced porphyrins released from Fe(III)-CBS, Mn(III)-CBS, and Co(III)-CBS. In each case a pyridine hemochrome spectrum of a protoporphyrin sample with a corresponding metal is shown. The reduced Fe(II)-PPXI spectrum (Fig. 7A) displays a peak at 416 and peaks at 521 and 554 nm in the α and β region, whereas the Fe(II)-CBS sample has peaks at 419, 524 and 556 nm. These peaks are within 1 nm of the pig liver CBS (H-450) hemochrome spectrum of Kim and Deal (31). The α and β peaks also agree with the hemochrome spectrum of heme b from myoglobin (32). The Mn(II)-PPXI Soret peak is red-shifted to 436 nm with the α and β peaks at 559 and 593 nm (Fig. 7B). Mn(II)-CBS absorbs at 434, 556, and 593 nm. In contrast, the Co(II)-PPXI shows a shift of the Soret peak to 423 nm and 534 and 567 nm, whereas Co(II)-CBS has very similar maxima at 423, 535, and 566 nm (Fig. 7C). These spectra demonstrate that the resulting purified proteins indeed contained the various metals and that the Mn(III)- and Co(III)-CBS did not contain any appreciable amount of iron.
FIGURE 6.
Absorption spectra of WT human CBS and GST-hemeless hCBS fusion protein. The proteins were diluted to final concentration 0.8 μg/ml in 20 mm HEPES, pH 7.4, 1 mm Tris-(2-carboxyethyl)phosphine, 0.01% Tween 20. Spectra were recorded on HP8453 UV-visible spectrophotometer.
FIGURE 7.
Reduced pyridine hemochromogen spectra of metalloporphyrins extracted from various heme-substituted CBS enzymes. Protoporphyrins were released from the respective CBS proteins (200 μg) in 30 mm NaOH and subsequently stabilized by adding 1 volume of pyridine to 3 volumes of the alkali mixture. Samples were reduced with a trace of solid Na2S2O4 and immediately measured. Solid line represents protoporphyrin released from GST-CBS fusion proteins. Dashed line corresponds to relevant protoporhyrin substance used as a standard. Stock solutions of metalloporhyrins were prepared at 2 mg/ml in 0.2 m sodium phosphate, pH 12. The standards were subsequently prepared the same way as the CBS proteins. A, iron; B, manganese; C, cobalt.
Activity assay on purified CBS enzymes showed interesting results (Table 2). Using our established procedure for expression and purification of wild type CBS we reproducibly get a fusion protein with specific activity of around 30 units/mg of protein. The CBS is expressed under aerobic conditions in a medium supplemented with the heme precursor, δ-ALA. The enzyme is fully responsive to AdoMet stimulation (2–4-fold) as well as fully saturated with PLP. However, the specific activity of CBS expressed under anaerobic conditions utilizing exogenous hemin was only 10.5 units/mg. The enzymes expressed in the presence of PPIX containing Co(III) and Mn(III) instead of iron showed similar properties except for somewhat lower specific activity: 6 and 3 units/mg, respectively. The heme-free GST-CBS fusion protein had a very low activity of about 1 unit/mg. All of the enzymes fully responded to AdoMet. With the exception of the heme-free enzyme the enzymes appear not to respond significantly to exogenous PLP suggesting that they exist predominantly as holoenzymes. These results suggest that the lack of a porphyrin-based cofactor has some effect on PLP binding as well as catalytic activity with no impact on AdoMet responsiveness. Thus, it appears that AdoMet responsiveness is independent of the presence of heme in CBS.
TABLE 2.
