Abstract
The primary bile acid receptor farnesoid X receptor (FXR) maintains lipid and glucose homeostasis by regulating expression of numerous bile acid-responsive genes, including an orphan nuclear receptor and metabolic regulator SHP. Using SHP as a model gene, we studied how FXR activity is regulated by p300 acetylase. FXR interaction with p300 and their recruitment to the SHP promoter and acetylated histone levels at the promoter were increased by FXR agonists in mouse liver and HepG2 cells. In contrast, p300 recruitment and acetylated histones at the promoter were not detected in FXR-null mice. p300 directly interacted with and acetylated FXR in vitro. Overexpression of p300 wild type increased, whereas a catalytically inactive p300 mutant decreased, acetylated FXR levels and FXR transactivation in cells. While similar results were observed with a related acetylase, CBP, GCN5 did not enhance FXR transactivation, and its recruitment to the promoter was not increased by FXR agonists, suggesting functional specificity of acetylases in FXR signaling. Down-regulation of p300 by siRNA decreased acetylated FXR and acetylated histone levels, and occupancy of FXR at the promoter, resulting in substantial inhibition of SHP expression. These results indicate that p300 acts as a critical coactivator of FXR induction of SHP by acetylating histones at the promoter and FXR itself. Surprisingly, p300 down-regulation altered expression of other metabolic FXR target genes involved in lipoprotein and glucose metabolism, such that beneficial lipid and glucose profiles would be expected. These unexpected findings suggest that inhibition of hepatic p300 activity may be beneficial for treating metabolic diseases.
Farnesoid X receptor (FXR)2 is a member of the nuclear receptor superfamily and the primary biosensor for endogenous bile acids (1-4). Upon activation by physiological concentrations of bile acids, FXR regulates expression of a number of bile acid-responsive genes in liver, kidney, and intestines (5). Previous studies utilizing synthetic FXR ligands and FXR-null mice have demonstrated that FXR plays a central role in maintaining whole body lipid and glucose homeostasis (6, 7). It has been established that bile acid-activated FXR regulates cholesterol and bile acid levels by induction of the orphan nuclear receptor and metabolic repressor, small heterodimeric partner (SHP) (8, 9). SHP inhibits the transcription of cholesterol 7α-hydroxylase (CYP7A1), a key enzyme in the conversion of cholesterol to bile acids in the classical bile acid biosynthetic pathway (10, 11). In addition to its crucial role in cholesterol and bile acid homeostasis, functions of FXR have recently been extended to glucose and fatty acid metabolism, prevention of gallstone formation, liver regeneration, and inhibition of intestinal bacterial growth (12-17). Although such important biological roles of FXR have now been established, molecular mechanisms by which FXR activity is modulated are largely unknown.
Nuclear receptors, including FXR, collaborate with a number of transcriptional cofactors to effectively modulate transcription of their target genes (18, 19). A recent study demonstrated that FXR activity is modulated by a metabolic coactivator, PGC-1α (PPARγ coactivator α) in response to fasting (20). PGC-1α enhances FXR gene transcription by coactivation of PPARγ and HNF-4 (hepatic nuclear factor 4) and also acts as a coactivator of FXR (20). Furthermore, histone arginine methyltransferases, CARM1 (coactivator-associated arginine methyltransferase 1, Ref. 21) and PRMT1 (protein arginine methyltransferase1, Ref. 22), and a transcriptional mediator, DRIP205, (23), have been shown to interact with FXR in vitro and coactivate FXR in cell-based reporter assays. Whether these cofactors identified from in vitro and cultured cell studies could regulate FXR activity in metabolic pathways in vivo needs to be established.
The transcription cofactor p300 functions in diverse biological pathways, including differentiation, development, and proliferation (24, 25) and expression of p300 is altered in human gastric, colorectal, and prostate carcinomas (26). Mice lacking the p300 gene die at early mid-gestation, suggesting that p300 is critical for embryonic development and organogenesis (27). p300 is a histone acetyl transferase (HAT) that catalyzes the acetylation of lysine residues not only in nucleosomal histones, but also in non-histone proteins, such as nuclear receptors, cofactors, and basal transcription factors, resulting in enhanced gene transcription (28, 29). Despite its functions in diverse biological processes, a role for p300 in metabolic regulation has not been reported.
Small heterodimer partner (SHP) is a well known FXR target and metabolic regulator (8, 9). SHP is an unusual orphan nuclear receptor, which lacks a DNA binding domain but contains a putative ligand binding domain (30). SHP interacts with and inhibits the activity of numerous nuclear receptors that are involved in regulation of diverse metabolic pathways (31-33). We recently reported that bile acid-induced SHP inhibits transcription of its target genes, including CYP7A1, by coordinately recruiting chromatin-modifying cofactors, such as mSin3A/HDACs corepressors, G9a histone lysine methyltransferase, and Swi/Snf-Brm remodeling complex to the promoter, resulting in chromatin remodeling and histone modification (32, 34). Marked alterations in cholesterol and bile acid levels in SHP-null mice have established a role for SHP in lipid homeostasis (35, 36). Interestingly, chronically elevated expression of SHP has been shown to associate with development of fatty liver and related metabolic disorders (37-39). Despite the established function of SHP in maintaining cholesterol and bile acid levels in health and disease states, how SHP is induced by bile acid-activated FXR remains relatively unknown.
From molecular, cellular, and mouse in vivo studies, we have obtained evidence indicating that p300 is critically involved in ligand-activated FXR signaling, particularly in SHP gene induction, by acetylating histones at the SHP promoter and FXR itself. Down-regulation of p300 substantially reduced SHP expression and further, altered expression of other hepatic FXR target genes, such that beneficial lipid and glucose profiles would be expected. We propose that inhibition of hepatic p300 activity may be beneficial for treating fatty liver disease and related metabolic disorders.
EXPERIMENTAL PROCEDURES
Cell Culture—Human hepatoma HepG2 cells (ATCC HB8065) were grown in phenol red-free Dulbecco's modified Eagle's medium (DMEM)/F12 (1:1). COS-1 cells were grown in DMEM media. Media were supplemented with 100 units/ml penicillin G-streptomycin sulfate and 10% heat-inactivated fetal bovine serum. For down-regulation of p300, HepG2 cells were infected with 5-25 MOI of Ad-empty or Ad-sip300, and 2 days later, cells were transfected with reporter and expression plasmids as indicated in the figure legends, and reporter assays were done.
