Abstract
Borrelia burgdorferi, the etiologic agent of Lyme disease, employs sophisticated means to evade killing by its mammalian hosts. One important immune escape mechanism is the inhibition of complement activation mediated by interactions of the host-derived immune regulators factor H (CFH) and factor H-like protein 1 (CFHL1) with borrelial complement regulator-acquiring surface proteins (BbCRASPs). BbCRASP-2 is a distinctive CFH- and CFHL1-binding protein that is produced by serum-resistant B. burgdorferi strains. Here we show that binding of CFH by BbCRASP-2 is due to electrostatic as well as hydrophobic forces. In addition, 14 individual amino acid residues of BbCRASP-2 were identified as being involved in CFH and CFHL1 binding. Alanine substitutions of most of those residues significantly inhibited binding of CFH and/or CFHL1 by recombinant BbCRASP-2 proteins. To conclusively define the effects of BbCRASP-2 residue substitutions on serum sensitivity in the bacterial context, a serum-sensitive Borrelia garinii strain was transformed with plasmids that directed production of either wild-type or mutated BbCRASP-2 proteins. Critical amino acid residues within BbCRASP-2 were identified, with bacteria producing distinct mutant proteins being unable to bind either CFH or CFHL1, showing high levels of complement components C3, C6, and C5b-9 deposited on their surfaces and being highly sensitive to killing by normal serum. Collectively, we mapped a structurally sensitive CFH/CFHL1 binding site within borrelial BbCRASP-2 and identified single amino acid residues potentially involved in the interaction with both complement regulators.
The ability of Borrelia burgdorferi sensu lato, the etiologic agent of Lyme disease, to infect immunocompetent mammalian hosts requires complex environmental sensing mechanisms and coordinated expression of proteins essential to overcome host defenses, in particular the innate and adaptive immune responses (1–4). As are many other blood-borne pathogens, most infectious isolates of B. burgdorferi sensu stricto (s.s.)2 are resistant to complement, a key effector system of the first line of defense (5–10). Resistance to the alternative pathway of complement activation is facilitated, in part, by attachment of host complement regulatory proteins such as factor H (CFH) and factor H-like protein 1 (CFHL1) to the spirochetal outer surface (11–14).
CFH and CFHL1, the two human immune regulators of the alternative pathway of the complement system, are structurally related plasma proteins. Both glycoproteins are composed of several homologous globular domains termed short consensus repeats (SCRs) (15). CFH is a 150-kDa glycoprotein composed of 20 SCR domains. CFHL1 is a 42-kDa glycoprotein and produced from an alternatively spliced transcript of the CFH gene, and consists of seven SCRs. The N-terminal seven SCRs of both complement regulators are identical, with the exception of four additional hydrophobic amino acids at the C terminus of CFHL1. Both molecules act as cofactors for factor I-mediated degradation of C3b, accelerate the decay of the C3 convertase of the alternative pathway by competing with Factor B for C3b-binding, and protect self surfaces from harmful attacks (16–18).
The interaction of CFH and/or CFHL1 with Borrelia species involves a variety of surface-exposed, outer membrane lipoproteins known as “CRASPs” (complement regulator-acquiring surface proteins) (19). Diverse Lyme disease genospecies can produce different CRASPs, so the proteins of B. burgdorferi s.s. are often referred to as “BbCRASPs,” those of Borrelia afzelii are called “BaCRASPs,” etc. (19, 20). Different B. burgdorferi strains express different numbers of CRASP proteins, each of which varies in relative affinity for the two host immune regulators. According to their ability to bind to CFH and/or CFHL1, the CRASPs produced by B. burgdorferi s.s. B. afzelii, and B. spielmanii are divided into (i) CFH- and CFHL1-binding proteins (Ba-, Bb-, BsCRASP-1 and Ba-, Bb-, BsCRASP-2), (ii) the CFHL1-binding protein BaCRASP-3 of B. afzelii, and (iii) molecules that soley interact with CFH (BbCRASP-3 to -5 from B. burgdorferi, BaCRASP-4 and BaCRASP-5 from B. afzelii, and BsCRASP-3 from B. spielmanii) (19, 20). CRASP-encoding genes belong to three genetically distinct classes (21, 22). BbCRASP-1, encoded by cspA, belongs to the formerly categorized pgf 54 family and is produced by B. burgdorferi (s.s.) only during tick-to-mammal and mammal-to-tick transmission stages (1, 2, 4, 22–24). Another class of proteins, including BbCRASP-3 (ErpP), BbCRASP-4 (ErpC), BbCRASP-5 (ErpA), and OspE, are encoded by the polymorphic erp gene family and produced at varying levels throughout mammalian infection (4, 14, 21, 25–30). The B. burgdorferi (s.s.) CFH/CFHL1 binding protein, BbCRASP-2, was recently identified as being encoded by the cspZ gene (31). Studies investigating its expression revealed that BbCRASP-2 is poorly produced by Borreliae during tick colonization, but is expressed at high levels during mammalian infection (1–2). Additionally, comparative analysis of the cspZ genes of diverse B. burgdorferi (s.s.) strains showed high degrees of sequence conservation, in particular within regions determined to be involved in binding CFH and/or CFHL1 (31–33). A recent study indicated that production of BbCRASP-2 is not essential for short term (up to 18 days) infection of mice, although the importance of BbCRASP-2 production for long term mammalian infection has yet to be examined (34).
In the present study we used site-directed mutagenesis to identify amino acid residues within BbCRASP-2 necessary for binding of CFH and CFHL1. A total of 31 residues were mutated, and the recombinant proteins were analyzed for CFH and CFHL1 binding. In addition, the effects on complement susceptibility and deposition of complement components were studied in Borreliae engineered to produce wild-type or mutated BbCRASP-2 proteins. The sensitivity of BbCRASP-2 interactions with CFH was also examined under varying salt and heparin concentrations. Collectively, these analyses defined several amino acids involved in binding of the immune regulators and showed that binding of CFH and CFHL1 contribute to serum resistance of Borreliae. The results presented in this study might further allow the definition of epitopes of BbCRASP-2 useful for vaccine development.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Condition—B. garinii strain G1 was originally isolated from the cerebrospinal fluid of a Lyme disease patient and is highly susceptible to complement-mediated killing in vitro (7). Borrelia were grown at 33 °C for 5–6 days up to cell densities of 1 × 107 ml-1 in modified Barbour-Stoenner-Kelly (BSK) medium as described previously (19). Escherichia coli JM109 was used as host for cloning and purification of recombinant proteins.
Site-directed Mutagenesis, Sequence Analysis, and Generation of Recombinant Proteins—To introduce single or double amino acid substitutions into the BbCRASP-2-encoding cspZ gene, a site-directed mutagenesis approach was conducted as described by the QuikChange™ protocol (Stratagene). The mutagenic high-performance liquid chromatography-purified primers employed in this study were purchased from Sigma-Aldrich (Steinheim, Germany) and listed in supplemental Table S1. Briefly, PCR reactions were carried out for 18 cycles (95 °C for 30 s, 55 °C for 30 s, and 68 °C for 13 min) using 100 ng of expression vector pGEX CSPZ, 125 ng of each mutagenic primer, and 2.5 units of Pfu DNA polymerase (Stratagene) in a final volume of 50 μl. For the introduction of site-specific mutations into shuttle vector pCSPZ (previously referred as pCRASP-2 (31)), thermal cycling was performed for 18 cycles at 95 °C for 30 s, 55 °C for 60 s, and 68 °C for 16 min. Before transformation of E. coli, the reactions were treated with 10 units of DpnI (New England Biolabs, Frankfurt, Germany) for 1 h at 37 °C. All mutations introduced into the cspZ gene were verified by DNA sequencing of both DNA strands. Proteins were purified using glutathione-Sepharose columns, with conditions as recommended by the manufacturer (GE Healthcare).
