Abstract
Interferon regulatory factor 3 (IRF-3) undergoes phosphorylation-induced activation in virus-infected cells and plays an important role in the antiviral innate immune response. The E3L protein encoded by vaccinia virus is known to impair phosphorylation and activation of IRF-3. Kinases in addition to IκB kinase-related kinases are implicated in the IRF-3-dependent antiviral response. To test in human cells the role of the protein kinase regulated by RNA (PKR) in IRF-3 activation, HeLa cells made stably deficient in PKR using an RNA interference strategy were compared with PKR-sufficient cells. Rapid phosphorylation and nuclear accumulation of IRF-3 were detected in PKR-sufficient cells following infection with E3L deletion mutant (ΔE3L) virus. By contrast, the full IRF-3 activation response was largely abolished in PKR-deficient cells. The ΔE3L virus-induced IRF-3 activation seen in PKR-sufficient cells was diminished by treatment with cytosine β-d-arabinofuranoside. Furthermore, the vaccinia mutant ts23, which displays increased viral double-stranded RNA production at 39 °C, induced PKR-dependent IRF-3 phosphorylation at 39 °C but not at 31 °C. Both IRF-3 phosphorylation and cell apoptosis induced by infection with ΔE3L virus were dependent upon RIG-I-like receptor signal transduction components, including the adapter IPS-1. These data suggest that PKR facilitates the host innate immune response and apoptosis in virus-infected cells by mediating IRF-3 activation through the mitochondrial IPS-1 signal transduction pathway.
In response to viral infection, host sensors of foreign nucleic acid, including cytoplasmic RIG (retinoic acid-inducible gene) I-like receptors (RLRs)2 and membrane-bound Toll-like receptors (TLRs), signal to induce the production of cytokines (1–3). These signaling cascades when triggered in response to double-stranded (ds) RNA act through adapter proteins, TIR domain-containing adapter-inducing IFN-β (TRIF) for the endosome membrane receptor TLR3 and IFN-β-promoter simulator 1 (IPS-1, also named as VISA, MAVS) for the RIG-I and mda-5 RLRs, to induce the synthesis of type I interferons (IFNs) (2–4). Type I IFN is an important component of the host antiviral innate immune response and subsequent defenses mounted against viral infection (2, 4). In addition, type I IFNs also serve as a link between innate and adaptive immunity (5).
Among the transcription factors important in the induction of type I interferons by the RLR and TLR signaling cascades is interferon regulatory factor 3 (IRF-3) (6, 7). Constitutively expressed IRF-3 resides in the cytosol in an inactive form. Following virus infection, IRF-3 is activated by phosphorylation, dimerization, translocation to the nucleus, and association with CREB-binding protein/p300 (6). Activation of IRF-3 is regulated by two IKK-related kinases, TBK-1 and IKKε, in a two-step manner (8, 9). The first-step phosphorylation of IRF-3 occurs at the C-terminal serine/threonine cluster between amino acids 396 and 405. This phosphorylation alleviates autoinhibition, thereby allowing interaction with CREB-binding protein, and facilitates the second-step phosphorylation at serines 385 and 386, which is required for IRF-3 dimerization (8). Activated IRF-3 together with NF-κB and activating transcription factor-2 (ATF-2)/c-Jun form a transcriptionally competent enhanceosome to induce the synthesis of type I IFNs, including IFN-β (4, 10). The importance of IRF-3 in the induction of IFNs is further illustrated by the large number of viruses that have evolved strategies to antagonize the production of IFNs through either inhibition of IRF-3 activation or by degradation of IRF-3 (11, 12).
Triphosphate-containing dsRNA represents one of the important viral triggers for activation of the host innate immune response. Production of dsRNA occurs during the life cycle of several viruses, both RNA viruses and DNA viruses (13, 14). dsRNA acts as an effector of multiple cellular enzymes that are part of the IFN response, including the protein kinase regulated by dsRNA (PKR) (4, 15–17); the family of dsRNA-dependent 2′,5′-oligoadenylate synthetases (4, 17); and ADAR1 (adenosine deaminase acting on dsRNA) (17, 18). Vaccinia virus (VV), a DNA virus, produces significant amounts of dsRNA in infected cells as a consequence of overlapping convergent transcription that occurs during the intermediate and late replication phases (13, 14). To counteract the cellular innate immune response triggered by viral dsRNA, vaccinia virus has evolved strategies to antagonize IFN signaling, IFN action, and apoptosis (17, 19). One viral gene important in the antagonism is E3L (14, 17). The E3L protein of VV is synthesized early during infection and functions as a key pathogenesis factor that affects the outcome of the virus-host interaction. E3L is an important virulence gene both in cultured cells and in the mouse model (14, 20). The E3L protein mediates IFN resistance, promotes virus growth, and impairs virus-mediated apoptosis. VV with the E3L gene deleted (ΔE3L) shows a restricted host range and induces apoptosis in cell culture (21, 22). Among the mechanisms by which E3L interferes with the host innate immune defense is by blocking the phosphorylation of IRF-3 and IRF-7 (23–25).
Two functional domains have been characterized in the E3L protein as follows: a C-terminal dsRNA-binding domain (RBM) identified by homology to the repeated RBM of PKR (15, 26, 27), and an N-terminal Z-DNA-binding domain (ZBM) identified by homology to the repeated ZBM of ADAR1 (28, 29). The dsRNA regulated kinase, PKR, is a principal cellular determinant of the phenotype characteristically seen in ΔE3L mutant virus-infected cells, including impaired virus multiplication and enhanced apoptotic response in human HeLa cells (30) and in Huh-7 cells (31). In addition to PKR's well established antiviral role through phosphorylation of eIF-2α and subsequent translation inhibition (4, 32), PKR also is implicated as an effector of multiple signaling transduction pathways, including the NF-κB and p38 MAPK pathways (18, 33). PKR was among the first pathogen recognition receptors identified and subsequently has been shown to be important in regulating IFN-β production in response to dsRNA or virus, as illustrated by West Nile virus (18, 33–35). However, the underlying mechanism as to how PKR mediates the host innate immune response and IFN production has not been elucidated.
The ability of E3L to impair the host innate immune response, including IRF-3 activation, coupled with the finding that loss of PKR expression in human HeLa cells complements the E3L deletion mutant phenotype of VV, led us to test whether PKR contributes to the activation of IRF-3 in VV-infected cells. Our results established that full IRF-3 phosphorylation and nuclear translocation are mediated in part by PKR in cells infected with vaccinia virus mutants that either lack E3L entirely (ΔE3L) or that overexpress dsRNA (ts23). Using an siRNA strategy, RIG-I, mda-5, and IPS-1 were found to affect the ΔE3L-induced IRF-3 activation in addition to PKR, but TRIF did not. Interestingly, suppression of IPS-1 and IRF-3 also impaired the ΔE3L-induced apoptotic response. These findings further establish PKR not only as a downstream effector of IFN action but also as an important upstream effector of signal transduction leading to IRF-3 activation.
