Abstract
The membrane type (MT) 6 matrix metalloproteinase (MMP) (MMP25) is a glycosylphosphatidylinositol-anchored matrix metalloproteinase (MMP) that is highly expressed in leukocytes and in some cancer tissues. We previously showed that natural MT6-MMP is expressed on the cell surface as a major reduction-sensitive form of Mr 120, likely representing enzyme homodimers held by disulfide bridges. Among the membrane type-MMPs, the stem region of MT6-MMP contains three cysteine residues at positions 530, 532, and 534 which may contribute to dimerization. A systematic site-directed mutagenesis study of the Cys residues in the stem region shows that Cys532 is involved in MT6-MMP dimerization by forming an intermolecular disulfide bond. The mutagenesis data also suggest that Cys530 and Cys534 form an intramolecular disulfide bond. The experimental observations on cysteines were also investigated by computational studies of the stem peptide, which validate these proposals. Dimerization is not essential for transport of MT6-MMP to the cell surface, partitioning into lipid rafts or cleavage of α-1-proteinase inhibitor. However, monomeric forms of MT6-MMP exhibited enhanced autolysis and metalloprotease-dependent degradation. Collectively, these studies establish the stem region of MT6-MMP as the dimerization interface, an event whose outcome imparts protease stability to the protein.
The MMP3 family of zinc-dependent endopeptidases includes both secreted and membrane-anchored proteases that can hydrolyze multiple substrates at distinct cellular and extracellular locations in physiological and pathological conditions (1). The membrane-anchored MMPs, known as membrane type-MMPs (MT-MMPs) evolved to specifically target substrates at the pericellular space and in the plasma membrane by incorporating two distinct anchoring motifs, namely a transmembrane domain as in the case of MT1-(MMP14), MT2-(MMP15), MT3-(MMP16), and MT5-MMP (MMP24) or a glycosylphosphatidylinositol (GPI) moiety as in the case of MT4-(MMP17) and MT6-MMP (MMP25) (2). The presence of membrane-anchoring domains confers unique properties and regulatory mechanisms to this subset of MMPs, which serve to tightly control the localization and amount of protease at the cell surface. These regulatory mechanisms include targeting to cell-matrix contacts, insertion into membrane microdomains, internalization and recycling, processing, oligomerization, and ectodomain shedding (3, 4). Although there is substantial information on the regulation of transmembrane MT-MMPs by these processes, in particular with MT1-MMP (3–5), little is known about the regulation of GPI-MT-MMPs, MT4- and MT6-MMP.
Structurally, the GPI-MT-MMPs comprise the classical domain organization of MMPs including an N-terminal prodomain, a zinc-containing catalytic domain, a hinge region, and a C-terminal hemopexin-like domain (6–8). Like all members of the MT-MMP family, the GPI-MT-MMPs also contain a stem (stalk) region downstream of the hemopexin-like domain which is followed by the anchoring motif. However, as opposed to the transmembrane MT-MMPs, the stem region of MT6- and MT4-MMP contains three and two unique cysteine residues, respectively, which may play a role in formation of disulfide bridges (8). Recent evidence from our laboratory has shown that under non-reducing conditions MT6-MMP is consistently detected as a major form of ∼120 kDa, likely representing dimeric enzyme complexes at the cell surface (9). Upon reduction, MT6-MMP is mostly found as a 57-kDa species. Based on these data, we hypothesized that the ∼120-kDa form of MT6-MMP represents enzyme homodimers held by intermolecular disulfide bridges within the stem region (9). In this study we examined the role of the cysteine residues located within the stem region in dimerization of MT6-MMP by site-directed mutagenesis and computational and functional studies. We show that that Cys532 is essential for dimer formation. These studies establish a unique role for the stem region of MT6-MMP in mediating disulfide-linked homodimerization.
EXPERIMENTAL PROCEDURES
Cell Culture—The human colon cancer cell line HCT-116 (CCL-247), the immortalized monkey kidney cell line COS-1 (CRL-1650), and the human promyelocytic cell line HL-60 (CCL-240) were all obtained from the American Type Culture Collection (ATCC). HCT-116 and COS-1 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 8% fetal calf serum and antibiotics. HL-60 cells were cultured in RPMI 1640 medium (Invitrogen) supplemented with 8% fetal calf serum and antibiotics (100 units/ml penicillin G sodium and 100 μg/ml streptomycin sulfate). All cell lines were maintained at 37 °C in an atmosphere of 95% air and 5% CO2.
HL-60 Differentiation—HL-60 cells were induced to differentiate into neutrophil-like cells by treatment with dimethyl sulfoxide (DMSO) as described (10, 11). Briefly, HL-60 cells at a density of 1.0 × 106cells/ml were treated with 1.25% DMSO in complete culture media for 7 days. The cells were then harvested, pelleted, and processed for immunoblot analyses as described below. Differentiation into neutrophil-like cells was monitored by mRNA expression of bcl-2 and bfl-1, as previously described (12).
Antibodies—The monoclonal antibody (mAb) MAB1142 to human MT6-MMP was purchased from R&D Systems (Minneapolis, MN). A rabbit pAb to human caveolin was from BD Biosciences. The mAb to the human transferrin receptor was purchased from BD Biosciences. The mAb to β-actin and the rabbit pAb to human α1-proteinase inhibitor (α1-PI) were purchased from Sigma.
cDNA Constructs and Transfections—The full-length human MT6-MMP cDNA was cloned into the expression plasmid pcDNA™3.1/myc-His (-) A (Invitrogen) to generate pcDNA3.1-MT6 for stable transfection, as described previously (9). Single, double, or triple Cys → Ala substitutions at Cys530, Cys532, and Cys534 present within the stem region of MT6-MMP were generated by the QuikChange® site-directed mutagenesis kit (Stratagene, La Jolla, CA) using sequence specific primers and wild type pcDNA3.1-MT6 DNA as the template. The Cys mutants were designated as depicted in Fig. 2. In addition, the single C532A mutant was further mutated by site-directed mutagenesis to substitute the catalytic Glu233 residue for alanine to generate catalytically inactive protease. The sequences of the inserts and junctions in the vectors were verified by DNA sequencing. COS-1 cells were transiently transfected with wild type, mutant, or chimeric MT6-MMP cDNAs using FuGENE™ 6 transfection reagent (Roche Diagnostics) as recommended by the manufacturer. 24–48 h after transfection the cells were harvested, and the lysates were subjected to immunoblot analyses as described below. To generate stable transfectants, HCT-116 cells were transfected with pcDNA3.1 vectors containing the single C532A or triple C530A/C2A/C4A MT6-MMP mutant cDNA using FuGENE™ 6 transfection reagent, and Geneticin® (G418 sulfate, Invitrogen)-resistant pooled populations were selected using conventional approaches. Stable pooled populations of HCT-116 cells expressing wild type MT6-MMP cDNA (referred to as MT6-HCT cells) or transfected with pcDNA3.1 without inserts (empty vector and, thus, referred to as EV-HCT cells) were generated as previously described (9).
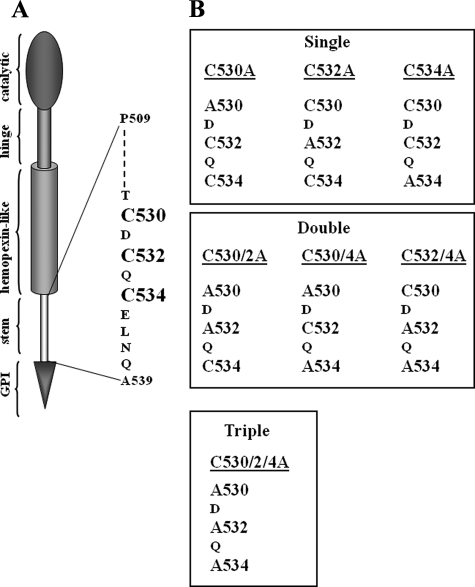
FIGURE 2.
A, schematic diagram of active MT6-MMP domains and cysteine residues located within the stem region. B, depiction of the Cys mutants generated in this study.