Characteristics of the various purified CBS enzymes
| Sample | Specific activity | AdoMet response | PLP response |
|---|---|---|---|
| units/mg protein | |||
| Aerobic WT hCBS fusion | 36 | YES (2.0x) | NO (1.1x) |
| Aerobic WT hCBS cleaved fusion | 30 | YES (1.7x) | NO (1.1x) |
| Anaerobic hemeless hCBS fusion | 1 | YES (2.1x) | YES (1.5x) |
| Anaerobic Fe(III) hCBS cleaved fusion | 10.5 | YES (2.3x) | NO (1.2x) |
| Anaerobic Mn(III) hCBS cleaved fusion | 3.0 | YES (3.5x) | NO (1.2x) |
| Anaerobic Co(III) hCBS cleaved fusion | 6.0 | YES (2.6x) | NO (1.1x) |
DISCUSSION
The heme is an essential molecule in living organisms from bacteria to humans, where it plays many critical roles ranging from redox sensing, a catalytic role as a prosthetic group of many hemeproteins to regulation of many molecular and cellular processes (33). Unlike other PLP-dependent enzymes catalyzing the production of cystathionine (i.e. cystathionine-γ-synthases, O-succinylhomoserine (thiol)-lyase, or yeast CBS), the human CBS contains in addition to PLP a second cofactor heme. The function of heme in human CBS enzyme is still, after many years of extensive research, a mystery. The crystal structure of truncated CBS revealed that the axial ligands of the heme are Cys52 and His65 (12). Thus the heme is located in the N-terminal region of CBS in a region that does not have any apparent structure. The heme of CBS is non-covalently cradled in a hydrophobic pocket ∼20 Å from the active site of the enzyme. As is typical for 6-coordinate heme proteins, CBS heme does not react readily with exogenous small molecule ligands. Oxidized Fe(III)-CBS does not react with any common heme-binding molecules (34). Heme-free enzyme is catalytically competent but the specific activity is far lower than that of the holo-enzyme (17, 35, 36). Two proposals have been made for the role of the CBS heme: 1) heme acts as a redox sensor (13, 18); and 2) heme functions as a structural support during protein folding (37, 38). Our previous data (20, 21) are inconsistent with the proposal that CBS activity is regulated by the redox state of the heme.
Our findings described here support the notion that heme in human CBS plays a structural role in subunit folding and/or native tetramer assembly rather than redox sensing or a regulatory role. In addition, we demonstrated that the iron protoporphyrin IX (heme) can be substituted with other analogs possessing different metal ions in their structure. Such substituted CBS enzymes retain the ability to form tetramers and are enzymatically active.
It was shown previously that the level of saturation of human CBS with heme substantially affects the yield and activity of the purified enzyme. The supplementation of cultures with δ-ALA, the heme biosynthesis precursor, had a dramatic impact on the yield, heme saturation as well as specific activity of human CBS expressed in E. coli (38). Moreover, the presence of heme moiety in CBS is necessary for PLP binding (30). PLP can be reversibly removed from CBS in the presence of heme. On the other hand, removal of heme is irreversible and leads to a substantial decrease in activity. Addition of heme to heme-deficient CBS could not restore either the activity or an absorption spectrum of the enzyme (30). Therefore, the presence of heme during CBS expression and folding is essential for preparation of highly active human CBS. The in vivo supplementation with hemin instead of its precursor is less effective. This could be due to a combination of several factors. First of all, heme is a substantially larger molecule than δ-ALA and its uptake may be significantly slower than that of δ-ALA. Second, the solubility of hemin as well as other porphyrins used in this study at neutral pH was very low. It increased after transformation of the protoporhyrin acids to salts by dissolving them in a strong base; however, this process is somewhat reversible depending on the final pH of the medium. Third, is it well known that hemin as the natural porphyrin possesses a significant antimicrobial activity that is augmented by the presence of hydrogen peroxide or a reducing agent (39).
