Abstract
C/EBP homology protein (CHOP), a stress-induced transcription factor, is involved in transcriptional regulation, cell cycle, and apoptosis. The present studies identified CHOP as an interacting partner of activating transcription factor (ATF) 4 in a yeast two-hybrid screen and confirmed their interaction in HEK293T cells. CHOP protein levels rose modestly and transiently during amino acid deprivation, whereas endoplasmic reticulum stress caused a much higher and sustained expression of CHOP protein. Exogenous CHOP expression enhanced the TRB3 gene induction by amino acid deprivation. Conversely, CHOP suppressed the induction of the endogenous asparagine synthetase (ASNS) gene and inhibited transcription from a reporter gene driven by the ASNS promoter following activation by ATF4 or amino acid deprivation. Short interfering RNA-mediated knockdown of CHOP further enhanced the induction of ASNS by either amino acid deprivation or endoplasmic reticulum stress. The CHOP-dependent repression of the ASNS gene required the entire CHOP protein, arguing against the possibility of simple sequestration of ATF4 by the CHOP leucine zipper domain, and chromatin immunoprecipitation analysis showed association of CHOP with the ASNS and TRB3 promoters. Interestingly, chromatin immunoprecipitation also showed that CHOP was associated with the C/EBP-ATF composite site regions of the SNAT2, VEGF, and CAT-1 genes, despite no significant effect on their expression after exogenous CHOP overexpression. Collectively, the results document that CHOP is a member of the transcription factor network that controls the stress-induced regulation of specific C/EBP-ATF-containing genes, such as ASNS.
Mammalian cells have evolved an array of pathways that mediate the cellular response to changes in the environment, including UV irradiation, endoplasmic reticulum (ER)2 stress, and deprivation of nutrients, such as amino acids and glucose (1). The cascade of events that is triggered by ER stress is referred to as the unfolded protein response (UPR), and the pathway initiated by amino acid limitation is called the amino acid response (AAR) pathway. These pathways initiate a wide array of adaptive mechanisms and ultimately, if necessary, programmed cell death (1, 2). Amino acid limitation leads to an increase in uncharged tRNA, which binds to and activates a kinase called GCN2 (general control nonderepressible 2) (3-5). ER stress, triggered by perturbation of calcium homeostasis, glucose deprivation, or other causes of misfolded protein accumulation in the ER lumen, activates a kinase called PERK (double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase) (6-8). Both GCN2 and PERK phosphorylate the eukaryotic initiation factor 2α on serine 51, which provokes a suppression of global protein synthesis, but a paradoxical increase in the translation of selected mRNAs containing short upstream opening reading frames (9). Among the proteins for which translation is increased is activating transcription factor 4 (ATF4) (10, 11), a member of the ATF subfamily of the basic leucine zipper (bZIP) transcription factor superfamily (12). ATF members are known to heterodimerize within the ATF family as well as with members of another bZIP subgroup, the CCAAT/enhancer-binding protein (C/EBP) family, which consists of six members: C/EBPα,-β,-γ,-δ,-ε, and CHOP (13). A C/EBPβ-ATF4 complex has been detected at cAMP-response elements (14, 15), as well as at C/EBP-ATF composite sites (16-18).
Among the ATF4 target genes are asparagine synthetase (ASNS) and CCAAT/enhancer-binding protein (C/EBP) homology protein (CHOP), also known as growth arrest and DNA damage protein 153 (GADD153). ASNS, for which the gene product catalyzes the glutamine and ATP-dependent conversion of aspartic acid to asparagine, is transcriptionally activated by ATF4 (19). The ASNS promoter contains two cis-acting sequences, Nutrient Sensing Response Elements-1 and -2 (NSRE-1 and NSRE-2), that together function as an enhancer element to mediate the transcriptional activation of the gene by either the AAR or the UPR pathway (20). The NSRE-1 sequence is a C/EBP-ATF composite site that has been shown by both electrophoretic mobility shift and ChIP assays to bind ATF4 following increased de novo synthesis of ATF4 following activation of either the AAR or the UPR (17, 19). C/EBP-ATF composite sites are composed of a half-site for C/EBP proteins and a half-site for ATF proteins (16, 21).
CHOP was originally isolated as a gene induced in response to DNA-damaging agents (22) and subsequently demonstrated to be induced by amino acid deprivation (23) and ER stress (24). The prototypic C/EBP protein contains a transcriptional activation/repression domain at its N terminus and a bZIP region at its C terminus, which consists of a basic DNA binding domain and a leucine zipper motif for dimerization. The basic region of CHOP contains proline and glycine substitutions in conserved residues believed to be essential to the interaction of these proteins with most C/EBP DNA-binding sites (25-27). Consequently, it was originally proposed that CHOP lacked DNA binding activity for prototypic C/EBP-binding sites and instead negatively regulated the activity of bZIP proteins by sequestering them through nonfunctional dimers, thereby inhibiting their ability to bind DNA (25). Indeed, CHOP-C/EBP heterodimers fail to bind several known C/EBP sequences in vitro, and when expressed in cells, CHOP attenuates the ability of other C/EBP proteins to activate promoter sites (25). However, subsequent studies revealed that a CHOP-C/EBP heterodimer could function as a positive trans-activator (28). More recently, TRB3 was also identified as a CHOP-inducible gene (29).
CHOP plays an essential role in the response to a wide variety of cell stresses and induces cell cycle arrest and apoptosis in response to ER stress (2). The basal expression level of CHOP is almost undetectable in most cell types, but expression is rapidly induced through ATF4-dependent transcription (25, 30). Therefore, CHOP is one of the most important components in the network of stress-inducible transcription factors, and deciphering the range of CHOP actions is critical for understanding the molecular mechanisms for the cellular response to stress. However, the number of investigations focused on interaction of CHOP with ATF transcription factors and the physiological role of those interactions is limited. Chen et al. (31) demonstrated that CHOP inhibits ATF3 function by forming a nonfunctional heterodimer, similar to the dominant negative effect on C/EBP proteins. Ohoka et al. (29) showed that CHOP and ATF4 cooperatively activate TRB3 expression during ER stress but did not investigate whether or not this response required direct interaction. Likewise, CHOP antagonizes the action of ATF4 and ATF5 at C/EBP-ATF composite sites, but not the action of CREB at cyclic AMP-response elements (32). The first published evidence for CHOP-ATF dimerization was from Gachon et al. (33) who showed that CHOP binds to ATF4 in vitro based on glutathione S-transferase pulldown assays and that in a reporter assay CHOP blocks the action of ATF4.
In this study, the interaction between ATF4 and CHOP was documented in intact cells, and the results also demonstrated that CHOP negatively regulates ATF4-dependent transcriptional activation of the ASNS gene. The inhibitory effect of CHOP on the ASNS gene was not observed when truncated proteins, encoding either the N- or C-terminal portion of the protein, were expressed. Despite its nonconventional C/EBP DNA binding domain, CHOP binds to the ASNS promoter, which contains a C/EBP-ATF composite site that mediates the response to amino acid deprivation or ER stress. Although the CHOP mRNA was elevated during amino acid deprivation, the increase in CHOP protein abundance was relatively modest and transient. In contrast, the increase in CHOP protein during the UPR was much greater and was sustained over 48 h. Consistent with the observed level of CHOP expression, amino acid deprivation did not cause cell apoptosis, whereas ER stress triggered the apoptotic pathway within 24 h.
MATERIALS AND METHODS
Antibodies and Plasmids—The CHOP monoclonal (sc-7351) and polyclonal (sc-575) antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). ATF4 rabbit polyclonal antibody was generated by Cocalico Biologicals (Reamstown, PA) using purified ATF4 protein. Caspase-3 antibody (9662) and cleaved caspase-3 antibody (9661S) were purchased from Cell Signaling Technology, Inc. (Danvers, MA). The following plasmids were generated for expression of specific proteins: human ATF4 cDNA was subcloned into pcDNA3.1/zeo+ (Invitrogen) to generate the ATF4 expression vector driven by the cytomegalovirus promoter (hATF4/pcDNA3.1-zeo+); human CHOP cDNA was generated by PCR from a human HepG2 cDNA library and then cloned into pcDNA3.1/zeo+ (hCHOP/pcDNA3.1-zeo+), which was then used to generate clones encoding the full-length protein (aa 1∼169), an N-terminal fragment (aa 1∼100), or a C-terminal fragment (aa 101∼169) that were subcloned into the pEF6-V5-His vector (CHOP-FL-V5/pEF6-V5-His, CHOP-N-Term-V5/pEF6-V5-His, and CHOP-C-Term-V5/pEF6-V5-His). The reporter plasmid (ASNS-173/+51/luciferase) was made by inserting an ASNS promoter fragment (nucleotides -173/+51) upstream of the Firefly luciferase reporter gene using the HindIII site of the pGL3 plasmid (Promega, Madison, WI). Hemagglutinin-tagged ATF4 (HA-ATF4/pCGN) and green fluorescent protein (GFP/pcDNA3.1) plasmids were used to express the corresponding proteins in HEK293T cells.