Semiquantitative Reverse Transcription-PCR—Total RNA of HL-60 cells treated with or without 1.25% DMSO was extracted with total RNA purification system (Invitrogen). Reverse transcription-PCR was performed with 2 μg each of total RNA sample using SuperScript™ III reverse transcriptase (Invitrogen) and subsequently Taq® DNA polymerase (Invitrogen) following the manufacturer's instructions. The sequences of the specific primers used for human MT6-MMP are as follows: forward, 5′-ATG GCC TGC AGC AAC TCT AT-3′; reverse, 5′-AGG GGC CTT TGA AGA AGA AA-3′. The sequences of human bcl-2 and bfl-1 primers are as follows: bcl-2 forward, 5′-AGA TGT CCA GCC AGC TGC ACC TGA C-3′; bcl-2 reverse, 5′-AGA TAG GCA CCC AGG GTG ATG CAA GCT-3′; bfl-1 forward, 5′-AGG CTG GCT CAG GAC TCT CTG C-3′; bfl-1 reverse, 5′-TTC TGG TCA ACA GTA TTG CTT CAG G-3′. Thirty cycles of PCR were performed with 30 s at 94 °C, 30 s at 52 °C, and 30 s at 72 °C. The housekeeping gene glyceraldehyde-3-phosphate dehydrogenase was also amplified and used as an internal control. The sequences of the human glyceraldehyde-3-phosphate dehydrogenase primers are as follows: forward, 5′-CCA CCC ATG GCA AAT TCC ATG GCA-3′; reverse, 5′-TCT AGA CGG CAG GTC AGG TCC ACC-3′. The amplified genes were resolved by 1.8% agarose gels and detected by ethidium bromide staining.
Preparation of Cell Lysates and Immunoblot Analyses—Cells were solubilized with cold lysis buffer (25 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1% IGEPAL CA-630, 5 mm EDTA, and 60 mm octyl glucoside) supplemented with 20 mm N-ethylmaleimide and protease inhibitor mixture without EDTA (Complete, Mini, EDTA-free, Roche Applied Science). Briefly, the cells were incubated with lysis buffer for 1 h on ice and centrifuged (13,000 rpm) for 15 min at 4 °C. The protein concentration in the lysates was determined by the BCA procedure (Pierce). An aliquot of each lysate was mixed with Laemmli SDS-sample buffer with or without 1% β-mercaptoethanol (β-ME), boiled (95 °C, 5 min), and resolved by SDS-PAGE followed by immunoblot analyses using various antibodies.
Cell Surface Biotinylation and Phosphatidylinositol-specific Phospholipase C (PI-PLC) Treatment—Cells (HCT-116 or COS-1) stably or transiently expressing wild type or mutant MT6-MMP in 6-well plates were surface-biotinylated with the cell-impermeable EZ-link-sulfo-NHS-biotin (Pierce), as described previously (9, 13). The cells were then lysed with 0.20 ml/well of cold lysis buffer on ice. After a brief centrifugation (15 min, 13,000 rpm), equal protein amounts of the supernatants were incubated with Neutravidin beads (Pierce) at 4 °C overnight. The beads were washed 5 times with harvest buffer (0.5% SDS, 60 mm Tris/HCl, pH 7.5, 2 mm EDTA) supplemented with 2.5% Triton X-100 (final concentration), and bound proteins were eluted with Laemmli SDS sample buffer, boiled, and resolved by reducing SDS-PAGE followed by transfer to a nitrocellulose membrane. The proteins were detected with MT6-MMP antibodies as described (9). In some experiments, 24 h after transient transfection with the appropriate cDNAs, COS-1 cells were washed with cold phosphate-buffered saline (PBS) and treated with 0.3 units/well of PI-PLC (Molecular Probes, Eugene, OR) in PBS for 30 min on ice. The supernatant was clarified and concentrated with Microcon® centrifugal filter devices (Millipore, Bedford, MA) in the presence of EDTA-free protease inhibitors mixture mix (Roche Applied Science). An aliquot of each concentrated supernatant was mixed with Laemmli SDS-sample buffer with or without β-ME, boiled and resolved by SDS-PAGE followed by immunoblot analysis.
Preparation of Crude Plasma Membrane Fraction and Successive Detergent Extraction of Lipid Rafts—Crude plasma membrane fraction and detergent extraction of lipid rafts were prepared as described previously (9).
Processing of α1-Proteinase Inhibitor by MT6-MMP Mutants—Crude plasma fractions of HCT-116 cells (25 and 50 μg/reaction) expressing wild type or MT6-MMP mutants (single C532A or triple C530A/C2A/C4A) were incubated with 25 ng of purified human α1-PI (Sigma) in 50 μl of collagenase buffer (50 mm Tris-HCl pH 7.5, 5 mm CaCl2, 150 mm NaCl, 0.02% Brij-35) supplemented with PI mixture (Roche Applied Science). After 16 h at 37 °C, the reactions were mixed with Laemmli SDS-sample buffer with 1% β-ME, boiled, and resolved by 10% SDS-PAGE followed by immunoblot analysis using anti-α1-PI pAb. The blots were reprobed with an anti-MT6-MMP MAB1142.
Computational Method—Sequence analysis was carried out to probe the secondary structure of the stem peptide of 31 amino acids (PAPSSGPRAPRPPKATPVSETCDCQCELNQA). The sequence homology method by SWISS-model failed to identify a template protein from the existing crystal structures. A more fundamental residue-by-residue analysis was applied by SDSC (San Diego Supercomputer Center) Biology Workbench and Garnier et al. (14) and Chou and Fasman (15) methods, both of which predicted α-helical conformation from Thr529 to Ala539. Secondary structure prediction was more difficult for the remainder of the sequence, but at least five amino acids were predicted to be involved in turn: Ser512, Pro515, Arg519, Pro521, and Glu528. The likelihood is that this 31-amino acid stretch is highly flexible. Model building started with construction of an α-helical structure within Sybyl (Tripos, 8.0) for Thr529 to Ala539 (434 atoms). The N-terminal residue Pro509 was protonated, and the C-terminal Ala539 was deprotonated in AMBER (Version 8.0). Solvent molecules (a total of 4378 TIP3P water molecules) were added to encompass the peptide. The entire complex, including the water molecules, was energy-minimized by the methodology described before (16). The 50,000-step energy minimization generated a low energy conformation (total energy change less than 0.2 kcal·mol-1). Subsequently, simulations were carried out using explicit-solvent molecular dynamics. Implicit-solvent simulation was also performed using the modified Generalized Born method (17), the findings of which were very similar to the explicit solvent method.
RESULTS
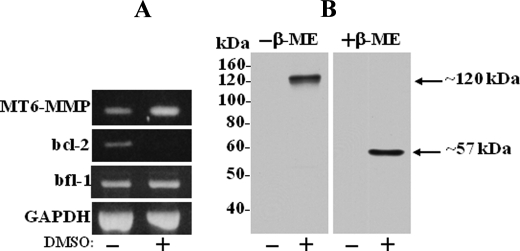
MT6-MMP Is Induced in Differentiated HL-60 Cells—MT6-MMP is known to be highly expressed in neutrophils (6, 18), where it is also detected as a major ∼120-kDa form, as we previously reported (9). To test whether human promyelocytic HL-60 cells express the ∼120-kDa MT6-MMP after differentiation into neutrophil-like cells, the cells were treated with DMSO, an agent known to induce their differentiation to the neutrophil lineage (19–21). After treatment, the cells were collected and examined for MT6-MMP expression by reverse transcription-PCR and immunoblot analyses. Expression of bcl-2 and bfl-1 mRNA was used as a marker of differentiation into neutrophil-like cells by DMSO as reported (12). As shown in Fig. 1A, untreated HL-60 cells express MT6-MMP mRNA. However, MT6-MMP protein was undetectable (Fig. 1B). DMSO treatment resulted in a significant increase (2.5-fold) in MT6-MMP mRNA (Fig. 1A), which correlated with detection of MT6-MMP protein (Fig. 1B). Under non-reducing conditions, MT6-MMP was detected as ∼120-kDa form, which after reduction was converted to ∼57-kDa species (Fig. 1B). Thus, differentiation of HL-60 cells into neutrophil-like cells correlates with expression of MT6-MMP, which is found as a reduction-sensitive form of ∼120 kDa, as reported for human neutrophils (9). These data confirm the ∼120-kDa form as the major species of MT6-MMP in leukocytes.
FIGURE 1.