Heme replacement with similar porphyrins is a powerful approach toward understanding its function in hemeproteins. Reconstitution of holoenzyme with non-iron metalloporphyrins has long been used in studies ranging from hemoglobin (40) to prostaglandin H2 synthase-1 (41). Recently, a novel way of introduction of heme cofactor analogs into proteins at the level of protein expression was presented (42). Substitution of iron protoporphyrin usually led to a decrease in enzymatic activity or even its loss (41, 43). Both the heme and the other metallo-PPIX-substituted enzymes have significantly lower activity and lower yield than the aerobically expressed WT CBS. This change may be due to the use of an E. coli strain with a mutation in a major pathway leading to heme biosynthesis. The enzymes with Co(III)-PPIX and Mn(III)-PPIX had somewhat lower activity than the Fe(III)-PPIX CBS (Table 2). Although the only difference between the heme and the studied porphyrins is in the centrally coordinated metal ion, the most likely reason of the diminished activity is altered coordination and subsequently induced perturbations in tertiary structure resulting in inefficient folding. This possibility may explain the presence of lower oligomers along with the active tetramers on native PAGE gels (result not shown).
On the other hand, CBS enzymes bearing various heme analogs retained their full responsiveness to AdoMet and ability to bind PLP. Even though responsiveness to AdoMet and heme binding occurred in CBS late in evolution they do not seem to be interdependent.
Improper protein folding resulting in its rapid degradation, aggregation, or altered trafficking usually due to point mutations can be in some cases restored by employing molecular, chemical, or pharmacological chaperones (44). Recently, the chemical chaperones such as TMAO, glycerol, or proline successfully partially rescued the enzymatic activity of three CBS mutants: I278T, T262M, and T353M (28). Moreover, as we documented here, the chaperone-like effect of TMAO ameliorated the CBS activity in the absence of heme. Heme-deficient CBS expressed in the presence of TMAO was fully responsive to AdoMet stimulation. Whether the effect of TMAO is direct by promoting better folding of heme-free CBS or indirect via alteration of activity of endogenous molecular chaperones, such as heat shock proteins, is unclear. However, in our previous work we demonstrated that addition of TMAO to an in vitro transcription/translation system could increase the specific activity of missense mutant CBS enzymes suggesting that it may work directly (28). The fact that Co(III)-PPIX or Mn(III)-PPIX substituted CBS remains active suggested that the protoporphyrin scaffold is required for proper folding of the enzyme and the nature of the central metal is not critical. The result obtained with TMAO suggests that the porphyrin itself is not essential for proper folding of the enzyme and that the chaperone-like function of the heme can be replaced by an unrelated chaperone. Moreover, rescue of CBS activity by a chemical chaperone in the absence of heme could have direct implications for some CBS-deficient homocystinuria patients having CBS, which are unable to bind or retain heme. Taken together, the presented results strongly support a structural role for heme in CBS.
Acknowledgments
We acknowledge Dr. Frank Frerman for help with the spectral analysis and many fruitful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant HL065217-06A2. This work was also supported by American Heart Association Grant AHA 2-5-80663, the Jerome Lejeune Foundation (to J. P. K.), and a generous gift from Margie McGlynn. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: CBS, cystathionine-β-synthase; PLP, pyridoxal-5′-phosphate; AdoMet, S-adenosyl-l-methionine; TMAO, trimethylamine-N-oxide; δ-ALA, δ-aminolevulinic acid; PPIX, protoporhyrin IX; GST, glutathione S-transferase; WT, wild type.