Binding and Inhibition Assay—Purified GST-BbCRASP-2, mutated GST-BbCRASP-2 proteins, and GST (2.5 ng/μl each) were immobilized overnight at 4 °C using Immobilizier™ glutathione microtiter plates (Nunc, Wiesbaden, Germany). Non-specific binding sites were blocked with 0.2% gelatin in PBS for 6 h at 4 °C. CFH (Calbiochem, Darmstadt, Germany) or purified CFHL1 (16) (5 μg/ml each) was added to the wells and left overnight at 4 °C. After addition of polyclonal anti-CFH antibodies (Calbiochem) for 2 h at room temperature, protein complexes were identified using a secondary peroxidase-conjugated anti-goat IgG antibody. Reactions were developed with 1,2-phenylenediamine dihydrochloride (Sigma-Aldrich).
Influences of heparin and salt on CFH binding were analyzed by ELISA. Purified GST-BbCRASP-2 (15 ng/μl) was immobilized onto wells of a microtiter plate (MaxiSorb, Nunc) overnight at 4 °C. Following three washing steps with PBS containing 0.05% Tween 20 (PBST), nonspecific binding sites were blocked with 0.2% gelatin in PBS for 6 h at 4 °C. Plates covered with BbCRASP-2 fusion protein were incubated overnight at 4 °C with CFH (0.25 ng/well) in PBST containing increasing concentrations of either low molecular weight heparin (Sigma-Aldrich, 0.5–16 μg/ml) or NaCl (29 mm to 1.16 m). After washing three times in PBS, binding of CFH was detected using mAb VIG8 antibody (35). The experiments were conducted at least three times, and the means ± S.D. were calculated.
SDS-PAGE, Ligand Affinity Blot, and Western Blot Analysis—Borrelial cell extracts (15 μg) were subjected to 10% Tris/Tricine-SDS-PAGE under reducing conditions and transferred to nitrocellulose. Binding of CFH and CFHL1 to borrelial proteins was assessed by ligand affinity blotting as previously described (19).
For Western blot analysis, membranes were incubated for 60 min at room temperature with either mAb L41 1C11 (FlaB) (36) or polyclonal anti-GST antibody (GE Healthcare, Germany) as described elsewhere (19).
Transformation of Serum-sensitive B. garinii and Characterization of Transformants—High passage, non-infectious B. garinii strain G1 was grown in 100 ml of BSK medium and harvested at mid-exponential phase (5 × 107 to 1 × 108 cells/ml). Electrocompetent cells were prepared as described previously (37) with slight modifications. Briefly, 50-μl aliquots of competent B. garinii strain G1 cells were electroporated at 12.5 kV/cm in 2-mm cuvettes with 10 μg of plasmid DNA. For control purpose B. garinii strain G1 cells also were transformed with pKFSS1 vector (38) alone. Cells were immediately diluted into 10 ml of BSK medium and incubated with antibiotic selection at 33 °C for 48–72 h. Bacteria were then diluted into 100 ml of BSK medium containing streptomycin (25 μg/ml), and 200-μl aliquots were plated into 96-well cell culture plates (Corning) for selection of transformants. Several clones selected were expanded in 1 ml of fresh BSK medium without antibiotic selection for 7 days, and then transferred into 10 ml of fresh BSK medium containing streptomycin (50 μg/ml).
The cspZ genes of transformed B. garinii G1 strains were detected by PCR with specific primers (Table S1). Spirochetes (100 μl) were sedimented by centrifugation, washed with PBS, and suspended in 50 μl of water. Five microliters of suspension was amplified by PCR using oligonucleotide primers at final concentrations of 100 nm each, plus 200 μm dNTPs. PCR was carried out for 25 cycles using the following parameters: denaturation at 94 °C for 1 min, annealing at 50 °C for 1 min, and extension at 72 °C for 1 min.
Plasmid DNA was prepared from the presumptive E. coli transformants with the Wizard® Plus SV Miniprep DNA purification system (Promega, Mannheim, Germany), and DNA inserts were sequenced by a commercial provider (GATC, Konstanz, Germany).
Immunofluorescence Assay for Detection of Surface-exposed BbCRASP-2—For indirect immunofluorescence assays, spirochetes (1 × 107) were incubated with 1:50 dilution of mouse polyclonal anti-BbCRASP-2 for 1 h at room temperature with gentle agitation. Following three washes with PBS containing 0.2% BSA, cells were collected by centrifugation at 14,000 × g for 10 min and resuspended in 100 μl of PBS containing 0.2% BSA. Aliquots of 10 μl were then spotted on coverslips and allowed to air dry overnight. After fixation with methanol, samples were dried for 90 min at room temperature and incubated for 90 min in a humidified chamber with a 1:500 dilution of Alexa 488-conjugated goat anti-mouse IgG (Molecular Probes). Slides were then washes four times with 0.2% BSA in PBS and mounted in ProLong® Gold Antifade reagent (Molecular Probes) before being sealed with glass slides. Slides were visualized at a magnification of × 1,000 using an Olympus CX40 fluorescence microscope mounted with a DS-5Mc charge-coupled device camera (Nikon).
Immunofluorescence Assay for Detection of Complement Proteins—Spirochetes (6 × 106) were incubated with 25% NHS for 30 min at 37 °C with gentle agitation, washed three times with PBS containing 1% BSA (PBS-BSA). Aliquots of 10 μl were then spotted on microscope slides and allowed to air dry overnight. After fixation, slides were incubated for 1 h in a humidified chamber with antibodies against complement components C3 (dilution of 1:1000), C6 (dilution of 1:200), and C5b-9 (dilution of 1:50). Following four washes with PBS, the slides were incubated for 1 h at room temperature with 1:2000 dilutions of appropriate Alexa 488-conjugated secondary antibodies (Molecular Probes, Leiden, The Netherlands). Slides were then washed, sealed, and visualized as described above.
Serum Adsorption Experiments—Spirochetes harvested by centrifugation were resuspended in 500 μl veronal-buffered saline (supplemented with 1 mm Mg2+, 0.15 mm Ca2+, 0.1% gelatin, pH 7.4) and a portion of 1 × 109 organisms were sedimented by centrifugation. The cell sediment was then resuspended in 750 μl of NHS supplemented with 34 mm EDTA and incubated for 1 h at room temperature with gentle agitation. After three washes with PBSA (0.15 m NaCl, 0.03 m phosphate, 0.02% sodium azide, pH 7.2) containing 0.05% Tween 20, the proteins bound to the spirochetes were eluted by incubation with 0.1 m glycine-HCl, pH 2.0, for 15 min. The bacterial cells were sedimented by centrifugation (14,000 × g, 20 min, 4 °C), and the proteins in the supernatant were analyzed by SDS-PAGE and Western blotting.