Neutrophil differentiation of HL-60 cells is associated with expression of the ∼120-kDa form of MT6-MMP. HL-60 cells were untreated or treated with 1.25% DMSO for 7 days. The cells were then harvested and processed for total RNA isolation and immunoblot analyses of cell lysates. A, reverse transcription-PCR analyses of MT6-MMP mRNA expression in untreated (-) and DMSO-treated (+) HL-60 cells. Expression of bcl-2 and bfl-1 mRNAs was also determined as markers of neutrophil-like differentiation (12). Expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA is shown as a loading control. B, immunoblot analysis of lysates of untreated (-) and DMSO-treated (+) HL-60 cells. Equal protein amounts (20 μg/lane) of lysates were mixed with Laemmli SDS sample buffer with (+) or without (-) β-ME and resolved by 10% SDS-PAGE followed by immunoblotting. MT6-MMP was detected with mAb MAB1142.
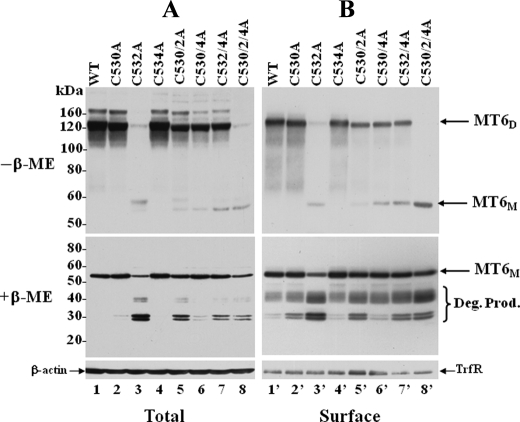
Cys532 in the Stem Region of MT6-MMP Is the Dimerization Site—The reduction sensitivity of the ∼120-kDa form of MT6-MMP suggested that this species may represent disulfide-linked homodimers mediated by cysteine residues within the stem region (Fig. 2A). Therefore, to map which of the three Cys residues of the stem region is involved in MT6-MMP dimerization, we performed single, double, and triple amino acid substitutions at the stem Cys residues at positions 530, 532, and 534 by site-directed mutagenesis, as depicted in Fig. 2B. The wild type and mutant MT6-MMP cDNAs were transiently expressed in COS-1 cells. Cells were lysed in the presence of N-ethylmaleimide to prevent oxidation of free thiols. The total cell lysates and the surface-biotinylated proteins of the transfected cells were then resolved by SDS-PAGE under reducing and non-reducing conditions and examined for the presence of MT6-MMP forms. As shown in Fig. 3, all the Cys mutants could be successfully expressed, as determined by the presence of the monomeric form of MT6-MMP in the cell lysates (Fig. 3A, +β-ME) and at the cell surface, as detected under reducing conditions (Fig. 3B, +β-ME). However, under non-reducing conditions, the single C532A (Fig. 3, A and B, lanes 3 and 3′) mutant exhibited a significant reduction in dimeric forms when compared with wild type MT6-MMP and the other single and double Cys mutants. The triple C530A/C2A/C4A (Fig. 3, A and B, lanes 8 and 8′) mutant showed no detectable dimers at the cell surface (Fig. 3B, lane 8′). However, a minor trace of a ∼120-kDa immunoreactive form was detected in the total cell lysate of the triple C530A/C2A/C4A mutant (Fig. 3A, lane 8, -β-ME), suggesting the existence of non-covalent interactions under these conditions. To further determine if the Cys mutants were membrane-anchored and exposed to the cell surface, the cells were treated with PI-PLC 24 h after transfection. This time was purposely selected to avoid excessive turnover of the proteins and, thus, evaluate the ability of the full-length wild type and mutant MT6-MMP proteins to traffic to the cell surface and be released by PI-PLC. Fig. 4 shows that overall all the recombinant MT6-MMP proteins were released by PI-PLC, consistent with GPI-anchoring and cell surface targeting, in agreement with the results of Fig. 3. However, the dimerization-deficient C532A and C530A/C2A/C4A mutants were detected in the cell surface in a monomeric form. Altogether, these results suggest a role for Cys532 in MT6-MMP dimerization.
FIGURE 3.
Expression of MT6-MMP Cys mutants. COS-1 cells were transiently transfected to express with type (wild type (WT)) (lanes 1 and 1′) or Cys mutant (lanes 2–8 and 2′–8′) MT6-MMP. Forty hours later the cells were surface-biotinylated as described under “Experimental Procedures.” The cells were then lysed in cold lysis buffer (supplemented with protease inhibitors and 20 mm N-ethylmaleimide), and protein concentrations were determined. Equal amount of lysates were subsequently used for analyses. One-third of the lysate from each transfectant was kept in the cold (total lysate), whereas the rest of the fraction was subjected to Neutravidin beads pulldown assays. The bound proteins were eluted with Laemmli SDS sample buffer and divided in two aliquots; one receivedβ-ME, and the other did not. The total lysates were also divided in two fractions, one with and the other without β-ME. The samples were resolved by 7.5% SDS-PAGE for reducing conditions and 12% SDS-PAGE for non-reducing conditions and then processed for immunoblot analysis. The blots were probed with mAb MAB1142 to MT6-MMP. The blots of reduced samples were also reprobed with antibodies toβ-actin (A) and transferrin receptor (TrfR, B). MT6D, MT6-MMP dimeric form; MT6M, MT6-MMP monomeric form. Deg. Prod, degradation products.
FIGURE 4.
PI-PLC release of MT6-MMP Cys mutants. COS-1 cells were transiently transfected to express with type (WT) MT6-MMP (lanes 1 and 1′) or Cys MT6-MMP mutants (lanes 2–5 and 2′–5′). Twenty-four hours later the cells were treated with 0.5 ml of PI-PLC (0.3 units/well) in phosphate-buffered saline for 30 min on ice. The supernatant was concentrated and mixed with Laemmli SDS-sample buffer with (+) or without (-) β-ME and resolved by 7.5% SDS-PAGE followed by immunoblot analysis with mAb MAB1142. MT6D, MT6-MMP dimeric form; MT6M, MT6-MMP monomeric form.
Although substitution of Cys532 inhibited dimerization (Fig. 3, A and B, lanes 3 and 3′), we found that all the double Cys mutants generated dimers (Fig. 3B, lanes 5′, 6′, and 7′) albeit at lower levels than wild type MT6-MMP (Fig. 3B, lane 1′) and the single C530A (Fig. 3B, lane 2′) and C534A (Fig. 3B, lane 4′) mutants. This result suggests that the double Cys mutants are capable of undergoing partial dimerization, likely via any of the remaining Cys residue. This is also supported by the low levels of the monomeric form detected under non-reducing conditions (Fig. 3, A and B, lanes 5–7 and 5′-7′). The observations that only substitution of Cys532 was sufficient to abrogate dimerization and that the double mutants (even those with mutated Cys532) could undergo partial dimerization suggested that in wild type MT6-MMP, Cys532 forms an intermolecular disulfide bond, whereas Cys530 and Cys534 form an intramolecular disulfide bond within the stem region.