Yeast Two-hybrid Screening—BD Matchmaker library construction and screening kits were purchased from Clontech. A human HepG2 library was generated as described in the manufacturer's protocols, using total RNA from HepG2 cells treated with 2 mm histidinol (HisOH) for 8 h. cDNAs corresponding to full-length or different domains of human ATF4 were generated by PCR, using the hATF4/pcDNA3.1-zeo+ plasmid as template, and subcloned into the pGBKT7 bait vector (Clontech). The procedures for culture media and plates for yeast growth and selection were as described in the manufacturer's protocols. Positive clones were sequenced and identified using National Center for Biotechnology Information databases.
Cell Culture—Human HepG2 hepatoma cells or HEK293T embryonic kidney cells were cultured in minimal essential medium (MEM, pH 7.4, Mediatech, Herndon, VA) or Dulbecco's modified Eagle's medium, pH 7.4 (Mediatech, Herndon, VA), respectively, each supplemented to contain 1× nonessential amino acids, 4 mm glutamine, 100 μg/ml streptomycin sulfate, 100 units/ml penicillin G, 0.25 μg/ml amphotericin B, and 10% (v/v) fetal bovine serum. Cells were maintained at 37 °C in a 5% CO2, 95% air incubator, and cultures were replenished with fresh MEM or Dulbecco's modified Eagle's medium and serum for 12 h prior to initiating all treatments to ensure that the cells were in the basal (“fed”) state. Amino acid deprivation was induced by transfer of cells to culture medium containing 2 mm HisOH, which blocks charging of histidine onto the corresponding tRNA and thus mimics histidine deprivation and triggers activation of the AAR cascade (34). ER stress was induced by transfer of cells to culture medium containing 300 nm thapsigargin (Tg), which activates the UPR pathway because of its inhibition of the ER resident Ca2+-ATPase (35).
RNA Isolation and Quantitative RT-PCR—Total RNA was isolated from HepG2 cells or HEK293T cells using the Qiagen RNeasy kit (Qiagen, Valencia, CA), including a DNase I treatment before the final elution to eliminate DNA contamination, according to the manufacturer's protocol. To measure the relative amount of ASNS mRNA, quantitative RT-PCR (qRT-PCR) analysis was performed using a DNA Engine Opticon 3 system (MJ Research, Reno, NV) and detection with SYBR Green I. To measure the steady state mRNA levels, the primers were as follows: for ASNS, sense, 5′-GCAGCTGAAAGAAGCCCAAGT-3′, and antisense, 5′-TGTCTTCCATGCCAATTGCA-3′; for CHOP, sense, 5′-CATCACCACACCTGAAAGCA-3′, and antisense, 5′-TCAGCTGCCATCTCTGCA-3′; for SNAT2, sense, 5′-GTGTCCTGTGGAAGCTGCTTTGA-3′, and antisense, 5′-CAGGTACAAGAGCTGTTGGCTGTGT-3′; for VEGF, sense, 5′-AGCTCCAGAGAGAAGTCGAGGAAGA-3′, and antisense, 5′-TCACTTTGCCCCTGTCGCTTT-3′; for TRB3, sense, 5′-TGGTACCAGCTCCTCTACG-3′, and antisense, 5′-GACAAAGCGACACAGCTTGA-3′; for CAT-1, sense, 5′-TGCCATTGTCATCTCCTTCCTG-3′, and antisense, 5′-AGCTCTCCAACGGTGACATAGC-3′; and for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), sense, 5′-TTGGTATCGTGGAAGGACTC-3′, and antisense, 5′-ACAGTCTTCTGGGTGGCAGT-3′. To measure the transcription rate from the ASNS gene, oligonucleotides derived from ASNS intron 12 and exon 13 were used to measure unspliced heteronuclear RNA as described previously (17), and the primers were as follows: sense, 5′-CCTGCCATTTTAAGCCATTTTGC-3′, and antisense, 5′-TGGGCTGCATTTGCCATCATT-3′. The PCRs were incubated at 48 °C for 30 min followed by 95 °C for 15 min to activate the Taq polymerase and amplification of 35 cycles of 95 °C for 15 s and 60 °C for 60 s. After PCR, melting curves were acquired by stepwise increase of the temperature from 55 to 95 °C to ensure that a single product was amplified in the reaction. PCR for each sample was done in duplicate with samples from at least three independent experiments, and the means ± S.E. between conditions were compared by Student's t test.
Immunoblotting—Total cell or nuclear protein extract (30 μg/lane) or co-immunoprecipitation (co-IP) eluate (45 μl/lane) was separated on a 10-14.5% Tris-HCl polyacrylamide gel (Bio-Rad) and then electrotransferred to a Protran nitrocellulose membrane (Schleicher & Schuell). The membrane was stained with Fast Green to check for equal loading and then incubated with 10% blocking solution (10% (w/v) Carnation nonfat dry milk, 30 mm Tris base, pH 7.5, 0.1% (v/v) Tween 20, and 200 mm NaCl) for 1 h at room temperature with mixing. Immunoblotting was performed using rabbit polyclonal antibodies against ATF4 or CHOP at an antibody concentration of 0.2-0.4 μg/ml in 10% dry milk blocking solution overnight at 4 °C. The blots were washed three times for 10 min in 5% blocking solution on a shaker and then incubated with peroxidase-conjugated goat anti-rabbit secondary antibody (Kirkegaard & Perry Laboratories, Gaithersburg, MD) at a 1:10,000 dilution for 2 h at room temperature. The blots were then washed two times for 10 min in 5% dry milk blocking solution and two times for 10 min in freshly made TBS/Tween (30 mm Tris base, 0.1% Tween 20, and 200 mm NaCl, pH 7.5). The bound secondary antibody was detected using an enhanced chemiluminescence kit (Amersham Biosciences) and exposing the blot to BioMax MR film (Eastman Kodak Co.). The films were quantified by densitometry using an UN-SCAN-IT software program (Silk Scientific, Orem, UT).
Transient Transfection—HepG2 cells (0.2 × 106 cells/well) were seeded on 24-well plates 18-24 h before transfection with Superfect reagent (Qiagen) at a ratio of 6 μl of Superfect to 1 μg of DNA. For each transfection, 0.5 μg of the ASNS-173/+51/luciferase plasmid was used along with the indicated amounts of the transcription factor expression plasmids. The total amount of transfected DNA was kept constant among experimental groups by the addition of empty pcDNA3.1 plasmid. At 12 h following transfection, the cells were transferred to fresh complete MEM for 12 h before all treatments. At 36 h following transfection, cellular extracts were prepared for analysis of luciferase activity, by washing the cells with phosphate-buffered saline (PBS) and incubating in 300 μl of Passive lysis buffer (Promega). The lysates were collected and stored at -80 °C until use. A 10-μl aliquot was used for Firefly luciferase assays using the luciferase reporter assay system (Promega, Madison, WI). The luciferase values were normalized to protein content for each sample. For each experimental condition, three assays were performed for each transfection, and at least two independent transfections were done.
