Abstract
c-Jun N-terminal kinases (JNKs) are part of the mitogen-activated protein kinase (MAPK) family and are important regulators of cell growth, proliferation, and apoptosis. Typically, a sequential series of events are necessary for MAPK activation: phosphorylation, dimerization, and then subsequent translocation to the nucleus. Interestingly, a constitutively active JNK isoform, JNK2α2, possesses the ability to autophosphorylate and has been implicated in several human tumors, including glioblastoma multiforme. Because overexpression of JNK2α2 enhances several tumorigenic phenotypes, including cell growth and tumor formation in mice, we studied the mechanisms of JNK2α2 autophosphorylation and autoactivation. We find that JNK2α2 dimerization in vitro and in vivo occurs independently of its autophosphorylation but is dependent on nine amino acids, known as the α-region. Alanine scanning mutagenesis of the α-region reveals that five specific mutants (L218A, K220A, G221A, I224A, and F225A) prevent JNK2α2 dimerization rendering JNK2α2 inactive and incapable of stimulating tumor formation. Previous studies coupled with additional mutagenesis of neighboring isoleucines and leucines (I208A, I214A, I231A, and I238A) suggest that a leucine zipper may play an important role in JNK2α2 homodimerization. We also show that a kinase-inactive JNK2α2 mutant can interact with and inhibit wild type JNK2α2 autophosphorylation, suggesting that JNK2α2 undergoes trans-autophosphorylation. Together, our results demonstrate that JNK2α2 differs from other MAPK proteins in two major ways; its autoactivation/autophosphorylation is dependent on dimerization, and dimerization most likely precedes autophosphorylation. In addition, we show that dimerization is essential for JNK2α2 activity and that prevention of dimerization may decrease JNK2α2 induced tumorigenic phenotypes.
The c-Jun N-terminal kinases (JNK),3 along with ERK1/2 and p38, comprise the mitogen-activated protein kinase (MAPK) family. By regulating AP-1 complexes and other transcription factors, JNK can regulate apoptosis, cellular proliferation, and differentiation (1, 2). Members of the JNK family have emerged as having important and diverse functions, depending on the isoform. Three distinct genes encoding JNKs are known: JNK1, JNK2, and JNK3, and at least 10 different splicing isoforms exist in mammalian cells. JNK1 and JNK2 are expressed in all tissues, whereas JNK3 expression is restricted to the brain, heart, and testis (3). JNK1 and JNK2 were initially believed to possess redundant functions; however, it was found that JNK1 preferentially mediates apoptosis (4), whereas JNK2 is associated with cellular proliferation (5). These findings suggest that individual JNK isoforms may play different roles in cellular transformation.
Recently, JNK2 has been implicated in tumorigenesis because JNK2-deficient mice were more resistant to the induction of papillomas, and silencing of JNK2 through RNA interference was shown to hinder the growth of the human T98G glioblastoma cell line (6, 7). We have also previously shown that 86% of primary glial tumors have increased activation of a specific JNK2 isoform, JNK2α2 (8, 9). In addition, we demonstrated that overexpression of JNK2α2 increased cell growth and increased tumor formation in nude mice, indicating that JNK2α2 may be important for tumor formation (9). JNK2α2 was also found to activate two crucial effectors of tumorigenesis: increased phosphorylation of AKT and increased eIF4E levels. AKT is a critical mediator for cell survival and proliferation, and eIF4E is a component of a cap-dependent translation initiation complex that is linked to breast, colon, and prostate tumors (9, 10).
Previous work argues that MAPK proteins, such as ERK1/2 and JNK, can dimerize and that dimerization may play an integral role in the activation of these MAPK proteins (11). Studies with ERK2 have shown that disruption of ERK homodimers reduces its ability to localize to the nucleus and that only homodimers are actively transported to the nucleus (12, 13). In addition, phosphorylation by upstream MAPK kinases is necessary for and precedes ERK2 dimerization (12). Unlike ERK2, the JNK2α2 isoform is capable of phosphorylating itself and downstream substrates, such as c-Jun, without the presence of upstream kinases (14). This ability to autophosphorylate can occur in either a cis- (intra-) or trans- (inter-) molecular mechanism. Domain analysis has revealed that JNK2α2 autophosphorylation activity and its ability to translocate to the nucleus is dependent on the alternatively spliced region known as the α-region (15). This 9-amino acid domain (LVKGCVIFQ) is critical for activation, because swapping this domain into the analogous region of the nonconstitutively active isoform, JNK1α2, renders the isoform active (15). Conversely, substitution of the JNK2α2 α-region with the analogous region of JNK1α2 renders JNK2α2 inactive. Despite knowing that the α-region is critical for activation, little is understood about how this region participates in JNK2α2 dimerization and autoactivation. Because JNK2α2 activity enhances tumor formation, it is important to fully understand its mechanism of autophosphorylation and autoactivation.
Our present findings demonstrate that JNK2α2 has a unique mechanism of autophosphorylation and autoactivation. We determined that dimerization is dependent on the α-region and occurs independently of phosphorylation. Also, disruption of JNK2α2 dimerization through specific mutations in the α-region resulted in loss of nuclear localization, loss of autophosphorylation, and decreased tumorigenicity. These results illustrate that JNK2α2 is unique from other MAPK proteins because its autophosphorylation/autoactivation is dependent on homodimerization and that this dimerization most likely precedes autophosphorylation. Characterization of the mechanism through which JNK2α2 is constitutively active will be of great value for designing and developing isoform specific inhibitors that could be used to help treat patients with glioblastoma multiforme.
EXPERIMENTAL PROCEDURES
Plasmid Construction of JNK2α2 Mutants—The cDNA encoding wild type JNK2α2 was cloned into BamHI/XhoI restriction sites of pcDNA 6A (Invitrogen). Point mutations were created by site-directed mutagenesis according to the QuikChange site-directed mutagenesis kit protocol (Stratagene). The oligonucleotide primer sequences are available upon request. The JNK2α2 mutant inserts were then subcloned into BamHI/XhoI restriction sites of pET42a (Novagen), pET28a (Novagen), or pEGFP-C1 (Clontech). The chimera constructs were generated as previously described (15).
Cell Culture and Cellular Transfection—Human embryonic kidney cell line HEK293 and U87-MG cells (American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mm-l-glutamine, and 100 units/ml penicillin/streptomycin. The cells grown to 30% confluence were transfected using FuGENE 6 (Roche Applied Science) according to the manufacturer's protocol.
Protein Expression and Purification—pET28 and pGEX constructs were expressed in the BL21(DE3)pLysS Escherichia coli strain (Novagen) and induced with 1 mm isopropyl β-d-1-thiogalactopyranoside for 4 h at 37 °C. The tagged proteins were purified according to the manufacturer's protocol using nickel-nitrilotriacetic acid-agarose beads (Qiagen). Protein was concentrated to 1 mg/ml using Microcon Ultracel YM-10 (Millipore) and stored in 20% glycerol at -80 °C.
Gel Filtration—2 mg of purified protein was applied to a 90-ml Sephacryl S-400 (GE Healthcare) gel filtration column equilibrated in phosphate-buffered saline. 1-ml fractions were collected in the same buffer, and the protein concentrations were determined by measuring the A280 nm. Relevant fractions were analyzed using SDS-PAGE and stained with Coomassie Blue. A standard curve was generated using the molecular mass gel filtration markers (Sigma) according to the manufacturer's protocol.