EXPERIMENTAL PROCEDURES
Cells and Viruses—HeLa, baby hamster kidney, and RK13 cells were maintained in Dulbecco's modified Eagle's medium complemented with 5 or 10% (v/v) fetal bovine serum (Hyclone), respectively, 100 μg/ml penicillin, and 100 units/ml streptomycin (Invitrogen) as described previously (36). HeLa cells in which PKR expression is stably knocked down (PKRkd) and a PKR knockdown control (PKRkd-con) were maintained in the above medium containing 1 μg/ml puromycin (Sigma). The Copenhagen strain (VC-2) of wild-type (WT) vaccinia virus and mutants deleted for the E3L gene (ΔE3L), the 83 N-terminal amino acids of E3L (Δ83N), or the 26 C-terminal amino acids of E3L (Δ26C) were provided by B. Jacobs (Arizona State University, Tempe). ts23 mutant that displays an increased dsRNA synthesis at (39 °C) was provided by R. Condit (University of Florida, Gainesville). Cytosine β-d-arabinofuranoside (Ara C, Sigma, C1768) treatment (10 μg/ml) was initiated 2 h before virus infection and was maintained throughout the 10-h incubation period. Vaccinia virus infections were carried out at a multiplicity of infection of 5. Virus titers were determined by plaque assay on RK13 cells (30).
siRNA Knockdown—The sequences targeted by validated chemically synthesized siRNAs (Dharmacon) in transient knockdown experiments were as follows: firefly luciferase, CTTACGCTGAGTACTTCGA (37); RIG-I, GGAAGAGGTGCAGTATATT (35, 38); mda-5, GGTGAAGGAGCAGATTCAG (38); IPS-1, TAGTTGATCTCGCGGACGA (39); TRIF, GACCAGACGCCACTCCAAC (38); IRF-3, CCGCAAAGAAGGGTTGCGT (40); and PKR, GCAGGGAGTAGTACTTAAATA (36).
A double-transfection approach was used to achieve maximal knockdown of the target proteins. Briefly, HeLa cells at 30–50% confluency were transfected with 50 pmol of siRNA per well (12-well plate) using Lipofectamine 2000 (Invitrogen). At 48 h after the first transfection, cells were re-seeded into 12-well plates at 1.5 × 105 cells per well. After overnight incubation, the second transfection with siRNA was performed; cells were infected with vaccinia virus (multiplicity of infection 5) at 48 h after the second transfection.
Western Immunoblot Analyses—Whole cell extracts were prepared in the presence of 1 mm phenylmethylsulfonyl fluoride and 1% (v/v) protease inhibitor mixture (Sigma) as described previously (36). For analysis of IRF-3 phosphorylation, 50 mm NaF and 2 mm Na2VO3 were included in the extract buffer. Proteins were fractionated by electrophoresis on SDS-10% polyacrylamide gels, transferred to nitrocellulose membranes, blocked, and then probed with an appropriate dilution of primary antibody in phosphate-buffered saline (PBS) containing 3% (w/v) skim milk. Antibodies used to detect human IRF-3 were from Santa Cruz Biotechnology, Inc. (sc-9082), or were generously provided by M. David (University of California, San Diego) and, against IRF-3 phosphorylated at Ser-396, by J. Hiscott (McGill University, Montreal). Rabbit polyclonal antibodies were used to detect human IPS-1 (Bethyl Laboratories, Inc.), human TRIF (Alexis Biochemicals Inc.), human PKR (Santa Cruz Biotechnology Inc.), phospho-PKR(Thr-446) (Santa Cruz Biotechnology Inc.), eIF-2α (Cell Signaling Technology), and phospho-eIF-2α(Ser-51) (Cell Signaling Technology); rabbit polyclonal antibody against purified vaccinia virus virions was as described previously (30). Mouse monoclonal antibodies were used to detect poly(ADP)-ribose polymerase (PARP) (Pharmingen), β-actin (Sigma), and α-tubulin (Sigma). Western immunoblot detection was performed with IRDye 800CW-conjugated anti-rabbit IgG or IRDye 680-conjugated anti-mouse IgG secondary antibody according to the manufacturer's protocols (LI-COR). Immunoreactive bands were visualized using an Odyssey infrared imaging system.
Indirect Immunofluorescence Microscopy—Cells were seeded onto coverslips (30–50% confluency) and infected with vaccinia virus (multiplicity of infection of 5) or left uninfected as indicated. Cells were fixed at 9 h after infection with 2% paraformaldehyde at 4 °C overnight and permeabilized with PBS containing 0.1% Triton X-100. Samples were incubated with antibody against IRF-3 (Santa Cruz Biotechnology, Inc., sc-9082) at 37 °C for 1 h, rinsed three times with PBS, and then incubated with secondary anti-rabbit antibody (1:500, Invitrogen) for 1 h at room temperature. Coverslips were mounted using ProLong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (Invitrogen); stained samples were visualized using an Olympus fluorescence microscope.
RESULTS
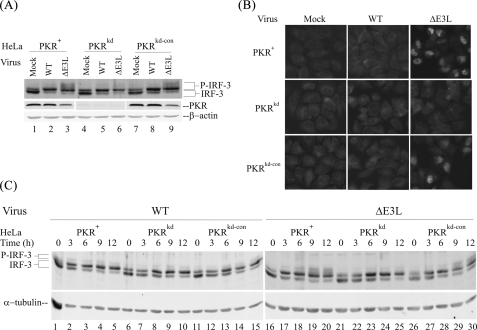
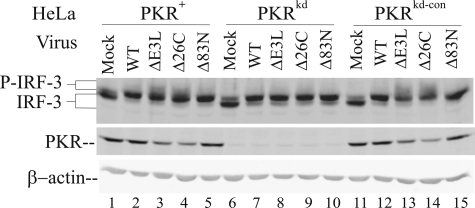
PKR Enhances and E3L Antagonizes IRF-3 Phosphorylation in Vaccinia Virus-infected Cells—To test whether PKR plays a role in the activation of transcription factor IRF-3, we first examined the phosphorylation of IRF-3 in PKR-deficient compared with PKR-sufficient HeLa cells following infection with either WT or ΔE3L mutant VV (Fig. 1). Phosphorylation of IRF-3 was assessed by a reduction in gel mobility on SDS-PAGE as described by Hiscott and co-workers (41, 42). One major lower band corresponding to unphosphorylated IRF-3 and a second more minor upper IRF-3 band corresponding to an N-terminal phosphorylated IRF-3 form were detected in uninfected cells, both PKR-sufficient and PKR-deficient cells (Fig. 1A, lanes 1, 4, and 7). This pattern of IRF-3 mobility is comparable with that seen earlier in uninfected cells (41, 42). The cells stable-deficient in PKR generated by a short hairpin RNA interference silencing strategy (PKRkd) possessed <5% of the PKR protein present in parental (PKR+) or puromycin-resistant knockdown control (PKRkd-con) cells (Fig. 1A), as described previously (30, 36).