ChIP Analysis—HepG2 cells were seeded at 1.5 × 107/150-mm dish with complete MEM and grown for 24 h. Cells were transferred to fresh MEM 12 h before transfer to either complete MEM, MEM containing 2 mm HisOH, or MEM containing 300 nm Tg for the time period indicated in each figure. ChIP analysis was performed as described previously (17). For double ChIP analysis, chromatin immunoprecipitated using CHOP antibody (sc-575, Santa Cruz Biotechnology) was eluted by incubation for 30 min at 37 °C in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8.0, 167 mm NaCl) containing 10 mm dithiothreitol. The eluate was diluted 100-fold with ChIP dilution buffer and divided evenly into two fractions. Each fraction was subjected again to the immunoprecipitation procedure using ATF4 antibody or normal rabbit IgG antibody (sc-2027, Santa Cruz Biotechnology), respectively. Purified, immunoprecipitated DNA was analyzed by qPCR, as described above. The results are expressed as the ratio to input DNA. Samples from three independent immunoprecipitations were analyzed in duplicate PCR assays, and the means ± S.E. between conditions were compared by the Student's t test. In the qPCR analysis, the primers 5′-TGGTTGGTCCTCGCAGGCAT-3′ and 5′-CGCTTATACCGACCTGGCTCCT-3′ were used to amplify the ASNS proximal promoter sequence (nucleotides -87 to -22, 5′-GGTTGGTCCTCGCAGGCATGATGAAACTTCCCGCACGCGTTACAGGAGCCAGGTCGGTATAAGCG-3′, NSRE-1 and NSRE-2 are underlined) and for ASNS exon 7 as a negative control (sense, 5′-GCAGCTGAAAGAAGCCCAAGT-3′ and 5′-TGTCTTCCATGCCAATTGCA-3′). As a positive control for CHOP-DNA interaction, primers designed to amplify the TRB3 promoter region, containing three sites reported to bind CHOP (29), were also used in the qPCR analysis. The primers for the TRB3 gene (nucleotides +205/+343) were as follows: sense, 5′-ATTAGCTCCGGTTTGCATCAC-3′, and antisense, 5′-TCCACTTCCGCTGCGAGTCTC-3′. CHOP binding to the C/EBP-ATF containing regions of other amino acid responsive genes was also tested. Those primers were as follows: for SNAT2, sense, 5′-GGGAAGACGAGTTGGGAACATTTG-3′, and antisense, 5′-CCCTCCTATGTCCGGAAAGAAAAC-3′; for VEGF, sense, 5′-CTCTGCCCAGTGCTAGGAGGAATT-3′, and antisense, 5′-CAGGGGCTTCTCTCCAGGCTAAA-3′; and for CAT-1, sense 5′-AGGTGTGAGCAGCGGGTCAACT-3′, and antisense, 5′-AGAGGAGTGGAGGCTGCGGTTA-3′. The qPCR analysis was performed using a DNA Engine Opticon 3 system, and the products were detected with SYBR Green I. Serial dilutions of input DNA were used to generate a standard curve for determining the relative amount of product. Duplicates for both the standards and the samples were simultaneously amplified using the same reaction master mixture. The reactions were incubated at 95 °C for 15 min to activate the polymerase, followed by amplification at 95 °C for 15 s and 60 °C for 60 s for 35 cycles. After PCR, melting curves were acquired by stepwise increases in the temperature from 55 to 95 °C to ensure that a single product was amplified in the reaction. The results are expressed as the ratio to input DNA. Samples from at least three independent immunoprecipitations were analyzed in duplicate PCR assays, and the means ± S.E. between conditions were compared by the Student's t test.
siRNA Transfection—The human CHOP siRNA (L-004819-00), siControl nontargeting siRNA (D-001210-02), and DharmaFECT 4 transfection reagent were purchased from Dharmacon, Inc. (Lafayette, CO). HepG2 cells were seeded in 12-well plates at a density of 2.5 × 105 cells/well in MEM and grown for 16 h. Transfection was performed according to the instructions of Dharmacon using 3 μl of DharmaFECT-4 and a 100 nm per well final siRNA concentration. At 24 h following transfection, cells were rinsed with PBS, given fresh MEM, and cultured for another 12 h. The medium was then removed and replaced with control MEM, MEM containing 2 mm HisOH, or MEM containing 300 nm Tg. After a 4- h incubation, total RNA or protein extracts were isolated and analyzed by RT-PCR or immunoblotting, respectively.
Co-immunoprecipitation—HEK293T cells were plated at 40% confluency in MEM medium 24 h before transfection. Cells were replenished with 3 ml/60-mm dish or 18 ml/150-mm dish of fresh MEM. CHOP-pcDNA3.1-zeo+, HA-ATF4-pCGN, or GFP-pcDNA3.1-zeo+ plasmids were transfected at 5 μg/60-mm dish or 20 μg/150-mm dish using a calcium phosphate-mediated transfection protocol. Transfection efficiency was evaluated by estimating the ratio of total cells to GFP-expressing cells. 36 h after transfection, cells were washed twice with PBS, harvested by scraping, and pelleted by centrifugation at 500 × g for 5 min. Cell pellets were resuspended in lysis buffer (20 mm HEPES, pH 7.6,10 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 20% glycerol, 0.5% Triton X-100) and incubated for 30 min at 4 °C with shaking. Lysates were cleared by centrifugation for 15 min at 14,000 × g. For immunoprecipitation of HA-ATF4, cell lysates were incubated with agarose beads conjugated with a monoclonal anti-HA antibody (A2095 Sigma) for 16 h at 4 °C, with shaking. The beads were precipitated by centrifugation for 5 min at 500 × g, washed with lysis buffer five times for 10 min, and the bound proteins were solubilized in 100 μl of sample dilution buffer (2% SDS, 125 mm Tris-HCl, pH 6.8, 20% glycerol, and 30 μg/ml bromphenol blue) at 65 °C. For immunoprecipitation of CHOP, cell lysates were incubated with anti-CHOP monoclonal antibody and Rec protein G-Sepharose 4B beads (Invitrogen) for 16 h at 4 °C, with shaking. The beads were precipitated by centrifugation for 5 min at 500 × g and washed with lysis buffer five times for 10 min, and the proteins were solubilized with 100 μl of sample dilution buffer at 65 °C. Eluates were brought to 5% β-mercaptoethanol and then subjected to immunoblotting (45 μl/lane).
RESULTS
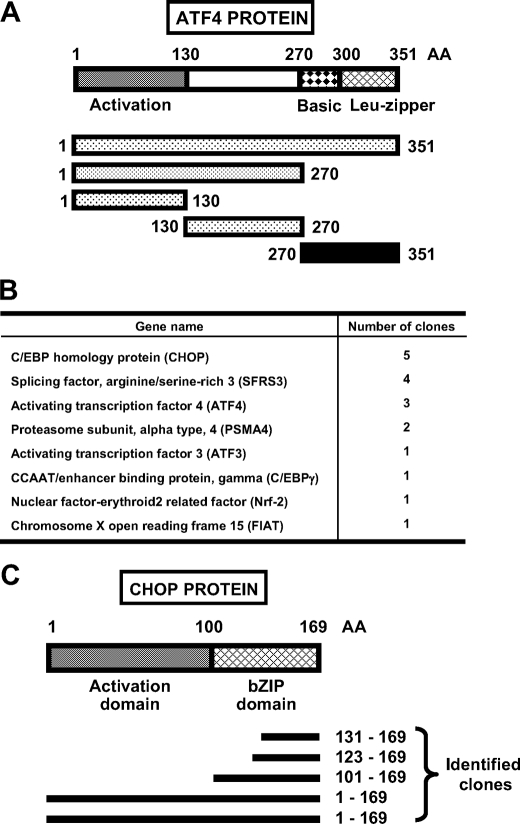
Yeast Two-hybrid Screen—Given the pivotal role of ATF4 in stress-responsive pathways, we performed a yeast two-hybrid screening to identify ATF4-interacting proteins. A cDNA encoding full-length ATF4 was cloned into the pGBKT7 vector and used as bait (Fig. 1A). However, the full-length ATF4 showed strong activation of the selection marker genes without prey. A deletion analysis was then performed, and different ATF4 domains were cloned into the bait vector. The ATF4 transcriptional activation domain (aa 1-130) and linker domain (aa 130-270) also showed strong self-activation activity. The ATF4 activation domain was further split into three fragments each containing 45 amino acids, but all three fragments still exhibited self-activation. The only ATF4 fragment that did not have self-activation activity was the bZIP domain (aa 270-351), and consequently, it was used as bait to screen a HepG2 cDNA library prepared using RNA from cells that had been incubated for 8 h in 2 mm HisOH. Positive clones included several bZIP transcription factors, such as ATF3, C/EBPγ, and ATF4 itself (Fig. 1B). Also identified were Nrf-2 (nuclear factorerythroid 2-related factor) and factor inhibiting ATF4-mediated transcription, two proteins known to bind ATF4 (36, 37). Among the positive clones obtained, five corresponded to the open reading frame of CHOP; two clones were the full-length CHOP sequence, one was the entire bZIP domain, and two were the leucine zipper domain (Fig. 1C).
FIGURE 1.
Identification of CHOP as an ATF4 binding partner by yeast two-hybrid screening. A, diagram shows the entire domain structure of the 351 amino acids (AA) that make up the ATF4 protein and the fragments used as bait for yeast two-hybrid screening. For the fragments, the stippled sections indicate the ATF4 regions that exhibited self-activation when tested. The black bar (aa 270-351) in A, corresponding to the basic leucine-zipper domain, was the sequence used to obtain the positive clones shown in B. C, diagram shows the domain structure of CHOP protein, and the black bars below indicate the amino acid sequence of different CHOP clones identified in the yeast two-hybrid screening. The indicated N-terminal (aa 1-100) and C-terminal (aa 100-169) are the CHOP fragments overexpressed to obtain the data in Fig. 7.