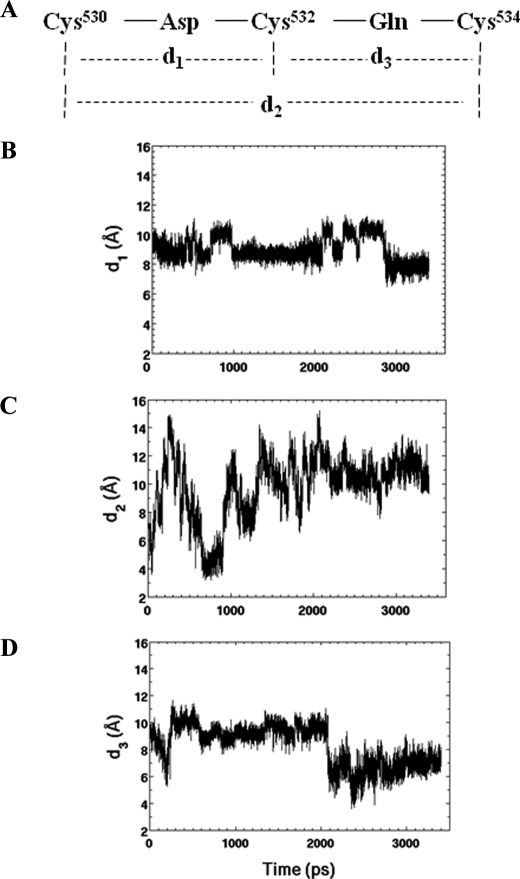
Analyses of MT6-MMP Stem Peptide by Computational Dynamics—The mutagenesis data (Fig. 3) suggested a model of the MT6-MMP stem region in which Cys530 and Cys534 form intramolecular disulfide bonds, whereas Cys532 mediates homodimerization. Although the existence of disulfide bonds can be determined by mass spectrometry, this approach is a challenging task with MT6-MMP due the close proximity of the cysteine residues in the stem region (22). Therefore, to gain further insight into the structural issues of cysteine residues within the stem region of MT6-MMP, we carried out computational dynamics simulations of the 31-amino acid stem peptide. Specifically, we looked at the propensity of the cysteine thiols to approach each other in the course of molecular dynamics, a prerequisite to oxidative disulfide bond formation. The simulations generated a 3.6-ns molecular dynamics trajectory which consists of 7200 conformational states. Distances between each two Sγ atoms are defined as d1 (Cys530 and Cys532), d2 (Cys530 and Cys534), and d3 Cys532 and Cys534) (Fig. 5A). Each was extracted with the ptraj program in AMBER. Fig. 5, B–D, shows that at the start of the simulations the geometry of the energy-minimized structure was defined by d1 = 9.6 Å, d3 = 9.2 Å, and d2 = 6.7 Å. In less than 0.1 ns, d1 held at ∼10 Å, d2 reached a minimum of 3.8 Å, and d3 approached 7 Å. After 0.5 ns, d2 was ∼3 Å, whereas d1 and d3 were about 9 Å. The closest approach for d2 occurred from 570 to 890 ps, which was 3.19 Å. This closest approach was appropriate for the ultimate disulfide bond formation with a length of the covalent S-S bond of 2.03 Å. The essence of this observation is that on the closest approach, the two thiols would be predisposed to experience oxidation and to experience the formation of the disulfide bond, which will tether Cys530 and Cys534 together.
FIGURE 5.
Explicit-solvent molecular dynamics simulations (3.6-ns duration) of the stem peptide of human MT6-MMP. A, the sequence in the cysteine-rich stretch is given, and the requisite distances d1, d2, and d3 define the positions of the side-chain sulfur atoms of the given cysteine residues. Changes of the distances d1 (B), d2 (C), and d3 (D) as a function of the simulation time are indicated.
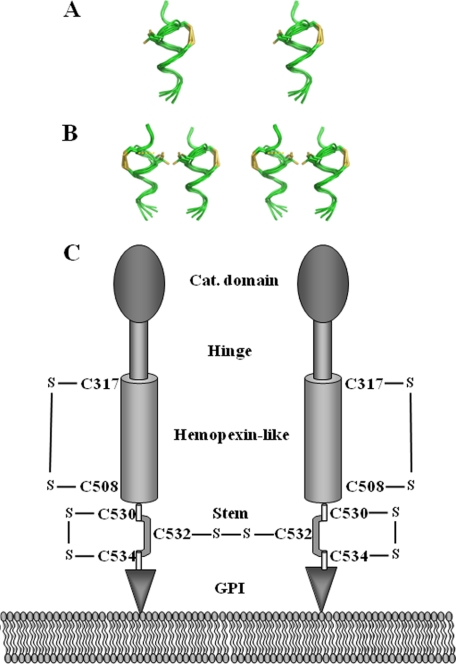
From the conformation data sampled in the simulations, six distinct conformations were extracted from the trajectory, which corresponded to the six smallest separations between the Cys530-Cys534 sulfur atoms. Coordinate transformation and superimposition were made in Sybyl. The shared conformational feature in all was the helicity for the segment Glu528 to Ala539. This conformational state predisposed the two proximal cysteine side chains to the formation of the disulfide bond between Cys530 and Cys534. Fig. 6A shows this arrangement graphically, after the formation of the disulfide bond within the stem region of MT6-MMP. On formation of the intramolecular disulfide bond within the stem region, Cys532 was now available for partnering with the stem region of another MT6-MMP molecule (Fig. 6B). Based on these data, we propose a model of MT6-MMP in which the homodimer is held by disulfide bonds mediated by Cys532, whereas Cys530 and Cys534 form an intramolecular disulfide bond, as depicted in the schematic of Fig. 6C.
FIGURE 6.
Stereo representation of six superimposed conformers of the stem peptide of MT6-MMP (A) and model of disulfide bridges in active MT6-MMP (B). A and B, the stem peptide backbone is shown in green. A, the conformers are shown within a single stem peptide in one molecule of MT6-MMP, depicting a sampling between 570 and 890 ps of the dynamics, where the sulfur atoms of the two distal cysteines (Cys530 and Cys534) move close to be able to form an intramolecular disulfide bond (shown in yellow). Cys532 (at 9 o'clock) is free in the loop created by the disulfide bond within the backbone of the peptide. B, availability of Cys532 predisposes two MT6-MMP stem peptides to dimer formation. The dashed yellow line shows where the intermolecular disulfide bond would exist for one set of the conformers. C, schematic indicating the location of disulfide bonds in active MT6-MMP. The existence of the Cys317-Cys508 intramolecular bond in the hemopexin-like domain is based on the conserved structure of this domain in MMPs (26, 28). The disulfide bonds of the Cys residues within the stem region are based on the data presented here.
Degradation of MT6-MMP Dimerization Mutants—The data of Fig. 3 indicated that mutations at the Cys residues of the stem region were also associated with enhanced degradation of the mutants to immunoreactive and surface-biotinylated fragments of ∼30–32 and ∼38 kDa (Fig. 3, A and B, +β-ME). However, the dimerization-deficient mutants C532A (Fig. 3, A and B, lanes 3 and 3′, and Fig. 7, lane 2) and C530A/C2A/C4A (Fig. 3, A and B, lanes 8 and 8′) yielded the highest levels of degradation fragments, consistent with a role for dimerization in MT6-MMP stability. Interestingly, mutations that abolish the intramolecular disulfide bond between Cys530 and Cys534 only slightly increased the amounts of degradation products (Fig. 3A and B, lanes 2, 2′, 4, and 4′). This suggests that the intramolecular Cys530-Cys534 disulfide bond (Figs. 6C) is not essential for MT6-MMP stability. Although the degradation products were detected at the cell surface by biotinylation (Fig. 3B), the PI-PLC-mediated released fraction (shown in Fig. 4, right panel) contained mostly the full-length enzymes. This discrepancy may be explained by the time elapsed after transfection for analyses (24 h for the PI-PLC treatment, as stated earlier, versus 40 h for surface biotinylation), which may affect the extent of degradation. Alternatively, the full-length enzyme and the degradation products may exhibit a differential sensitivity to PI-PLC cleavage due to changes in conformation or accessibility. Interestingly, a previous study from Kang et al. (18) in human neutrophils also showed that the degradation fragments of MT6-MMP (>40 kDa) escaped PI-PLC-mediated release.
FIGURE 7.
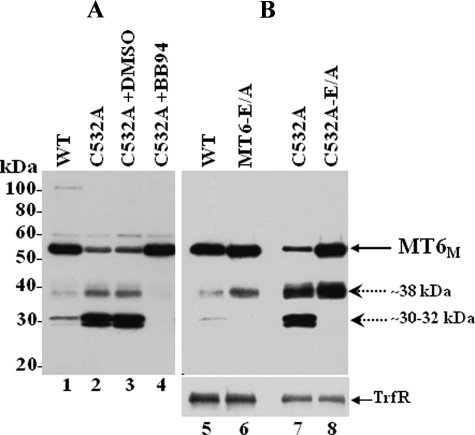
Degradation of MT6-MMP monomers. A, COS-1 cells were transiently transfected to express wild type (WT; lane 1) or C532A (lanes 2–4) MT6-MMP. Six hours after transfection, the cells expressing C532A (lanes 2–4) were incubated without (lane 2) or with 10 μm BB-94 (lane 4) or vehicle (DMSO, lane 3) in complete media. Forty hours post-transfection the cells were harvested, and the lysates were resolved by reducing 10% SDS-PAGE followed by immunoblot analyses with MT6-MMP antibodies. B, COS-1 cells were transiently transfected to express wild type (lane 5), MT6-E/A (lane 6), C532A (lane 7), or C532A-E/A (lane 8) MT6-MMP. Forty-hours later the cells were surface-biotinylated as described under “Experimental Procedures.” The cells were then lysed in cold lysis buffer, and protein concentrations were determined. Equal amounts of protein were subjected to Neutravidin beads pulldown assays. The bound biotinylated proteins were eluted with Laemmli SDS-sample buffer containing β-ME and resolved by 10% SDS-PAGE followed by immunoblot analyses with mAb MAB1142 to MT6-MMP. The blot was reprobed with an antibody to the transferrin receptor (TrfR). MT6M, MT6-MMP monomeric form. Dashed arrows indicate immunoreactive degradation fragments.