FIGURE 1.
PKR-dependent phosphorylation and nuclear localization of IRF-3 in vaccinia virus-infected cells is impaired by E3L. A, whole cell extracts were prepared from mock-infected cells (lanes 1, 4, and 7) or cells infected for 10 h with the either wild-type (WT)(lanes 2, 5, and 8) or the E3L deletion mutant (ΔE3L) (lanes 3, 6, and 9) vaccinia virus and analyzed by Western immunoblot assay with antibody against IRF-3 and PKR. β-Actin was used as a loading control. Cells used were as follows: PKR-sufficient parental (PKR+) and knockdown control (PKRkd-con); PKR-deficient knockdown (PKRkd). B, subcellular distribution of IRF-3. Cells were mock-infected or infected with either wild-type (WT) or ΔE3L mutant virus and analyzed for IRF-3 subcellular localization at 8 h after infection by indirect immunofluorescence. C, time course of IRF-3 phosphorylation in parental PKR+, PKR-deficient knockdown PKRkd, and PKR-sufficient knockdown control PKRkd-con cells. Cells were mock-infected (0) or infected for 3, 6, 9, or 12 h with either WT (left) or ΔE3L mutant (right) virus, and whole cell extracts were prepared, and immunoblot analyses were performed with antibodies against IRF-3 and α-tubulin as a loading control.
By contrast, in cells infected for 10 h with WT virus (Fig. 1A, lanes 2, 5, and 8), the ratios of the two IRF-3 bands were reversed; the upper band corresponding to the N-terminal phosphorylated form of IRF-3 was more abundant than the unphosphorylated lower band in VV-infected PKR-sufficient and -deficient cells, again the anticipated pattern of IRF-3 mobility under conditions of virus infection (41, 42). However, in cells infected with the ΔE3L mutant, two additional IRF-3 forms, corresponding to the C-terminal phosphorylation of IRF-3 (41, 42), were seen in the two PKR-sufficient cell lines (PKR+ and PKRkd-con) (Fig. 1A, lanes 3 and 9). By contrast, in the PKRkd cells these phosphorylated IRF-3 species were substantially reduced (Fig. 1A, lane 6). The slower mobility of the C-terminally phosphorylated IRF-3 (41, 42) was confirmed using antibody against IRF-3 phosphorylated at serine 396 (data not shown, see Fig. 5A). These findings indicate that PKR plays a role in the process of vaccinia virus-induced IRF-3 C-terminal phosphorylation and that the viral E3L protein impairs this PKR-dependent IRF-3 modification.
FIGURE 5.
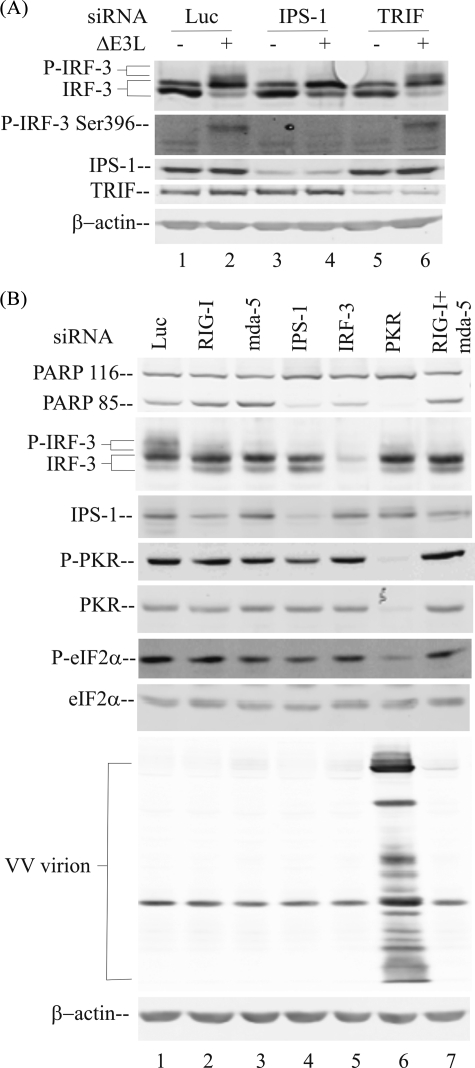
PKR-mediated IRF-3 phosphorylation occurs within the RIG-like receptor-IPS adapter signal transduction pathway. A, whole cell extracts were prepared from parental PKR+ cells, either uninfected (–) or ΔE3L virus-infected (+) for 10 h following transient knockdown as described under “Experimental Procedures,” utilizing chemically synthesized siRNAs against luciferase as a control (lanes 1 and 2), IPS-1 (lanes 3 and 4), or TRIF (lanes 5 and 6). Western immunoblot analyses were carried out with antibodies against IRF-3, phospho-IRF-3(Ser-396), TRIF, IPS-1, and β-actin as a loading control. B, whole cell extracts were prepared from parental PKR+ cells infected with ΔE3L mutant virus following transient knockdown utilizing siRNAs against the following targets: lane 1, luciferase control; lane 2, RIG-I; lane 3, mda-5; lane 4, IPS-1; lane 5, IRF-3; lane 6, PKR; lane 7, RIG-I and mda-5. Western immunoblot analyses were carried out with extracts prepared at 10 h after infection with antibodies against IRF-3, IPS-1, phospho-PKR (P-PKR), PKR, phospho-eIF-2α (P-eIF-2α), and eIF-2α or 24 h after infection to measure PARP cleavage (PARP 85), vaccinia virus protein expression, and β-actin.
IRF-3 Phosphorylation and Nuclear Localization Are Dependent upon PKR in ΔE3L Mutant-infected Cells—The phosphorylation of IRF-3 on Ser and Thr in the C-terminal region, which corresponds to the most slowly migrating forms of IRF-3 on SDS-PAGE, leads to IRF-3 activation and the cytoplasmic to nuclear translocation of the factor (41, 43). To further verify that the ΔE3L virus-induced IRF-3 phosphorylation was modulated by the PKR protein, we carried out indirect immunofluorescence microscopy to assess the subcellular distribution of IRF-3. As shown in Fig. 1B, the latent IRF-3 protein showed a cytoplasmic distribution in the mock-infected cells, both PKR-sufficient and -deficient (Fig. 1B, left panel). Infection with WT VV did not alter the diffuse IRF-3 cytosolic localization pattern (Fig. 1B, middle panel). However, infection with the ΔE3L mutant virus resulted in the nuclear accumulation of IRF-3 protein in PKR-sufficient (PKR+ or PKRkd-con) but not in PKR-deficient (PKRkd) cells (Fig. 1B, right panel). These images suggest that the nuclear translocation of IRF-3 induced by infection with ΔE3L mutant virus, in addition to the C-terminal phosphorylation, was impaired in the absence of PKR.