Protein Analysis and Immunoprecipitation—Protein extracts from total cells were harvested as previously described (16). 50 μg of total protein was separated by denaturing electrophoresis. The following antibodies were used for immunoblotting: mouse anti-GFP (catalog number 11814460001; Roche Applied Science), rabbit anti-phosphorylated JNK that recognizes the phosphorylated T-P-Y motif (98F2; Cell Signaling Technology), rabbit anti-phosphorylated c-Jun (9261; Cell Signaling Technology), mouse anti-JNK2 (sc-7345; Santa Cruz), mouse anti-actin (MAB1501R; Chemicon), mouse ant-GST (sc-138; Santa Cruz), and mouse anti-FLAG (Sigma; F3165). Immunoprecipitations for 3×FLAG constructs were conducted in radioimmunoprecipitation assay buffer using 2 μg of mouse anti-FLAG antibody (F3165; Sigma) bound to Gammabind G-Sepharose (GE Healthcare). The protein extracts were harvested and immunoblotted as described previously (16). The protein levels were quantitated using National Institutes of Health Image J.
Immunofluorescence—Immunofluorescence was performed on formaldehyde-fixed cells as previously described (17). The images were taken using a Leica SP2 AOBS confocal laser scanning microscope. Images that compared protein levels were collected using equal exposure times and processed in a similar fashion.
In Vitro Kinase Assay—1 μg of the fusion protein was incubated in 25 μl of the kinase buffer (25 mm HEPES, pH 7.4, 25 mm MgCl2, 2 mm dithiothreitol, 0.1 mm NaVO4, and 25 mm β-glycerophosphate) containing 30 μm ATP at 30 °C for 30 min. The reactions were terminated by adding protein loading buffer and boiling for 5 min (8). Radioactive kinase assays were conducted in a similar manner except 2 μm ATP and 5 μCi of [32P]ATP was added. Western blots were analyzed using anti-phospho-JNK antibody previously described (98F2; Cell Signaling Technology).
BS3 Cross-linking Assay—The experiments were carried out with 1 mg of protein (250 nm) in phosphate-buffered saline. A 50 mm stock solution of BS3 (Pierce) was serially diluted, added to each protein sample, and then incubated at room temperature for 30 min. The reactions were terminated by adding protein loading buffer and boiling the samples for 5 min (18). The samples were then analyzed by SDS-PAGE followed by staining using Coomassie Blue. The final concentration of BS3 ranged between 30 μm and 5 mm.
Retroviral Infections—The retrovirus pMXIH-YPet was generated by subcloning YPet from the pCEP4-YPet plasmid into the pMXIH backbone (19, 20). Retroviral infections were carried out as previously described except that the Phoenix-Ampho cell lines were used instead of HEK293 cells (16). 36 h after infection of the U87-MG cells, the infected cells were selected by culturing in selective medium containing 500 μg/ml of hygromycin-B for 2 days.
Cell Growth Analysis—U87-MG-infected cells were plated in 6-well plates (5.0 × 105 cells/well) and cultured in DMEM supplemented with 1% fetal bovine serum, 2 mm l-glutamine, and 100 units/ml penicillin/streptomycin. The numbers of live cells were counted daily by means of trypan blue exclusion assay. The experiments were done in triplicate, and the results were expressed as the means ± S.D.
Soft Agar Assay—U87-MG-infected cells were plated in 6-well plates (3 × 105 cells/well), suspended in DMEM containing 0.3% of agarose and 1% fetal bovine serum, and overlaid onto a bottom layer of solidified 0.7% agarose in DMEM containing 10% fetal bovine serum, 2 mm l-glutamine, and 100 units/ml penicillin/streptomycin. The presence of colonies was scored after 10 days using Genetools (Syngene). The experiments were done in triplicate, and the results were expressed as the means ± S.D.
RESULTS
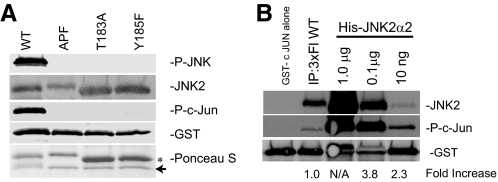
Bacterially Expressed JNK2α2 Can Autophosphorylate and Autoactivate—We previously have observed strong autophosphorylation and constitutive activation of the JNK2α2 isoform, which is unique from the JNK1 and JNK3 genes (8). Understanding the mechanism by which JNK2α2 becomes activated would be of significant interest, because it would assist in the creation of inhibitors for the treatment of relevant diseases. Two hallmarks of JNK activity are the dual phosphorylation of the T-P-Y motif and the ability to phosphorylate downstream substrates such as c-Jun. To determine whether bacterially expressed JNK2α2 possesses these two hallmarks of activity, we conducted an in vitro kinase assay with bacterially expressed His-JNK2α2 with GST-c-Jun. We used an anti-active JNK antibody that only recognizes JNK when it is phosphorylated at both Thr183 and Tyr185. This reagent demonstrated that His-JNK2α2 could autophosphorylate itself, in the absence of an upstream kinase, on the T-P-Y motif (Fig. 1A). We verified that the antibody could only recognize a molecule phosphorylated at both sites by generating JNK2α2 mutants that blocked phosphorylation at each individual site (T183A and Y185F) or at both sites (APF). This showed that only the intact wild type (WT) JNK2 could be recognized by this antibody. Consistently, only the dually phosphorylated WT JNK2α2 protein can phosphorylate the GST-c-Jun substrate (Fig. 1A). Because our assay used bacterially expressed protein, we wished to determine its activity relative to JNK2α2 isolated from cells. To compare JNK2α2 activity, we conducted an in vitro kinase assay with GST-c-Jun and either immunoprecipitated 3×FLAG JNK2α2, which was transiently transfected in U87-MG cells, or the bacterially expressed His-JNK2α2. We used an antibody specific for phosphorylated c-Jun to measure the relative JNK2α2 activity. Although there was a greater amount of JNK2α2 in the immunoprecipitation than in 10 ng of fusion protein, Western analysis revealed that the bacterially expressed His-JNK2α2 had a ∼2–3-fold higher level of c-Jun phosphorylation compared with the cellular JNK2α2, suggesting that bacterially expressed JNK2α2 has a specific activity greater than cellular JNK2α2 (Fig. 1B).
FIGURE 1.
Bacterially expressed His-JNK2α2 can autophosphorylate itself at the T-P-Y motif and is constitutively active. A, in vitro kinase reactions with GST-c-Jun and recombinant His-JNK2α2 WT, a mutant incapable of being phosphorylated at the T-P-Y motif (APF), or individual mutants of the T-P-Y motif, T183A or Y185F. The samples were immunoblotted with an antibody specific for JNK phosphorylated at the T-P-Y motif (P-JNK), phosphorylated c-Jun (P-c-Jun), anti-JNK2, or anti-GST. A Ponceau S stain was conducted to monitor relative protein expression (arrow designates GST-c Jun and the asterisk represents His-JNK2α2). B, Western analysis comparing the relative activity of bacterially expressed JNK2α2 versus cellular JNK2α2. In vitro kinase reactions with GST c-Jun and immunoprecipitated 3×FLAG JNK2α2(IP:3×Fl WT) or decreasing concentrations of bacterially expressed His-JNK2α2. The samples were separated using a 4–20% gradient SDS-PAGE and immunoblotted with anti-JNK2, an antibody specific for phosphorylated c-Jun, and anti-GST. The signals from JNK2 and phosphorylated c-Jun were quantitated, and the ratio of phosphorylated c-Jun/JNK2 was determined. These ratios were normalized relative to the IP sample (designated 1.0) to obtain the fold increase. The 1.0-μg sample levels were too high to obtain an accurate measurement (N/A).