References
- 1.Mudd, S. H., Levy, H. L., and Kraus, J. P. (2001) in The Metabolic and Molecular Bases of Inherited Disease (Scriver, C. R., Beaudet, A. L., Sly, W. S., Valle, D., Childs, B., Kinzler, K., and Vogelstein, B., eds) pp. 2007–2056, 8 Ed., McGraw-Hill, New York
- 2.Stampfer, M. J., Malinow, M. R., Willett, W. C., Newcomer, L. M., Upson, B., Ullmann, D., Tishler, P. V., and Hennekens, C. H. (1992) J. Am. Med. Assoc. 268 877–881 [PubMed] [Google Scholar]
- 3.Kruman, I. I., Kumaravel, T. S., Lohani, A., Pedersen, W. A., Cutler, R. G., Kruman, Y., Haughey, N., Lee, J., Evans, M., and Mattson, M. P. (2002) J. Neurosci. 22 1752–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mills, J. L., Scott, J. M., Kirke, P. N., McPartlin, J. M., Conley, M. R., Weir, D. G., Molloy, A. M., and Lee, Y. J. (1996) J. Nutr. 126 756S–760S [DOI] [PubMed] [Google Scholar]
- 5.Seshadri, S., Beiser, A., Selhub, J., Jacques, P. F., Rosenberg, I. H., D'Agostino, R. B., Wilson, P. W., and Wolf, P. A. (2002) N. Engl. J. Med. 346 476–483 [DOI] [PubMed] [Google Scholar]
- 6.Miles, E. W., and Kraus, J. P. (2004) J. Biol. Chem. 279 29871–29874 [DOI] [PubMed] [Google Scholar]
- 7.Scott, J. W., Hawley, S. A., Green, K. A., Anis, M., Stewart, G., Scullion, G. A., Norman, D. G., and Hardie, D. G. (2004) J. Clin. Investig. 113 274–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank, N., Kery, V., Maclean, K. N., and Kraus, J. P. (2006) Biochemistry 45 11021–11029 [DOI] [PubMed] [Google Scholar]
- 9.Janosik, M., Kery, V., Gaustadnes, M., Maclean, K. N., and Kraus, J. P. (2001) Biochemistry 40 10625–10633 [DOI] [PubMed] [Google Scholar]
- 10.Shan, X., Dunbrack, R. L., Jr., Christopher, S. A., and Kruger, W. D. (2001) Hum. Mol. Genet. 10 635–643 [DOI] [PubMed] [Google Scholar]
- 11.Kery, V., Poneleit, L., and Kraus, J. P. (1998) Arch. Biochem. Biophys. 355 222–232 [DOI] [PubMed] [Google Scholar]
- 12.Meier, M., Janosik, M., Kery, V., Kraus, J. P., and Burkhard, P. (2001) EMBO J. 20 3910–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taoka, S., Lepore, B. W., Kabil, O., Ojha, S., Ringe, D., and Banerjee, R. (2002) Biochemistry 41 10454–10461 [DOI] [PubMed] [Google Scholar]
- 14.Jhee, K. H., McPhie, P., and Miles, E. W. (2000) J. Biol. Chem. 275 11541–11544 [DOI] [PubMed] [Google Scholar]
- 15.Maclean, K. N., Janosik, M., Oliveriusova, J., Kery, V., and Kraus, J. P. (2000) J. Inorg. Biochem. 81 161–171 [DOI] [PubMed] [Google Scholar]
- 16.Nozaki, T., Shigeta, Y., Saito-Nakano, Y., Imada, M., and Kruger, W. D. (2001) J. Biol. Chem. 276 6516–6523 [DOI] [PubMed] [Google Scholar]
- 17.Oliveriusova, J., Kery, V., Maclean, K. N., and Kraus, J. P. (2002) J. Biol. Chem. 277 48386–48394 [DOI] [PubMed] [Google Scholar]
- 18.Taoka, S., Ojha, S., Shan, X., Kruger, W. D., and Banerjee, R. (1998) J. Biol. Chem. 273 25179–25184 [DOI] [PubMed] [Google Scholar]
- 19.Frank, N., Kent, J. O., Meier, M., and Kraus, J. P. (2008) Arch. Biochem. Biophys. 470 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pazicni, S., Lukat-Rodgers, G. S., Oliveriusova, J., Rees, K. A., Parks, R. B., Clark, R. W., Rodgers, K. R., Kraus, J. P., and Burstyn, J. N. (2004) Biochemistry 43 14684–14695 [DOI] [PubMed] [Google Scholar]