Serum Susceptibility Testing of Borrelia Strains—Serum susceptibility of B. garinii strains G1, G1/pKFSS1, G1/pCSPZ, and G1 containing shuttle vector pCSPZ that harbor point mutations in the cspZ gene was assessed by using a growth inhibition assay as described previously (7). Each experiment was conducted at least three times, and means ± S.D. were calculated.
Human Sera, CFH, CFHL1, and Monoclonal and Polyclonal Antibodies—NHS obtained from 20 healthy human blood donors without known history of spirochetal infections was used as the source for CFH. Purified human CFH was purchased from Calbiochem. CFHL1 were expressed in Spodoptera frugiperda Sf9 insect cells infected with recombinant baculovirus (16, 39).
Generation of mAb L41 1C11 against FlaB was described elsewhere (35). To detect GST fusion proteins a goat anti-GST antibody (GE Healthcare, Germany) was used. Polyclonal rabbit αSCR1–4 antiserum was used for detection of CFHL1 (16), and the mAb VIG8 was applied to specifically detect CFH (35). For detection of both complement regulators a goat anti-human CFH antiserum (Merck Biosciences, Bad Soden, Germany) was used. The goat anti-human C3 and C6 antibodies were purchased from Calbiochem, and the monoclonal anti-human C5b-9 antibody was from Quidel (San Diego, CA). Polyclonal mouse anti-BbCRASP-2 sera were generated by injection (intraperitoneal) of recombinant BbCRASP-2 into Balb/c mice (31).
RESULTS
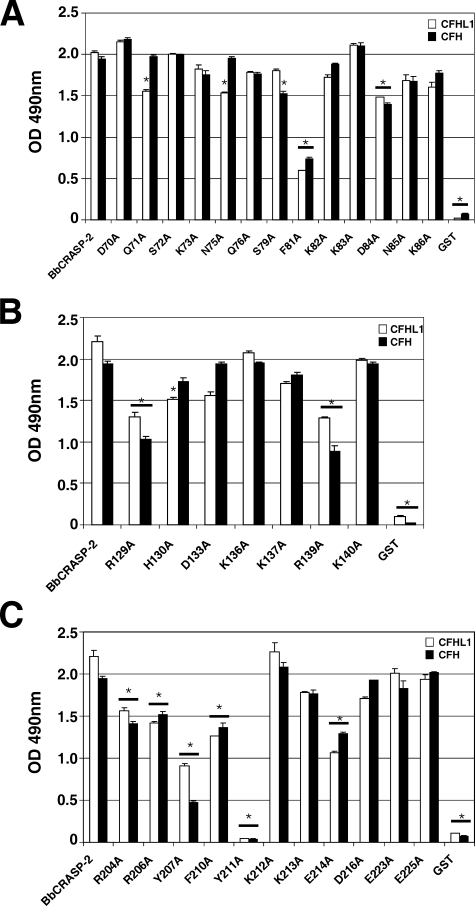
Identification of Amino Acid Residues within BbCRASP-2 Region 2 Required for CFH and CFHL1 Binding—To identify amino acid residues potentially involved in the interaction of BbCRASP-2 with complement regulators CFH and CFHL1, a comprehensive alanine-scanning approach was conducted (Fig. 1). Charged, hydrophobic, or polar amino acids within the putative CFH and/or CFHL1 binding region 2, which encompasses amino acid positions 70–88 (31), were systematically replaced by alanine (Fig. 1). Initially, binding of each recombinant mutant BbCRASP-2 protein to CFH and CFHL1 was quantified using ELISA (Fig. 2). Mutant proteins D70A, S72A, K73A, Q76A, K82A, K83A, N85A, and K86A all bound CFH and CFHL1 with intensities comparable to the wild-type BbCRASP-2 protein. Mutant S79A showed reduced binding to CFH, and binding of CFHL1 was decreased in mutants Q71A and N75A. Replacement of aspartate with alanine at position 84 (mutant D84A) impaired binding of both CFH and CFHL1. The strongest effect on binding of both immune regulators was observed when phenylalanine was replaced by alanine at position 81 (mutant F81A). These results indicate that several amino acid residues within region 2 mediate BbCRASP-2 binding to CFH and CFHL1 and suggest somewhat different interactions with each host protein.
FIGURE 1.
Schematic representation of three binding regions of the BbCRASP-2 protein. The putative binding regions 2–4 previously identified by pepspot analysis (31), and their encompassing amino acid residues are indicated. Amino acid residues changed to alanine during the present studies are underlined.
FIGURE 2.
Analysis of CFHL1 and CFH binding to BbCRASP-2 mutants. Purified GST-BbCRASP-2 (positive control), GST-BbCRASP-2 mutants as well as purified GST protein (negative control) were immobilized onto microtiter plates and incubated with purified CFH or CFHL1 as described under “Experimental Procedures.” Binding of CFH or CFHL1 was then assessed by an ELISA together with a polyclonal anti-CFH antibody. A, BbCRASP-2 mutants representing binding region 2, B, BbCRASP-2 mutants representing binding region 3; C, BbCRASP-2 mutants representing binding region 4. Experiments were performed at least three times, and one representative experiment is shown. Bars represent the mean of triplicates within one experiment ± S.D. One-way analysis of variance was performed to analyze statistical significance in CFH/CFHL1 binding between BbCRASP-2 and mutant proteins, and significant differences from the wild-type (p < 0.001) are indicated by asterisks.
Identification of Amino Acid Residues within Region 3 of BbCRASP-2 That Are Involved in CFH and CFHL1 Binding—Charged amino acid residues within proposed binding region 3 were also systematically mutated and replaced by alanine (Fig. 1). Individual substitutions of charged amino acids Asp-133, Lys-136, Lys-137, and Lys-140 with a neutral alanine did not affect binding of either complement regulator (Fig. 2B). However, weaker binding to both complement regulators was observed for mutants R129A and R139A. In addition the mutant H130A bound CFH but showed reduced binding of CFHL1.
Identification of Amino Acid Residues within BbCRASP-2 Region 4 Involved in CFH and CFHL1 Binding—To identify specific amino acid residues within the C terminus required for binding of complement regulators, 11 positions were systematically replaced by alanine (Fig. 1). ELISA revealed significantly reduced ligand binding by mutants R204A, R206A, Y207A, F210A, and E214A (Fig. 2C). Replacement of tyrosine 211 by alanine (Y211A) completely abolished binding of both complement regulators. Mutant Y207A showed no detectable binding to CFH, but did bind CFHL1 with weaker intensity. Mutant Y211A did bind either of the human complement regulators. Taken together, single mutations in residues 204–211 significantly impaired binding of CFH and CFHL1, suggesting that this region forms a contiguous binding domain.