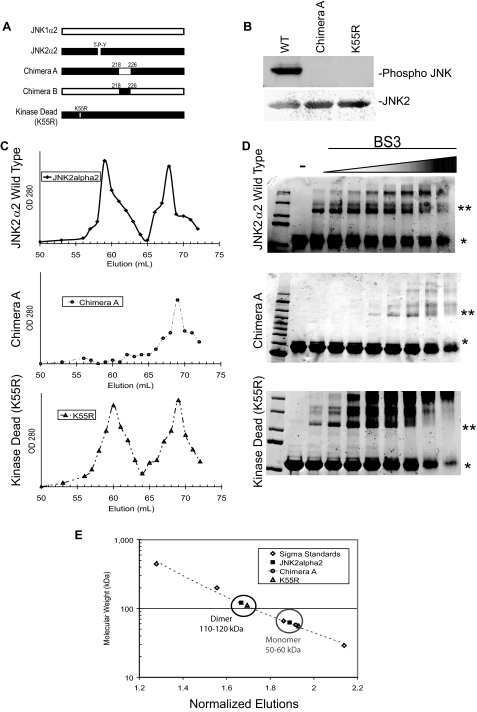
JNK2α2 Dimerization Is Dependent on the α-Region, but Not Phosphorylation—Some MAPK family members, such as ERK1/2, dimerize to achieve full activation, and typically phosphorylation occurs prior to dimerization (11, 12). We constructed a series of mutants to dissect the order in which JNK2α2 autoactivates. To determine whether phosphorylation is necessary for dimerization, we generated a kinase dead JNK2α2 mutant, K55R, in which the lysine that is critical for phosphotransfer in the ATP-binding motif was mutated to an arginine (Fig. 2A). In vitro kinase assays using purified recombinant protein confirmed that K55R does not autophosphorylate (Fig. 2B). To directly analyze the importance of the α-region, we used a JNK2α2 chimera (Chimera A) in which the JNK2α2 α-region was exchanged for the α-region from the JNK1α2 isoform, which was previously shown not to possess autophosphorylation activity (15) (Fig. 2A). Consistently, Chimera A showed no autophosphorylation activity in these assays (Fig. 2B).
FIGURE 2.
JNK2α2 dimerization in vitro is dependent on the α-region and is independent of phosphorylation. A, schematic of JNK2α2 chimeras and mutants. JNK1α2 and JNK2α2 sequences are indicated by white bars and black bars, respectively. The α-region is defined as the amino acids 218–226. The kinase dead JNK2α2 mutation (K55R) and the position of the T-P-Y motif are also shown. B, in vitro kinase reactions with purified JNK2α2 constructs. Western analysis using an antibody specific for phosphorylated T-P-Y in JNK (top panel) or JNK2 (bottom panel). WT, wild type JNK2α2. K55R, kinase dead mutant. C, gel filtration of JNK2α2 wild type and JNK mutants. JNK2α2 wild type (top panel), Chimera A (middle panel), and kinase dead mutant-K55R (bottom panel) were applied to a Sephacryl S-400 gel filtration column. 1-ml fractions were collected, and the A280 nm value was measured. D, cross-linking analysis of purified JNK2α2 protein was incubated with increasing concentrations of the homo-bifunctional cross-linker BS3. The samples were analyzed using SDS-PAGE and then stained with Coomassie Blue. -, samples not treated with BS3. * indicates the JNK2α2 monomer. ** indicates the JNK2α2 dimer. E, Sigma Protein Standards were applied to the gel filtration column to generate a standard curve in which the molecular mass of each standard protein was plotted against its normalized fraction (see “Experimental Procedures”). The black circle indicates fractions that are ∼110–120 kDa (dimer), and the gray circle shows fractions that are ∼50–60 kDa (monomer).
JNK2α2 Dimerization in Vitro—To analyze JNK2α2 dimerization in its native state, we used size exclusion gel filtration chromatography. Previous research has shown that JNK2α2 exists both as a dimer and a monomer (11). Gel filtration of recombinant wild type JNK2α2 resulted in two distinct peaks at 59 and 67 ml (Fig. 2C). To determine which peak contained dimeric or monomeric forms, a standard curve was generated using gel filtration protein markers (Fig. 2E). We determined that the 59-ml fraction contained proteins of ∼110–120 kDa in size, consistent with a JNK2α2 dimer, whereas the 67-ml fraction contained proteins of 50–60 kDa in size, consistent with a JNK2α2 monomer (Fig. 2, C and E). Interestingly, the kinase dead (K55R) mutant revealed two distinct peaks at 59 and 66 ml, and the column profile was similar to the wild type JNK2α2 (Fig. 2C). Application of the APF mutant also yielded a similar profile as WT JNK2α2 (supplemental Fig. S1). These findings suggested that JNK2α2 dimerization is not dependent upon autophosphorylation. Chromatography of Chimera A yielded only one single protein peak at 59 ml, indicating that the α-region is necessary for JNK2α2 to form dimers.
To verify the importance of the α-region, we also conducted a cross-linking assay. We used a primary amine reactive cross-linking reagent (BS3) that has been shown to cross-link dimerized protein, allowing the identification of protein dimers (18). Recombinant wild type JNK2α2 protein was incubated with increasing amounts of the BS3 cross-linker. We found that the lowest concentration of BS3 (30 μm) was sufficient to visualize a 120-kDa JNK2α2 dimer (Fig. 2D). When we extended our studies to Chimera A, we found that a 16-fold higher concentration of BS3 was necessary to visualize the 120-kDa JNK2α2 dimer. The Chimera A dimer is most likely an artifact because a 100-fold molar excess of BS3 was needed to visualize the dimer. In addition, the presence of the 200-kDa band is likely an artifact because of high BS3 concentration resulting in two dimers being cross-linked together. The kinase dead mutant behaved similarly to wild type JNK2α2, verifying that dimerization is phosphorylation-independent (Fig. 2D). Taken together, our findings suggest that the α-region, but not phosphorylation, is necessary for JNK2α2 dimerization.