To further examine the proteolytic nature of the degradation process of MT6-MMP, we treated COS-1 cells expressing C532A with the broad spectrum metalloproteinase inhibitor, BB-94. As shown in Fig. 7A, BB-94 completely inhibited the generation of both the ∼30–32- and ∼38-kDa fragments (Fig. 7A, lane 4) and consistently caused higher levels of the monomeric 57-kDa form. Thus, generation of both the ∼30–32- and ∼38-kDa fragments is a metalloprotease-dependent event. To determine whether autolysis was partly responsible, we substituted the catalytic Glu233 with an alanine residue (E/A mutation) in both wild type and C532A MT6-MMP. The E/A substitution abrogates MMP catalytic activity and, thus, permits determination of the involvement of autocatalysis in enzyme degradation. These studies showed that an E/A substitution in both wild type (Fig. 7B, lane 6) and C532A (Fig. 7B, lane 8) MT6-MMP caused a disappearance of the ∼30–32-kDa fragment and an increase of the ∼38-kDa fragment in transiently transfected COS-1 cells. The C532A-E/A mutant also showed higher levels of the monomeric form (Fig. 7B, lane 8). These data are consistent with the ∼30–32-kDa fragment being the product of autolysis, whereas formation of the ∼38-kDa fragment is due to a metalloprotease. These data suggest that lack of dimerization enhances degradation of MT6-MMP by both autocatalytic and non-autocatalytic metalloprotease-dependent processes.
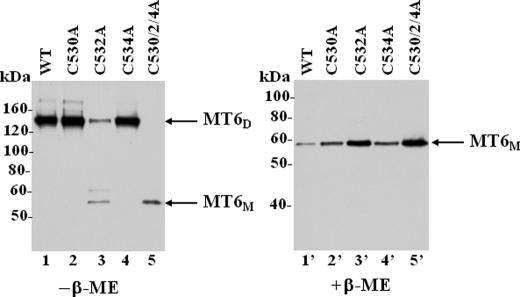
Dimerization Is Not Required for Partition of MT6-MMP into Lipid Rafts—We previously demonstrated that the ∼120-kDa form of MT6-MMP is present in lipid rafts (9). Therefore, we examined whether dimerization is required for the distribution of MT6-MMP into lipid rafts. To this end, we generated pooled populations of HCT-116 colon cancer cells stably expressing single C532A or triple C530A/C2A/C4A MT6-MMP mutants. As expected, only the monomeric form of MT6-MMP was displayed at the surface in cells expressing these mutants (Fig. 8, A and B, lanes 3 and 4 and 3′ and 4′). The cells were then lysed, and the lysates were fractionated into detergent-resistant fractions by differential fractionation into Triton X-100-soluble (referred to as S fraction) and Triton X-100-insoluble/β-octylglucoside-soluble (referred to as I fraction) fractions, as described (9). Under these conditions lipid raft components such as GPI-anchored proteins and members of the caveolin family partition into the insoluble I fraction (23, 24). The resultant S and I fractions were resolved by SDS-PAGE under reducing conditions followed by immunoblot analyses. As shown in Fig. 8C, the dimerization mutants C532A and C530A/C2A/C4A, like wild type MT6-MMP, partitioned completely into the insoluble fraction. Taken together these data suggest that trafficking to the cell surface (Figs. 3 and 4) and lipid raft insertion (Fig. 8C) of MT6-MMP are not dependent on dimerization.
FIGURE 8.
Lack of dimerization does not affect partitioning of MT6-MMP into lipid rafts. A and B, HCT-116 colon cancer cells were stably transfected to express wild type (WT) MT6-MMP (lanes 2 and 2′), or single C532A (lanes 3 and 3′) or triple C530A/C2A/C4A (lanes 4 and 4′) MT6-MMP mutants. Control cells were stably transfected with pcDNA vector without insert (EV) (lanes 1 and 1′). Lysates of stable-pooled populations were resolved by non-reducing (-β-ME) or reducing (+β-ME) 10% SDS-PAGE and examined for expression of MT6-MMP forms by immunoblot analyses. The blots were reprobed with an antibody to the transferrin receptor (TrfR). C, HCT-116 cells stably expressing wild type MT6-MMP (WT) or single C532A or triple C530A/C2A/C4A MT6-MMP mutants were harvested, and cellular proteins were separated into Triton X-100 soluble (S) and Triton X-100-insoluble/β-octylglucoside-soluble (I) fractions. Equal amounts of protein (10 μg/lane) from each fraction were mixed with Laemmli SDS-sample buffer with β-ME and resolved by 7.5% SDS-PAGE followed by immunoblot analysis with mAb MAB1142. The blots were reprobed with an anti-caveolin pAb to detect the ∼22–24-kDa caveolin (Cav). MT6D, MT6-MMP dimeric form; MT6M, MT6-MMP monomeric form.
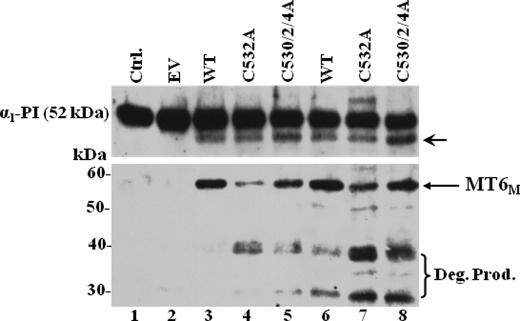
Proteolytic Activity of Dimerization-deficient MT6-MMP Mutants—Nie and Pei (25) demonstrated that α1-PI is a substrate of MT6-MMP. In agreement, we recently showed that isolated plasma membranes of HCT-116 cells stably expressing MT6-MMP cleave α1-PI, and this process is inhibited by TIMP-1 and TIMP-2 (8). Therefore, we used α1-PI cleavage to assess the role of dimerization in proteolytic activity of MT6-MMP. To this end, purified human α1-PI was incubated with crude plasma membranes isolated from HCT-116-expressing recombinant wild type MT6-MMP or the C532A or C530A/C2A/C4A MT6-MMP mutants. Because determination of the precise amount of active MT6-MMP in each plasma membrane extract was not plausible due to the lack of a quantitative assay, we estimated the relative amounts of MT6-MMP by immunoblot analyses in a separate experiment. Then, comparable levels of MT6-MMP-containing plasma membranes were incubated with α1-PI. To further compensate for differences in MT6-MMP expression, α1-PI was incubated with two concentrations of plasma membranes. Cleavage of α1-PI was verified by immunoblot analyses of the reaction mixtures, as described under “Experimental Procedures.” As shown in Fig. 9, uncleaved α1-PI exhibits a molecular mass of ∼52 kDa (Fig. 9, lanes 1 and 2). Incubation of α1-PI with plasma membranes isolated from EV-HCT cells did not result in cleavage (Fig. 9, lane 2), as expected. In contrast, plasma membranes of HCT-116 cells enriched with either wild type (Fig. 9, lanes 3 and 6) or dimerization-deficient (Fig. 9, lanes 4, 5, 7, and 8) MT6-MMP resulted in the appearance of the cleaved product of α1-PI, as reported (8, 25). Under these conditions, cleavage of α1-PI by wild type and mutant MT6-MMP was also completely inhibited by exogenously added recombinant TIMP-1 or TIMP-2 (100 nm, data not shown). Immunoblot analyses of the reaction mixtures to detect MT6-MMP forms (Fig. 9, lower panel) revealed higher amounts of wild type MT6-MMP than mutant MT6-MMP, possibly due to degradation of the mutants during the incubation time. Indeed, both C532A and C530A/C2A/C4A mutants exhibited increased amounts of the ∼38- and ∼30-kDa degradation fragments (Fig. 9, lanes 7 and 8, lower panel). Based on these semiquantitative data we conclude that lack of dimerization does not impair the ability of MT6-MMP to accomplish the cleavage of α1-PI or to be inhibited by TIMPs. Furthermore, the relatively lower levels of the dimerization-deficient mutants in the reaction mixtures suggest that the monomeric form may be somewhat more effective in processing α1-PI than the dimeric form.