Mouse in Vivo Experiments—Eight-week-old male mice were maintained on a 12-h light and 12-h dark cycle. Cholic acid (CA) feeding was started at 5:00 PM to reduce variability between experiments, and food intake was monitored. For GW4064 experiments, mice were treated with GW4064 (2 mg/20 g mouse in 1% Tween 80 and 1% methylcellulose) or vehicle using oral gavage and 3 h later, livers were collected for further studies. For adenoviral experiments, mice were injected with about 0.5 × 109 active viral particles (Ad-empty, Ad-3Flag-FXR) in 200 μl of phosphate-buffered saline via the tail vein, and 4-7 days after infection, the mice were fed normal chow or chow supplemented with 0.5% CA for 3-24 h. All the animal use and adenoviral protocols were approved by the Institutional Animal Care and Use and Institutional Biosafety Committees at University of Illinois at Urbana-Champaign and were in accordance with National Institutes of Health guidelines.
Plasmids and Adenoviral Vectors—Expression vectors for Gal4-FXR and 3Flag-FXR (40) were kindly provided by R. Sato, for p300 wild type and mutant (41) by M. Stallcup, and for GCN5 wild type and mutant by P. Puigserver. Reporter plasmids, SHP promoter-luc (42) and FXRE-tk-luc (21), were provided by Y. Lee and M. Ananthanarayanan, respectively. An adenoviral vector expressing p300 siRNA (Ad-sip300) was kindly provided by Dr. P. Rotwein (44). Adenoviral vectors were amplified, purified, and titered as described (34, 45).
Real-time RTPCR—Total RNA was isolated using TRIzol reagent, and cDNA was synthesized and RTPCR was performed with an iCycler iQ (Bio-Rad). The amount of PCR product for each mRNA was normalized by dividing by the amount of 36B4 PCR product. Sequences of the primers are available upon request.
In Vitro and in Cell Acetylation Assays—For in vitro acetylation assays, p300, CBP, pCAF, and GCN5, were purified from Sf9 insect cells infected with baculovirus encoding each of these proteins, as described (43). 1 μg of purified GST, GST-FXR, or core histones were incubated with each of the purified HATs in the presence of [3H]acetyl-CoA (0.25 μCi) in acetylation buffer (50 mm Hepes, pH 7.9, 10% glycerol, 1 μm GW4064). After incubation at 30 °C for 1 h, the proteins were separated by SDS-PAGE, proteins were detected by Coomassie Blue staining, and radioactivity was detected by fluorography.
To detect acetylated FXR in cells, HepG2 or COS-1 cells were transfected with expression plasmids for p300 (or infected with Ad-p300 wild type), along with Ad-Flag-FXR (5 MOI). Cells were treated with histone deacetylase inhibitors such as 0.5 μm trichostatin A (TSA) and 5 mm nicotinamide (Nam), in the presence of 200 nm GW4064 for 5 h and collected for co-IP assays as described (46-51). Briefly, 3× Flag-FXR was immunoprecipitated in post-translational modification (PTM) buffer (50 mm Tris-HCl, pH 8.0, 5 mm EDTA, 10% glycerol, 150 mm NaCl, 1% Nonidet P-40, 0.1% SDS, protease inhibitors, 1 μm TSA, 10 mm sodium butyrate, 10 mm Nam, 1 mm dithiothreitol, and phosphatase inhibitors) with 1 μg of either M2 antibody (Sigma, Inc) or goat FXR antibody (Santa Cruz Biotechnology, sc-1204) and immunoprecipitates were stringently washed with PTM buffer. Acetylated Flag-FXR in the immunoprecipitates was detected by Western blotting using acetyl lysine antibody (Cell Signaling, Inc). Membranes were stripped and Flag-FXR was detected by Western blotting using M2 or rabbit FXR (sc-13063) antibody.
Coimmunoprecipitation (Co-IP) Assays—To examine protein-protein interactions in mouse liver, co-IP assays were performed essentially as described (32, 34, 45). Briefly, 0.25-0.5 mg of mouse liver nuclear extracts was incubated with 1 μg of p300 antibody (sc-584) at 4 °C overnight. The immune complex was isolated by incubation with protein G-agarose, and proteins in the immunoprecipitates were detected by Western blotting.
Chromatin Immunoprecipitation (ChIP) Assays—ChIP assays in HepG2, normal mice, and FXR-null mice were carried out essentially as described (32, 34, 45, 52). In re-ChIP assays, chromatin was first immunoprecipitated with antisera to FXR and then eluted with 100 μl of elution buffer with 10 mm dithiothreitol at 37 °C for 30 min and then, diluted (25-fold) with dilution buffer (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 2 mm EDTA, 1% Triton X-100), and re-immunoprecipitated with IgG or p300 antibody. ChIP experiments were repeated at least three times with reproducible results. Sequences of the ChIP primers for mouse and human SHP promoters are available upon request.
RESULTS
Cholic Acid Feeding Increases the Interaction of p300 with FXR in Mouse Liver—To determine if the p300 acetylase is involved in bile acid signaling in vivo, we first examined the effects of cholic acid (CA), a primary bile acid and natural FXR agonist, on p300 interaction with FXR in mouse liver. Mice were infected with Ad-Flag-FXR or control Ad-empty and then, fed normal chow or 0.5% CA-supplemented chow for 6 h. In Western analysis of liver extracts, Flag-FXR was not detected in the Ad-empty-infected mice and was present at similar levels in the two Ad-Flag-FXR groups (supplemental Fig. S1). p300 was immunoprecipitated from liver nuclear extracts, and Flag-FXR in the immunoprecipitates was detected by Western blotting. The levels of Flag-FXR were similar in input samples, but were substantially increased in the anti-p300 immunoprecipitates after CA feeding and not detected in the IgG control precipitates (Fig. 1A).
FIGURE 1.
CA feeding increases p300 interaction with FXR in mouse liver. A, mice were tail vein-injected with Ad-3Flag-FXR or control Ad-empty and 5 days later, were fed normal (-) or 0.5% CA-supplemented (+) chow for 6 h. p300 was immunoprecipitated from liver nuclear extracts, and Flag-FXR in the immunoprecipitates was detected by Western blotting. B and C, uninfected mice were fed normal or 0.5% CA chow for 6 h, and livers were collected for further studies. B, the Shp and Cyp7a1 mRNA levels were determined by q-RTPCR, and normalized to those of 36B4. The S.E. was calculated using the Student's t test (n = 3); *, p < 0.05. C, interaction between endogenous p300, and FXR was detected by coimmunoprecipitation. FXR was immunoprecipitated from liver nuclear extracts, and p300 in the immunoprecipitates were detected by Western blotting. p300 and IgG heavy chain are indicated by an arrow and asterisk, respectively.