We next examined the phosphorylation of IRF-3 in the three cells lines after increasing times following vaccinia virus infection (Fig. 1C). Similar to the results shown in Fig. 1A at 10 h after infection, the IRF-3 phosphorylation events were marginally detectable in mock or WT virus-infected HeLa cells regardless of PKR expression level (Fig. 1C, left panel, lanes 1–15). However, in ΔE3L virus-infected PKR-sufficient HeLa cells, the phosphorylated IRF-3 proteins were detected as early as 3–6 h after infection (Fig. 1C, lanes 17 and 18 and 27 and 28). Furthermore, the IRF-3 phosphorylation products increased in abundance with time after infection of PKR-sufficient cells (Fig. 1C, lanes 19 and 20 and 29 and 30). In ΔE3L mutant-infected PKR-deficient PKRkd cells, the C-terminal region phosphorylation of IRF-3 was effectively abolished, even at 12 h after infection (Fig. 1C, lanes 22–25).
Vaccinia Virus Induction of IRF-3 Phosphorylation Is Modulated by Ara C Treatment and Displays a ts23 Phenotype—Because PKR knockdown in HeLa cells abrogated IRF-3 activation induced by the infection with the ΔE3L mutant virus, and because a common feature shared between the viral E3L and cellular PKR proteins is their ability to bind dsRNA, we hypothesized that viral dsRNA is likely a principal inducer of IRF-3 activation in vivo in ΔE3L-infected PKR-sufficient cells. Also, the time after infection when the phosphorylation of IRF-3 significantly increased, between 6 and 9 h, paralleled the time course for viral dsRNA production (13, 14) as measured by PKR activation (30).
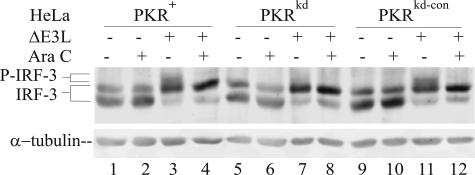
To test this hypothesis, we examined the relationship between viral dsRNA and IRF-3 phosphorylation utilizing two established approaches to modulate dsRNA levels (21). One strategy was to minimize the dsRNA produced during VV infection by using the pharmacologic agent, Ara C, which inhibits the DNA replication and reduces by about 85% viral dsRNA production (44). Treatment with Ara C abolished the PKR-dependent phosphorylation of IRF-3 seen in ΔE3L-infected PKR-sufficient HeLa cells (Fig. 2, lanes 4 and 12) compared with untreated but infected cells (Fig. 2, lanes 3 and 11). The IRF-3 protein present in ΔE3L-infected PKRkd HeLa cells did not display the PKR-dependent hyperphosphorylation state, and treatment with Ara C did not have a detectable effect (Fig. 2, lanes 7 and 8). Treatment of uninfected cells with Ara C, either PKR-sufficient or PKR-deficient, also had no discernable effect on either cell morphology (data not shown) or the phosphorylation pattern of IRF-3 (Fig. 2, lanes 2, 6, and 10).
FIGURE 2.
Ara C blocks PKR-dependent phosphorylation in ΔE3L mutant virus-infected cells. Whole cell extracts were prepared from uninfected (–) or ΔE3L virus-infected (+) cells at 10 h after infection, either untreated (–) or treated (+) with Ara C, and analyzed by Western immunoblot assay with antibody against IRF-3. α-Tubulin was used as a loading control. Cells used were as follows: PKR-sufficient parental (PKR+) and knockdown control (PKRkd-con); PKR-deficient knockdown (PKRkd).
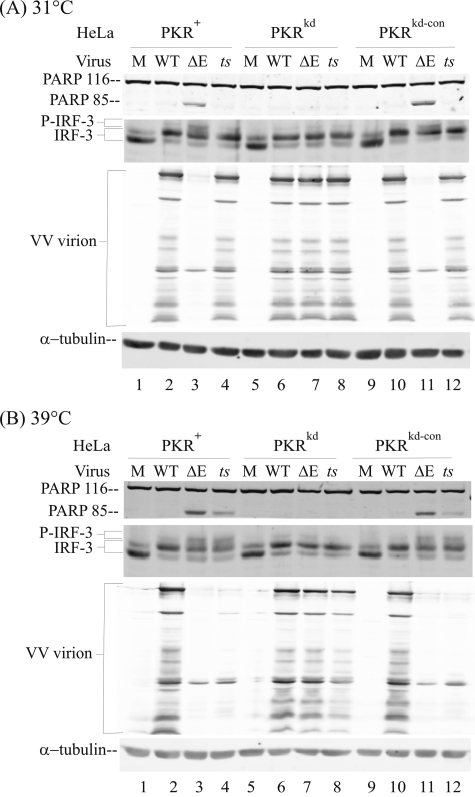
The second strategy used to assess the role of dsRNA was to increase the production of viral dsRNA in infected cells in the presence of E3L protein but without the use of pharmacologic agents. We utilized a temperature-sensitive (ts) mutant vaccinia virus, ts23, which produces a significantly increased amount of dsRNA because of aberrant transcription at restrictive (39 °C) but not at permissive (31 °C) temperatures (21, 45). As shown in Fig. 3A, when infection was carried out at permissive temperature (31 °C), the ts23 mutant displayed a phenotype indistinguishable from WT virus as follows: no PARP cleavage indicative of minimal apoptosis; no gel mobility shift of IRF-3 characteristic of PKR-dependent modification; and, importantly, robust production of viral structural proteins indicative of late gene expression.
FIGURE 3.
PKR-dependent IRF-3 phosphorylation and inhibition of viral capsid protein expression displays a temperature-sensitive phenotype in ts23 mutant vaccinia virus-infected cells. Whole cell extracts were prepared from mock-infected (M) cells or cells infected with either WT, ΔE3L mutant (ΔE), or with ts23 mutant (ts) vaccinia virus at 31 °C (A) or 39 °C (B). Cell used were as follows: PKR-sufficient parental (PKR+) and knockdown control (PKRkd-con); PKR-deficient knockdown (PKRkd). Western immunoblot analyses were carried out with antibodies against IRF-3, PARP, and vaccinia virus proteins, and α-tubulin as a loading control. PARP 116, intact PARP; PARP 85, cleavage product.