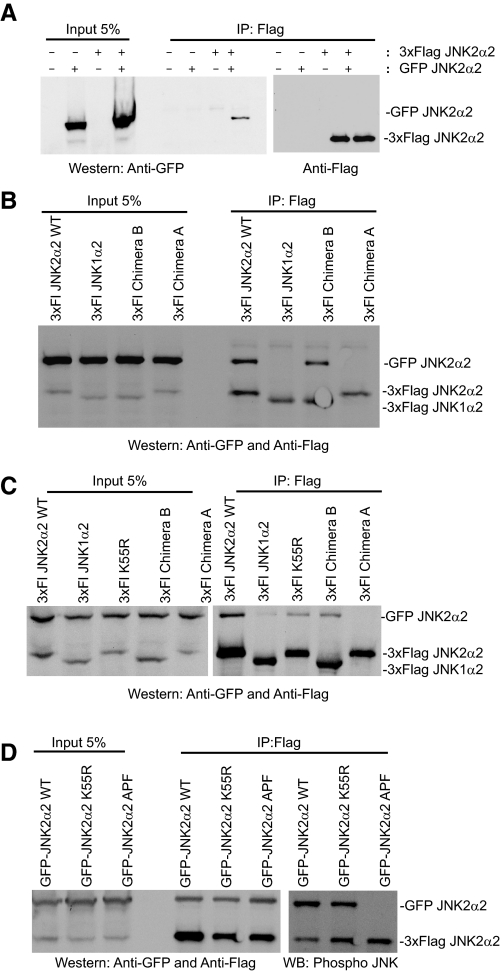
JNK2α2 Dimerization in Vivo—To determine whether the α-region is necessary for JNK2α2 dimerization in vivo, we co-transfected GFP-JNK2α2 and 3×FLAG JNK2α2 in HEK293 cells and performed co-immunoprecipitation assays. We discovered that the two tagged JNK2α2 proteins co-immunoprecipitated, suggesting that JNK2α2 does dimerize in vivo (Fig. 3A). When we extended our studies to 3×FLAG-JNK1α2, 3×FLAG Chimera A, and 3×FLAG Chimera B (a JNK1α2 mutant containing the α-region from JNK2α2; Fig. 2A), we found that only the 3×FLAG WT and 3×FLAG Chimera B could be immunoprecipitated with GFP-JNK2α2 (Fig. 3B; note Chimera B and JNK1α2 migrate similarly). These results suggest that the α-region from JNK2α2 is necessary for dimerization in vivo. The assays were then performed in a glioblastoma cell line, U87-MG. Consistently, we found that the wild type 3×FLAG JNK2α2 could interact with the GFP-JNK2α2, whereas Chimera A did not interact (Fig. 3C). Analysis with Chimera B revealed that 8-fold more GFP-JNK2α2 precipitated than JNK1α2, showing that it is the addition of the α-region from JNK2α2 that enhances the binding of JNK2α2 to JNK1α2. In addition, the 3×FLAG K55R mutant co-immunoprecipitated with GFP-JNK2α2, although it appeared that less 3×FLAG K55R co-immunoprecipitated compared with the 3×FLAG WT, suggesting that phosphorylation may contribute to JNK2α2 dimerization in vivo. However, this apparent decrease was likely due to variations in the immunoprecipitation technique because a duplicate assay revealed similar amounts of precipitated K55R mutant to the WT protein (Fig. 3D). To further show that dimerization does occur independently of phosphorylation, we also analyzed the GFP JNK2α2 APF mutant, which is incapable of being phosphorylated at the T-P-Y motif (Figs. 1A and 3D). We found that the JNK2α2 APF mutant was co-immunoprecipitated at levels similar to the GFP-WT protein, suggesting that phosphorylation does not affect JNK2α2 dimerization (Fig. 3D). Together these findings suggest that although JNK2α2 may form heterodimers with other JNK isoforms in vivo, it is the α-region, and not phosphorylation, which strongly promotes homodimers within cells.
FIGURE 3.
Co-immunoprecipitation indicates that the α-region is necessary for JNK2α2 dimerization in vivo. A, 3×FLAG-JNK2α2 and GFP-JNK2α2 constructs were cotransfected in HEK293 cells. The immunoprecipitates (IP) obtained with anti-FLAG were separated by SDS-PAGE and detected by immunoblotting with anti-GFP or anti-FLAG. B, 3×FLAG JNK2α2 wild type (3×Fl JNK2α2WT), 3×FLAG JNK1α2, 3×FLAG Chimera B, or 3×FLAG Chimera A constructs were cotransfected with GFP-JNK2α2 in HEK293 cells. The immunoprecipitates were analyzed in a similar fashion as in A. C, 3×FLAG JNK constructs were cotransfected with GFP-JNK2α2 in U87-MG cells and analyzed similarly as described above. D, GFP-JNK2α2 WT, GFP-JNK2α2 K55R, or GFP-JNK2α2 APF were cotransfected with 3×FLAG JNK2α2 WT in U87-MG cells and analyzed similarly as described above.
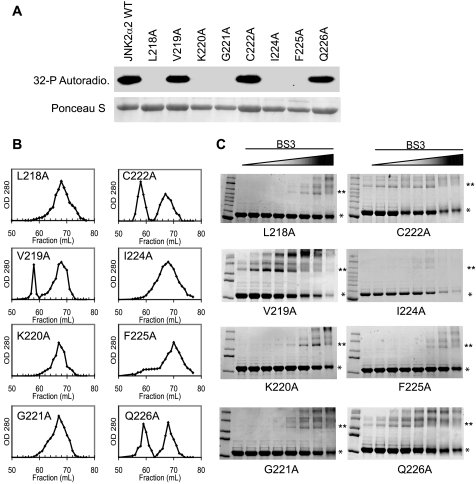
Inhibition of JNK2α2 Dimerization Prevents Autophosphorylation—To enhance our understanding of the role that the α-region plays in JNK2α2 activity, we conducted an alanine mutagenesis scan of the α-region (LVKGCVIFQ). Eight of the amino acids were individually mutated to an alanine residue to determine which amino acid(s) are necessary for JNK2α2 autophosphorylation and/or dimerization. We mutated only one valine (Val219) because valine and alanine are structurally very similar. In vitro kinase assays using radioactively labeled [32P]ATP demonstrated that five mutants (L218A, K220A, G221A, I224A, and F225A) could no longer autophosphorylate, whereas only three mutants (V219A, C222A, and Q226A) retained their autophosphorylation activity (Fig. 4A). To determine whether the loss of autophosphorylation activity correlated with an inability to dimerize, we analyzed each mutant using size exclusion chromatography and the aforementioned cross-linking assay. We observed that each autophosphorylation “inactive” mutant (L218A, K220A, G221A, I224A, and F225A) existed mainly as a monomer and required 16–32-fold more BS3 to form the 120-kDa dimer (Fig. 4, B and C). Consistently, the mutants that retained the ability to autophosphorylate (V219A, C222A, and Q226A) existed both as dimers and monomers and required only the lowest concentration of cross-linker to identify the JNK2α2 dimer (Fig. 4, B and C). These findings taken together with the K55R mutant data suggest that JNK2α2 autophosphorylation is dependent on dimerization and that the amino acids Leu218, Lys220, Gly221, Ile224, and Phe225 play an important role in dimerization.
FIGURE 4.
Alanine Scanning Mutagenesis of the JNK2α2 α-region. A, in vitro kinase assay using radioactive [32P]ATP with JNK2α2 wild type and the alanine mutants. The indicated amino acid in JNK2α2 was mutated to alanine. 1 μg of purified fusion protein was then used in an in vitro kinase/autophosphorylation assay. Top panel, autoradiogram of autophosphorylation reactions. In vitro kinase reactions were electrophoresed in SDS-PAGE and the subsequent Western blot used for autoradiography. Bottom panel, Ponceau S stain of the same Western blot to demonstrate equal loading of the respective JNK mutants. B, gel filtration of alanine mutants. Purified fusion proteins for the same mutants were used in size exclusion chromatography. 1-ml fractions were collected, and the A280 nm value was measured. C, cross-linking analysis using increasing concentrations of the homo-bifunctional cross-linker BS3. * indicates the JNK2α2 monomer. ** indicates the JNK2α2 dimer.