We confirmed these results with endogenous FXR without overexpression of Flag-FXR. First, to confirm that CA feeding is operating in vivo, we measured the mRNA levels of Shp, a well known FXR target and metabolic corepressor, and Cyp7a1, a key bile acid biosynthetic enzyme and inhibited by Shp. The mRNA levels of Shp were significantly increased, whereas those of Cyp7a1 were decreased, after 6 h of CA feeding (Fig. 1B). Endogenous FXR was immunoprecipitated with FXR antisera or IgG and p300 was detected. While p300 was not detected in the IgG precipitates, the amount of p300 in the anti-FXR immunoprecipitates was increased by CA feeding (Fig. 1C). Endogenous p300 levels were not changed after 6 h of CA feeding (supplemental Fig. S2). These results indicate that CA feeding increases the interaction of p300 with FXR in mouse liver, suggesting that p300 may be involved in bile acid-activated FXR signaling in vivo.
p300 Enhances FXR Transactivation—To determine whether the p300/FXR interaction is functionally relevant, we tested the effects of down-regulation of p300 on FXR activity. Infection with Ad-sip300 decreased the endogenous p300 mRNA levels by 80% in HepG2 cells (supplemental Fig. S3). It was shown that infection with this Ad-sip300 did not reduce mRNA levels of the acetylase CBP, which is highly conserved and functionally related with p300 (44). Expression of FXR and RXRα activated the FXRE reporter in the presence of chenodeoxy cholic acid (CDCA), a primary bile acid, or GW4046, a synthetic FXR agonist (Fig. 2A, lanes 2, 9, 16). Infection with increasing amounts of Ad-sip300 progressively and significantly inhibited FXR transactivation (Fig. 2A, lanes 9-22). These results indicate that p300, at endogenous levels, functions as a coactivator of ligand-activated FXR.
FIGURE 2.
p300 enhances FXR transactivation. HepG2 cells were infected with increasing amounts (5-25 MOI, indicated by triangles) of Ad-sip300 or Ad-empty, and 2 days later, the cells were transfected with 200 ng of (FXRE)3-luc (A) or SHP promoter-luc (B) vectors along with expression plasmids for FXR and RXR. Cells were treated with ligands overnight and luciferase and β-galactosidase activities. The values for firefly luciferase activities were normalized by dividing by the β-galactosidase activities. The S.E. was calculated using the Student's t test (n = 3), *, p < 0.05 and NS, not statistically significant, compared with control lanes 9 or 16.
Because SHP is a well known direct FXR target, we also tested if p300 coactivates FXR transactivation of the natural SHP promoter. Treatment with GW4064 or CDCA substantially increased FXR activity on the SHP promoter (Fig. 2B, lanes 2, 9, 16). Ad-sip300 significantly reduced the FXR activity in a dosedependent manner, whereas Ad-empty had little effect (lanes 9-22). These results indicate that p300 functions as a coactivator for FXR transactivation of the SHP promoter.
CA Feeding Induces Recruitment of p300 to the Shp Promoter in Mouse Liver—To further determine whether p300 is involved in ligand-activated FXR signaling, we examined whether p300 can be recruited to the SHP promoter after CA feeding. In ChIP assays, FXR was associated with the Shp promoter in livers of control mice, and CA feeding significantly increased FXR occupancy at the promoter (Fig. 3, A and B). CA feeding also substantially increased recruitment of p300 to the promoter (Fig. 3A). Consistent with p300 recruitment, acetylation of histone H3 at K9/K14, a gene activation histone mark, was increased. These effects were not observed for the control Gapdh sequence (Fig. 3A).
FIGURE 3.
CA feeding increases association of p300, FXR, and acetylated histone H3 K9/K14 with the Shp promoter. A and B, mice were fed normal or CA chow for 6 h, and livers were collected for ChIP analyses. B, the FXR band intensity was measured by densitometry, and the band intensity from control samples was set as 1. The S.E. was calculated using the Student's t test (n = 3). C, mice were fed CA chow for indicated times, and livers were collected for ChIP assays. D, FXR knockout (k/o) mice or normal mice were fed normal or CA chow for 6 h and association of indicated factors or acetylated histones with the Shp promoter were determined by ChIP analyses. For ChIP assays, sheared chromatin was precipitated with the indicated antibodies, and SHP promoter DNA was amplified by PCR. DNA from samples before immunoprecipitation was used for amplification in the total sample.
To monitor temporal association of FXR and p300 with the Shp promoter, time course ChIP assays were done. To confirm that CA feeding effectively altered expression of the Shp gene, mRNA levels were monitored by q-RTPCR. The Shp mRNA levels were increased by about 2-fold as early as 6 h of CA feeding (supplemental Fig. S4). Association of FXR with the promoter was detected in livers of untreated mice and was increased by 6 h after CA feeding (Fig. 3C). p300 was recruited to the Shp promoter at 3 h of CA feeding and reached a maximum at 6 h. Consistent with p300 recruitment, acetylated histone H3 levels were increased after CA feeding (Fig. 3C).
To determine whether the p300 recruitment was dependent on FXR, we performed ChIP assays in FXR-null mice and normal mice in parallel. It has been demonstrated that Shp expression was substantially reduced in FXR-null mice fed either control chow or CA-supplemented chow (7). In FXR-null mice, association of FXR was not observed, as expected, and the association of p300, acetylated histones, and RNA polymerase II was also largely eliminated (Fig. 3D). These results demonstrate that recruitment of p300 to the Shp promoter is FXR-dependent. Interestingly, association of RXR was observed in wild type and FXR-null mice independent of CA feeding, suggesting that another nuclear receptor that can form a heterodimer with RXR was present at the promoter.
Ligand-regulated FXR Induction of SHP in HepG2 Cells—The mechanism of p300-mediated coactivation of FXR signaling was studied in more detail in HepG2 cells. The effects of FXR ligands on the levels of acetylated histones at the SHP promoter were first studied to determine if FXR activation resulted in the same histone modification in HepG2 cells as in vivo. Acetylated histone H3 K9/K14 was detected in untreated cells, and the levels were increased after treatment with CDCA, and even more dramatically by treatment with GW4046 (Fig. 4A). Interestingly, co-treatment with the reported FXR antagonist, gugglesterone (53), blocked the CDCA-mediated increase in histone H3 acetylation and reduced acetylation to lower than basal levels (Fig. 4A). These results suggest that ligand-regulated FXR signaling in HepG2 cells is similar to that observed in mouse liver in vivo.
FIGURE 4.
FXR agonists increase association of p300, FXR, and acetylated histones at the SHP promoter in HepG2 cells. A, HepG2 cells were treated for 6 h with FXR ligands as indicated and acetylated histone H3 K9/14 levels at the SHP promoter. GS denotes gugglesterone. B-E, HepG2 cells were treated with GW4064 (B) or CDCA (C) for 3 h. HepG2 cells were treated with CDCA for indicated times (D). A-D, samples were analyzed by ChIP assay as described in the legend to Fig. 3. E, in re-ChIP assays, chromatin was first immunoprecipitated with antisera to FXR and then, re-immunoprecipitated with IgG or p300 antibody.