By contrast, when infection of PKR-sufficient (PKR+, PKRkd-con) cells was done at the restrictive temperature (39 °C) where the ts23 virus produces excess dsRNA (45), the ts23 virus displayed a phenotype characteristic of ΔE3L mutant virus as shown in Fig. 3B. The ts23 mutant and the ΔE3L mutant both mediated activation of PKR-dependent IRF-3 phosphorylation, cleavage of PARP, and inhibition of late viral protein synthesis at 39 °C (Fig. 3B). Finally, the phenotypes of all three viruses, WT, ΔE3L mutant, and ts23 mutant, were similar to each other in PKR-deficient cells, both at 31 °C (Fig. 3A) and at 39 °C (Fig. 3B). Importantly, the phenotype of ts23 mutant at restrictive temperature (39 °C) in PKR-deficient cells was ΔE3L-like, i.e. rescued by the PKR knockdown. These two experimental tests, use of Ara C to inhibit DNA synthesis and reduce viral dsRNA production (Fig. 2) and use of ts23 to increase aberrant transcription and dsRNA production in a temperature-dependent manner (Fig. 3), taken together further indicate VV-produced dsRNA as a principal viral mediator of the PKR-dependent IRF-3 activation.
dsRNA Binding Domain Is Essential for E3L to Block IRF-3 Activation—To further probe the mechanism as to how the vaccinia virus E3L protein blocks PKR-dependent phosphorylation, we utilized two domain-selective E3L deletion mutants, Δ26C and Δ83N, that respectively lack either the C-terminal dsRNA binding domain (Δ26C) or the N-terminal Z-DNA binding domain (Δ83N) of E3L protein. As shown in Fig. 4, infection with the Δ26C mutant that lacks the ability to bind and sequester dsRNA triggered the phosphorylation of IRF-3 in PKR-sufficient HeLa cells (lanes 4 and 14) at a level comparable with that of the ΔE3L mutant (lanes 3 and 13), whereas infection with the Δ83N mutant had no effect on the IRF-3 activation in PKR-sufficient or -deficient cells was seen with WT virus. In the absence of PKR, none of the four viruses triggered IRF-3 hyperphosphorylation to a detectable level (Fig. 4, lanes 7–10). These results suggest that the C-terminal dsRNA-binding domain of the E3L protein is indispensable for E3L to interfere with IRF-3 activation and signaling, and further indicate that viral dsRNA is the critical mediator of IRF-3 phosphorylation (Figs. 1A, 2, and 3) and activation (Fig. 1B).
FIGURE 4.
IRF-3 phosphorylation following infection of PKR+, PKRkd, and PKRkd-con with mutant viruses expressing truncated E3L proteins lacking the ZBM or RBM domain. Whole cell extracts were prepared from uninfected cells (Mock) or infected cells at 10 h post-infection with either WT or the following E3L deletion mutant viruses: ΔE3L deletion mutant; Δ26C mutant lacking RBM; Δ83N mutant lacking ZBM. Cells used were as follows: PKR-sufficient parental (PKR+) and knockdown control (PKRkd-con); PKR-deficient knockdown (PKRkd). Western immunoblot analysis was carried out with antibody against IRF-3, PKR, and β-actin as a loading control.
PKR Mediates IRF-3 Phosphorylation through an IPS-dependent Signaling Pathway—To test whether PKR mediates IRF-3 activation in response to infection with the ΔE3L mutant VV through the RIG-I-like receptor signal transduction pathway or the TLR3 receptor that also senses dsRNA (4), we examined the effect of transient knockdown of the cognate adapter proteins, IPS-1 for the RIG-I-like receptors, and TRIF for TLR3. As shown in Fig. 5A, siRNA-mediated knockdown of IPS-1 abolished the virus-induced C-terminal phosphorylation of IRF-3 measured by either mobility shift of IRF-3 or with phospho-IRF-3(Ser-396)-specific antibody (lane 4), whereas the TRIF knockdown had no effect (lane 6). Infection with the ΔE3L mutant led to an increased and comparable phosphorylation of IRF-3 in PKR+ cells transfected with siRNA against either luciferase or TRIF (Fig. 5A, lanes 2 and 6), similar to that seen in virus-induced cells not transfected with any siRNA (Figs. 1, 2, 3).
We next examined the effect of transient siRNA knockdown of individual RLR signaling pathway components, including the two dsRNA sensors, RIG-I and mda-5, and their adapter protein IPS-1, IRF-3, and PKR. As shown in Fig. 5B, siRNAs targeting mda-5 (lane 3) or IPS-1 (lane 4), or RIG-I and mda-5 together (lane 7), largely abolished the C-terminal phosphorylation of IRF-3 in parental PKR+ HeLa cells following infection with ΔE3L virus (Fig. 5B). siRNA knockdown of IRF-3 also eliminated detectable PKR-dependent phosphorylated form as expected (Fig. 5B, lane 5).
Although Western immunoblot analyses (Fig. 5) revealed that the siRNA-mediated silencing caused greater than 80% loss of the target proteins IPS-1 (Fig. 5, A, lanes 3 and 4, and B, lane 4), TRIF (Fig. 5A, lanes 5 and 6), IRF-3 (Fig. 5B, lane 5), and PKR (Fig. 5B, lane 6), inadequate antibody reagents were available to assess the knockdown of endogenous RIG-I and mda-5 in human cells. However, functional evidence consistent with mda-5 loss, in addition to the validated IPS-1 loss, was obtained as described above (Fig. 5B). Reduced but not abolished phosphorylation of IRF-3 was seen in the RIG-I (Fig. 5B, lane 2) compared with the mda-5 (Fig. 5B, lane 3) transient knockdown cells, possibly because of insufficient knockdown of these proteins to mediate a full functional change. Alternatively, mda-5 may be the key sensor of the VV dsRNAs. However, taken together, these knockdown results suggest that the PKR-mediated IRF-3 phosphorylation occurs by the RIG-I-like receptor pathway and requires the sensor mda-5, together with the adapter IPS-1, to signal in a PKR-dependent manner following vaccinia virus infection.
We also measured the growth rate of ΔE3L mutant virus in these PKR+ HeLa cells following siRNA-mediated knockdown of individual RLR signaling pathway components. The yield of ΔE3L virus was rescued by transient knockdown of PKR (data not shown), similar to our earlier finding with cells made stably deficient in PKR by RNA interference (30). However, ΔE3L virus growth was not significantly increased by transient knockdown of mda-5, RIG-I, IPS-1, or IRF-3. The rescue of ΔE3L virus growth correlated with the impairment of eIF-2α phosphorylation, which was seen only in the cells transiently knocked down for PKR (Fig. 5B, lane 6).