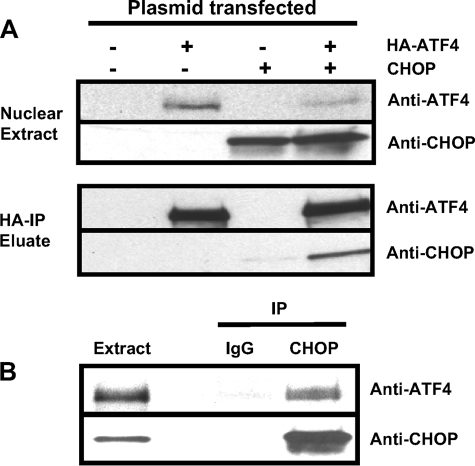
CHOP Interacts with ATF4 in Vivo—To further investigate the interaction of CHOP and ATF4 in intact cells, the HA-ATF4 fusion protein and CHOP were expressed either alone or in combination in human HEK293T cells. Whole cell extracts were subjected to immunoprecipitation (IP) with HA antibody-conjugated agarose beads, and the presence of CHOP was detected by immunoblot analysis (Fig. 2A). CHOP was only detected in the immunoprecipitate when HA-ATF4 and CHOP were co-expressed but not when HA-ATF4 or CHOP was expressed alone indicating that the co-IP of CHOP with ATF4 was specific. In a reciprocal co-IP experiment, whole cell extracts from HEK293T cells co-expressing CHOP and HA-ATF4 were subjected to IP with anti-CHOP antibody or, as a negative control, normal mouse IgG antibody (Fig. 2B). ATF4 was absent from the immunoprecipitate after IP with normal mouse IgG (nonspecific IgG), but ATF4 specifically co-immunoprecipitated with CHOP. Taken together, these experiments demonstrate that ATF4 and CHOP interact with each other in vivo and are consistent with the previous in vitro glutathione S-transferase pulldown data by Gachon et al. (33).
FIGURE 2.
Interaction of CHOP and ATF4 in vivo. HA-ATF4 and CHOP were expressed either alone or in combination in HEK293T cells. Whole cell extracts were collected and subjected to immunoprecipitation using HA antibody-conjugated agarose beads. Bound protein was eluted and subjected to immunoblot analysis to detect the presence of ATF4 and CHOP (A). In a reciprocal experiment, extracts were subjected to immunoprecipitation (IP) using CHOP antibody or normal mouse IgG antibody (IgG). Bound protein was eluted and subjected to immunoblot analysis to detect the presence of ATF4 and CHOP (B).
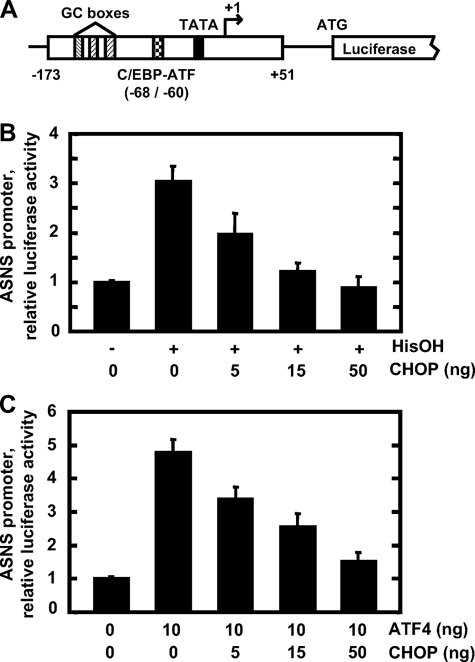
CHOP Inhibits Transcriptional Activation from ASNS Promoter—Given that the induction of the ASNS gene by amino acid limitation is mediated by ATF4 (17, 19, 38), a possible role for CHOP was tested. HepG2 cells were co-transfected with the ASNS-173/+51 promoter/luciferase reporter plasmid (Fig. 3A) and with a CHOP expression vector and then incubated in MEM or MEM containing 2 mm HisOH for 12 h. ASNS promoter activity as measured by luciferase activity was induced by HisOH treatment, and overexpression of CHOP negatively regulated the induction in a concentration-dependent manner (Fig. 3B). To determine whether CHOP directly antagonizes ATF4 activity on the ASNS promoter, HepG2 cells were co-transfected with the ASNS-173/+51 promoter/luciferase reporter plasmid and expression vectors for ATF4 and CHOP (Fig. 3C). When HepG2 cells were transfected with 10 ng/well of ATF4 expression plasmid, ASNS promoter activity was induced to a level approximately comparable with that by HisOH treatment (Fig. 3C). Co-expression of CHOP and ATF4 revealed a concentration-dependent CHOP repression of the promoter activity induced by ATF4. Exogenous expression of ATF4 and CHOP had no significant effect on the ASNS promoter activity if the NSRE-1 (C/EBP-ATF site), and NSRE-2 sequences were mutated (data not shown). Taken together, these experiments demonstrate that CHOP negatively regulates the ATF4-driven transcription mediated by the NSRU within the ASNS promoter.
FIGURE 3.
Effect of CHOP on ASNS-driven transcription activated by HisOH or ATF4. A diagram showing the structure of the ASNS-173/+51 promoter/luciferase reporter (A). HepG2 cells were co-transfected with 0.5 μg/well of the ASNS-173/+51 promoter/luciferase reporter plasmid and the CHOP-pcDNA3.1 expression plasmid at the amounts indicated. To activate ATF4-dependent transcription, half of the cells were treated with 2 mm HisOH for 12 h (B) and the other half were co-transfected with 10 ng/well of ATF4-pcDNA3.1 expression plasmid (C). The total amount of transfected DNA was kept constant among experimental groups by the addition of pcDNA3.1 plasmid. Cell extracts were assayed for luciferase activity as described under “Materials and Methods.” Each experiment was repeated with three different batches of cells, and the results shown represent the means ± S.E.
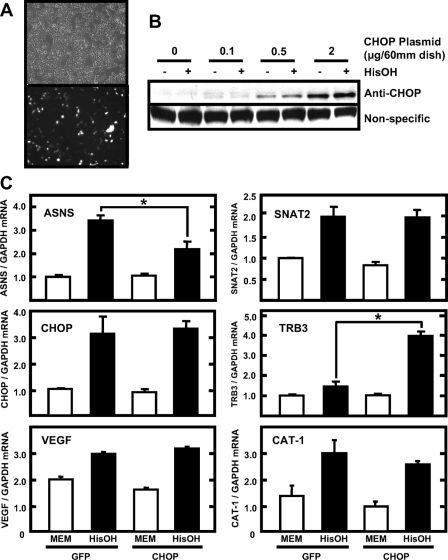
Effect of CHOP Overexpression on Endogenous Gene Expression—To examine whether CHOP overexpression affects the endogenous expression of ASNS and other amino acid-responsive genes, HEK293T cells were transfected with CHOP or green fluorescent protein (GFP) (negative control) expression plasmids. An estimated transfection efficiency of 50-60% was achieved as shown by the ratio of GFP fluorescent cells to total cells (Fig. 4A). Monitoring exogenous CHOP protein production by immunoblotting showed a concentration-dependent expression from a 0-2-μg plasmid/60-mm dish (Fig. 4B). Incubation of transfected cells in HisOH for 8 h did not alter the exogenous CHOP expression, and the amount of endogenous CHOP expression under this condition was relatively small (Fig. 4B, see Fig. 5B for comparison). CHOP overexpression did not affect the basal level of the endogenous ASNS mRNA, but the induction of ASNS expression by HisOH treatment was decreased by ∼40% (Fig. 4C). Given the transfection efficiency of only 50-60%, the repressive effect of CHOP on the endogenous ASNS gene is likely to be quite strong. In contrast, other genes that are activated by amino acid limitation, SNAT2 (39), VEGF (40), CAT-1 (41), and CHOP itself (23), were largely unaffected by CHOP overexpression (Fig. 4C). To determine whether the overexpressed CHOP was functioning in a manner consistent with previously published reports, the regulation of the CHOP-inducible gene TRB3 was monitored (Fig. 4C). Jousse et al. (42) have shown that TRB3 expression is increased following amino acid limitation, and Ohoka et al. (29) reported that the TRB3 gene contains AARE-like genomic elements that are responsive to activation by ATF4 and CHOP. In the HepG2 cells, TRB3 mRNA content was modestly increased by HisOH treatment, but the increase was substantially enhanced by overexpression of CHOP. These results are consistent with TRB3 as a CHOP-inducible gene and in direct contrast to the CHOP-mediated repression of ASNS gene expression.
FIGURE 4.
Effect of CHOP overexpression on endogenous gene expression. HEK293T cells were transfected with GFP-pcDNA3.1 or CHOP-pcDNA3.1 plasmids as described under “Materials and Methods.” A shows phase contrast fluorescence images of the GFP-transfected cells (2 μg/60-mm dish). At 24 h post-transfection, cells were incubated for 8 h in MEM or in MEM containing 2 mm HisOH, and then RNA and protein extracts were isolated. Protein extracts from GFP- or CHOP-transfected cells were subjected to immunoblot analysis to detect the presence of CHOP protein (B). The RNA was subjected to qRT-PCR analysis for the mRNA content of the indicated genes, as well as GAPDH mRNA as an internal control. The results are shown as the means ± S.E. for three independent experiments (C). An asterisk indicates that the induction of expression was significantly changed by CHOP expression (p < 0.05).