In ChIP assays, FXR association with the SHP promoter and p300 recruitment were markedly increased after GW4064 (Fig. 4B) or CDCA treatment (Fig. 4C). Consistent with the p300 recruitment, acetylation of histone H3 K9/K14 was also increased. An increase in the association of FXR and p300 was detected as early as 1 h after CDCA treatment reaching a maximum at 3-6 h (Fig. 4D). These ChIP studies indicate that p300 is recruited to the SHP promoter in a FXR ligand-dependent manner.
ChIP studies from FXR-null mice indicate that p300 recruitment to the SHP promoter is FXR-dependent (Fig. 3D). To further determine if p300 and FXR are simultaneously associated with the promoter, chromatin immunoprecipitated with FXR antisera was re-precipitated with p300 antisera. SHP promoter sequences were detected in CDCA-treated samples that had been re-precipitated with p300 antibody (Fig. 4E). These results suggest that both p300 and FXR are simultaneously associated with the promoter.
p300 Down-regulation Decreases Acetylated Histone Levels and FXR Binding at the SHP Promoter—To directly determine if increased histone H3 K9/K14 acetylation and FXR occupancy at the SHP promoter is dependent on p300, p300 was down-regulated over 70% by infection with Ad-sip300 (Fig. 5, A and C). GW4064 treatment increased SHP mRNA levels about 3-fold, and this increase was largely inhibited by p300 siRNA (Fig. 5A). Acetylated histone H3 K9/K14 levels and association of FXR and RNA polymerase II with the promoter were increased by GW4064, and these increases were largely eliminated in cells infected with Ad-sip300 (Fig. 5B). Similar effects of p300 siRNA were observed with CDCA treatment (Fig. 5D). These results demonstrate that increased histone H3 K9/K14 acetylation and enhanced FXR binding to the SHP promoter are dependent on p300, suggesting that p300 coactivates FXR-mediated SHP induction by increasing acetylation of promoter histones and FXR binding to the promoter chromatin.
FIGURE 5.
Increased FXR binding and histone acetylation at the SHP promoter after treatment with FXR agonists are p300-dependent. HepG2 cells were infected with Ad-sip300 (sip300) or Ad-empty (empty) and 3 days later, cells were treated with GW4064 (A and B) or CDCA (C and D). Cells were harvested for q-RTPCR (A and C) or ChIP (B and D) assays as described in the legends to Figs. 1 and 3, respectively. The S.E. was determined by the Student's t test; *, p < 0.05 (n = 3).
FXR Is Acetylated by p300 in Vitro and in Cells—The binding of transcriptional factors to promoter chromatin is increased, not only by acetylation of histones, but also by acetylation of the DNA binding factors (54, 55). Because FXR binding to the SHP promoter was inhibited by down-regulation of p300 (Fig. 5, B and D), we further asked whether FXR can be acetylated by p300. To test this possibility, we examined first whether p300 can directly interact with and acetylate FXR in vitro. GST pull-down assays showed that FXR directly interacted with p300 in vitro (supplemental Fig. S5). To determine whether FXR can be acetylated by p300, GST-FXR, or control GST were incubated with purified p300 in the presence of [3H]acetyl-CoA. Acetylation of GST-FXR as well as GST-FXR fragment was detected, whereas acetylation of GST was not (Fig. 6A). In both reactions, auto-acetylation of p300 was detected.
FIGURE 6.
FXR is acetylated by p300 in vitro and in cells. A, GST-FXR or control GST was incubated with purified p300 and [3H]acetyl-CoA, and acetylated proteins were detected by fluorography after SDS-PAGE. Acetylated FXR bands are indicated by arrows and auto-acetylated p300 is indicated by an asterisk. The dotted arrow denotes a degraded fragment of FXR, which was confirmed by Western analysis using FXR antibody. B, isolated Flag-FXR from HepG2 cells were incubated with p300 and acetyl-CoA in vitro, and acetylated FXR and FXR were detected by Western blotting as indicated. C and D, COS-1 cells were transfected with plasmids for Flag-FXR and p300 wild type (WT). The cells were treated with 200 nm GW4064 and HDAC inhibitors, 500 nm TSA and 5 mm Nam, for 5 h, and cell extracts were prepared. Flag-FXR was immunoprecipitated with M2 and acetylated FXR in the immunoprecipitates were detected by Western blotting using acetyl lysine antibody (upper panel). The membrane was stripped, and FXR was detected by FXR antibody (lower panel). IgG heavy chain is indicated by an asterisk. E, HepG2 cells were infected with Ad-3Flag-FXR and adenoviral vectors expressing p300 WT, p300 siRNA (sip300), or a catalytically inactive p300 mutant (MT), as indicated. Flag-FXR was immunoprecipitated with goat FXR antibody and acetylated FXR in the immunoprecipitates were detected by Western blotting using acetyl lysine (upper panel). The use of goat FXR antibody increases separation of the Flag-FXR from the nonspecific IgG heavy chain compared with the mouse M2 antibody. FXR in the immunoprecipitates were detected with rabbit FXR antibody (lower panel). Above the upper panel, the numbers in parentheses are the intensities for the acetylated protein bands relative to the corresponding total FXR (lower panel) with control sample (lane 1) set to 1.0. F, HepG2 cells were cotransfected with plasmids of 200 ng of Gal4-TATA-luc reporter and 5 ng of Gal4-DBD or Gal4-FXR in the presence of p300 wild type or mutant as indicated. Cells were treated with vehicle or 200 nm GW4064 overnight and harvested for reporter assays. The values for firefly luciferase activities were normalized by dividing by the β-galactosidase activities. The S.E. was calculated using the Student's t test (n = 3); *, p < 0.05.
To further confirm these results, Flag-FXR was expressed in HepG2 cells by infection with Ad-Flag-FXR. Purified Flag-FXR was incubated with the p300 acetylase and acetylated FXR was detected by Western blotting using acetyl lysine antibody. Flag-FXR was robustly acetylated by p300 in vitro (Fig. 6B).
To examine whether FXR can be acetylated in cells, COS-1 cells were transfected with plasmids for Flag-FXR and treated with deacetylase inhibitors, and then, acetylated FXR levels were detected. Acetylated FXR levels were increased by deacetylase inhibitor treatment (Fig. 6C), and similar results were observed in HepG2 cells (not shown). These results suggest that FXR is an acetylated protein that undergoes dynamic acetylation and deacetylation in cells.
To determine if p300 acetylates FXR in cells, COS-1 cells were transfected with plasmids for Flag-FXR and p300 wild type and acetylated FXR levels were detected. Acetylated FXR levels were elevated in cells overexpressing p300 (Fig. 6D). Similar results were observed after immunoprecipitation with FXR antibody instead of M2 antibody (supplemental Fig. S6). These results suggest that p300 increases FXR acetylation levels in cells.