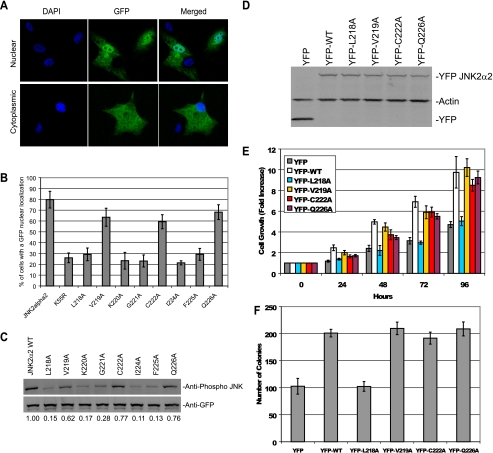
Dimerization Is Necessary for JNK2α2 Translocation to the Nucleus—If dimerization is necessary for JNK2α2 autophosphorylation, it may also be important for other components of JNK2α2 activity. One hallmark of JNK2α2 activity is the ability to translocate to the nucleus. To determine whether the autophosphorylation and dimer “inactive” mutants were also deficient in the ability to localize to the nucleus, we transfected GFP-tagged alanine mutants into U87-MG cells. GFP-JNK2α2 WT had a strong nuclear localization signal in which 80% of the GFP-positive cells had nuclear staining, consistent with previous reports (15). Analysis of the alanine mutants revealed that each autophosphorylation/dimerization “inactive” mutant (L218A, K220A, G221A, I224A, and F225A) showed only 20–30% of the GFP-positive cells with a clear nuclear localization, whereas the three autophosphorylation/dimer “active” mutants (V219A, C222A, and Q226A) had 60–70% cells with a strong nuclear staining (Fig. 5, A and B). In addition, analysis of the kinase dead mutant revealed that only 25% of the cells had a strong GFP nuclear localization, demonstrating that phosphorylated dimers are preferentially translocated to the nucleus (Fig. 5B). Consistently, Western analysis using an antibody specific for phosphorylated JNK revealed that each autophosphorylation “inactive” mutant exhibited a 6–7-fold reduction in phosphorylation, whereas the “active” mutants had only a 1.2–1.6-fold reduction (Fig. 5C). These results indicate that nuclear localization is dependent on dimerization.
FIGURE 5.
Activity of JNK2α2 alanine mutants in U87-MG cells. A, direct immunofluorescence of GFP-tagged alanine mutants in U87-MG cells. Top panels, example of nuclear localization of GFP-tagged wild type JNK2α2 construct. Bottom panels, example of cytoplasmic localization. B, percentage of positive cells that possessed a nuclear localization for the respective GFP-tagged alanine mutants. C, GFP-tagged alanine mutants transfected in U87-MG cells. Western analysis using an antibody specific for phosphorylated JNK or GFP. The relative amount of the phosphorylated JNK is normalized to wild type JNK2α2 and is designated 1.00. D, Western analysis of the U87-MG cells stably expressing YFP tagged JNK2α2 constructs. The blot was probed with anti-YFP and anti-actin. E, cell growth analysis. Each cell line was cultured in DMEM supplemented with 1% fetal bovine serum, and viable cells were counted daily. The results are from two separate experiments, each done in triplicate. F, anchorage-independent growth of each cell lines in soft agar. Each sample was done in triplicate, and the number of colonies was counted after 10 days.
JNK2α2 Dimerization Is Important for Tumorigenicity—Overexpression of JNK2α2 in U87-MG cells has been previously shown to increase cellular proliferation and enhance anchorage-independent cell growth, whereas overexpression of the inactive JNK2α2 APF mutant does not enhance these properties (9). We tested whether dimerization plays a role in the tumorigenic phenotype. To this end, we used a retrovirus to generate U87-MG cells that stably expressed YFP-JNK2α2 WT, YFP-V219A, YFP-C222A, YFP-Q226A, and YFP-L218A (Fig. 5D). We analyzed the cellular growth of the stable cell lines in DMEM supplemented with 1% fetal bovine serum. This concentration of fetal bovine serum was previously shown to maintain the cellular growth of U87-MG cells while eliminating the growth factors that activate endogenous JNK (9). Consistent with previous findings, the YFP-WT cells had a significantly faster growth rate compared with the cells expressing YFP alone (9) (Fig. 5E). The cell lines stably expressing YFP-V219A, YFP-C222A, and YFP-Q226A behaved similarly to the YFP-WT cells and also had a dramatically faster growth rate. On the other hand, the YFP-L218A cells were found to proliferate at a much slower rate and had a similar growth profile as YFP alone control cells. Analysis of anchorage-independent growth of the stable cell lines yielded similar results. Compared with the YFP alone cells, the YFP-WT, YFP-V219A, YFP-C222A, and YFP-Q226A cell lines all produced ∼2-fold more colonies, whereas the YFP-L218A cells yielded a similar number of colonies to the control cell line (Fig. 5F). Together, these findings demonstrate that JNK2α2 dimerization is necessary for its cellular activity as well as its ability to enhance tumorigenesis.
A Leucine Zipper Motif May Be Responsible for JNK2α2 Dimerization—To help elucidate the structural mechanism of JNK dimerization, we reapplied the dimer-containing fraction from our initial gel filtration analysis of WT JNK2α2 (Fig. 6A). If JNK2α2 dimerization is the result of a monomer-dimer equilibrium, two peaks should result. If the dimer peak is a stable chemical form, then one peak should be obtained. Analysis of the dimer peak yielded two peaks in which the protein sizes corresponded to a JNK2α2 dimer and monomer (Fig. 6A). This finding suggests that JNK2α2 dimers are not stably bound and therefore do not occur through a covalent interaction such as a disulfide bond occurring between intermolecular cysteines. This finding is supported by the discovery that the C222A mutation does not influence JNK2α2 dimerization or activity (Fig. 5, B–E).
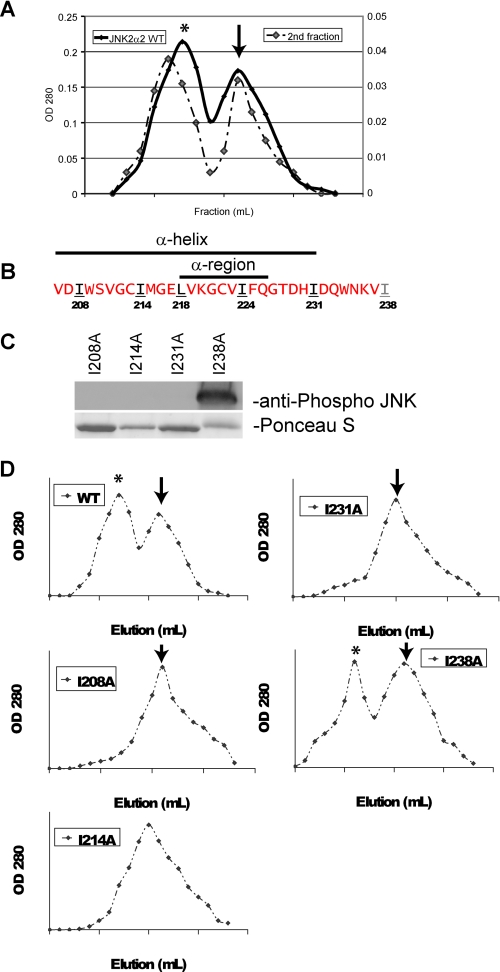
FIGURE 6.