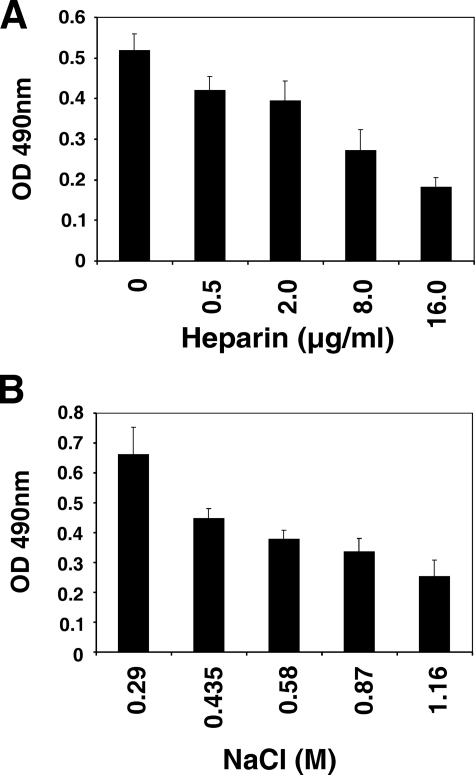
Effects of Heparin and Salt on Ligand Binding by BbCRASP-2—To further characterize the nature of interactions between BbCRASP-2 and complement regulators, we first tested whether binding of CFH is influenced by polyanions such as heparin or by salt. To this end, purified CFH was incubated with BbCRASP-2 immobilized to the wells of microtiter plates. The buffers used were supplemented with increasing concentrations of either heparin or NaCl. Binding of BbCRASP-2 to CFH was inhibited in dose-dependent manners by both heparin and salt (Fig. 3). These data indicate that electrostatic forces are involved in BbCRASP-2 and CFH binding.
FIGURE 3.
Effect of heparin and salt on the binding of CFH to BbCRASP-2. Purified CFH was added to microtiter plates covered with GST-BbCRASP-2 and incubated with buffer supplemented with increasing concentrations of heparin (A) or NaCl concentrations (B). After incubation at 4 °C overnight, the plates were washed, and bound CFH was detected by using mAb VIG8. Bars represent the mean of triplicates within one experiment ± S.D.
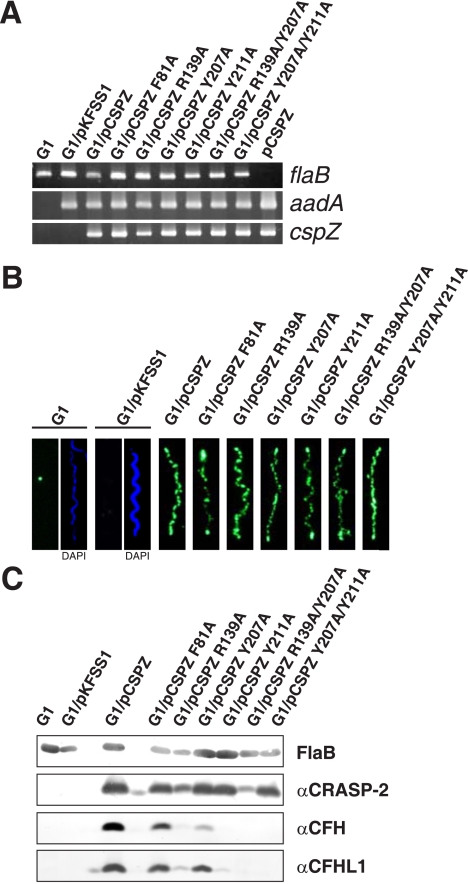
Generation and Analysis of B. garinii Strains Expressing Mutated BbCRASP-2 Proteins—To confirm the results of the mutagenesis studies in vivo and to assess the relevance of CFH/CFHL1 binding in evasion of complement-mediated killing, we engineered a serum-sensitive strain of B. garinii to produce either wild-type or a mutant BbCRASP-2 protein. To this end the shuttle vector pCSPZ, which encodes the wild-type cspZ gene of serum-resistant B. burgdorferi s.s. strain B31, was used as the template for substitution of residues. Plasmid pCSPZ containing either mutations at one site (F81A, R139A, Y207A, and Y211A), or at two sites (R139A/Y207A and Y207A/Y211A) were generated. Each mutagenized pCSPZ plasmid, pCSPZ, or the empty shuttle vector pKFSS1 were then introduced into the highly serum-sensitive B. garinii strain G1. This strain does not produce any CFH and CFHL1 binding CRASP proteins. Transformants were tested by PCR amplification of the streptomycin resistance cassette and the cspZ gene (Fig. 4A). First we evaluated heterologous expression of the various BbCRASP-2 proteins on the surface of B. garinii by immunofluorescence microscopy (31). All cspZ-transformed bacteria expressed a BbCRASP-2 protein at their surface, whereas the parental strain and the bacteria transformed with the empty shuttle vector pKFSS1 did not (Fig. 4B). Cells showed a punctate staining of the various BbCRASP-2 proteins, which is in agreement with the surface distribution of the wild-type protein in the serum-resistant B. burgdorferi s.s. strain (31). Those results indicated that heterologously expressed BbCRASP-2 protein was translocated to the surface of B. garinii in a manner comparable to that of B. burgdorferi s.s.
FIGURE 4.
Characterization of B. garinii G1 complemented with BbCRASP-2 and BbCRASP-2 mutants. A, B. garinii G1, G1/pKFSS1, G1/pCSPZ, and G1 strains expressing mutated BbCRASP-2 proteins were characterized by PCR amplification of the flaB, cspZ, and aadA genes using primers listed in supplemental Table S1. B, surface expression of BbCRASP-2 as well as mutated BbCRASP-2 proteins were assessed by indirect immunofluorescence microscopy of intact borrelial cells. Spirochetes were incubated with mouse polyclonal anti-BbCRASP-2 antiserum before fixation. Slides were then incubated with an Alexa 488-conjugated anti-mouse antibody. For counterstaining, the DNA-binding dye 4′,6-diamidino-2-phenylindole were used to identify cells within a given field. For a clearer view only 4′,6-diamidino-2-phenylindole stain of strains G1 and G1/pKFSS1 was presented (right windows of panel 1 and 2). Slides were visualized at a magnification of ×1000. C, expression of BbCRASP-2 proteins in all strains was assessed by ligand affinity blotting. Whole cell lysates were separated by 10% Tris/Tricine-SDS-PAGE and transferred to nitrocellulose. The membranes were incubated with either purified CFHL1 or NHS, and binding of proteins was detected with the indicated antisera, e.g. polyclonal rabbit αSCR1–4 antiserum specific to CFHL1 and monoclonal antibody VIG8 specific for SCR20 of CFH. A monoclonal antibody, L41 1C11, specific for the flagellin protein, FlaB, was used to show similar loading of the borrelial cell lysates.
Binding of CFH and CFHL1 to all transformants was assessed by ligand affinity blot analysis (40). Membrane-exposed BbCRASP-2 proteins obtained from borrelial cell lysates showed the same binding to CFH and CFHL1 as did the recombinant counterparts expressed in E. coli (Fig. 4C and data not shown). These results indicate that recombinant BbCRASP-2 proteins synthesized by E. coli exhibit similar characteristics as do native proteins produced by Borreliae, confirming the validity both the in vitro and in vivo studies.