FIGURE 9.
Cleavage of α1-PI by MT6-MMP wild type and Cys mutants. HCT-116 cells stably expressing wild type (WT) MT6-MMP or single C532A or triple C530A/C2A/C4A MT6-MMP mutants or transfected with vector without insert (EV) were harvested, and plasma membranes were isolated as described under “Experimental Procedures.” Equal amounts of the plasma membrane fractions (25 μg/reaction in lanes 2–5 and 50 μg/reaction in lanes 6–8) resuspended in 50 μl/reaction of collagenase buffer were incubated (16 h, 37 °C) with α1-PI (25 ng/reaction, lanes 2–8). As control (Ctrl.), α1-PI was incubated in collagenase buffer without plasma membranes (lane 1). The mixtures were resolved by reducing 10% SDS-PAGE followed by immunoblot analysis using anti-α1-PI pAb. The blots were reprobed with mAb MAB1142. The arrow in the upper panel indicates the cleaved α1-PI. MT6M, MT6-MMP monomeric form. The bracket in the lower panel indicates the degradation products of MT6-MMP. Deg. Prod, degradation products.
DISCUSSION
Previously we showed that MT6-MMP is displayed at the cell surface as a ∼120-kDa species (9). The reduction and SDS sensitivity of the 120-kDa form of MT6-MMP was suggestive of covalent dimerization of active monomers via disulfide linkages (8). Analysis of the primary sequence of MT6-MMP reveals six cysteine residues, some of which are conserved with members of the MT-MMP subfamily (6). Conserved cysteines include the cysteine residue located within the pro-domain (Cys90 in MT6-MMP) and the two cysteines at the beginning and end of the hemopexin-like domain (Cys317 and Cys508 in MT6-MMP). Cys90 in the pro-domain of MT6-MMP is predicted to interact with the catalytic zinc and, thus, to play a role in the cysteine switch mechanism of pro-MMP activation after furin cleavage (26, 27). Cys317 and Cys508 are predicted to form the disulfide bond stabilizing the four-bladed propeller structure of the hemopexin-like domain (26, 28) and, thus, precluded from playing a role in dimerization. The stem region of MT6-MMP also contains three cysteine residues, Cys530, Cys532, and Cys534, of unknown functions (6), which were good candidates for mediating dimerization. To assess their putative role(s) in dimerization, we generated single, double, and triple substitutions at these cysteine residues and analyzed the profile of MT6-MMP forms after expression in COS-1 and HCT-116 cells by immunoblot analyses under reducing and non-reducing conditions. Furthermore, we used computational analyses to examine the predisposition of the stem cysteine residues to undergo disulfide bond formation. Collectively, these data clearly showed that Cys532 is responsible for MT6-MMP homodimerization, whereas Cys530 and Cys534 are involved in the formation of intramolecular disulfide bonds within the stem region. This conclusion is supported by the following findings; (i) substitution of only Cys532 was sufficient to abrogate dimerization, (ii) substitutions at both Cys530 and Cys534 did not prevent dimerization, and (iii) Cys530 and Cys534 were predicted to be at a molecular distance that favors disulfide bond formation, as determined in the computational study. In agreement, individual substitutions of these cysteines had no effect on dimerization. The ability of the double Cys mutants to dimerize, including those with a mutation at Cys532, can be explained by disulfide bonds generated by any of the remaining Cys residue, which can be made available for oxidization under the experimental conditions used here. In contrast, rearrangement of disulfide bonds and, consequently, MT6-MMP dimerization is not possible in the single Cys532 mutant, consistent with the presence of a Cys530-Cys534 disulfide bond.
The computational study of the stem peptide validated the experimental findings, with the prediction that a sequential disulfide bond formation in which Cys530 and Cys534 generate the first bond. We searched the Research Collaboratory for Structural Bioinformatics data base for examples of disulfide bonds in proteins of known crystal structures that share the aforementioned CXXXC motif (Cys530 to Cys534) of MT6-MMP stem peptide or motifs that come close to it. Three structures were found which are relevant for discussion here. The disulfide bonds in each case are between the first cysteine in the short sequence and the one that follows it, two (CXC), three (CXXC), or five (CXXXXC) residues to the C terminus. However, there are no examples of CXXXC, which is the case for MT6-MMP. We call these sequential disulfide bonds as opposed to through-space disulfides. The first is the structure of thiol oxidase Erv2p. This protein has the motifs CXC and CXXC (PDB code 1JR8) (29). The second is flavoenzyme Ero1p (PDB code 2HI7), which exhibits the motifs CXXC and CXXXXC (30). The third example is the structure of the complex of DsbB-DsbA, with the motif CXXC (31). In all these motifs one sees either a loop or a helical turn that brings the side chains of the two cysteines into close proximity for the oxidative disulfide bond formation. Whereas we have not been able to find an example for the CXXXC motif in the crystallographic data base of proteins, the principle that we just defined for bringing the two cysteines to close spatial proximity is the same in all, including the case of MT6-MMP (i.e. the CXXXC motif). These observations strengthen the case for the sequential disulfide bond formation in MT6-MMP, as described in this report. It also worth noticing that from the three cysteines of the stem region of MT6-MMP, only the first two cysteines are conserved in mammals and zebrafish (ClustalW Version 1.82). This observation suggests that the intramolecular Cys530-Cys534 disulfide bond appeared at a later stage of evolution. Functionally, the data suggest that the intramolecular Cys530-Cys534 disulfide bond is not a perquisite for dimerization because MT6-MMP molecules bearing either single or double mutations at Cys530 and Cys534 generated homodimers. However, these mutants generated reduced levels of homodimers when compared with wild type MT6-MMP (Fig. 3). This result and the computational study suggest that the intramolecular Cys530-Cys534 disulfide bond predisposes MT6-MMP dimerization via Cys532, resulting in a more efficient dimerization process.
Previous studies showed that oligomerization permits stabilization of GPI-anchored proteins into lipid rafts, a process that leads to their apical sorting in polarized epithelial cells (32, 33). MT6-MMP also partitions exclusively into detergent-resistant lipid rafts, where it is detected in a dimeric form (9). However, here we have found that the C532A and C530A/C2A/C4A mutants, similar to the case of the wild type MT6-MMP, associate into detergent-resistant fractions, and their monomeric forms are displayed at the cell surface. Thus, covalent homodimerization is not a requirement for MT6-MMP partitioning to lipid rafts or cell surface targeting. In this regard, because MT6-MMP dimers are mediated by intermolecular disulfide bridges likely generated in the endoplasmic reticulum (34), the mechanisms regulating MT6-MMP partitioning into lipid rafts and trafficking may be different from those exhibited by GPI-anchored proteins that undergo oligomerization via non-covalent interactions in the medial Golgi (32, 33, 35). Regardless, more studies are warranted to understand the cellular distribution of MT6-MMP dimers in normal and cancer cells.