JNK2α2 dimerization may occur through a leucine zipper motif. A, gel filtration of wild type JNK2α2. The left axis represents the initial filtration of WT JNK2α2 in which 1-ml fractions were collected, and the A280 nm was measured. *, JNK2α2 dimer; arrow, monomer. The right axis is the subsequent gel filtration of the dimer fraction. B, amino acid sequence of JNK2α2 indicating the location of the α-region and α-helix. Red letters indicate amino acids that were not mutated. Black/underlined letters indicate amino acids that were mutated to an alanine and resulted in loss of JNK2α2 autophosphorylation. The gray/underlined letter indicates an amino acid that was mutated to an alanine but retained autophosphorylation capability. C, in vitro kinase reactions with the mutant His-JNK2α2. Western blots were probed with an antibody specific for phosphorylated JNK at the T-P-Y motif. Ponceau S stain of the same Western blot demonstrated equal loading of the respective JNK mutants. D, gel filtration of JNK2α2 wild type and JNK2α2 mutants. 1-ml fractions were collected, and the A280 nm values were measured. *, JNK2α2 dimer; arrow, monomer.
Recent studies suggest that JNK dimerization may be dependent on a leucine zipper. A leucine zipper is composed of two α-helical segments that have leucine/isoleucine generally separated by 6 amino acids. The leucine/isoleucine side chains extend from one α-helix to a similar α-helix on a second polypeptide. ERK2 and c-Jun were found to homodimerize through a leucine zipper (11, 21). In addition, the crystal structure of JNK2α revealed that an α-helix may encompass the α-region, indicating that the leucines/isoleucines within the α-region may be a component of a leucine zipper (Fig. 6B) (22, 23). Supporting this hypothesis is our finding that the mutations L218A and I224A prevent dimerization, autophosphorylation, and JNK2α2 activity (Fig. 5, B–E).
To provide additional evidence for the importance of a leucine zipper in JNK2α2 dimerization, we mutated isoleucines and leucines that surround the α-region and reside within the α-helix (I208A, I214A, and I231A). As a control we also mutated I238A because it resides outside the α-helix and the potential leucine zipper (Fig. 6B) (23). In vitro kinase assays revealed that each mutant within the α-helix did not autophosphorylate or form dimers (Fig. 6, C and D). Consistently, the I238A mutation was found not to alter the ability of JNK2α2 to autophosphorylate or form dimers. Together our results suggest that JNK2α2 dimerization occurs in a reversible fashion and that it may be due to a leucine zipper, although a conclusive determination awaits a crystal structure of the JNK2α2 dimer.
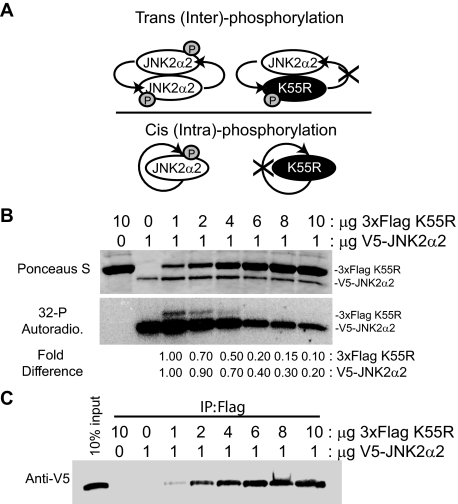
JNK2α2 Autophosphorylates in a trans-Molecular Manner—There are two possibilities for how JNK2α2 undergoes autophosphorylation: 1) there is a conformational change that facilitates a cis- (or intra-) autophosphorylation or 2) the bound kinases of the JNK2α2 homodimer phosphorylate each other resulting in trans- (or inter-) autophosphorylation (Fig. 7A). To address this issue, we incubated wild type JNK2α2 with the K55R kinase-inactive mutant. To differentiate between the two proteins, a 3×FLAG tag was added to the K55R mutant, resulting in a 5-kDa size shift, whereas a V5 tag was added to the wild type JNK2α2 protein (Fig. 7B). Increasing amounts of the 3×FLAG K55R was added to 1 μg of purified wild type V5-JNK2α2. If JNK2α2 autophosphorylation occurs in cis, then increasing amounts of 3×FLAG K55R should not affect the phosphorylation of wild type JNK2α2. However, if there is a trans-molecular phosphorylation event, then the kinase dead mutant should prevent wild type phosphorylation (Fig. 7A). In vitro kinase assays using radioactively labeled [32P]ATP showed that a 6-fold ratio of 3×FLAG K55R compared with wild type JNK2α2 caused a 60% decrease in wild type phosphorylation, and a 10-fold higher concentration resulted in an 80% decrease (Fig. 7B). This finding suggests that 3×FLAG K55R behaves in a dominant negative manner consistent with a trans-molecular reaction. To verify that JNK2α2 autophosphorylation occurs in trans, we also looked at the ability of wild type JNK2α2 to phosphorylate the kinase dead mutant. If JNK2α2 autophosphorylation occurs in cis, then only the wild type JNK2α2 will be phosphorylated, but if there is a trans-molecular phosphorylation, then both the 3×FLAG K55R mutant and wild type protein will be phosphorylated. We discovered that both the kinase dead mutant and wild type protein were phosphorylated when the proteins were at a 1:1 ratio, demonstrating a trans-molecular interaction. Surprisingly, a 6-fold higher concentration of 3×FLAG K55R compared with wild type JNK2α2 resulted in an 80% decrease in 3×FLAG K55R phosphorylation (Fig. 7B). One possible explanation for these results is that the kinase dead mutant is incapable of forming heterodimers with wild type JNK2α2 at higher concentrations. To address this issue, we conducted immunoprecipitation assays for the different concentrations of the 3×FLAG K55R mutant. We determined that increasing concentrations of 3×FLAG K55R was able to precipitate more wild type JNK2α2, suggesting that more heterodimers are formed at higher K55R concentrations (Fig. 7C). Because increasing concentrations of K55R yielded more heterodimers, another secondary phosphorylation mechanism may be occurring. Overall, our findings support a JNK2a2 trans-autophosphorylation model where dimerization precedes phosphorylation. However, they also indicate a possible secondary mechanism of JNK2α2 phosphorylation (see “Discussion”).
FIGURE 7.
JNK2α2 autophosphorylation occurs by a trans-molecular mechanism. A, schematic of trans- or cis-autophosphorylation with JNK2α2 wild type (white oval) and the kinase dead JNK2α2 (K55R) (black ovals). B, in vitro kinase assay using radioactive [32P]ATP. Autophosphorylation/kinase reactions were performed using the indicated amounts of a 3× FLAG-tagged K55R mutant and V5-tagged wild type JNK2α2. Top panel, Ponceau S stain as a loading control. Middle panel, autoradiogram of the in vitro kinase reaction. Bottom panel, fold change in JNK phosphorylation when normalized to the 1:1 ratio sample. The first two lanes (3×FLAG K55R alone and V5-JNK2α2 alone) are controls for JNK2α2 autophosphorylation. C, co-immunoprecipitation using anti-FLAG were separated by SDS-PAGE and detected by immunoblotting with anti-V5. The load control is on the far left (10% input).