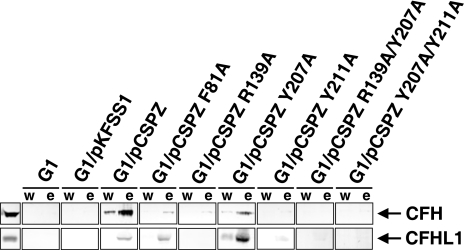
Analysis of CFH and CFHL1 Binding Capabilities of B. garinii Strains Expressing Mutated BbCRASP-2 Proteins—To examine whether the transformants are capable of binding CFH and/or CFHL1 to their surfaces, serum adsorption experiments were performed. To this end, borrelial cells were incubated with NHS supplemented with EDTA to prevent complement activation. Consistent with previous results, transformants showed different binding capabilities for these complement regulators. B. garinii G1/pCSPZ expressing the wild-type BbCRASP-2 protein bound both complement regulators, and negative controls G1 and G1/pKFSS1 did not (Fig. 5). Strains G1/pCSPZ Y211A, G1/pCSPZ R139A/Y207A, and G1/pCSPZ Y207A/Y211A showed no detectable binding of CFH or CFHL1. Reduced binding of CFH was observed for transformants G1/pCSPZ F81A, G1/pCSPZ R139A, and G1/pCSPZ Y207A. The latter strain also showed binding of CFHL1 but with weaker intensity than did G1/pCSPZ. A faint band indicative of weak binding to CFHL1 was observed for G1/pCSPZ F81A.
FIGURE 5.
Binding of complement regulator CFH and CFHL1 by various B. garinii transformants. B. garinii strains G1, G1/pKFSS1, G1/pCSPZ, G1/pCSPZ F81A, G1/pCSPZ R139A, G1/pCSPZ Y207A, G1/pCSPZ Y211A, G1/pCSPZ R139A/Y207A, and G1/pCSPZ Y207A/Y211A incubated in NHS-EDTA were extensively washed with PBSA containing 0.05% Tween 20, and bound proteins were eluted using 0.1 m glycine (pH 2.0). Both the last wash (w) and the eluate (e) fractions obtained from each strain were separated in a non-reducing conditions 12.5% SDS-PAGE gel, transferred to nitrocellulose, and probed with either mAb VIG8 specific for SCR 20 of CFH or αSCR1–4 antiserum specific for the N terminus of CFHL1. As controls purified CFH and CFHL1 (500 ng each) were loaded on the gel.
Serum Susceptibility of B. garinii Strains Expressing Mutant BbCRASP-2 Proteins—CRASP-mediated binding of complement regulators to the bacterial surface directly correlates with serum resistance of human pathogenic B. burgdorferi s.s. B. afzelii, and B. spielmanii, whereas B. garinii that lacks CRASP proteins is highly susceptible to complement (11, 13, 14, 20, 41, 42). We therefore examined each B. garinii transformant for complement activation and serum resistance under physiological conditions. Wild-type strains G1 and G1/pKFSS1 and each BbCRASP-2-complemented strain were incubated in 50% NHS or in 50% heat-inactivated NHS, and survival was monitored (7). Different levels of serum susceptibility were observed among the various strains (Fig. 6). Strains G1/pCSPZ, G1/pCSPZ F81A, G1/pCSPZ R139A, and G1/pCSPZ Y207A were resistant to complement-mediated lysis, whereas growth of wild-type G1, G1/pKFSS1, G1/pCSPZ R139A/Y207A and G1/pCSPZ Y207A/Y211A was strongly inhibited under the same conditions. Strain G1/pCSPZ Y211A displayed an intermediate serum-resistant phenotype. Using heat-inactivated NHS instead of NHS, growth of all strains analyzed was unaffected and led to a continuous decrease of absorbance (Fig. 6).
FIGURE 6.
Serum susceptibility of B. garinii G1 strains expressing diverse BbCRASP-2 proteins. A growth inhibition assay was used to investigate susceptibility to human serum of G1, G1/pKFSS1, G1/pCSPZ, G1/pCSPZ F81A, G1/pCSPZ R139A, G1/pCSPZ Y207A, G1/pCSPZ Y211A, G1/pCSPZ R139A/Y207A, and G1/pCSPZ Y207A/Y211A. Spirochetes were incubated in either 50% NHS (filled triangles) or 50% heat-inactivated NHS (open triangles) over a cultivation period of 9 days at 33 °C, respectively. Color changes were monitored by measurement of the absorbance at 562/630 nm. All experiments were performed three times in which each test was done at least 3-fold with very similar results. For clarity only data from representative experiments are shown. Error bars represent ± S.D.
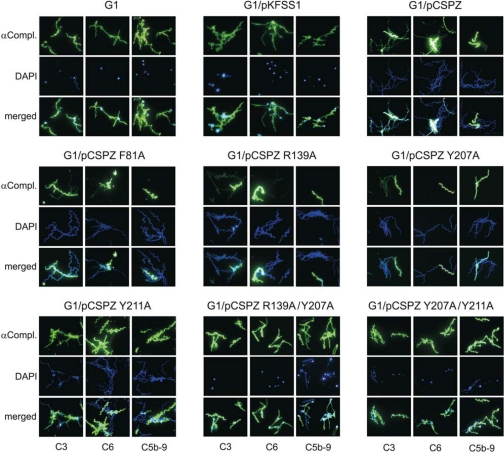
Deposition of Complement Components on the Surface of Transformed B. garinii Strains—Because the B. garinii transformants exhibit differential serum susceptibility, we also assessed complement activation on the surface by analyzing deposition of C3 and late activation products C6 and C5b-9 (terminal complement complex, TCC). Spirochetes were incubated in 25% NHS, and binding of complement components was analyzed by immunofluorescence microscopy. C3 deposition with variable staining with few strongly and many weakly stained cells was observed for strains G1/pCSPZ, G1/pCSPZ F81A, G1/pCSPZ R139A, and G1/pCSPZ Y207A (Fig. 7). In contrast, C3 bound strongly to wild-type strain G1 and transformants G1/pKFSS1, G1/pCSPZ Y211A, G1/pCSPZ R139A/Y207A, and G1/pCSPZ Y207A/Y211A (Fig. 7). Similarly, the serum-resistant transformants showed relatively weak signals, whereas all other strains displayed prominent signals for late complement components C6 and C5b-9 (TCC). Spirochetes that were fully covered with complement components exhibited blebs of various sizes and showed membrane damage and alterations in cell morphology (Fig. 7). In addition, most of the complement-affected cells stained negatively with 4′,6-diamidino-2-phenylindole, indicating that these spirochetes might represent cell ghosts.
FIGURE 7.
Deposition of complement components C3, C6, and C5b-9 on the surface of B. garinii G1 expressing diverse BbCRASP-2 proteins. Complement components deposited on G1, G1/pKFSS1, G1/pCSPZ, G1/pCSPZ F81A, G1/pCSPZ R139A, G1/pCSPZ Y207A, G1/pCSPZ Y211A, G1/pCSPZ R139A/Y207A, and G1/pCSPZ Y207A/Y211A were detected by indirect immunofluorescence microscopy. Spirochetes were incubated with either 25% NHS for 30 min at 37 °C with gentle agitation, and bound C3, C6, and C5b-9 were analyzed with specific antibodies against each component and appropriate Alexa 488-conjugated secondary antibodies. For visualization of the spirochetes in a given microscopic field, the DNA-binding dye 4′,6-diamidino-2-phenylindole (DAPI) was used. Slides were visualized at a magnification of ×1000. Panels shown are representative for at least 20 microscope fields.