- 21.Pazicni, S., Cherney, M. M., Lukat-Rodgers, G. S., Oliveriusova, J., Rodgers, K. R., Kraus, J. P., and Burstyn, J. N. (2005) Biochemistry 44 16785–16795 [DOI] [PubMed] [Google Scholar]
- 22.Cherney, M. M., Pazicni, S., Frank, N., Marvin, K. A., Kraus, J. P., and Burstyn, J. N. (2007) Biochemistry 46 13199–13210 [DOI] [PubMed] [Google Scholar]
- 23.Shan, X., and Kruger, W. D. (1998) Nat. Genet. 19 91–93 [DOI] [PubMed] [Google Scholar]
- 24.Kruger, W. D., and Cox, D. R. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 6614–6618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kraus, J. P. (1987) Methods Enzymol. 143 388–394 [DOI] [PubMed] [Google Scholar]
- 26.Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951) J. Biol. Chem. 193 265–275 [PubMed] [Google Scholar]
- 27.Laemmli, U. K. (1970) Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- 28.Singh, L. R., Chen, X., Kozich, V., and Kruger, W. D. (2007) Mol. Genet. Metab. 91 335–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, J. M., Umanoff, H., Proenca, R., Russell, C. S., and Cosloy, S. D. (1988) J. Bacteriol. 170 1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kery, V., Bukovska, G., and Kraus, J. P. (1994) J. Biol. Chem. 269 25283–25288 [PubMed] [Google Scholar]
- 31.Kim, I. C., and Deal, W. C., Jr. (1976) Biochemistry 15 4925–4930 [DOI] [PubMed] [Google Scholar]
- 32.Berry, E. A., and Trumpower, B. L. (1987) Anal. Biochem. 161 1–15 [DOI] [PubMed] [Google Scholar]
- 33.Mense, S. M., and Zhang, L. (2006) Cell Res. 16 681–692 [DOI] [PubMed] [Google Scholar]
- 34.Vadon-Le Goff, S., Delaforge, M., Boucher, J. L., Janosik, M., Kraus, J. P., and Mansuy, D. (2001) Biochem. Biophys. Res. Commun. 283 487–492 [DOI] [PubMed] [Google Scholar]
- 35.Bruno, S., Schiaretti, F., Burkhard, P., Kraus, J. P., Janosik, M., and Mozzarelli, A. (2001) J. Biol. Chem. 276 16–19 [DOI] [PubMed] [Google Scholar]
- 36.Evande, R., Ojha, S., and Banerjee, R. (2004) Arch. Biochem. Biophys. 427 188–196 [DOI] [PubMed] [Google Scholar]
- 37.Janosik, M., Oliveriusova, J., Janosikova, B., Sokolova, J., Kraus, E., Kraus, J. P., and Kozich, V. (2001) Am. J. Hum. Genet. 68 1506–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kery, V., Elleder, D., and Kraus, J. P. (1995) Arch. Biochem. Biophys. 316 24–29 [DOI] [PubMed] [Google Scholar]
- 39.Stojiljkovic, I., Evavold, B. D., and Kumar, V. (2001) Expert Opin. Investig. Drugs 10 309–320 [DOI] [PubMed] [Google Scholar]
- 40.Fabry, T. L., Simo, C., and Javaherian, K. (1968) Biochim. Biophys. Acta 160 118–122 [DOI] [PubMed] [Google Scholar]
- 41.Gupta, K., Selinsky, B. S., and Loll, P. J. (2006) Acta Crystallogr. Sect. D Biol. Crystallogr. 62 151–156 [DOI] [PubMed] [Google Scholar]
- 42.Woodward, J. J., Martin, N. I., and Marletta, M. A. (2007) Nat. Meth. 4 43–45 [DOI] [PubMed] [Google Scholar]
- 43.Bender, A. T., Kamada, Y., Kleaveland, P. A., and Osawa, Y. (2002) J. Inorg. Biochem. 91 625–634 [DOI] [PubMed] [Google Scholar]
- 44.Chaudhuri, T. K., and Paul, S. (2006) FEBS J. 273 1331–1349 [DOI] [PubMed] [Google Scholar]
- 45.Umanoff, H., Russell, C. S., and Cosloy, S. D. (1988) J. Bacteriol. 170 4969–4971 [DOI] [PMC free article] [PubMed] [Google Scholar]