DISCUSSION
In this study, we have examined the mechanisms leading to the constitutive activity of JNK2α2. Using size exclusion chromatography, cross-linking assays, and co-immunoprecipitations, we demonstrated that a 9-amino acid region (LVKGCIVFQ), known as the α-region, is necessary for JNK2α2 dimerization. To determine which amino acids in the α-region are important for dimerization, we conducted an alanine mutagenesis scan. Eight different mutants were analyzed and through the use of size exclusion chromatography and cross-linking assays, we discovered that five mutants (L218A, K220A, G221A, I224A, and F225A) abolished dimerization. Each of these mutants also lost its autophosphorylation activity as well as its ability to localize to the nucleus. These findings strongly suggest that JNK2α2 activity is dependent on dimerization. Additionally, a U87-MG cell line stably expressing L218A did not stimulate cell proliferation or increase anchorage-independent growth, indicating that dimerization is also necessary for JNK2α2 induced tumorigenesis. Careful dissection of the mechanism of JNK2α2 dimerization revealed that: 1) dimerization occurs independently of autophosphorylation; 2) JNK2α2 dimers exists in a dimer-monomer equilibrium, suggesting that the dimers are not constitutively bound; and 3) in vitro kinase assays using [32P]ATP with wild type JNK2α2 and a kinase dead mutant (K55R) demonstrate that wild type JNK2α2 can phosphorylate the K55R mutant, revealing that JNK2α2 autophosphorylation occurs in a trans-mechanism, rather than a cis-mechanism. Lastly, mutagenesis of isoleucines and leucines surrounding the α-region (I208A, I214A, and I231A) resulted in loss of JNK2α2 dimerization, indicating that homodimerization may occur through a leucine zipper. Other mutations within the α-region could also affect dimerization, and we speculate that these mutations disrupted the secondary structure of the α-helix. Overall, we have enhanced our understanding of the mechanisms of JNK2α2 autoactivation/autophosphorylation, and we have shown that dimerization is important for JNK2α2-induced tumorigenic phenotypes.
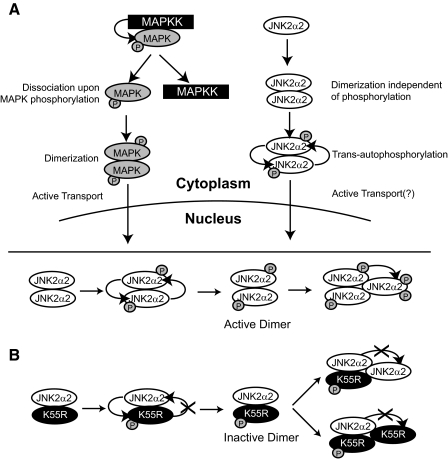
Possible Secondary Mechanism of JNK2α2 Autophosphorylation—Our findings suggest that JNK2α2 undergoes a trans-autophosphorylation mechanism because: 1) wild type JNK2α2 phosphorylates the K55R mutant and 2) K55R acts as a dominant negative mutant. We consistently noted that increasing concentrations of K55R generated more heterodimers with wild type JNK2α2. This provides a mechanism for the dose-dependent manner in which K55R inhibited JNK2α2 phosphorylation activity. Surprisingly, higher concentrations of K55R also reduced the K55R phosphorylation, suggesting a second mechanism for JNK2α2 phosphorylation. To explain this finding we hypothesize that the trans-autophosphorylation event could “activate” the JNK2α2 homodimer. The “active” phosphorylated homodimer could subsequently phosphorylate other JNK2α2 molecules leading to increased levels of JNK2α2 phosphorylation. Consequently, increasing concentrations of K55R would generate more “inactive” heterodimers and hence would explain the dose-dependent decrease in wild type and K55R phosphorylation (Fig. 8B). This potential trimer would most likely be very unstable because our gel filtration analysis only identified JNK2α2 dimers and monomers.
FIGURE 8.
Models of the mechanism of JNK2α2 autophosphorylation/activation. A, model comparing the known mechanism of MAPK activation versus JNK2α2 autophosphorylation/autoactivation. B, possible secondary mechanism of JNK2α2 autophosphorylation. Top, the “active” homodimer of wild type JNK2α2. Bottom, the “inactive” heterodimer of wild type JNK2α2 and the kinase dead mutant (K55R). See text for details.
The Role of MAPK Dimerization—The majority of our knowledge concerning MAPK dimerization is based on ERK2 studies. Extensive studies in Xenopus and human cell lines have shown that ERK2 exists both as a dimer and a monomer, but upon phosphorylation ERK2 will dissociate from the MAPK kinase and form homodimers (12, 13). Surprisingly, in vitro studies have demonstrated that ERK2 kinase activity is not dependent on dimerization because its kinase activity is concentration-independent, and dimerization-defective mutants have similar kinase activity as the wild type protein (24). However, in vivo reports have illustrated the importance of dimerization, because only phosphorylated ERK2 homodimers are actively transported to the nucleus, and disruption of ERK2 dimerization by mutagenesis reduces its nuclear localization (13). Without proper nuclear localization, MAPK proteins are unable to properly regulate gene expression. Another MAPK family member, p38, was also shown to homodimerize, but it is still unknown whether its nuclear localization is dimerization-dependent (13). Both p38 and JNK do not contain an obvious nuclear localization sequence, but when activated by UV light or osmotic stress, both p38 and JNK will translocate to the nucleus, demonstrating that both MAPK proteins are also actively transported (25, 26). Studies using the fission yeast Schizosaccharomyces pombe, indicated that the yeast homologue of JNK, Spc1, is actively transported to the nucleus by Pim1, a homologue of the guanine nucleotide exchange factor RCC1 (27). Here we show that JNK2α2 dimerization is necessary for activation both in vitro and in vivo and that nuclear translocation is dependent on homodimerization. However, future studies are needed to determine which protein chaperones, such as RCC1, are responsible for transporting JNK2α2 to the nucleus.
JNK2α2 Inhibitor—Since the discovery of JNKs, many researchers have attempted to design JNK-selective inhibitors for potential use as therapeutic drugs. Many of the commercially available JNK inhibitors are small molecule, ATP competitive inhibitors such as SP600125. This particular inhibitor was found to reduce lung injury caused by ischemia/reperfusion and alleviate damage from smoke inhalation, illustrating the potential of JNK inhibitors as therapeutic drugs (28). Although there has been some success with the ATP competitive inhibitors, some problems have arisen because of their promiscuous nature. For example, SP600125 was shown to inhibit 13 of 30 tested protein kinases, suggesting that possible deleterious effects could occur because of inhibition of other kinases (29). Consequently, very specific kinase inhibitors to JNK must be developed to ensure proper therapeutic targeting of these drugs. Our work here suggests that targeting JNK2α2 dimerization will not only inhibit JNK2α2 autophosphorylation and autoactivation, but it may also enable the generation of a more selective JNK inhibitor because each MAPK family member has unique domains necessary for their own dimerization (11, 30). Peptide inhibitors of protein kinases have been generated to act in a protein substrate competitive manner, implying that a properly designed peptide inhibitor could inhibit JNK2α2 homodimerization.