DISCUSSION
Lyme disease spirochetes have developed sophisticated strategies to evade destruction by host immune defenses (3, 43, 44). Active recruitment of immune regulators CFH and CFHL1 represents an important escape strategy to overcome host complement attack. Pathogenic B. burgdorferi s.s., B. afzelii, and B. spielmanii isolates resist complement-mediated killing due to the production of multiple CRASPs proteins, which bind CFH and CFHL1 (13, 14, 20, 22, 25, 27, 41). All serum-resistant B. burgdorferi s.s. isolates examined so far produce as many as five distinct CRASPs, belonging to three genetically distinct classes. Complementation of serum-sensitive Borreliae with either BbCRASP-1 or BbCRASP-2 increases resistance to complement-mediated lysis (Refs. 23, 31 and this work). The recently identified CFH- and CFHL1-binding BbCRASP-2 protein (also referred as CspZ) is highly conserved among B. burgdorferi s.s. isolates, and infected humans and laboratory animals produce robust antibody responses against this protein (31, 32, 34).
In our earlier analyses, through use of a peptide-based approach, we identified four linear regions within BbCRASP-2 capable of binding of CFH and CFHL1 (31). Until cocrystals of BbCRASP-2 with its ligands become available, the identification of residues acting upon CFH/CFHL1 binding has to be sought by other means, such as the site-directed mutagenesis approach used here. We decided to systematically substitute residues within the putative binding region -2, -3, and -4 with a neutral alanine residue, to limit side-effects and to best allow maintenance of the native conformation of BbCRASP-2 molecules. Substitutions of 14 distinct amino acid residues within the putative binding regions affected CFH and CFHL1 binding. In addition, some substitutions affected binding of CFH (S79A) or only CFHL1 (N75A, H130A, and D133A). The binding profiles show that the N-terminal part of region 4 appears to represent the most sensitive CFH/CFHL1 binding site, as changes at any position between 204 and 211 decreased binding of complement regulators. In addition, several amino acid residues within binding region 2 (Gln-71, Asn-75, Ser-79, Phe-81, and Asp-84) and binding region 3 (Arg-129, His-130, and Arg-139) also influence binding of the complement regulators. These data confirm the concept that BbCRASP-2 uses multiple contact sites for CFH/CFHL1 binding. ELISA experiments indicated that several charged residues, e.g. Asp-84, Arg-129, Arg-139, Arg-204, Lys-206, and Glu-214 as well as hydrophobic residues Phe-81, Tyr-207, Phe-210, and Tyr-211 are involved in these interactions. These results suggest that interactions between BbCRASP-2 and immune regulators are dictated by two different binding mechanisms, one dependent on electrostatic forces and the other being favored by hydrophobic contacts and hydrogen bonding. This is in line with the general conclusion that the predominant driving forces in protein-protein interactions include covalent and non-covalent interactions, e.g. van der Waals contacts, electrostatic forces, and hydrogen bonds (45, 46). Effects of increasing salt concentrations on CFH binding to BbCRASP-2 suggest that long range electrostatic forces and ion pairings play a subordinate role. In general, salt concentrations in the range of 0.1–1 m reduce the formation of salt bridges and drastically influence protein-protein interactions that are primarily mediated by electrostatic forces, such as the interactions between CFH and host C3b (26, 45, 47). The relative influence of the electronegative polysaccharide heparin on BbCRASP-2-CFH interactions also suggests that other forces beside electrostatic are involved in this interaction. Alternatively, heparin and CFH may share overlapping but not identical binding sites on BbCRASP-2. In contrast, CFH binding to the borrelial proteins BbCRASP-1, BbCRASP-3, and OspE is completely inhibited by heparin (21, 22, 48). Thus, the type of interaction of the individual CRASP proteins with the same host complement regulator ligands is different. Long range electrostatic forces together with hydrophobic interactions might also be required for the stabilization of the complex formed by BbCRASP-2 and CFH/CFHL1.
Human pathogenic B. burgdorferi s.s. B. afzelii, and B. spielmanii evading complement-mediated killing by binding CFH and/or CFHL1 (7, 11, 12, 14, 20, 41). As a corollary, loss of CRASP function or expression leads to a highly serum-susceptible phenotype, as is observed for many B. garinii strains (11, 13). The importance of CFH and CFHL1 binding for mediating serum resistance was demonstrated by transformation of an initial serum-sensitive B. garinii strain with a shuttle vector that allows exogenous production of BbCRASP-2. Here we clearly show that production of the CFH/CFHL1-binding BbCRASP-2 protein converts the parental serum-sensitive strain to a serum-resistant phenotype (Fig. 6C). Thus, serum resistance directly correlated with binding of CFH and CFHL1 and significant reduction of complement activation products deposited on the cell surface. Moreover, we showed that some exchanges of single amino acid residues (in mutants F81A, R139, and Y207A) did not always result in higher serum susceptibility, whereas mutations at two positions had additive effects to increase susceptibility to complement (mutants R139A/Y207A and Y207A/Y211A). Our findings suggest that several amino acid residues directly participate in the interaction with CFH and CFHL1 and that the binding region 4 formed a highly sensitive binding site on BbCRASP-2.
The BbCRASP-2-encoding cspZ gene is highly conserved within a wide variety of geographically dispersed B. burgdorferi s.s. isolates (32, 33). The amino acid residues relevant for CFH and CFHL1 binding are conserved in all isolates examined. In contrast, BbCRASP-2 orthologs from other Borrelia species, including B. garinii, B. afzelii, and B. spielmanii did not bind either CFH and CFHL1 (32, 33). Because the N-terminal insertion found in those BbCRASP-2 orthologs was not responsible for the lack of CFH binding, it is tempting to speculate that the sequence variations within binding regions 2 and 4 are responsible for the non-binding features. The consecutive mutational analysis of BbCRASP-2 revealed that seemingly subtle changes of amino acid residues can have significant impacts on CFH and CFHL1 binding. Similarly, single amino acid substitutions within the C terminus of B. burgdorferi BbCRASP-1 also abrogate binding to CFH and CFHL1 (49). These lines of evidence suggest that distinct amino acid residues play pivotal roles for interactions of BbCRASP-2 with CFH and CFHL1. In addition to the Lyme disease Borrelia species a number of other important human pathogenic bacteria produce proteins utilize host immune regulators CFH and/or CFHL1 for immune and complement evasion and for adherence to human cells (44, 50–60). The underlying mechanism(s) of the molecular protein-protein interaction are largely unclear, but it appears that various bacterial proteins bind to human immune regulators by different modes. Involvement of conformational binding structures and linear determinants is uncertain regarding the CFH-binding proteins of relapsing fever Borrelia (61, 62, 63) and various pneumococci strains (51, 53, 56). Thus, elucidating the structural underpinnings of this aspect of host-pathogen interactions will have major impacts upon developing new specific therapeutics against human pathogens.
In summary, we mapped a structurally sensitive CFH/CFHL1 binding site within BbCRASP-2 and identified single amino acid residues, which are involved in interactions with the two host immune regulators. Furthermore, binding of CFH and CFHL1 to Borrelia producing BbCRASP-2 on their surface is sufficient to facilitate serum resistance in this pathogen. To our knowledge, this is the first report analyzing mutated proteins on the surface of live spirochetes under physiological conditions. Altogether, this study represents a major step toward a full understanding of the molecular interactions between serum-resistant Borreliae and immune regulators of the human host.