The data presented here indicate that homodimerization of MT6-MMP contributes to the stabilization of the active protease. Indeed, disruption of the Cys532 disulfide bond caused the resultant monomeric form of MT6-MMP to be partially converted into lower molecular weight fragments by a process involving autocatalysis and the action of one or more metalloprotease(s). In contrast, mutations at Cys530 and Cys534 had minimal effect, suggesting that the intramolecular disulfide bridge is not critical for MT6-MMP stability. We surmise that the homodimeric structure of MT6-MMP evolved to exhibit a structural conformation that is more resistant to degradation. The precise nature of the degradation products of MT6-MMP (38 and 32 kDa) is presently unknown. However, based on their relative molecular mass and on preliminary immunoblot data, we surmise that both species lack the catalytic domain and, thus, are inactive species. The 38-kDa species is a natural product of MT6-MMP turnover because it is also detected in neutrophil lysates, as we reported earlier (9), and is also present in differentiated HL-60 cells (data not shown). Moreover, Kang et al. (18) were the first to report similar degradation products of MT6-MMP in human neutrophils. As shown here, formation of the 38-kDa species in transfected COS-1 cells is not autocatalytic, yet it is sensitive to metalloproteinase inhibition (Fig. 7). Furthermore, lack of dimerization enhances formation of the 38-kDa form. Thus, we surmise that dimerization protects MT6-MMP against the proteolytic attack by a putative metalloproteinase. As opposed to the 38-kDa species, the 32-kDa form is readily detected under conditions of recombinant protein overexpression in COS-1 cells and is particularly noticeable in cells expressing the dimerization deficient mutants (Figs. 3 and 7). Formation of the 32-kDa form is an autocatalytic event as it is inhibited in the E/A mutant. We have not yet detected this species in human neutrophils or HL-60 cells, suggesting that its formation may be a consequence of overexpression.
Interestingly, although the homodimeric form of MT6-MMP constitutes the major pool of active MT6-MMP at the cell surface, monomeric forms are also observed. The low levels of monomer may be explained in part by its enhanced sensitivity to proteolytic attack and/or by its propensity toward dimerization. Indeed, transport to the cell surface does not appear to be regulated by homodimerization, as discussed earlier. It is important to recognize that the presence of biotinylated degradation fragments of MT6-MMP does not rule out the possibility that the degradation of monomers occurs intracellularly after endocytosis. Preliminary evidence from our laboratory using antibody cross-linking-mediated internalization assays indicates that MT6-MMP undergoes endocytosis and recycling.4 Therefore, it will be important to determine the extent by which homodimerization or lack thereof regulates the endocytosis of MT6-MMP.
Using a semiquantitative assay of α1-PI cleavage, we found that both the wild type and dimerization-deficient mutants of MT6-MMP accomplished a similar extent of α1-PI cleavage under similar experimental conditions. These data suggest that the proteolytic activity of MT6-MMP in plasma membranes is not completely compromised by lack of dimerization, at least when tested with α1-PI as a substrate. In fact, considering that the dimerization-deficient mutants are prone to degradation, the monomer may be more effective than the dimeric form in accomplishing α1-PI cleavage. Furthermore, both TIMP-1 and TIMP-2 inhibited α1-PI cleavage under these conditions, suggesting that both the monomeric and the dimeric forms of MT6-MMP are sensitive TIMP inhibition. However, the precise kinetics of enzyme activity and inhibition of monomeric versus dimeric MT6-MMP needs to be evaluated under experimental conditions in which the net amount of active site monomers and dimers can be accurately determined on the cell surface, a challenging task when studying enzyme kinetics of membrane proteases. Furthermore, the relative importance of MT6-MMP dimerization in regulating substrate recognition and hydrolysis needs to be determined with a variety of targets. Currently, the information available on MT6-MMP substrates is only limited to proteins that are cleaved by the isolated catalytic domain (8), and there is little information on the functional consequences of MT6-MMP activity in cells. We previously showed that ectopic expression of MT6-MMP in human colon cancer cell lines enhances tumor growth when the cells are implanted in nude mice (9). However, how MT6-MMP promotes this effect in vivo remains unknown. MT6-MMP did not promote in vitro cell migration and invasion of collagen or Matrigel-coated filters (9) nor facilitated the degradation of DQ™ collagen I or gelatin substrates of the colon cancer cell lines.4 In agreement, MT6-MMP did not promote in vitro invasion of a peritoneal basement membrane when expressed in COS-1 cells (36). Furthermore, we showed that MT6-MMP is not an activator of pro-gelatinases regardless of TIMP presence (9). In this regard it is worth mentioning that the dimerization mutants of MT6-MMP, like the wild type enzyme, showed no ability to activate pro-gelatinases (data not shown). In all events, understanding the role of dimerization in MT6-MMP catalytic activity and biological function awaits the determination of its substrate profile. However, considering the physical constraints that disulfide-linked homodimers may impose to the structural conformation of MT6-MMP catalytic site, the physiological relevance of newly identified MT6-MMP substrates by degradomic approaches will have to be validated with dimeric and lipid raft-inserted MT6-MMP forms.
Formation of homodimers has been reported with other members of the MMP family (37). MMP-9 forms homodimers that are SDS-resistant and -sensitive to reduction (38, 39). Dimerization of MMP-9 is thought to involve non-covalent associations mediated by the hemopexin-like domain (40) and the hinge region (41). Functionally, the dimeric form of MMP-9 may regulate activation (38); however, a full understanding of the function of MMP-9 dimers still remains elusive. The transmembrane MT1-MMP (MMP-14) has also been shown to generate homodimers (42–45). However, under similar experimental conditions used here to detect MT6-MMP dimers, we were unable to detect MT1-MMP homodimers in a variety of cells. This suggests that MT1-MMP dimerization may be a transient process and/or that the dimeric forms represent a small fraction of the total enzyme at the cell surface. Consistently, the relative importance of MT1-MMP dimers for enzyme function both in vitro and in vivo has been questioned (36, 46, 47). There is also a lack of consensus on the nature (covalent and/or non-covalent) of MT1-MMP dimers and the domains involved in dimerization as both the hemopexin-like domain (42, 43) and the anchoring apparatus (transmembrane and cytosolic tail) (44, 48) were all claimed to play a role. In the case of MT6-MMP the mutagenesis data presented here do not support a key role for the hemopexin-like domain in MT6-MMP dimerization. Indeed, the single C532A and the triple C530A/C2A/C4A mutant, which maintain the hemopexin-like domain, show a dramatic decrease in dimerization when compared with the wild type enzyme. From these data however, we cannot completely rule out secondary contributions of the hemopexin-like domain to dimerization, possibly in stabilization or folding of the dimeric form. In view of this information, the emerging picture suggests that the GPI-anchored MT-MMPs are unique in regard to their ability to generate oligomeric forms. First, the GPI-anchored MT-MMPs contain a stem region with unique Cys residues, which we have shown here to mediate protease dimerization in MT6-MMP. Second, the disulfide-linked homodimers of MT6-MMP are readily detected in cells expressing natural MT6-MMP, such as differentiated neutrophil-like HL-60 cells and human neutrophils, indicating that this form represents the physiological organization of MT6-MMP. Preliminary evidence from our laboratory shows that MT4-MMP (MMP17) is also detected in oligomeric forms.5 Altogether, these observations further establish oligomerization as a general characteristic of the human GPI-anchored MT-MMPs.
This work was supported, in whole or in part, by National Institutes of Health Grant CA-61986 (to R. F.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: MMP, matrix metalloproteinase; MT, membrane type; GPI, glycosylphosphatidylinositol; TIMP, tissue inhibitor of metalloproteinase; pAb, polyclonal antibody; mAb, monoclonal antibody; PI-PLC, phosphatidylinositol-specific phospholipase C; β-ME, β-mercaptoethanol; α1-PI, α1-proteinase inhibitor.
Bernardo, M. M., H. Zhao, A. Sohail, Q. Sun, and R. Fridman, unpublished data.