High phosphorylation of both PKR and eIF-2α was seen in ΔE3L virus-infected cells following the luciferase, RIG-I, mda-5, IPS-1, or IRF-3 knockdown (Fig. 5B, lanes 1–5 and 7). But these phosphorylations were impaired by transient PKR knockdown that correlated with reduced eIF-2α phosphorylation (Fig. 5B, lane 6). Consequently virion protein accumulation in the PKR-sufficient cells was far less abundant compared with cells in which PKR was knocked down and eIF-2α phosphorylation was reduced. These results, taken together, suggest that in cell culture the impairment of eIF-2α phosphorylation rather than IRF-3 activation is the major determinant for the rescue of viral late protein production and ΔE3L mutant virus growth.
DISCUSSION
Activation of the IRF-3 transcription factor, a key component of the antiviral innate immune response, is well established to occur by phosphorylation (6, 12). Although genetic disruption and biochemical analysis have firmly established the importance of two IKK-related kinases, TBK-1 and IKKε,inthe phosphorylation of IRF-3 in mouse cells (6, 8, 9, 46), additional kinases have been implicated for RNA and virus-induced phosphorylation of IRF-3. These include phosphatidylinositol 3-kinase and protein kinase Cα (47, 48). In the results reported herein, we utilized human cells in which >95% of the PKR protein was stably depleted (36) to investigate the role of PKR in the phosphorylation and activation of IRF-3 in vaccinia virus-infected cells. We demonstrate that PKR plays an important role in mediating full IRF-3 phosphorylation and subsequent nuclear translocation triggered through an IPS-1-dependent pathway. These effects were seen following infection with vaccinia virus mutants, either the ΔE3L mutant that does not express the E3L protein or the ts23 mutant that expresses increased aberrant RNA transcripts at nonpermissive temperature, but not with wild-type vaccinia virus.
Two RNA interference strategies were used to knock down PKR protein expression in HeLa cells as follows: generation of cells stably deficient in PKR by a short hairpin RNA interference approach and transient knockdown of PKR using a chemically synthesized siRNA. In cells in which PKR knockdown was achieved both stably (Fig. 1) and transiently (Fig. 5B), the phosphorylation of IRF-3 and the nuclear translocation of IRF-3 were greatly impaired in the PKR-deficient cells following VV infection with the ΔE3L mutant. Several lines of evidence point toward viral dsRNA as the key inducer of IRF-3 activation in response to VV infection of PKR-sufficient cells. These include the significant activation of IRF-3 in cells infected with E3L deletion mutants that do not express the dsRNA-binding protein E3L or express a truncated E3L protein lacking the dsRNA binding domain; activation of IRF-3 in cells infected with the ts23 mutant whereby at the restricted temperature increased amounts of dsRNA accumulate as a result of aberrant transcription (45), but not at the permissive temperature; and minimal activation in cells infected with wild-type vaccinia virus that expresses the E3L protein. The finding that Ara C treatment impairs activation of IRF-3 in ΔE3L mutant-infected cells suggests that late transcription following DNA replication facilitates production of RNA in sufficient concentration and with sufficient structure necessary to mediate activation of IRF-3.
What mechanistic role does PKR play as a nucleic acid sensor for IRF-3 activation in vaccinia virus-infected cells? Because vaccinia virus is a DNA virus that multiplies in the cytoplasm of infected cells (49) and because dsRNA has been identified in situ in vaccinia virus-infected cells (13, 45), we considered the possibility that PKR functioned within the RIG-I-like receptor signal transduction pathway for sensing cytosolic viral dsRNA (2) or TLR3 for sensing endosomal dsRNA (50). The RIG-I and mda-5 RNA helicases that signal through the mitochondrial IPS-1 adapter constitute a key pathway for sensing foreign RNA and triggering an antiviral innate immune response. We found that transient knockdown of IPS-1 almost completely abolished the PKR-dependent phosphorylation of IRF-3 induced by ΔE3L mutant virus infection, but TRIF knockdown had no effect. Likewise, transient knockdown of both RIG-I and mda-5 together essentially completely abolished the PKR-dependent IRF-3 phosphorylation, whereas knockdown of either alone had a partial effect. These results, taken together, suggest that the recognition of the intracellular vaccinia virus dsRNA was predominantly if not exclusively by the cytoplasmic helicases RIG-I and mda-5 (2, 51) and not by the membrane-bound sensor TLR3 (50).
The PKR protein possesses two putative TNF receptor-associated factor (TRAF)-interacting motifs and physically interacts in vivo with TRAF proteins, a family of adapter molecules linking different pathways with IKK activation (52, 53). TRAF3, a key component linking IPS-1 signal transduction to two downstream IKK-related kinases (TBK-1 and IKKε) in the IRF-3 signaling pathway, is reported to associate with PKR (54). Thus, it is tempting to speculate that PKR mediates the IRF-3 activation through interaction with TRAF3. However, the detailed mechanism of the PKR dependence for full activation of IRF-3 is presently unresolved, including whether the catalytic activation of PKR by dsRNA is required.
Our studies using cultured human HeLa cells further establish the importance of the viral E3L protein in antagonism of IRF-3 phosphorylation in vaccinia virus-infected cells, consistent with earlier studies with mouse embryo fibroblast cells (23). However, the mechanism by which E3L interferes with the IRF-3 activation likely differs between mouse and human cells. In the ΔE3L mutant-infected PKR–/– mouse embryo fibroblast cells, dsRNA and PKR were reportedly not involved in mediating IRF-3 phosphorylation, suggesting that E3L acted through a PKR-independent mechanism (23). By contrast, our data indicate that the E3L protein, specifically the dsRNA binding domain region of E3L (Fig. 3), impaired the IRF-3 activation signal following infection. Furthermore, dsRNA has been demonstrated in vaccinia virus-infected cells (13, 45). Furthermore, we found that with the ts23 mutant that expresses E3L but also produces greatly increased amounts of dsRNA (45), the PKR-dependent phenotype for IRF-3 activation is displayed.
Our observations that the signal transduction adapter protein IPS-1 plays an essential role in two PKR-mediated cellular responses to ΔE3L mutant infection, the IRF-3 activation pathway and the cell death signaling pathway, are intriguing. IPS-1 is located on the outer membrane of mitochondria, an organelle where apoptosis-associated proteins such as Bcl-2 family members are localized (55). IPS-1 interacts with the Fas-associated protein with death domain via its C-terminal effector domain to mediate the NF-κB activation (56). Previously PKR has been shown to be a key mediator in the apoptosis triggered by the ΔE3L mutant virus infection and dsRNA (21, 30). Although how PKR mediates apoptosis is not fully resolved, several effector proteins, including eIF-2α, NF-κB, ATF-3, and p53, are implicated in PKR-induced apoptosis (17, 57), with the apoptotic signals transmitted through Fas-associated protein with death domain and caspase-8 (58–60). These findings taken together suggest that IPS-1 and PKR may cooperatively function in the cellular responses to virus infection. Our observations obtained with the ΔE3L mutant VV are consistent with recent findings showing both IPS-1 and IRF-3 are required for efficient apoptosis induced by infection with reovirus Dearing-type 3 (T3D), a dsRNA virus (61). In Sendai virus-infected cells, the apoptotic response is impaired by siRNA knockdown of IRF-3 or by blocking the IRF-3 activation pathway via RIG-I (62).