To directly determine whether p300 acetylates FXR, HepG2 cells were infected with Ad-3Flag-FXR and either Ad-empty or Ad-p300 wild type and further infected with Ad-sip300 or catalytically inactive Ad-p300 mutant. Acetylated FXR levels were markedly increased in cells infected with Ad-p300 wild type (Fig. 6E, lanes 1 and 2). Elevated acetylated FXR levels by p300 were decreased in cells co-infected with Ad-sip300 (lane 3) or a higher dose of Ad-p300 mutant (lane 5). In control experiments, infection of Ad-sip300 markedly decreased the p300 protein levels (supplemental Fig. S7). Consistent results were observed in COS-1 cells transfected with plasmids for p300 wild type and mutant (not shown). These results indicate that FXR is acetylated by p300 in cells.
p300 Acetylase Activity Is Important for FXR Transactivation—To determine whether the p300 acetylase activity is important in enhancing FXR activity, we examined the effect of overexpression of p300 wild type or a catalytically inactive p300 mutant on FXR transactivation in HepG2 cells. Exogenous expression of p300 wild type significantly enhanced FXR transactivation in GW4064-dependent manner but overexpression of a catalytically inactive p300 mutant did not increase FXR transactivation (Fig. 6F). Consistent results were observed in cells treated with CDCA (not shown). These results with the p300 mutant indicate that p300 acetylase activity is required for p300-mediated coactivation of FXR, further suggesting that p300 enhances FXR activity by increasing acetylation of both histones at the target gene promoter and probably FXR.
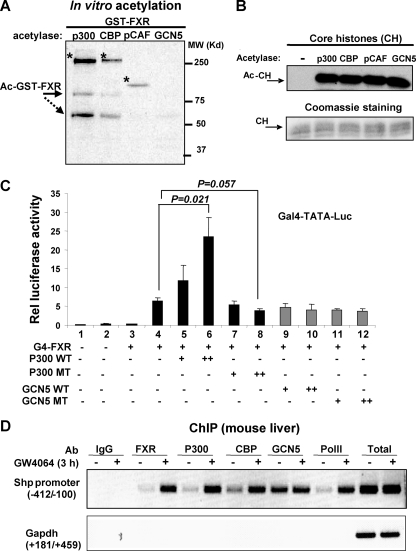
CBP, but Not GCN5, Coactivates FXR Induction of SHP—To test if FXR can be acetylated by other HATs, in vitro acetylation studies were done with CBP, pCAF, and GCN5 as well as p300. Core histones or GST were also incubated with HATs to serve positive or negative controls, respectively. GST-FXR was acetylated by p300 and to a lesser extent by CBP, but not efficiently acetylated by pCAF or GCN5 (Fig. 7A). In contrast, histones were robustly acetylated by all these acetylases (Fig. 7B). GST protein was not acetylated by any of these HATs (not shown).
FIGURE 7.
Functional specificity of acetylases in ligand-activated FXR signaling. A, purified p300, CBP, pCAF, or GCN5, were incubated with GST-FXR and [3H]acetyl-CoA. The positions of acetylated FXR and FXR fragment are indicated by solid and dotted arrows, respectively. Auto-acetylated p300, CBP, and pCAF are indicated by asterisks. B, as a control, core histones (CH) were also incubated with each of acetylases and subjected to fluorography (upper panel) or Coomassie Blue staining (lower panel) after SDS-PAGE. C, HepG2 cells were cotransfected with Gal4-TATA-luc reporter and Gal4-DBD or Gal4-FXR in the presence of p300 or GCN wild type (WT) or catalytically inactive mutants (MT). Cells were treated with vehicle or 200 nm GW4064 overnight and harvested for reporter assays. The values for firefly luciferase activities were normalized by dividing by the β-galactosidase activities. The S.E. was calculated using the Student's t test (n = 3). D, mice were treated with GW4064 using oral gavage and 3-h later, liver were collected for ChIP assays as described in the legend to Fig. 3.
In functional reporter assays, overexpression of p300 significantly increased FXR transactivation in HepG2 cells, whereas p300 mutant did not (Fig. 7C, lanes 4-8). In contrast, overexpression of GCN5 wild type or catalytically inactive mutant did not alter FXR transactivation (Fig. 7C, lanes 9-12). As a control, expression of GCN5 proteins in transfected cells was confirmed by Western analyses (not shown).
To further determine the functional specificity of acetylases in ligand-activated FXR signaling in vivo, the recruitment of p300, CBP, and GCN5, to the SHP promoter was examined in mouse liver after treatment with GW4064. GW4064 treatment increased the Shp mRNA levels by 6-fold.3 In ChIP studies, promoter occupancy of FXR, p300, or RNA polymerase II was substantially increased after GW4064 treatment (Fig. 7D). Recruitment of CBP was also markedly increased. In contrast, GCN5 was associated with the promoter and GW4064 treatment did not increase the promoter occupancy. Similar associations of these acetylases with the SHP promoter were observed in mouse liver after CA feeding (not shown). These results suggest that p300 and possibly CBP, but not GCN5, are directly involved in ligand-activated FXR induction of SHP, suggesting functional specificity of acetylases in FXR signaling.
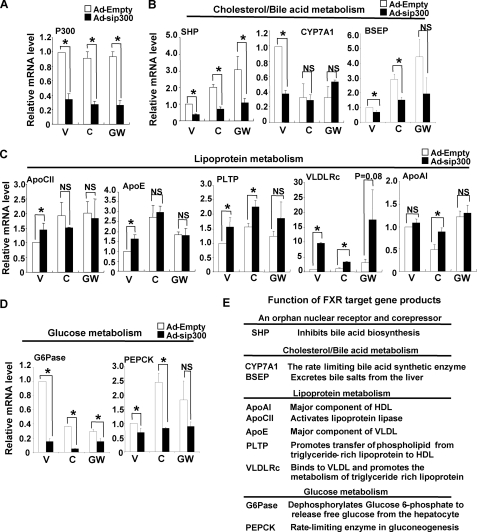
Effects of p300 Down-regulation on Expression of other Metabolic FXR Target Genes—Function of p300 in metabolic pathways was analyzed by testing the effects of down-regulation of p300 on expression of FXR target genes. The p300 mRNA levels were reduced by 70% by Ad-sip300 infection and were not altered by CDCA or GW4064 treatment in HepG2 cells (Fig. 8A). The functions of representative FXR target gene products that are examined in this study are briefly summarized in Fig. 8E. Down-regulation of p300 resulted in substantial decreases in SHP mRNA levels and significant decreases in BSEP mRNA levels in cells treated with vehicle or FXR agonists (Fig. 8B). CDCA or GW4064 treatment reduced CYP7A1 mRNA levels about 70% as expected and while infection with Ad-sip300 reduced basal levels of CYP7A1, a further reduction was not observed with agonist treatment (Fig. 8B). The mRNA levels of ApoCII or ApoE were not changed by Ad-sip300 infection, and interestingly, PLTP and VLDLR mRNA levels were increased (Fig. 8C). The mRNA levels of ApoA1 were reduced by CDCA treatment and down-regulation of p300 reversed the inhibition of ApoA1 expression (Fig. 8C). Strikingly, down-regulation of p300 reduced mRNA levels of G-6-Pase and reduced those of PEPCK (Fig. 8D).