FIGURE 5.
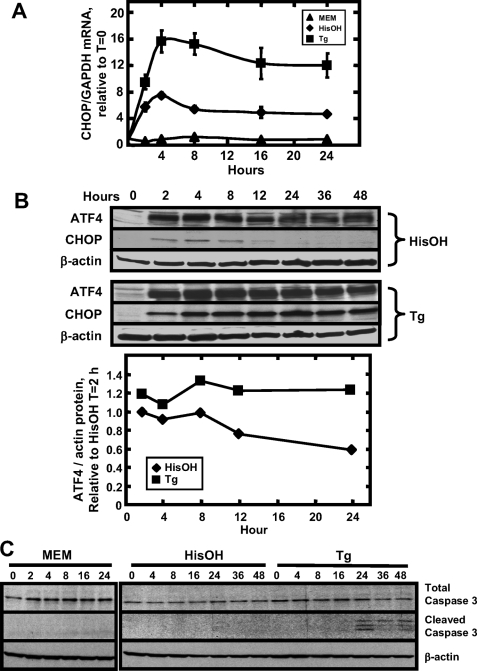
Time course of CHOP expression and apoptosis following amino acid limitation and ER stress. HepG2 cells were incubated in MEM, MEM containing 2 mm HisOH, or MEM containing 300 nm Tg. At the times indicated, RNA was isolated and analyzed by qRT-PCR analysis for CHOP or GAPDH mRNA content (A). The graph illustrates the means ± S.E. for three independent experiments. Where not shown, the error bars are contained with the symbol. In a second series of experiments, protein extracts were collected and subjected to immunoblot analysis for ATF4, CHOP, and β-actin content (B). The blot shown is a single experiment, and the ATF4 quantification below it represents the average of two independent experiments. To collect the data for C, HepG2 cells were incubated for 0-48 h in MEM, MEM containing 2 mm HisOH, or MEM containing 300 nm Tg. At the times indicated, protein extracts were collected and subjected to immunoblot analysis for caspase 3 (uncleaved and cleaved forms) and β-actin content. The blot shown is representative of two independent experiments.
Regulation of CHOP Expression by the AAR and UPR Pathways—The induction pattern of CHOP expression by amino acid deprivation and ER stress was established in HepG2 cells by incubation in MEM, MEM containing 2 mm HisOH, or MEM containing 300 nm Tg for 0-24 h (Fig. 5). After either HisOH or Tg treatment, CHOP mRNA expression was rapidly induced within 2 h and remained elevated throughout the 24-h time course for both treatments, but the magnitude was quite different for the two pathways. Compared with the AAR, activation of the UPR caused a greater induction of CHOP mRNA by about 2-fold (Fig. 5A). Although the basal level of CHOP protein was below detection, after HisOH or Tg treatment, CHOP protein was induced within 2 h (Fig. 5B). Surprisingly, in the presence of HisOH the protein level of CHOP peaked at about 4 h and then decreased after 8-12 h. In striking contrast, during Tg treatment the protein level of CHOP peaked at 4 h, at a level much higher than that for HisOH, and remained at that level throughout the entire 48 h (Fig. 5B). The transient and relatively low level of CHOP protein expression following amino acid deprivation may be an important aspect of its cellular actions in this context. As a positive control, the ATF4 protein abundance was also measured, and there was less of a difference between the two pathways with both producing a robust and prolonged induction of ATF4 protein (Fig. 5B). Harding et al. (43) demonstrated that the PERK/ATF4 pathway is the dominant pathway for the CHOP induction by ER stress. To determine whether the level of ATF4 correlated with the degree of CHOP induction by amino acid deprivation or ER stress, the ATF4 immunoblots from two independent experiments were quantified relative to the actin control (Fig. 5B) and compared with the CHOP induction (Fig. 5A). The initial induction (2-4 h) of ATF4 by Tg is slightly higher than that by HisOH. During the later phase of these stress responses (8-24 h), the ATF4 protein level was maintained in the Tg-treated cells but gradually declined in the HisOH-treated cells. Indeed, higher CHOP mRNA induction in the Tg-treated cells is correlated with higher ATF4 levels during the later phase of the response (8-24 h). However, additional factors may also contribute to the CHOP induction at the early stage (2-4 h) of Tg treatment, as the induction of ATF4 was only slightly higher than that by HisOH treatment, but the CHOP induction is larger (∼2-fold). These data suggest that during the AAR there may be translational or post-translational mechanisms that control the CHOP protein abundance.
ER Stress, but Not Amino Acid Deprivation, Triggers Cell Apoptosis—As noted above, the magnitude and time course of CHOP protein expression differs significantly following activation of the AAR and UPR pathways. Given that CHOP is reported to be an inducer of cell cycle arrest and apoptosis, experiments were designed to determine whether these two pathways have different kinetics in terms of apoptosis and if those kinetics correlate with CHOP protein expression. HepG2 cell were incubated in MEM, MEM containing 2 mm HisOH, or MEM containing 300 nm Tg for 0-24 h. Whole cell extracts were collected at specific times and subjected to immunoblot analysis to probe for cleavage of caspase 3, which is an indicator of cell apoptosis (44). Cells cultured in MEM containing HisOH did not exhibit detectable levels of caspase 3 cleavage up to 48 h of incubation, whereas significant cleavage was observed after 24 h of Tg treatment (Fig. 5C).
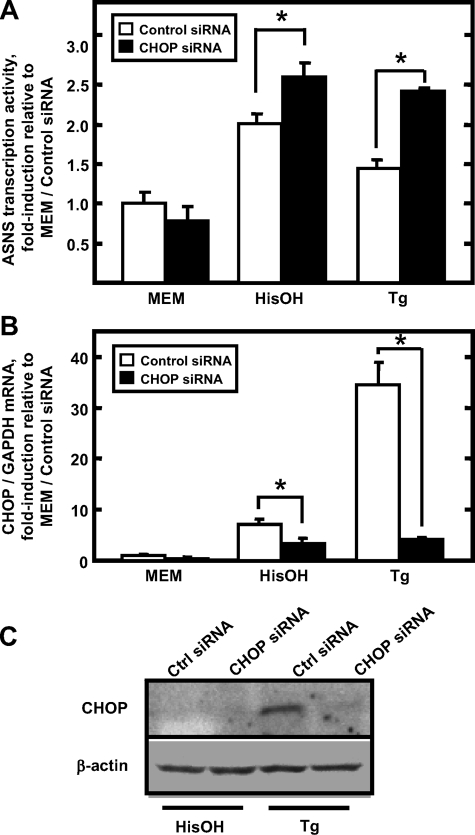
Induction of ASNS Expression Is Enhanced by CHOP Knockdown—Having demonstrated CHOP induction by the AAR and UPR pathways, siRNA was used to knockdown the CHOP expression and then the induction of ASNS by these stress pathways was assessed. HepG2 cells were transfected with either control siRNA or CHOP siRNA, and 36 h later the cells were then incubated in MEM, MEM containing 2 mm HisOH, or MEM containing 300 nm Tg for 4 h. As documented by qRT-PCR and immunoblot analysis, both mRNA (Fig. 6B) and protein (Fig. 6C) expression of CHOP were efficiently reduced. In the face of reduced CHOP expression, the induction of ASNS transcriptional activity by either HisOH or Tg treatment was further enhanced (Fig. 6A). Consistent with the observation that more CHOP protein is produced during Tg treatment, the additional enhancement of ASNS transcription during the UPR was larger than that during HisOH treatment. These experiments further support the proposal that CHOP functions as a repressor of ASNS transcription in response to the AAR and UPR stress pathways.
FIGURE 6.
Effect of siRNA-mediated CHOP knockdown on endogenous ASNS expression. HepG2 cells were transfected with either control siRNA or CHOP siRNA. At 36 h post-transfection, cells were incubated in MEM, MEM containing 2 mm HisOH, or MEM containing 300 nm Tg for 4 h. Total RNA was isolated and subjected to qRT-PCR analysis for ASNS heteronuclear RNA content to assess ASNS transcriptional activity (A). The data are presented as the ratio of ASNS heteronuclear RNA to the GAPDH control. The CHOP mRNA and GAPDH mRNA content were also measured (B). The graph illustrates the means ± S.E. for three independent experiments. Protein extracts were subjected to immunoblot analysis for CHOP and β-actin content (C). Ctrl, control.