In summary, we have established that in vaccinia virus infected human HeLa cells infected with either the ΔE3L mutant or the ts23 mutant, the full activation of IRF-3 occurs in a PKR-dependent manner. Furthermore, the PKR-dependent signal transduction appears to occur through an IPS-1-dependent RNA sensor response. PKR, in the context of innate immunity, thus functions in two conceptually different roles as follow: one as an IFN-induced protein that alters translational patterns in virus-infected cells through phosphorylation of eIF-2α to inhibit virus growth, and the other as a signal transducer leading to the full activation of the IRF-3 factor following virus infection. Although PKR is one of many IFN-inducible genes and, as such, most likely plays a redundant role in the antiviral actions of interferon (12, 17), the results presented herein suggest that PKR may play a more unique role in the full activation of IRF-3 following infection with mutant vaccinia viruses. It is now of utmost importance to establish the precise mechanism by which PKR functions in signal transduction, together with IPS-1 and TBK-1 and IKKε, to mediate the full activation of IRF-3.
Acknowledgments
We thank R. Condit, M. David, J. Hiscott, and B. Jacobs for generously providing reagents and materials.
This work was supported, in whole or in part, by National Institutes of Health Grants AI-12520 and AI-20611 (NIAID research grants from USPHS). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: RLR, retinoic acid-inducible gene (RIG) I-like receptor; dsRNA, double-stranded RNA; IPS-1, IFN-β-promoter simulator 1; IRF-3, interferon regulatory factor 3; PKR, the RNA-dependent protein kinase inducible by interferon; TLR, Toll-like receptor; VV, vaccinia virus; CREB, cAMP-response element-binding protein; PBS, phosphate-buffered saline; WT, wild type; siRNA, short interfering RNA; TRAF, TNF receptor-associated factor; PARP, poly(ADP)-ribose polymerase; IFN, interferon; Ara C, cytosine β-d-arabinofuranoside.
References
- 1.Akira, S., and Hemmi, H. (2003) Immunol. Lett. 85 85–95 [DOI] [PubMed] [Google Scholar]
- 2.Yoneyama, M., and Fujita, T. (2007) J. Biol. Chem. 282 15315–15318 [DOI] [PubMed] [Google Scholar]
- 3.Onomoto, K., Yoneyama, M., and Fujita, T. (2007) Curr. Top. Microbiol. Immunol. 316 193–205 [DOI] [PubMed] [Google Scholar]
- 4.Borden, E. C., Sen, G. C., Uze, G., Silverman, R. H., Ransohoff, R. M., Foster, G. R., and Stark, G. R. (2007) Nat. Rev. Drug Discov. 6 975–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Bon, A., and Tough, D. F. (2002) Curr. Opin. Immunol. 14 432–436 [DOI] [PubMed] [Google Scholar]
- 6.Hiscott, J. (2007) J. Biol. Chem. 282 15325–15329 [DOI] [PubMed] [Google Scholar]
- 7.Tamura, T., Yanai, H., Savitsky, D., and Taniguchi, T. (2008) Annu. Rev. Immunol. 26 535–584 [DOI] [PubMed] [Google Scholar]
- 8.Panne, D., McWhirter, S. M., Maniatis, T., and Harrison, S. C. (2007) J. Biol. Chem. 282 22816–22822 [DOI] [PubMed] [Google Scholar]
- 9.Fitzgerald, K. A., McWhirter, S. M., Faia, K. L., Rowe, D. C., Latz, E., Golenbock, D. T., Coyle, A. J., Liao, S. M., and Maniatis, T. (2003) Nat. Immunol. 4 491–496 [DOI] [PubMed] [Google Scholar]
- 10.Panne, D., Maniatis, T., and Harrison, S. C. (2007) Cell 129 1111–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haller, O., Kochs, G., and Weber, F. (2006) Virology 344 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Randall, R. E., and Goodbourn, S. (2008) J. Gen. Virol. 89 1–47 [DOI] [PubMed] [Google Scholar]
- 13.Weber, F., Wagner, V., Rasmussen, S. B., Hartmann, R., and Paludan, S. R. (2006) J. Virol. 80 5059–5064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs, B. L., and Langland, J. O. (1996) Virology 219 339–349 [DOI] [PubMed] [Google Scholar]
- 15.McCormack, S. J., Thomis, D. C., and Samuel, C. E. (1992) Virology 188 47–56 [DOI] [PubMed] [Google Scholar]
- 16.Samuel, C. E. (1979) Proc. Natl. Acad. Sci. U. S. A. 76 600–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samuel, C. E. (2001) Clin. Microbiol. Rev. 14 778–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toth, A. M., Zhang, P., Das, S., George, C. X., and Samuel, C. E. (2006) Prog. Nucleic Acids Res. Mol. Biol. 81 369–434 [DOI] [PubMed] [Google Scholar]
- 19.Seet, B. T., Johnston, J. B., Brunetti, C. R., Barrett, J. W., Everett, H., Cameron, C., Sypula, J., Nazarian, S. H., Lucas, A., and McFadden, G. (2003) Annu. Rev. Immunol. 21 377–423 [DOI] [PubMed] [Google Scholar]
- 20.Brandt, T. A., and Jacobs, B. L. (2001) J. Virol. 75 850–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kibler, K. V., Shors, T., Perkins, K. B., Zeman, C. C., Banaszak, M. P., Biesterfeldt, J., Langland, J. O., and Jacobs, B. L. (1997) J. Virol. 71 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beattie, E., Paoletti, E., and Tartaglia, J. (1995) Virology 210 254–263 [DOI] [PubMed] [Google Scholar]
- 23.Smith, E. J., Marie, I., Prakash, A., Garcia-Sastre, A., and Levy, D. E. (2001) J. Biol. Chem. 276 8951–8957 [DOI] [PubMed] [Google Scholar]