FIGURE 8.
Down-regulation of p300 results in differential effects on the expression of FXR target genes. A-D, HepG2 cells were infected with Ad-sip300 or Ad-empty, 3 days later, cells were treated with ligands overnight, and the mRNA levels of FXR target genes were determined by q-RTPCR. V, C, and GW indicate vehicle, 50 μm CDCA, and 100 nm GW4064, respectively. Statistical significance was determined by the Student's t test; *, p < 0.05; NS indicates not statistically significant (n = 3). E, representative FXR target genes examined in this study are listed with a brief description of the function of their gene products.
These data indicate that down-regulation of p300 has differential effects on the expression of hepatic FXR target genes involved in lipoprotein and glucose metabolism. Interestingly, the changes in expression of genes involved in lipid metabolism are in directions that would lead to improved lipid profiles, and those in glucose metabolism would lead to decreased serum glucose levels.
DISCUSSION
We present evidence indicating that the p300 acetylase acts as a critical regulator in hepatic SHP gene induction, by coactivating ligand-activated FXR via acetylation of histones at the SHP promoter and probably FXR itself (Fig. 9). Several lines of evidence support this conclusion. First, the p300 interaction with FXR was increased, p300 was recruited to the SHP promoter, and histone H3 K9/K14 acetylation (a gene activation histone mark) at the promoter was increased after treatment with FXR agonists. Further, the time course of increased acetylation at the promoter after ligand treatment correlated with that of p300 in both HepG2 cells and mouse liver in vivo. Most importantly, down-regulation of p300 by siRNA nearly completely blocked the increased histone acetylation and substantially inhibited expression of the SHP gene. These results strongly suggest that p300 coactivates FXR induction of SHP by acetylating histones at the promoter.
FIGURE 9.
A proposed molecular mechanism by which p300 coactivates ligand-activated FXR induction of SHP. Unliganded FXR/RXR heterodimer is associated with the SHP promoter and acetylated histone H3 levels are minimal, resulting in basal levels of SHP expression. Activation of FXR signaling by bile acids or GW4064 increases the FXR interaction with p300 and recruits p300 to the SHP promoter. The recruited p300 increases both histone H3 acetylation at K9/14 (a gene activation histone mark) and FXR acetylation, which would increase the association of FXR, coactivator complexes, and RNA polymerase II with the SHP promoter (the solid circle denotes increased FXR binding, compared with the dotted circle in the basal state). These processes at the promoter lead to the SHP gene induction in hepatocytes in response to ligand-activated FXR signaling.
p300 may also coactivate ligand-activated FXR by acetylation of FXR itself (Fig. 9). We observed that FXR is acetylated by p300, and acetylated FXR levels were increased by p300 wild type and decreased by a catalytically inactive p300 mutant or siRNA for the p300. Importantly, increased occupancy of FXR and RNA polymerase II at the SHP promoter after treatment with FXR agonists was largely eliminated by down-regulation of p300. These results, taken together, suggest that p300 coactivates ligand-activated FXR induction of SHP by acetylating both histone at the promoter and FXR itself, which may contribute to increased association of FXR and RNA polymerase II with the promoter. Consistent with these, in functional reporter assays, overexpression of the p300 mutant blocked FXR transactivation, while p300 wild type significantly increased FXR activity. To directly demonstrate functional roles of FXR acetylation in the modulation of FXR activity, lysine residues in FXR acetylated by p300 remain to be determined.
We also observed that endogenous FXR was acetylated at a low level, and acetylated FXR levels were not markedly increased after CA feeding or treatment with GW4064 in mouse liver (not shown). It is possible that acetylation and deacetylation of FXR are dynamic processes under normal physiological conditions so that acetylated FXR levels are transiently elevated in response to activated FXR signaling pathway in hepatocytes. Whether constitutively elevated FXR acetylation is associated with pathological states will be an interesting question.
In vitro acetylation studies showed that FXR can be acetylated by p300 and CBP, but to a much lesser extent by GCN5 and pCAF. Also, overexpression of GCN5 did not increase FXR transactivation, whereas p300 significantly increased FXR activity. Consistent with these results, association of GCN5 with the SHP promoter was not increased, whereas recruitment of p300 and CBP was substantially increased in mouse liver after GW4064 treatment. These results suggest a functional specificity for p300 and probably CBP, but not GCN5, in coactivating ligand-activated FXR induction of SHP.
We observed unexpected intriguing results that down-regulation of p300 altered the expression of metabolic FXR target genes such that beneficial metabolic profiles would be expected. The expected effects of the down-regulation are complex because direct targets of FXR/p300, like SHP, would be down-regulated and indirect targets of FXR via SHP induction would be up-regulated. Furthermore, if these metabolic genes are also induced by other transcription factors that are coactivated by p300, decreased expression would be expected. Nevertheless, for the genes involved in lipoprotein metabolism, down-regulation of p300 resulted in a dramatic increase in the expression of the VLDL receptor gene, which should increase the metabolism of triglyceride-rich lipoproteins and, therefore, decrease the levels of triglycerides, which would have a beneficial cardiovascular result. Down-regulation of p300 also increased the basal levels of ApoCII, ApoE, and PLTP. While modest, these changes should result in increased synthesis of an HDL component, transfer of phospholipid to HDL, and increased metabolism of chylomicrons and VLDL. Each of these changes would result in improved lipid profiles that may reduce the probability of cardiovascular disease. For enzymes related to glucose metabolism, p300 reduced expression of G6Pase and PEPCK, both of which are involved in the production of glucose. Therefore, p300 down-regulation should contribute to decreased serum glucose levels and increased insulin sensitivity, beneficial effects in diabetes.
These potentially beneficial outcomes from p300 down-regulation and subsequent inhibition of SHP expression are in-line with recent reports that elevation of SHP expression is implicated in development of the fatty liver phenotype and diabetes (37, 38, 60). For instance, liver-specific prolonged overexpression of SHP was shown to result in depleted bile acid pools and accumulation of hepatic lipids by promoting lipogenesis in transgenic mice (37). Also, decreased lipid levels and increased insulin sensitivity were detected in SHP-null mice (60). Ablation of the SHP gene in the ob/ob mice resulted in decreases in liver and serum lipid levels and beneficial changes in metabolic profiles (38). Our studies, taken together with these reports, suggest that suppression of SHP expression by inhibiting hepatic p300 activity in metabolic disease states may result in beneficial metabolic outcomes that would be useful for treating fatty liver and related metabolic diseases.