Supplementary Material
Acknowledgments
We thank Steffi Hälbich, Christa Hanssen-Hübner, and Jane Herrlich for skillful and expert technical assistance. We are indebted to Scott Samuels for generously providing the B. burgdorferi shuttle vector pKFSS1.
This work was funded by the Deutsche Forschungsgemeinschaft (Grant Kr3383/1-1 to P. K.). This work forms part of the doctoral theses of C.S. and J.S. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Author's Choice—Final version full access.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
Footnotes
The abbreviations used are: s.s., sensu stricto; CFH, complement factor H; CFHL1, complement factor H-like protein 1; CRASP, complement regulator-acquiring surface protein; NHS, normal human serum; SCR, short consensus repeat; GST, glutathione S-transferase; BbCRASP, borrelial complement regulator-acquiring surface protein; BSK, Barbour-Stoenner-Kelly medium; PBS, phosphate-buffered saline; ELISA, enzyme-linked immunosorbent assay; mAb, monoclonal antibody; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine; BSA, bovine serum albumin.
References
- 1.Bykowski, T., Woodman, M. E., Cooley, A. E., Brissette, C. A., Brade, V., Wallich, R., Kraiczy, P., and Stevenson, B. (2007) Infect. Immun. 75 4227-4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bykowski, T., Woodman, M. E., Cooley, A. E., Brissette, C. A., Wallich, R., Brade, V., Kraiczy, P., and Stevenson, B. (2007) Int. J. Med. Microbiol. 298 Suppl. 1, 249-256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hovius, J. W. R., van Dam, A. P., and Fikrig, E. (2007) Trends Parasitol. 23 434-438 [DOI] [PubMed] [Google Scholar]
- 4.Stevenson, B., von Lackum, K., Riley, S. P., Cooley, A. E., Woodman, M. E., and Bykowski, T. (2007) Wien. Klin. Wochenschr. 118 643-652 [DOI] [PubMed] [Google Scholar]
- 5.Brade, V., Kleber, I., and Acker, G. (1992) Immunobiology 185 453-465 [DOI] [PubMed] [Google Scholar]
- 6.Breitner-Ruddock, S., Würzner, R., Schulze, J., and Brade, V. (1997) Med. Microbiol. Immunol. (Berl.) 185 253-260 [DOI] [PubMed] [Google Scholar]
- 7.Kraiczy, P., Hunfeld, K.-P., Breitner-Ruddock, S., Würzner, R., Acker, G., and Brade, V. (2000) Immunobiology 201 406-419 [DOI] [PubMed] [Google Scholar]
- 8.Kurtenbach, K., Sewell, H.-S., Ogden, N. H., Randolph, S. E., and Nuttall, P. A. (1998) Infect. Immun. 66 1248-1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurtenbach, K., De Michaelis, S., Etti, S., Schäfer, S. M., Sewell, H.-S., Brade, V., and Kraiczy, P. (2002) Trends Microbiol. 10 74-79 [DOI] [PubMed] [Google Scholar]
- 10.Van Dam, A. P., Oei, A., Jaspars, R., Fijen, C., Wilske, B., Spanjaard, L., and Dankert, J. (1997) Infect. Immun. 65 1228-1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alitalo, A., Meri, T., Rämö, L., Jokiranta, T. S., Heikkilä, T., Seppälä, I. J. T., Oksi, J., and Meri, S. (2001) Infect. Immun. 69 3685-3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hellwage, J., Meri, T., Heikkilä, A., Panelius, J., Lahdenne, P., Seppälä, I., and Meri, S. (2001) J. Biol. Chem. 276 8427-8435 [DOI] [PubMed] [Google Scholar]
- 13.Kraiczy, P., Skerka, C., Brade, V., and Zipfel, P. F. (2001) Eur. J. Immunol. 31 1674-1684 [DOI] [PubMed] [Google Scholar]
- 14.McDowell, J. V., Tran, E., Hamilton, D., Wolfgang, J., Miller, K., and Marconi, R. T. (2003) J. Clin. Microbiol. 41 3905-3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zipfel, P. F., and Skerka, C. (1999) Immunol. Today 20 135-140 [DOI] [PubMed] [Google Scholar]
- 16.Kühn, S., Skerka, C., and Zipfel, P. F. (1995) J. Immunol. 155 5663-5670 [PubMed] [Google Scholar]
- 17.Kühn, S., and Zipfel, P. F. (1995) Gene 162 225-229 [DOI] [PubMed] [Google Scholar]
- 18.Pangburn, M. K., Schreiber, G. M., and Müller-Eberhard, H. J. (1977) J. Exp. Med. 146 257-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraiczy, P., Skerka, C., Brade, V., and Zipfel, P. F. (2001) Infect. Immun. 69 7800-7809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herzberger, P., Siegel, C., Skerka, C., Fingerle, V., Schulte-Spechtel, U., van Dam, A., Wilske, B., Brade, V., Zipfel, P. F., Wallich, R., and Kraiczy, P. (2007) Infect. Immun. 75 4817-4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraiczy, P., Hellwage, J., Skerka, C., Kirschfink, M., Brade, V., Zipfel, P. F., and Wallich, R. (2003) Eur. J. Immunol. 33 697-707 [DOI] [PubMed] [Google Scholar]
- 22.Kraiczy, P., Hellwage, J., Skerka, C., Becker, H., Kirschfink, M., Simon, M. M., Brade, V., Zipfel, P. F., and Wallich, R. (2004) J. Biol. Chem. 279 2421-2429 [DOI] [PubMed] [Google Scholar]
- 23.Brooks, C. S., Vuppala, S. R., Jett, A. M., Alitalo, A., Meri, S., and Akins, D. R. (2005) J. Immunol. 175 3299-3308 [DOI] [PubMed] [Google Scholar]
- 24.von Lackum, K., Miller, J. C., Bykowski, T., Riley, S. P., Woodmann, M. E., Brade, V., Kraiczy, P., Stevenson, B., and Wallich, R. (2005) Infect. Immun. 73 7398-7405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alitalo, A., Meri, T., Lankinen, H., Seppälä, I. J. T., Lahdenne, P., Hefty, P. S., Akins, D. R., and Meri, S. (2002) J. Immunol. 169 3847-3853 [DOI] [PubMed] [Google Scholar]
- 26.Hellwage, J., Jokiranta, T. S., Friese, M. A., Wolk, T. U., Kampen, E., Zipfel, P. F., and Meri, S. (2002) J. Immunol. 169 6935-6944 [DOI] [PubMed] [Google Scholar]
- 27.Metts, M. S., McDowell, J. V., Theisen, M., Hansen, P. R., and Marconi, R. T. (2003) Infect. Immun. 71 3587-3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, J. C., von Lackum, K., Babb, K., McAlister, J. D., and Stevenson, B. (2003) Infect. Immun. 71 6943-6952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, J. C., and Stevenson, B. (2006) Int. J. Med. Microbiol. 296 S1 185-194 [DOI] [PubMed] [Google Scholar]