H. Zhao, A. Sohail, and R. Fridman, manuscript in preparation.
References
- 1.Nagase, H., Visse, R., and Murphy, G. (2006) Cardiovasc. Res. 69 562-573 [DOI] [PubMed] [Google Scholar]
- 2.Zucker, S., Pei, D., Cao, J., and Lopez-Otin, C. (2003) Curr. Top. Dev. Biol. 54 1-74 [DOI] [PubMed] [Google Scholar]
- 3.Osenkowski, P., Toth, M., and Fridman, R. (2004) J. Cell. Physiol. 200 2-10 [DOI] [PubMed] [Google Scholar]
- 4.Itoh, Y., and Seiki, M. (2006) J. Cell. Physiol. 206 1-8 [DOI] [PubMed] [Google Scholar]
- 5.Barbolina, M. V., and Stack, M. S. (2008) Semin. Cell Dev. Biol. 19 24-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pei, D. (1999) Cell Res. 9 291-303 [DOI] [PubMed] [Google Scholar]
- 7.Puente, X. S., Pendas, A. M., Llano, E., Velasco, G., and Lopez-Otin, C. (1996) Cancer Res. 56 944-949 [PubMed] [Google Scholar]
- 8.Sohail, A., Sun, Q., Zhao, H., Bernardo, M. M., Cho, J. A., and Fridman, R. (2008) Cancer Metastasis Rev. 27 289-302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun, Q., Weber, C. R., Sohail, A., Bernardo, M. M., Toth, M., Zhao, H., Turner, J. R., and Fridman, R. (2007) J. Biol. Chem. 282 21998-22010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trayner, I. D., Bustorff, T., Etches, A. E., Mufti, G. J., Foss, Y., and Farzaneh, F. (1998) Leuk. Res. 22 537-547 [DOI] [PubMed] [Google Scholar]
- 11.Wang, F., Herzmark, P., Weiner, O. D., Srinivasan, S., Servant, G., and Bourne, H. R. (2002) Nat. Cell Biol. 4 513-518 [DOI] [PubMed] [Google Scholar]
- 12.Santos-Beneit, A. M., and Mollinedo, F. (2000) J. Leukocyte Biol. 67 712-724 [DOI] [PubMed] [Google Scholar]
- 13.Zhao, H., Bernardo, M. M., Osenkowski, P., Sohail, A., Pei, D., Nagase, H., Kashiwagi, M., Soloway, P. D., DeClerck, Y. A., and Fridman, R. (2004) J. Biol. Chem. 279 8592-8601 [DOI] [PubMed] [Google Scholar]
- 14.Garnier, J., Osguthorpe, D. J., and Robson, B. (1978) J. Mol. Biol. 120 97-120 [DOI] [PubMed] [Google Scholar]
- 15.Chou, P. Y., and Fasman, G. D. (1974) Biochemistry 13 222-245 [DOI] [PubMed] [Google Scholar]
- 16.Zhang, W., Shi, Q., Meroueh, S. O., Vakulenko, S. B., and Mobashery, S. (2007) Biochemistry 46 10113-10121 [DOI] [PubMed] [Google Scholar]
- 17.Onufriev, A., Bashford, D., and Case, D. A. (2000) J. Phys. Chem. B 104 3712-3720 [Google Scholar]
- 18.Kang, T., Yi, J., Guo, A., Wang, X., Overall, C. M., Jiang, W., Elde, R., Borregaard, N., and Pei, D. (2001) J. Biol. Chem. 276 21960-21968 [DOI] [PubMed] [Google Scholar]
- 19.Birnie, G. D. (1988) Br. J. Cancer Suppl. 9, 41-45 [PMC free article] [PubMed]
- 20.Collins, S. J., Ruscetti, F. W., Gallagher, R. E., and Gallo, R. C. (1978) Proc. Natl. Acad. Sci. U. S. A. 75 2458-2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacob, C., Leport, M., Szilagyi, C., Allen, J. M., Bertrand, C., and Lagente, V. (2002) Int. Immunopharmacol. 2 1647-1656 [DOI] [PubMed] [Google Scholar]
- 22.Gorman, J. J., Wallis, T. P., and Pitt, J. J. (2002) Mass Spectrom. Rev. 21 183-216 [DOI] [PubMed] [Google Scholar]
- 23.Solomon, K. R., Mallory, M. A., and Finberg, R. W. (1998) Biochem. J. 334 325-333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhuang, L., Lin, J., Lu, M. L., Solomon, K. R., and Freeman, M. R. (2002) Cancer Res. 62 2227-2231 [PubMed] [Google Scholar]
- 25.Nie, J., and Pei, D. (2004) Exp. Cell Res. 296 145-150 [DOI] [PubMed] [Google Scholar]
- 26.Maskos, K., and Bode, W. (2003) Mol. Biotechnol. 25 241-266 [DOI] [PubMed] [Google Scholar]
- 27.Nagase, H. (1997) Biol. Chem. 378 151-160 [PubMed] [Google Scholar]
- 28.Visse, R., and Nagase, H. (2003) Circ. Res. 92 827-839 [DOI] [PubMed] [Google Scholar]
- 29.Han, G. W., Kopka, M. L., Langs, D., Sawaya, M. R., and Dickerson, R. E. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 9214-9219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gross, E., Kastner, D. B., Kaiser, C. A., and Fass, D. (2004) Cell 117 601-610 [DOI] [PubMed] [Google Scholar]
- 31.Inaba, K., Murakami, S., Suzuki, M., Nakagawa, A., Yamashita, E., Okada, K., and Ito, K. (2006) Cell 127 789-801 [DOI] [PubMed] [Google Scholar]
- 32.Paladino, S., Sarnataro, D., Pillich, R., Tivodar, S., Nitsch, L., and Zurzolo, C. (2004) J. Cell Biol. 167 699-709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paladino, S., Sarnataro, D., Tivodar, S., and Zurzolo, C. (2007) Traffic 8 251-258 [DOI] [PubMed] [Google Scholar]
- 34.Anelli, T., and Sitia, R. (2008) EMBO J. 27 315-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cunningham, O., Andolfo, A., Santovito, M. L., Iuzzolino, L., Blasi, F., and Sidenius, N. (2003) EMBO J. 22 5994-6003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hotary, K., Li, X. Y., Allen, E., Stevens, S. L., and Weiss, S. J. (2006) Genes Dev. 20 2673-2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maskos, K. (2005) Biochimie (Paris) 87 249-263 [DOI] [PubMed] [Google Scholar]
- 38.Olson, M. W., Bernardo, M. M., Pietila, M., Gervasi, D. C., Toth, M., Kotra, L. P., Massova, I., Mobashery, S., and Fridman, R. (2000) J. Biol. Chem. 275 2661-2668 [DOI] [PubMed] [Google Scholar]
- 39.Wilhelm, S. M., Collier, I. E., Marmer, B. L., Eisen, A. Z., Grant, G. A., and Goldberg, G. I. (1989) J. Biol. Chem. 264 17213-17221 [PubMed] [Google Scholar]
- 40.Cha, H., Kopetzki, E., Huber, R., Lanzendorfer, M., and Brandstetter, H. (2002) J. Mol. Biol. 320 1065-1079 [DOI] [PubMed] [Google Scholar]
- 41.Van den Steen, P. E., Van Aelst, I., Hvidberg, V., Piccard, H., Fiten, P., Jacobsen, C., Moestrup, S. K., Fry, S., Royle, L., Wormald, M. R., Wallis, R., Rudd, P. M., Dwek, R. A., and Opdenakker, G. (2006) J. Biol. Chem. 281 18626-18637 [DOI] [PubMed] [Google Scholar]
- 42.Lehti, K., Lohi, J., Juntunen, M. M., Pei, D., and Keski-Oja, J. (2002) J. Biol. Chem. 277 8440-8448 [DOI] [PubMed] [Google Scholar]
- 43.Itoh, Y., Takamura, A., Ito, N., Maru, Y., Sato, H., Suenaga, N., Aoki, T., and Seiki, M. (2001) EMBO J. 20 4782-4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rozanov, D. V., Deryugina, E. I., Ratnikov, B. I., Monosov, E. Z., Marchenko, G. N., Quigley, J. P., and Strongin, A. Y. (2001) J. Biol. Chem. 276 25705-25714 [DOI] [PubMed] [Google Scholar]
- 45.Galvez, B. G., Genis, L., Matias-Roman, S., Oblander, S. A., Tryggvason, K., Apte, S. S., and Arroyo, A. G. (2005) J. Biol. Chem. 280 1292-1298 [DOI] [PubMed] [Google Scholar]
- 46.Cho, J. A., Osenkowski, P., Zhao, H., Kim, S., Toth, M., Cole, K., Aboukameel, A., Saliganan, A., Schuger, L., Bonfil, R. D., and Fridman, R. (2008) J. Biol. Chem. 283 17391-17405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li, X. Y., Ota, I., Yana, I., Sabeh, F., and Weiss, S. J. (2008) Mol. Biol. Cell 19 3221-3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Itoh, Y., Ito, N., Nagase, H., and Seiki, M. (2008) J. Biol. Chem. 283 13053-13062 [DOI] [PMC free article] [PubMed] [Google Scholar]