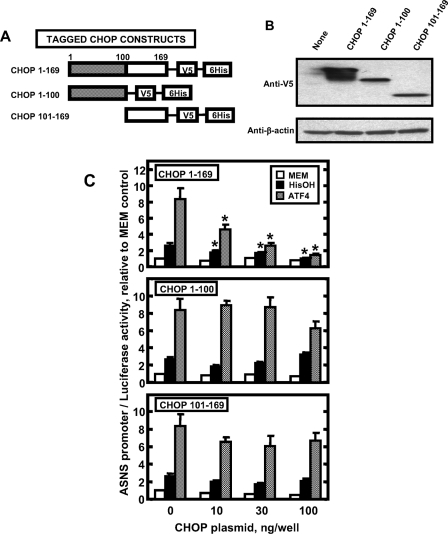
Repressive Function of CHOP Requires Both N- and C-terminal Portions of the Protein and Is Associated with DNA Binding—Two distinct mechanisms have been proposed for CHOP action, both of which involve the formation of heterodimers between CHOP and its binding partners. The first mechanism arises from data showing that CHOP forms nonfunctional heterodimers with its binding partners and sequesters them from binding DNA (25), whereas the second mechanism is based on the observation that CHOP heterodimers can bind to specific DNA sequences (29). Given the fact that the leucine zipper domain of CHOP is sufficient to bind ATF4, as suggested by the yeast two-hybrid screening, if CHOP inhibits ATF4 activity on the ASNS gene by sequestering it from binding to the ASNS promoter, the bZIP domain alone should be able to inhibit ATF4 activity. To investigate this possibility for CHOP inhibition of ATF4 action on the ASNS gene, full-length CHOP, the N terminus (aa 1-100, transcriptional activation/repression domains), or the C terminus (aa, 101-169 bZIP domain) of CHOP were cloned into the pEF6-V5/His expression vector (Fig. 7A), and their expression was confirmed by transfecting these constructs into HEK293T cells followed by immunoblot analysis (Fig. 7B). Co-transfection of the CHOP fragments with the ASNS173/+51 promoter/luciferase reporter plasmid was performed in combination with either HisOH treatment or ATF4 overexpression, in HepG2 cells (Fig. 7C). As expected, the full-length CHOP protein repressed the induction of ASNS promoter activity by HisOH treatment or ATF4 overexpression. However, both the N-terminal and the C-terminal portions of CHOP had little or no effect on the induction of ASNS-driven transcription (Fig. 7C).
FIGURE 7.
The effect of truncated CHOP proteins on transcription driven by the ASNS promoter. A diagram showing the constructs that express full-length or truncated CHOP proteins labeled with a V5 epitope tag (A). The CHOP constructs were transfected into HEK293T cells, and 12 h later protein extracts were subjected to immunoblot analysis for V5-tagged proteins and β-actin (B). The CHOP constructs were also transfected into HepG2 cells along with the ASNS-173/+51 promoter/luciferase reporter plasmid in combination with either 2 mm HisOH treatment or ATF4 overexpression, as indicated (C). Cell extracts were assayed for luciferase activity as described under “Materials and Methods.” Each value represents three assays, and each experiment was repeated with two different batches of cells. The results shown represent the means ± S.E. An asterisk indicates that the value is significantly different (p < 0.05) from the control (0 ng/well CHOP).
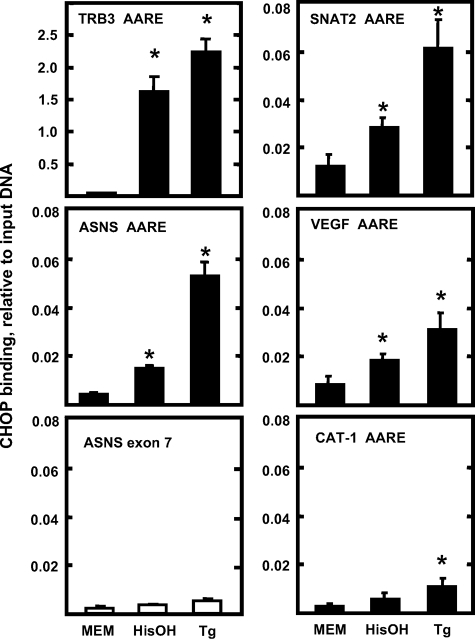
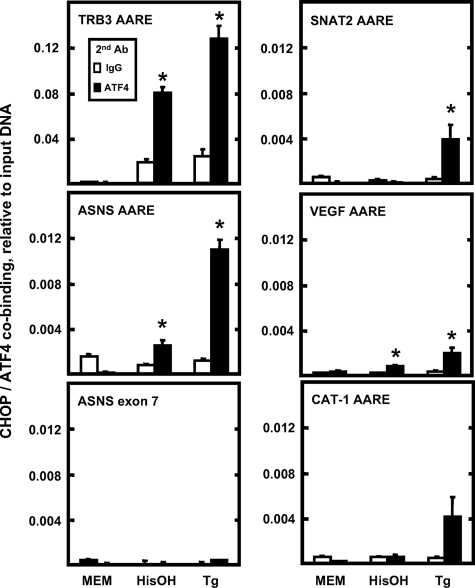
Chromatin Immunoprecipitation to Monitor CHOP Binding to AARE-containing Genes—The fact that CHOP inhibition of ATF4 action requires the entire protein led to the hypothesis that DNA binding is associated with CHOP action on the ASNS gene. To investigate CHOP binding directly to AARE sequences, ChIP analysis was performed in HepG2 cells incubated in MEM, MEM containing 2 mm HisOH, or MEM containing 300 nm Tg for 4 h. The TRB3 promoter contains three AARE-like sequences in tandem that were shown to bind CHOP (29). As a positive control for CHOP binding, CHIP analysis of the TRB3 promoter was investigated. The binding of CHOP to the TRB3 promoter was dramatically increased following HisOH or Tg treatment (Fig. 8). Following activation of the AAR and UPR pathways, the association of CHOP with the ASNS promoter was also significantly increased, and the magnitude was greater in Tg-treated cells compared with that in the HisOH-treated cells (Fig. 8), consistent with the observation that Tg induces CHOP protein abundance to a higher level than does HisOH (Fig. 5B). As a negative control, the association of CHOP with ASNS exon 7 (coding region) was minimal and exhibited little change after HisOH or Tg treatment, supporting the interpretation that the association of CHOP with the ASNS promoter is specific (Fig. 8). These data, in conjunction with the data of Fig. 7C, indicate that direct binding of CHOP to the ASNS promoter is a mechanistic step in the CHOP inhibition of ASNS transcription. To determine the extent of CHOP binding to C/EBP-ATF4 composite sites that have amino acid-responsive function, ChIP analysis was also performed on the SNAT2, VEGF, and CAT-1 genes (Fig. 8). For SNAT2 and VEGF, there was a low basal level of binding that was significantly increased in response to HisOH or Tg treatment. For CAT-1, although the increased CHOP association reached statistical significance after Tg treatment, the absolute amount of CHOP association was quite small (Fig. 8).
FIGURE 8.
The association of CHOP with genes containing a C/EBP-ATF composite site. HepG2 cells were incubated in MEM control medium or treated with either 2 mm HisOH or 300 nm Tg for 4 h. ChIP assays were performed as described under “Materials and Methods.” DNA fragments were immunoprecipitated with CHOP antibody, and the enrichment of CHOP protein at the indicated gene was analyzed by qPCR, using primer sets specific for the regions of interest. Data were plotted as the ratio to the value obtained with a 1:20 dilution of input DNA. Each condition was analyzed in triplicate, and each point represents the means ± S.E. for two independent experiments. An asterisk indicates that the value is significantly different from the MEM control (p < 0.05).
To determine whether CHOP and ATF4 are bound to the same DNA fragment, a “double ChIP” was performed in HepG2 cells incubated in MEM, MEM containing 2 mm HisOH, or MEM containing 300 nm Tg for 4 h. Formaldehyde-cross-linked chromatin was first immunoprecipitated with CHOP antibody and then subjected to a second immunoprecipitation with ATF4 antibody or normal rabbit IgG antibody. Following activation of the AAR or UPR pathway, genomic fragments containing the AARE regions from the TRB3, ASNS, and VEGF genes were significantly enriched in the ATF4-immunoprecipitated chromatin compared with the control IgG (Fig. 9). The degree of enrichment on each gene roughly paralleled that observed in the CHOP ChIP experiment (Fig. 8). Significant enrichment of CHOP/ATF4 on the SNAT2 AARE region was only observed during UPR but not AAR. The co-occupancy of CHOP and ATF4 did not reach statistic significance on the CAT-1 AARE region, which is consistent with the weak binding of CHOP on that region (Fig. 8). As a negative control, the association of CHOP/ATF4 with ASNS exon 7 (coding region) was minimal (Fig. 9).
FIGURE 9.