Supplementary Material
Acknowledgments
We thank R. Sato for providing plasmids Gal4-FXR and 3Flag-FXR, P. Rotwein for Ad-sip300, P. Edwards for FXR deletion constructs, B. Forman for CMX-FXR, R. Evans for CMX-RXRα, P. Puigserver for the GCN5 plasmids, H. Lu for Ad-p300 wild type and mutant, M. Stallcup for p300 wt and the HAT mutant, M. Ananthanarayanan for FXRE-tk-luc, and Y. Lee and D. Moore for SHP-luc reporter plasmid. We thank F. Gonzalez for permission to use FXR-null mice. We give special thanks to B. Kemper for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants CA103867 and CA124760 (to C. M. C.) and DK062777 and AHA 0756028Z (to J. K. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S7.
Footnotes
The abbreviations used are: FXR, farnesoid X receptor; SHP, small heterodimer partner; CYP7A1, cytochrome P450 7A1; CA, cholic acid; CDCA, chenodeoxy cholic acid; ChIP, chromatin immunoprecipitation; HDAC, histone deacetylase; HAT, histone acetylase; TSA, trichostatin A; Nam, nicotine amide; Ad, adenovirus; HDL, high density lipoprotein; VLDL, very low density lipoprotein; WT, wild type; MOI, multiplicity of infection; GST, glutathione S-transferase.
J. Y. Lee and J. K. Kemper, unpublished data.
References
- 1.Makishima, M., Okamoto, A. Y., Repa, J. J., Tu, H., Learned, M., Luk, A., Hull, M. V., Lustig, K. D., Mangelsdorf, D. J., and Shan, B. (1999) Science 284 1362-1365 [DOI] [PubMed] [Google Scholar]
- 2.Mangelsdorf, D. J., and Evans, R. M. (1995) Cell 83 841-850 [DOI] [PubMed] [Google Scholar]
- 3.Wang, H., Chen, J., Hollister, K., Sowers, L., and Forman, B. M. (1999) Mol. Cell 3 543-553 [DOI] [PubMed] [Google Scholar]
- 4.Parks, D. J., Blanchard, S. G., Bledsoe, R. K., Chandra, G., Consler, T. G., Kliewer, S. A., Stimmel, J. B., Willson, T. M., Zavacki, A. M., Moore, D. D., and Lehmann, J. M. (1999) Science 284 1365-1368 [DOI] [PubMed] [Google Scholar]
- 5.Kalaany, N. Y., and Mangelsdorf, D. J. (2006) Annu. Rev. Physiol. 68 159-191 [DOI] [PubMed] [Google Scholar]
- 6.Willson, T. M., Jones, S. A., Moore, J. T., and Kliewer, S. A. (2001) Med. Res. Rev. 21 513-522 [DOI] [PubMed] [Google Scholar]
- 7.Sinal, C., Tohkin, M., Miyata, M., Ward, J., Lambert, G., and Gonzalez, F. J. (2000) Cell 102 731-744 [DOI] [PubMed] [Google Scholar]
- 8.Lu, T. T., Makishima, M., Repa, J. J., Schoonjans, K., Kerr, T. A., Auwerx, J., and Mangelsdorf, D. J. (2000) Mol. Cell 6 507-515 [DOI] [PubMed] [Google Scholar]
- 9.Goodwin, B., Jones, S. A., Price, R. R., Watson, M. A., McKee, D. D., Moore, L. B., Galardi, C., Wilson, J. G., Lewis, M. C., Roth, M. E., Maloney, P. R., Wilson, T. M., and Kliewer, S. A. (2000) Mol. Cell 6 517-526 [DOI] [PubMed] [Google Scholar]
- 10.Chiang, J. Y. L. (1998) Front. Biosci. 3 D176-D193 [DOI] [PubMed] [Google Scholar]
- 11.Russell, D. W. (1999) Cell 97 539-542 [DOI] [PubMed] [Google Scholar]
- 12.Ma, K., Saha, P. K., Chan, L., and Moore, D. D. (2006) J. Clin. Investig. 116 1102-1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe, M., Houten, S. M., Wang, L., Moschetta, A., Mangelsdorf, D. J., Heyman, R. A., Moore, D. D., and Auwerx, J. (2004) J. Clin. Investig. 113 1408-1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, W., Ma, K., Zhang, J., Qatanani, M., Cuvillier, J., Liu, J., Dong, B., Huang, X., and Moore, D. D. (2006) Science 312 233-236 [DOI] [PubMed] [Google Scholar]
- 15.Inagaki, T., Moschetta, A., Lee, Y. K., Peng, L., Zhao, G., Downes, M., Yu, R. T., Shelton, J. M., Richardson, J. A., Repa, J. J., Mangelsdorf, D. J., and Kliewer, S. A. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 3920-3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang, Y., Lee, F. Y., Barrera, G., Lee, H., Vales, C., Gonzalez, F. J., Willson, T. M., and Edwards, P. A. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 1006-1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moschetta, A., Bookout, A. L., and Mangelsdorf, D. J. (2004) Nat. Med. 10 1352-1358 [DOI] [PubMed] [Google Scholar]
- 18.Rosenfeld, M., and Glass, C. (2001) J. Biol. Chem. 276 36865-36868 [DOI] [PubMed] [Google Scholar]
- 19.Lazar, M. A. (2003) Nucl. Recept. Signal. 1 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang, Y., Castellani, L. W., Sinal, C. J., Gonzalez, F. J., and Edwards, P. A. (2004) Genes Dev. 18 157-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ananthanarayanan, M., Li, S., Balasubramaniyan, N., Suchy, F. J., and Walsh, M. J. (2004) J. Biol. Chem. 279 54348-54357 [DOI] [PubMed] [Google Scholar]
- 22.Rizzo, G., Renga, B., Antonelli, E., Passeri, D., Pellicciari, R., and Fiorucci, S. (2005) Mol. Pharmacol. 68 551-558 [DOI] [PubMed] [Google Scholar]
- 23.Pineda Torra, I., Freedman, L. P., and Garabedian, M. J. (2004) J. Biol. Chem. 279 36184-36191 [DOI] [PubMed] [Google Scholar]
- 24.Ogryzko, V. V., Schiltz, R. L., Russanova, V., Howard, B. H., and Nakatani, Y. (1996) Cell 87 953-959 [DOI] [PubMed] [Google Scholar]
- 25.Yao, T. P., Oh, S. P., Fuchs, M., Zhou, N. D., Ch'ng, L. E., Newsome, D., Bronson, R. T., Li, E., Livingston, D. M., and Eckner, R. (1998) Cell 93 361-372 [DOI] [PubMed] [Google Scholar]
- 26.Muraoka, M., Konishi, M., Kikuchi-Yanoshita, R., Tanaka, K., Shitara, N., Chong, J. M., Iwama, T., and Miyaki, M. (1996) Oncogene 12 1565-1569 [PubMed] [Google Scholar]