- 30.Stevenson, B., El-Hage, N., Hines, M., Miller, J. C., and Babb, K. (2002) Infect. Immun. 70 491-497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartmann, K., Corvey, C., Skerka, C., Kirschfink, M., Karas, M., Brade, V., Miller, J. C., Stevenson, B., Wallich, R., Zipfel, P. F., and Kraiczy, P. (2006) Mol. Microbiol. 61 1220-1236 [DOI] [PubMed] [Google Scholar]
- 32.Kraiczy, P., Seling, A., Brisette, C. A., Rossmann, E., Hunfeld, H.-P., Bykowski, T., Burns, L. H., Troese, M. J., Cooley, A. E., Miller, J. C., Brade, V., Wallich, R., Casjens, S., and Stevenson, B. (2008) Clin. Vaccine Immunol. 15 484-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogers, E. A., and Marconi, R. T. (2007) Infect. Immun. 75 5272-5281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colemann, A. S., Yang, X., Kumar, M., Zhang, X., Promnares, K., Shroder, D., Kenedy, M. R., Anderson, J. F., Akins, D. R., and Pal, U. (2008) PLoS One 3 3010e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prodinger, W., Hellwage, J., Spruth, M., Dierich, M. P., and Zipfel, P. F. (1998) Biochem. J. 31 41-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hauser, U., Lehnert, G., and Wilske, B. (1999) J. Clin. Microbiol. 37 2241-2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samuels, S. D. (1995) Electroporation Protocols for Microorganismen, Vol. 47, Humana Press. Totowa, NJ
- 38.Frank, K. L., Bundle, S. F., Kresge, M. E., Eggers, C. H., and Samuels, D. S. (2003) J. Bacteriol. 185 6723-6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kühn, S., and Zipfel, P. F. (1996) Eur. J. Immunol. 26 2383-2387 [DOI] [PubMed] [Google Scholar]
- 40.Phizicky, E. M., and Fields, S. (1995) Microbiol. Rev. 59 94-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallich, R., Pattathu, J., Kitiratschky, V., Brenner, C., Zipfel, P. F., Brade, V., Simon, M. M., and Kraiczy, P. (2005) Infect. Immun. 73 2351-2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siegel, C., Herzberger, P., Skerka, C., Brade, V., Fingerle, V., Schulte-Spechtel, U., Wilske, B., Zipfel, P. F., Wallich, R., and Kraiczy, P. (2008) Int. J. Med. Microbiol. 298 Suppl. 1, 292-294 [DOI] [PubMed] [Google Scholar]
- 43.Embers, M. E., Ramamoorthy, R., and Phillip, M. T. (2004) Microbes Infect. 6 312-318 [DOI] [PubMed] [Google Scholar]
- 44.Kraiczy, P., and Würzner, R. (2006) Mol. Immunol. 43 31-44 [DOI] [PubMed] [Google Scholar]
- 45.Dill, K. A. (1990) Biochemistry 29 7133-7155 [DOI] [PubMed] [Google Scholar]
- 46.Kollman, P. A. (1994) Curr. Opin. Struct. Biol. 4 240-245 [Google Scholar]
- 47.Giannakis, E., Jokiranta, T. S., Male, D. A., Ranganathan, S., Ormsby, R. J., Fischetti, V. A., Mold, C., and Gordan, D. L. (2003) Eur. J. Immunol. 33 962-969 [DOI] [PubMed] [Google Scholar]
- 48.Alitalo, A., Meri, T., Chen, T., Lankinen, H., Cheng, Z.-Z., Jokiranta, T. S., Seppälä, I. J. T., Lahdenne, P., Hefty, P. S., Akins, D. R., and Meri, S. (2004) J. Immunol. 172 6195-6201 [DOI] [PubMed] [Google Scholar]
- 49.Cordes, F. S., Roversi, P., Kraiczy, P., Simon, M. M., Brade, V., Jahrhaus, O., Wallis, R., Skerka, C., Zipfel, P. F., Wallich, R., and Lea, S. (2005) Nat. Struct. Mol. Biol. 12 276-277 [DOI] [PubMed] [Google Scholar]
- 50.Ram, S., Mackinnon, F. G., Gulati, S., McQuillen, D. P., Vogel, U., Frosch, M., Elkins, C., Guttormsen, H. K., Wetzler, L. M., Oppermann, M., Pangburn, M. K., and Rice, P. A. (1999) Mol. Immunol. 36 915-928 [DOI] [PubMed] [Google Scholar]
- 51.Jarva, H., Janulczyk, R., Hellwage, J., Zipfel, P. F., Björck, L., and Meri, S. (2002) J. Immunol. 168 1886-1894 [DOI] [PubMed] [Google Scholar]
- 52.Jarva, H., Hellwage, J., Jokiranta, T. S., Lehtinen, M. J., Zipfel, P. F., and Meri, S. (2004) J. Immunol. 172 3111-3118 [DOI] [PubMed] [Google Scholar]
- 53.Hammerschmidt, S., Kunert, A., Agarwal, V., Skerka, C., and Zipfel, P. F. (2007) J. Immunol. 178 5848-5858 [DOI] [PubMed] [Google Scholar]
- 54.Johnsson, E., Berggard, K., Kotarsky, H., Hellwage, J., Zipfel, P. F., Sjöbring, U., and Lindahl, G. (1998) J. Immunol. 161 4894-4901 [PubMed] [Google Scholar]
- 55.Kotarsky, H., Hellwage, J., Johnsson, E., Skerka, C., Svensson, H. G., Lindahl, G., Sjobring, U., and Zipfel, P. F. (1998) J. Immunol. 160 3349-3354 [PubMed] [Google Scholar]
- 56.Lu, L., Ma, Y., and Zhang, J.-R. (2007) J. Biol. Chem. 281 15464-15474 [DOI] [PubMed] [Google Scholar]
- 57.Verma, A., Hellwage, J., Artiushin, S., Zipfel, P. F., Kraiczy, P., Timoney, J. F., and Stevenson, B. (2006) Infect. Immun. 74 2659-2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kunert, A., Losse, J., Gruzin, C., Hühn, M., Kaendler, K., Mikkat, S., Volke, D., Hoffmann, R., Jokiranta, T. S., Seeberger, H., Moellmann, U., Hellwage, J., and Zipfel, P. F. (2007) J. Immunol. 179 2979-2988 [DOI] [PubMed] [Google Scholar]
- 59.Stevenson, B., Choy, H. A., Pinne, M., Rotondi, M. L., Miller, M. C., De-Moll, E., Kraiczy, P., Cooley, A. E., Creamer, T. P., Suchard, M. A., Brissette, C., Verma, A., and Haake, D. A. (2007) PLoS ONE 2 e1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lambris, J. D., Ricklin, D., and Geisbrecht, B. V. (2008) Nat. Rev. Microbiol. 6 132-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hovis, K. M., Jones, J. P., Sadlon, T., Raval, G., Gordon, D. L., and Marconi, R. T. (2006) Infect. Immun. 74 2007-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hovis, K. M., Freedman, J. C., Zhang, H., Forbes, J. L., and Marconi, R. T. (2008) Infect. Immun. 76 2113-2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rossmann, E., Kraiczy, P., Herzberger, P., Skerka, C., Kirschfink, M., Simon, M. M., Zipfel, P. F., and Wallich, R. (2007) J. Immunol. 178 7292-7301 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.