- 24.Xiang, Y., Condit, R. C., Vijaysri, S., Jacobs, B., Williams, B. R., and Silverman, R. H. (2002) J. Virol. 76 5251–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langland, J. O., Kash, J. C., Carter, V., Thomas, M. J., Katze, M. G., and Jacobs, B. L. (2006) J. Virol. 80 10083–10095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang, H. W., and Jacobs, B. L. (1993) Virology 194 537–547 [DOI] [PubMed] [Google Scholar]
- 27.Ho, C. K., and Shuman, S. (1996) J. Virol. 70 2611–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, Y. G., Muralinath, M., Brandt, T., Pearcy, M., Hauns, K., Lowenhaupt, K., Jacobs, B. L., and Rich, A. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 6974–6979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patterson, J. B., and Samuel, C. E. (1995) Mol. Cell. Biol. 15 5376–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, P., Jacobs, B. L., and Samuel, C. E. (2008) J. Virol. 82 840–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arsenio, J., Deschambault, Y., and Cao, J. (2008) Virology 377 124–132 [DOI] [PubMed] [Google Scholar]
- 32.Samuel, C. E. (1993) J. Biol. Chem. 268 7603–7606 [PubMed] [Google Scholar]
- 33.Williams, B. R. (1999) Oncogene 18 6112–6120 [DOI] [PubMed] [Google Scholar]
- 34.Diebold, S. S., Montoya, M., Unger, H., Alexopoulou, L., Roy, P., Haswell, L. E., Al-Shamkhani, A., Flavell, R., Borrow, P., and Reis e Sousa, C. (2003) Nature 424 324–328 [DOI] [PubMed] [Google Scholar]
- 35.Gilfoy, F. D., and Mason, P. W. (2007) J. Virol. 81 11148–11158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, P., and Samuel, C. E. (2007) J. Virol. 81 8192–8200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouxsein, N. F., McAllister, C. S., Ewert, K. K., Samuel, C. E., and Safinya, C. R. (2007) Biochemistry 46 4785–4792 [DOI] [PubMed] [Google Scholar]
- 38.Li, K., Chen, Z., Kato, N., Gale, M., Jr., and Lemon, S. M. (2005) J. Biol. Chem. 280 16739–16747 [DOI] [PubMed] [Google Scholar]
- 39.Kawai, T., Takahashi, K., Sato, S., Coban, C., Kumar, H., Kato, H., Ishii, K. J., Takeuchi, O., and Akira, S. (2005) Nat. Immunol. 6 981–988 [DOI] [PubMed] [Google Scholar]
- 40.Uno, T., Hirabayashi, K., Murai, M., Yano, J., and Ozato, K. (2005) Mol. Cancer Ther. 4 799–805 [DOI] [PubMed] [Google Scholar]
- 41.Lin, R., Heylbroeck, C., Pitha, P. M., and Hiscott, J. (1998) Mol. Cell. Biol. 18 2986–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Servant, M. J., ten Oever, B., LePage, C., Conti, L., Gessani, S., Julkunen, I., Lin, R., and Hiscott, J. (2001) J. Biol. Chem. 276 355–363 [DOI] [PubMed] [Google Scholar]
- 43.Wathelet, M. G., Lin, C. H., Parekh, B. S., Ronco, L. V., Howley, P. M., and Maniatis, T. (1998) Mol. Cell 1 507–518 [DOI] [PubMed] [Google Scholar]
- 44.Colby, C., Jurale, C., and Kates, J. R. (1971) J. Virol. 7 71–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bayliss, C. D., and Condit, R. C. (1993) Virology 194 254–262 [DOI] [PubMed] [Google Scholar]
- 46.Sharma, S., tenOever, B. R., Grandvaux, N., Zhou, G. P., Lin, R., and Hiscott, J. (2003) Science 300 1148–1151 [DOI] [PubMed] [Google Scholar]
- 47.Sarkar, S. N., Peters, K. L., Elco, C. P., Sakamoto, S., Pal, S., and Sen, G. C. (2004) Nat. Struct. Mol. Biol. 11 1060–1067 [DOI] [PubMed] [Google Scholar]
- 48.Johnson, J., Albarani, V., Nguyen, M., Goldman, M., Willems, F., and Aksoy, E. (2007) J. Biol. Chem. 282 15022–15032 [DOI] [PubMed] [Google Scholar]
- 49.Moss, B. (2001) in Fields Virology (Knipe, D. M., Howley, P. M., Griffin, D. E., Lamb, R. A., Martin, M. A., Roizman, B., and Straus, S. E., eds) 4th Ed., pp. 2849–2883, Lippincott Williams & Wilkins, Philadelphia
- 50.Uematsu, S., and Akira, S. (2007) J. Biol. Chem. 282 15319–15323 [DOI] [PubMed] [Google Scholar]
- 51.Takeuchi, O., and Akira, S. (2008) Curr. Opin. Immunol. 20 17–22 [DOI] [PubMed] [Google Scholar]
- 52.Arch, R. H., Gedrich, R. W., and Thompson, C. B. (1998) Genes Dev. 12 2821–2830 [DOI] [PubMed] [Google Scholar]
- 53.Gil, J., Garcia, M. A., Gomez-Puertas, P., Guerra, S., Rullas, J., Nakano, H., Alcami, J., and Esteban, M. (2004) Mol. Cell. Biol. 24 4502–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oganesyan, G., Saha, S. K., Guo, B., He, J. Q., Shahangian, A., Zarnegar, B., Perry, A., and Cheng, G. (2006) Nature 439 208–211 [DOI] [PubMed] [Google Scholar]
- 55.Hockenbery, D., Nunez, G., Milliman, C., Schreiber, R. D., and Korsmeyer, S. J. (1990) Nature 348 334–336 [DOI] [PubMed] [Google Scholar]
- 56.Seth, R. B., Sun, L., Ea, C. K., and Chen, Z. J. (2005) Cell 122 669–682 [DOI] [PubMed] [Google Scholar]
- 57.Garcia, M. A., Gil, J., Ventoso, I., Guerra, S., Domingo, E., Rivas, C., and Esteban, M. (2006) Microbiol. Mol. Biol. Rev. 70 1032–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Balachandran, S., Kim, C. N., Yeh, W. C., Mak, T. W., Bhalla, K., and Barber, G. N. (1998) EMBO J. 17 6888–6902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gil, J., and Esteban, M. (2000) Oncogene 19 3665–3674 [DOI] [PubMed] [Google Scholar]
- 60.Iordanov, M. S., Ryabinina, O. P., Schneider, P., and Magun, B. E. (2005) Apoptosis 10 153–166 [DOI] [PubMed] [Google Scholar]
- 61.Holm, G. H., Zurney, J., Tumilasci, V., Leveille, S., Danthi, P., Hiscott, J., Sherry, B., and Dermody, T. S. (2007) J. Biol. Chem. 282 21953–21961 [DOI] [PubMed] [Google Scholar]
- 62.Peters, K., Chattopadhyay, S., and Sen, G. C. (2008) J. Virol. 82 3500–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]