We demonstrated the therapeutic potential of preventing JNK2α2 dimerization by studying the ability of our active and inactive mutants to stimulate tumorigenesis. Each active mutant (V219A, C222A, and Q226A) behaved similarly to wild type JNK2α2 and enhanced two tumorigenic phenotypes: cellular proliferation and anchorage-independent growth 2-fold (Fig. 5, D and E). Interestingly, the inactive mutant (L218A), which was found to exist as a monomer and is not constitutively active, did not enhance these phenotypes. These results suggest that blocking or inhibiting dimerization may decrease the tumorigenic ability of JNK2α2. Overall, given the oncogenic role of JNK2α2 in glioblastoma multiforme, designing a specific inhibitor that blocks JNK2α2 dimerization could be very important in improving the prognosis of brain tumor patients.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant CA69495, CA96539 and CA124832 (to A. J. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: JNK, c-Jun N-terminal kinase; MAPK, mitogen-activated protein kinase; ERK, extracellular signal-regulated kinase; GFP, green fluorescent protein; DMEM, Dulbecco's modified Eagle's medium; GST, glutathione S-transferase; WT, wild type; YFP, yellow fluorescent protein; BS3, bis(sulfosuccinimidyl) suberate.
References
- 1.Kyriakis, J. M., Liu, H., and Chadee, D. N. (2004) Methods Mol. Biol. 250 61-88 [DOI] [PubMed] [Google Scholar]
- 2.Johnson, G. L., and Lapadat, R. (2002) Science 298 1911-1912 [DOI] [PubMed] [Google Scholar]
- 3.Bode, A. M., and Dong, Z. (2007) Mol. Carcinog. 46 591-598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butterfield, L., Storey, B., Maas, L., and Heasley, L. E. (1997) J. Biol. Chem. 272 10110-10116 [DOI] [PubMed] [Google Scholar]
- 5.Arbour, N., Naniche, D., Homann, D., Davis, R. J., Flavell, R. A., and Oldstone, M. B. (2002) J. Exp. Med. 195 801-810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, N., Nomura, M., She, Q. B., Ma, W. Y., Bode, A. M., Wang, L., Flavell, R. A., and Dong, Z. (2001) Cancer Res. 61 3908-3912 [PubMed] [Google Scholar]
- 7.Potapova, O., Gorospe, M., Bost, F., Dean, N. M., Gaarde, W. A., Mercola, D., and Holbrook, N. J. (2000) J. Biol. Chem. 275 24767-24775 [DOI] [PubMed] [Google Scholar]
- 8.Tsuiki, H., Tnani, M., Okamoto, I., Kenyon, L. C., Emlet, D. R., Holgado-Madruga, M., Lanham, I. S., Joynes, C. J., Vo, K. T., and Wong, A. J. (2003) Cancer Res. 63 250-255 [PubMed] [Google Scholar]
- 9.Cui, J., Han, S. Y., Wang, C., Su, W., Harshyne, L., Holgado-Madruga, M., and Wong, A. J. (2006) Cancer Res. 66 10024-10031 [DOI] [PubMed] [Google Scholar]
- 10.Avdulov, S., Li, S., Michalek, V., Burrichter, D., Peterson, M., Perlman, D. M., Manivel, J. C., Sonenberg, N., Yee, D., Bitterman, P. B., and Polunovsky, V. A. (2004) Cancer Cell 5 553-563 [DOI] [PubMed] [Google Scholar]
- 11.Wilsbacher, J. L., Juang, Y. C., Khokhlatchev, A. V., Gallagher, E., Binns, D., Goldsmith, E. J., and Cobb, M. H. (2006) Biochemistry 45 13175-13182 [DOI] [PubMed] [Google Scholar]
- 12.Adachi, M., Fukuda, M., and Nishida, E. (1999) EMBO J. 18 5347-5358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khokhlatchev, A. V., Canagarajah, B., Wilsbacher, J., Robinson, M., Atkinson, M., Goldsmith, E., and Cobb, M. H. (1998) Cell 93 605-615 [DOI] [PubMed] [Google Scholar]
- 14.Pimienta, G., Ficarro, S. B., Gutierrez, G. J., Bhoumik, A., Peters, E. C., Ronai, Z., and Pascual, J. (2007) Cell Cycle 6 1762-1771 [DOI] [PubMed] [Google Scholar]
- 15.Cui, J., Holgado-Madruga, M., Su, W., Tsuiki, H., Wedegaertner, P., and Wong, A. J. (2005) J. Biol. Chem. 280 9913-9920 [DOI] [PubMed] [Google Scholar]
- 16.Nitta, R. T., Jameson, S. A., Kudlow, B. A., Conlan, L. A., and Kennedy, B. K. (2006) Mol. Cell. Biol. 26 5360-5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennedy, B. K., Barbie, D. A., Classon, M., Dyson, N., and Harlow, E. (2000) Genes Dev. 14 2855-2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliver, A. W., Paul, A., Boxall, K. J., Barrie, S. E., Aherne, G. W., Garrett, M. D., Mittnacht, S., and Pearl, L. H. (2006) EMBO J. 25 3179-3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nitta, R. T., Smith, C. L., and Kennedy, B. K. (2007) PLoS ONE 2 e963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen, A. W., and Daugherty, P. S. (2005) Nat. Biotechnol. 23 355-360 [DOI] [PubMed] [Google Scholar]
- 21.Lively, T. N., Nguyen, T. N., Galasinski, S. K., and Goodrich, J. A. (2004) J. Biol. Chem. 279 26257-26265 [DOI] [PubMed] [Google Scholar]
- 22.Xie, X., Gu, Y., Fox, T., Coll, J. T., Fleming, M. A., Markland, W., Caron, P. R., Wilson, K. P., and Su, M. S. (1998) Structure 6 983-991 [DOI] [PubMed] [Google Scholar]
- 23.Shaw, D., Wang, S., Villasenor, A. G., Tsing, S., Walter, D., Browner, M., Barnett, J., and Kuglstatter, A. (2008) J. Mol. Biol. 383 885-893 [DOI] [PubMed] [Google Scholar]
- 24.Robbins, D. J., Zhen, E., Cheng, M., Xu, S., Vanderbilt, C. A., Ebert, D., Garcia, C., Dang, A., and Cobb, M. H. (1993) J. Am. Soc. Nephrol. 4 1104-1110 [DOI] [PubMed] [Google Scholar]
- 25.Cheng, H. L., and Feldman, E. L. (1998) J. Biol. Chem. 273 14560-14565 [DOI] [PubMed] [Google Scholar]
- 26.Kondoh, K., Torii, S., and Nishida, E. (2005) Chromosoma 114 86-91 [DOI] [PubMed] [Google Scholar]
- 27.Gaits, F., and Russell, P. (1999) Mol. Biol. Cell 10 1395-1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishii, M., Suzuki, Y., Takeshita, K., Miyao, N., Kudo, H., Hiraoka, R., Nishio, K., Sato, N., Naoki, K., Aoki, T., and Yamaguchi, K. (2004) J. Immunol. 172 2569-2577 [DOI] [PubMed] [Google Scholar]
- 29.Bain, J., McLauchlan, H., Elliott, M., and Cohen, P. (2003) Biochem. J. 371 199-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson, D. J., Fortner, K. A., Lynch, D. H., Mattingly, R. R., Macara, I. G., Posada, J. A., and Budd, R. C. (1996) Eur. J. Immunol. 26 989-994 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.