Co-occupancy of CHOP and ATF4 at C/EBP-ATF composite sites. HepG2 cells were incubated in MEM control medium or treated with either 2 mm HisOH or 300 nm Tg for 4 h. Double-ChIP assays were performed as described under “Materials and Methods.” DNA fragments were first immunoprecipitated with CHOP antibody, eluted, and then subjected to the second immunoprecipitation with ATF4 antibody or normal rabbit IgG antibody. The enrichment of CHOP/ATF4 proteins at the indicated gene was analyzed by qPCR, using primer sets specific for the regions of interest. Data are plotted as the ratio to the value obtained with a 1:200 dilution of input DNA. Each point represents the means ± S.E. for three independent experiments. An asterisk indicates that the value is significantly different from both the MEM control and the IgG control (p < 0.05).
DISCUSSION
The results described in this study document several novel observations and illustrate the counterbalancing function of CHOP on ATF4-dependent transcriptional activation of human ASNS gene during amino acid deprivation or ER stress. 1) The association of CHOP-ATF4 in vivo was documented in mammalian cells by co-immunoprecipitation analysis. 2) CHOP negatively regulates ATF4-mediated transcriptional activation and consequently represses the induction of endogenous ASNS expression by amino acid deprivation. 3) Although CHOP mRNA content is elevated for up to 24 h during prolonged amino acid deprivation, its protein induction is relatively modest and transient, peaking at 4 h and then declining. In contrast, the CHOP mRNA and protein abundance is much greater and more sustained during ER stress. 4) siRNA-mediated CHOP knockdown results in enhanced induction of endogenous ASNS expression by either amino acid deprivation or ER stress. 5) The inhibition effect of CHOP on ATF4 function requires the entire protein, arguing against a mechanism by which CHOP sequesters its binding partners from binding to DNA through its bZIP domain only. 6) ChIP analysis documents recruitment of both ATF4 and CHOP to the ASNS promoter region, which contains the C/EBP-ATF-binding site for ATF4. 7) The degree of apoptosis triggered by ER stress or amino acid deprivation correlated with the degree of CHOP protein induction.
The responses of mammalian cells to stress conditions are a robust yet intricately regulated array of cellular processes. The transcription factor ATF4 is translationally induced by many stress conditions, such as amino acid deprivation (43), ER stress (45), the presence of long double-strand RNA (46), and heme deficiency (47). ATF4 is the functional ortholog of the yeast transcription factor GCN4, which is thought to be a “master regulator” of genes responsive to nutrient deprivation in yeast (48). Although ATF4 and GCN4 share little or no similarity in their protein sequence, when artificially recruited to DNA in the yeast two-hybrid screening, even small fragments (45 aa) of the ATF4 trans-activation domain strongly activated the yeast reporter genes, all in the absence of mammalian co-factors. The strong induction of ATF4 by stress and its potency in activating target genes raises the question of whether or not ATF4 activity is subject to counter-regulation by other proteins, perhaps to provide gene specificity to the ATF4 response. Indeed, several negative regulators of ATF4 have been identified, such as factor inhibiting ATF4-mediated transcription (37), ATF3 (16, 21, 38), and TRB3 (42, 50). After 8-12 h of amino acid limitation, ATF3 and C/EBPβ were shown to feedback repress the ATF4 induction of ASNS gene transcription in a self-limiting process during which ATF4 up-regulates the synthesis of ATF3 and C/EBPβ (17, 34). The current discovery of CHOP as a repressor of ATF4-dependent regulation of the ASNS gene provides another example of such a regulator.
The interaction of CHOP with other bZIP transcription factors and the physiological role of those interactions are poorly investigated. Chen et al. (31) demonstrated that CHOP interacts with ATF3 and inhibits ATF3 function by forming a nonfunctional heterodimer, a mechanism similar to that by which CHOP inhibits the activity of C/EBP proteins. Ohoka et al. (29) showed that CHOP and ATF4 cooperatively activate TRB3 expression during ER stress. Given the fact that TRB3 inhibits ATF4-dependent transcriptional activation of several stress-regulated genes, including ASNS (42, 50), CHOP appears to play an important role, both directly and indirectly, in the network of negative regulators of stress-inducible genes. Our data are consistent with the observations of Al Sarraj et al. (49) who reported CHOP antagonism of either ATF4 or ATF5 action when they were co-expressed exogenously. The present ChIP analysis demonstrated that CHOP is associated with the ASNS promoter region during amino acid deprivation and ER stress. Furthermore, a double-ChIP approach documented that both AFT4 and CHOP are bound to the same DNA fragment. These results suggest that the ASNS C/EBP-ATF element serves as an ATF4-CHOP-binding site. This result appears to hold for other C/EBP-ATF element-containing genes as well. Although different primer sequences do not allow for direct quantitative comparison of CHOP association among these genes, it is interesting to note that the absolute amount of increased binding for the TRB3 gene, which has three tandem copies of the C/EBP-ATF sequence, was an order of magnitude greater than the values for the genes containing a single binding site.
Another intriguing question is why CHOP cooperates with ATF4 to activate the TRB3 gene, whereas it inhibits others. Ohoka et al. (29) demonstrated that the flanking sequence of the TRB3 AARE might contain one or more binding sites for additional factors that contribute to the distinct function of the ATF4/CHOP heterodimer at a particular gene. The stress response unit (nucleotides +201 to +312) of the TRB3 gene contains three identical tandem repeats each consisting of 33 bp. Each of the repeats contains a CCAAT-like element, a CHOP-binding site, and an AARE, which partially overlaps with the CHOP-binding site. Mutagenesis analysis showed that not only were the CHOP-binding site and the AARE essential for the TRB3 induction by ER stress, but the CCAAT-like element was also required (29). The ASNS promoter does not have the CCAAT-like element and is repressed by CHOP overexpression, leading to the possibility that the CCAAT-like element may be involved in the discrimination between activation or repression by the ATF4/CHOP heterodimer. In a microarray study by Harding et al. (1), using ATF4-/- mouse embryo fibroblast cells, many genes were identified to require ATF4 for their induction by ER stress. In a separate microarray study by Marciniak et al. (51), using CHOP-/- mouse embryo fibroblast cells, 26 genes were identified to have impaired induction following ER stress. Interestingly, comparison of the two sets of identified genes, only the chloride channel 3 (CLCN3) gene appeared in both studies. Additional studies need to be completed with arrays covering the entire genome.
CHOP is also involved in the induction of cell cycle arrest and apoptosis (2). In the HepG2 cells used in this study, CHOP protein induction was transient during amino acid deprivation, and no apoptosis was observed, whereas CHOP protein was induced and maintained at a much higher level during ER stress, and under this circumstance apoptosis was triggered. Collectively, the results presented in this study document new transcriptional regulatory roles for CHOP. Further investigation of the mechanisms by which CHOP contributes to the cellular stress pathways will provide additional insight into these important processes.
Acknowledgments
We thank other members of the laboratory for technical advice, reagents, and helpful discussion.
This work was supported, in whole or in part, by National Institutes of Health Grants DK-52064 and DK70647 (NIDDK) (to M. S. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ER, endoplasmic reticulum; AAR, amino acid response; AARE, amino acid response element; ASNS, asparagine synthetase; ATF, activating transcription factor; CAT-1, cationic amino acid transporter 1; C/EBP, CCAAT/enhancer-binding protein; CHOP, C/EBP homology protein; ChIP, chromatin immunoprecipitation; HisOH, histidinol; NSRE, nutrient sensing response element; qPCR, quantitative real time PCR; RT, reverse transcriptase; Tg, thapsigargin; TRB3, tribbles 3; SNAT2, system A transporter 2; VEGF, vascular endothelial growth factor; MEM, minimal essential medium; PERK, protein kinase-like endoplasmic reticulum kinase; aa, amino acids; siRNA, short interfering RNA; PBS, phosphate-buffered saline; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; UPR, unfolded protein response; IP, immunoprecipitation; co-IP, co-immunoprecipitation; GFP, green fluorescent protein; HA, hemagglutinin; bZIP, basic leucine zipper.