- 27.Shikama, N., Lutz, W., Kretzschmar, R., Sauter, N., Roth, J. F., Marino, S., Wittwer, J., Scheidweiler, A., and Eckner, R. (2003) EMBO J. 22 5175-5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu, M., Rao, M., Wang, C., Sakamaki, T., Wang, J., Di Vizio, D., Zhang, X., Albanese, C., Balk, S., Chang, C., Fan, S., Rosen, E., Palvimo, J. J., Janne, O. A., Muratoglu, S., Avantaggiati, M. L., and Pestell, R. G. (2003) Mol. Cell. Biol. 23 8563-8575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kraus, W. L., and Wong, J. (2002) Eur. J. Biochem. 269 2275-2283 [DOI] [PubMed] [Google Scholar]
- 30.Seol, W., Choi, H., and Moore, D. D. (1996) Science 272 1336-1339 [DOI] [PubMed] [Google Scholar]
- 31.Lee, Y., and Moore, D. D. (2002) J. Biol. Chem. 277 2463-2467 [DOI] [PubMed] [Google Scholar]
- 32.Kemper, J., Kim, H., Miao, J., Bhalla, S., and Bae, Y. (2004) Mol. Cell. Biol. 24 7707-7719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bavner, A., Sanyal, S., Gustafsson, J. A., and Treuter, E. (2005) Trends Endocrinol. Metab. 16 478-488 [DOI] [PubMed] [Google Scholar]
- 34.Fang, S., Miao, J., Xiang, L., Ponugoti, B., Treuter, E., and Kemper, J. K. (2007) Mol. Cell. Biol. 27 1407-1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerr, T. A., Saeki, S., Schneider, M., Schaefer, K., Berdy, S., Redder, T., Shan, B., Russell, D. W., and Schwarz, M. (2002) Dev. Cell 2 713-720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, L., Lee, Y., Bundman, D., Han, Y., Thevananther, S., Kim, C., Chua, S., Wei, P., Heyman, R., Karin, M., and Moore, D. (2002) Dev. Cell 2 721-731 [DOI] [PubMed] [Google Scholar]
- 37.Boulias, K., Katrakili, N., Bamberg, K., Underhill, P., Greenfield, A., and Talianidis, I. (2005) EMBO J. 24 2624-2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang, J., Iqbal, J., Saha, P. K., Liu, J., Chan, L., Hussain, M. M., Moore, D. D., and Wang, L. (2007) Hepatology 46 147-157 [DOI] [PubMed] [Google Scholar]
- 39.Wang, L., Huang, J., Saha, P., Kulkarni, R. N., Hu, M., Kim, Y. D., Park, K. G., Chan, L., Rajan, A. S., Lee, I., and Moore, D. D. (2006) Mol. Endocrinol. 11 2671-2681 [DOI] [PubMed] [Google Scholar]
- 40.Nakahara, M., Furuya, N., Takagaki, K., Sugaya, T., Hirota, K., Fukamizu, A., Kanda, T., Fujii, H., and Sato, R. (2005) J. Biol. Chem. 280 42283-42289 [DOI] [PubMed] [Google Scholar]
- 41.Lee, Y., Koh, S., Zhang, X., Cheng, X., and Stallcup, M. (2002) Mol. Cell. Biol. 22 3621-3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, Y., Dell, H., Dowhan, D. H., Hadzopoulou-Cladaras, M., and Moore, D. D. (2000) Mol. Cell. Biol. 20 187-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas, M. C., and Chiang, C. M. (2005) Mol. Cell 17 251-264 [DOI] [PubMed] [Google Scholar]
- 44.Kuninger, D., Stauffer, D., Eftekhari, S., Wilson, E., Thayer, M., and Rotwein, P. (2004) Hum. Gene Ther. 15 1287-1292 [DOI] [PubMed] [Google Scholar]
- 45.Ponugoti, B., Fang, S., and Kemper, J. K. (2007) Mol. Endocrinol. 11 2698-2712 [DOI] [PubMed] [Google Scholar]
- 46.Gu, W., and Roeder, R. G. (1997) Cell 90 595-606 [DOI] [PubMed] [Google Scholar]
- 47.Martinez-Balbas, M. A., Bauer, U. M., Nielsen, S. J., Brehm, A., and Kouzarides, T. (2000) EMBO J. 19 662-671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lerin, C., Rodgers, J. T., Kalume, D. E., Kim, S. H., Pandey, A., and Puigserver, P. (2006) Cell Metab. 3 429-438 [DOI] [PubMed] [Google Scholar]
- 49.Fu, M., Liu, M., Sauve, A. A., Jiao, X., Zhang, X., Wu, X., Powell, M. J., Yang, T., Gu, W., Avantaggiati, M. L., Pattabiraman, N., Pestell, T. G., Wang, F., Quong, A. A., Wang, C., and Pestell, R. G. (2006) Mol. Cell. Biol. 26 8122-8135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lagouge, M., Argmann, C., Gerhart-Hines, Z., Meziane, H., Lerin, C., Daussin, F., Messadeq, N., Milne, J., Lambert, P., Elliott, P., Geny, B., Laakso, M., Puigserver, P., and Auwerx, J. (2006) Cell 127 1109-1122 [DOI] [PubMed] [Google Scholar]
- 51.Baur, J. A., Pearson, K. J., Price, N. L., Jamieson, H. A., Lerin, C., Kalra, A., Prabhu, V. V., Allard, J. S., Lopez-Lluch, G., Lewis, K., Pistell, P. J., Poosala, S., Becker, K. G., Boss, O., Gwinn, D., Wang, M., Ramaswamy, S., Fishbein, K. W., Spencer, R. G., Lakatta, E. G., Le Couteur, D., Shaw, R. J., Navas, P., Puigserver, P., Ingram, D. K., de Cabo, R., and Sinclair, D. A. (2006) Nature 444 337-342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miao, J., Fang, S., Bae, Y., and Kemper, J. K. (2006) J. Biol. Chem. 281 14537-14546 [DOI] [PubMed] [Google Scholar]
- 53.Urizar, N. L., Liverman, A. B., Dodds, D. T., Silva, F. V., Ordentlich, P., Yan, Y., Gonzalez, F. J., Heyman, R. A., Mangelsdorf, D. J., and Moore, D. D. (2002) Science 296 1703-1706 [DOI] [PubMed] [Google Scholar]
- 54.Soutoglou, E., Katrakili, N., and Talianidis, I. (2000) Mol. Cell 5 745-751 [DOI] [PubMed] [Google Scholar]
- 55.Luo, J., Li, M., Tang, Y., Laszkowska, M., Roeder, R. G., and Gu, W. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2259-2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deleted in proof
- 57.Deleted in proof
- 58.Deleted in proof
- 59.Deleted in proof
- 60.Wang, L., Liu, J., Saha, P., Huang, J., Chan, L., Spiegelman, B., and Moore, D. D. (2005) Cell Metab. 2 227-238 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.