References
- 1.Harding, H. P., Zhang, Y., Zeng, H., Novoa, I., Lu, P. D., Calfon, M., Sadri, N., Yun, C., Popko, B., Paules, R., Stojdl, D. F., Bell, J. C., Hettmann, T., Leiden, J. M., and Ron, D. (2003) Mol. Cell 11 619-633 [DOI] [PubMed] [Google Scholar]
- 2.Zinszner, H., Kuroda, M., Wang, X. Z., Batchvarova, N., Lightfoot, R. T., Remotti, H., Stevens, J. L., and Ron, D. (1998) Gene. Dev. 12 982-995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sood, R., Porter, A. C., Olsen, D. A., Cavener, D. R., and Wek, R. C. (2000) Genetics 154 787-801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang, P., McGrath, B. C., Reinert, J., Olsen, D. S., Lei, L., Gill, S., Wek, S. A., Vattem, K. M., Wek, R. C., Kimball, S. R., Jefferson, L. S., and Cavener, D. R. (2002) Mol. Cell. Biol. 22 6681-6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinnebusch, A. G. (2005) Annu. Rev. Microbiol. 59 407-450 [DOI] [PubMed] [Google Scholar]
- 6.Okada, T., Yoshida, H., Akazawa, R., Negishi, M., and Mori, K. (2002) Biochem. J. 366 585-594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harding, H. P., Zhang, Y., Bertolotti, A., Zeng, H., and Ron, D. (2000) Mol. Cell 5 897-904 [DOI] [PubMed] [Google Scholar]
- 8.Liang, S. H., Zhang, W., McGrath, B. C., Zhang, P., and Cavener, D. R. (2006) Biochem. J. 393 201-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wek, R. C., and Cavener, D. R. (2007) Antioxid. Redox. Signal. 9 2357-2371 [DOI] [PubMed] [Google Scholar]
- 10.Vattem, K. M., and Wek, R. C. (2004) Proc. Natl. Acad. Sci. U. S. A 101 11269-11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu, P. D., Harding, H. P., and Ron, D. (2004) J. Cell Biol. 167 27-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ameri, K., and Harris, A. L. (2008) Int. J. Biochem. Cell Biol. 40 14-21 [DOI] [PubMed] [Google Scholar]
- 13.Ramji, D. P., and Foka, P. (2002) Biochem. J. 365 561-575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vallejo, M., Ron, D., Miller, C. P., and Habener, J. F. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 4679-4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang, G., and Hai, T. (1997) J. Biol. Chem. 272 24088-24095 [DOI] [PubMed] [Google Scholar]
- 16.Fawcett, T. W., Martindale, J. L., Guyton, K. Z., Hai, T., and Holbrook, N. J. (1999) Biochem. J. 339 135-141 [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, H., Pan, Y. X., Dudenhausen, E. E., and Kilberg, M. S. (2004) J. Biol. Chem. 279 50829-50839 [DOI] [PubMed] [Google Scholar]
- 18.Lopez, A. B., Wang, C., Huang, C. C., Yaman, I., Li, Y., Chakravarty, K., Johnson, P. F., Chiang, C. M., Snider, M. D., Wek, R. C., and Hatzoglou, M. (2007) Biochem. J. 402 163-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siu, F., Bain, P. J., LeBlanc-Chaffin, R., Chen, H., and Kilberg, M. S. (2002) J. Biol. Chem. 277 24120-24127 [DOI] [PubMed] [Google Scholar]
- 20.Barbosa-Tessmann, I. P., Chen, C., Zhong, C., Siu, F., Schuster, S. M., Nick, H. S., and Kilberg, M. S. (2000) J. Biol. Chem. 275 26976-26985 [DOI] [PubMed] [Google Scholar]
- 21.Wolfgang, C. D., Chen, B. P., Martindale, J. L., Holbrook, N. J., and Hai, T. (1997) Mol. Cell. Biol. 17 6700-6707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fornace, A. J., Jr., Alamo, I., Jr., and Hollander, M. C. (1988) Proc. Natl. Acad. Sci. U. S. A. 85 8800-8804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruhat, A., Jousse, C., Wang, X.-Z., Ron, D., Ferrara, M., and Fafournoux, P. (1997) J. Biol. Chem. 272 17588-17593 [DOI] [PubMed] [Google Scholar]
- 24.Wang, X. Z., Lawson, B., Brewer, J. W., Zinszner, H., Sanjay, A., Mi, L. J., Boorstein, R., Kreibich, G., Hendershot, L. M., and Ron, D. (1996) Mol. Cell. Biol. 16 4273-4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ron, D., and Habener, J. F. (1992) Gene. Dev. 6 439-453 [DOI] [PubMed] [Google Scholar]
- 26.Vinson, C. R., Sigler, P. B., and McKnight, S. L. (1989) Science 246 911-916 [DOI] [PubMed] [Google Scholar]
- 27.Landschulz, W. H., Johnson, P. F., and McKnight, S. L. (1989) Science 243 1681-1688 [DOI] [PubMed] [Google Scholar]
- 28.Wang, X.-Z., Kuroda, M., Sok, J., Batchvarova, N., Kimmel, R., Chung, P., Zinszner, H., and Ron, D. (1998) EMBO J. 17 3619-3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohoka, N., Yoshii, S., Hattori, T., Onozaki, K., and Hayashi, H. (2005) EMBO J. 24 1243-1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Averous, J., Bruhat, A., Jousse, C., Carraro, V., Thiel, G., and Fafournoux, P. (2004) J. Biol. Chem. 279 5288-5297 [DOI] [PubMed] [Google Scholar]
- 31.Chen, B. P., Wolfgang, C. D., and Hai, T. (1996) Mol. Cell. Biol. 16 1157-1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon, S., Govind, C. K., Qiu, H., Kim, S. J., Dong, J., and Hinnebusch, A. G. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 11713-11718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gachon, F., Gaudray, G., Thebault, S., Basbous, J., Koffi, J. A., Devaux, C., and Mesnard, J. (2001) FEBS Lett. 502 57-62 [DOI] [PubMed] [Google Scholar]
- 34.Thiaville, M. M., Dudenhausen, E. E., Zhong, C., Pan, Y. X., and Kilberg, M. S. (2008) Biochem. J. 410 473-484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Treiman, M., Caspersen, C., and Christensen, S. B. (1998) Trends Pharmacol. Sci. 19 131-135 [DOI] [PubMed] [Google Scholar]
- 36.He, C. H., Gong, P., Hu, B., Stewart, D., Choi, M. E., Choi, A. M., and Alam, J. (2001) J. Biol. Chem. 276 20858-20865 [DOI] [PubMed] [Google Scholar]
- 37.Yu, V. W., Ambartsoumian, G., Verlinden, L., Moir, J. M., Prud'homme, J., Gauthier, C., Roughley, P. J., and St-Arnaud, R. (2005) J. Cell Biol. 169 591-601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan, Y.-X., Chen, H., Siu, F., and Kilberg, M. S. (2003) J. Biol. Chem. 278 38402-38412 [DOI] [PubMed] [Google Scholar]
- 39.Palii, S. S., Chen, H., and Kilberg, M. S. (2004) J. Biol. Chem. 279 3463-3471 [DOI] [PubMed] [Google Scholar]
- 40.Abcouwer, S. F., Marjon, P. L., Loper, R. K., and Vander Jagt, D. L. (2002) Investig. Ophthalmol. Vis. Sci. 43 2791-2798 [PubMed] [Google Scholar]
- 41.Fernandez, J., Lopez, A. B., Wang, C., Mishra, R., Zhou, L., Yaman, I., Snider, M. D., and Hatzolgou, M. (2003) J. Biol. Chem. 278 50000-50009 [DOI] [PubMed] [Google Scholar]
- 42.Jousse, C., Deval, C., Maurin, A. C., Parry, L., Cherasse, Y., Chaveroux, C., Lefloch, R., Lenormand, P., Bruhat, A., and Fafournoux, P. (2007) J. Biol. Chem. 282 15851-15861 [DOI] [PubMed] [Google Scholar]
- 43.Harding, H. P., Novoa, I., Zhang, Y., Zeng, H., Wek, R., Schapira, M., and Ron, D. (2000) Mol. Cell 6 1099-1108 [DOI] [PubMed] [Google Scholar]
- 44.Mazumder, S., Plesca, D., and Almasan, A. (2008) Methods Mol. Biol. 414 13-21 [DOI] [PubMed] [Google Scholar]
- 45.Costa-Mattioli, M., Gobert, D., Stern, E., Gamache, K., Colina, R., Cuello, C., Sossin, W., Kaufman, R., Pelletier, J., Rosenblum, K., Krnjevic, K., Lacaille, J. C., Nader, K., and Sonenberg, N. (2007) Cell 129 195-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barber, G. N. (2005) Cell Death. Differ. 12 563-570 [DOI] [PubMed] [Google Scholar]
- 47.Lu, L., Han, A. P., and Chen, J. J. (2001) Mol. Cell. Biol. 21 7971-7980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hinnebusch, A. G., and Natarajan, K. (2002) Eukaryot. Cell 1 22-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Al Sarraj, J., Vinson, C., and Thiel, G. (2005) Biol. Chem. 386 873-879 [DOI] [PubMed] [Google Scholar]
- 50.Ord, D., and Ord, T. (2003) Exp. Cell Res. 286 308-320 [DOI] [PubMed] [Google Scholar]
- 51.Marciniak, S. J., Yun, C. Y., Oyadomari, S., Novoa, I., Zhang, Y., Jungreis, R., Nagata, K., Harding, H. P., and Ron, D. (2004) Genes Dev. 18 3066-3077 [DOI] [PMC free article] [PubMed] [Google Scholar]