Abstract
Chimaerins are a family of GTPase activating proteins (GAPs) for the small G-protein Rac that have gained recent attention due to their important roles in development, cancer, neuritogenesis, and T-cell function. Like protein kinase C isozymes, chimaerins possess a C1 domain capable of binding phorbol esters and the lipid second messenger diacylglycerol (DAG) in vitro. Here we identified an autoinhibitory mechanism in α2-chimaerin that restricts access of phorbol esters and DAG, thereby limiting its activation. Although phorbol 12-myristate 13-acetate (PMA) caused limited translocation of wild-type α2-chimaerin to the plasma membrane, deletion of either N- or C-terminal regions greatly sensitize α2-chimaerin for intracellular redistribution and activation. Based on modeling analysis that revealed an occlusion of the ligand binding site in the α2-chimaerin C1 domain, we identified key amino acids that stabilize the inactive conformation. Mutation of these sites renders α2-chimaerin hypersensitive to C1 ligands, as reflected by its enhanced ability to translocate in response to PMA and to inhibit Rac activity and cell migration. Notably, in contrast to PMA, epidermal growth factor promotes full translocation of α2-chimaerin in a phospholipase C-dependent manner, but not of a C1 domain mutant with reduced affinity for DAG (P216A-α2-chimaerin). Therefore, DAG generation and binding to the C1 domain are required but not sufficient for epidermal growth factor-induced α2-chimaerin membrane association. Our studies suggest a role for DAG in anchoring rather than activation of α2-chimaerin. Like other DAG/phorbol ester receptors, including protein kinase C isozymes, α2-chimaerin is subject to autoinhibition by intramolecular contacts, suggesting a highly regulated mechanism for the activation of this Rac-GAP.
Signaling via the small GTPase Rac has been implicated in a wide range of cell processes such as cytoskeleton reorganization, migration, gene expression, and cell cycle progression. Rac is a molecular switch that cycles between GTP-loaded “on” and GDP-loaded “off” states. Tight mechanisms exist to fine-tune Rac responses by controlling the cycling between Rac active and inactive states. In the off state, cytosolic GDP-bound Rac is associated with Rho family-specific guanine nucleotide dissociation inhibitors (RhoGDIs) (1). In response to an activating signal, such as the stimulation of tyrosine kinase or G protein-coupled receptors, Rac translocates to the plasma membrane where guanine nucleotide exchange factors promote the exchange of GDP for GTP, rendering Rac active. On the other hand, termination of Rac signal is mediated by GTPase activating proteins (GAPs),3 which interact with GTP-bound Rac and accelerate GTP hydrolysis, leading to Rac inactivation (2, 3). Although the mechanisms by which Rac-guanine nucleotide exchange factor promote Rac activation have been extensively studied, much less is known about the mechanistic basis of Rac inactivation by Rac-GAPs.
The chimaerin family of Rac-GAPs (α1-, α2-, β1-, and β2-chimaerin) gained significant attention in the last years due to their unique regulatory properties, as they represent the only Rac-GAPs regulated directly by the lipid second messenger diacylglycerol (DAG). Chimaerins have a single C1 domain, a 50-amino acid motif originally identified in protein kinase C (PKC) isozymes and involved in DAG binding. Chimaerin Rac-GAPs bind DAGs and DAG-mimetics such as phorbol esters in vitro with similar affinities to PKC isozymes, an indication that DAG can signal through pathways independent of PKC (4, 5). It is now well established that DAG can also signal through other C1-domain containing proteins, such as protein kinase D isozymes, RasGRPs, and DAG kinases (6). Beside the C1 domain and the C-terminal GAP domain with high specificity toward Rac, α2- and β2-chimaerin have a poorly characterized N-terminal region containing a SH2 domain possibly involved in protein-protein interactions. Recent studies established that activation of tyrosine kinase and G protein-coupled receptors leads to β2-chimaerin activation and translocation to the plasma membrane, a mechanism that depends on DAG generated by phospholipase C (PLC) (7, 8). This redistribution places the Rac-GAP in close proximity to activated Rac and serves the purpose of limiting the intensity and duration of Rac activation, arguing for a key role for DAG in the termination of Rac signaling in response to receptor activation.
Among the members of the chimaerin family, α2-chimaerin has gained recent attention due to its important roles in development. α2-Chimaerin modulates neuritogenesis and plays a central role in motor circuit formation and growth cone collapse mediated by EphA4 forward signaling (9-13). We found that the related zebrafish homologue chn1 plays an important modulatory role in cell migration during gastrulation (14). Despite this emerging functional information, little is known regarding the mechanisms that lead to α2-chimaerin activation. Although α2-chimaerin may interact with tyrosine kinase receptors via the SH2 domain in a tyrosine phosphorylation-dependent manner and probably through the C terminus (10, 11, 13), it is unclear whether its localization and activation are modulated by ligand binding to the C1 domain. Moreover, whether receptors coupled to PLC and DAG generation can modulate α2-chimaerin localization and activity is unknown. Addressing this issue would be relevant to define potential cross-talks between DAG and small G-protein signaling and more importantly, it may define a novel DAG effector unrelated to PKC.
In this study we report that α2-chimaerin adopts a conformation in which the C1 and Rac-GAP domains are not fully exposed. Using a series of deletions and point mutants we revealed that release of this autoinhibition facilitates the access of C1 domain ligands, which favors the relocalization of α2-chimaerin to the cell periphery to render the Rac-GAP active. Our analysis led to the identification of amino acids that play key roles in stabilizing the inactive conformation of α2-chimaerin. Furthermore, we established for the first time that α2-chimaerin is an effector of the epidermal growth factor receptor (EGFR), suggesting a role for this Rac-GAP in limiting Rac activation in response to physiological stimuli.
EXPERIMENTAL PROCEDURES
Materials—Phorbol 12-myristate 13-acetate (PMA) and GF109203X were purchased from LC Laboratories (Woburn, MA). Epidermal growth factor (EGF) was purchased from Roche Molecular Biochemicals. Cell culture reagents were obtained from Invitrogen. U73122 was purchased from Calbiochem.
Cell Culture and Transfections—COS-1 and HeLa cells (ATCC) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin in a humidified 5% CO2 atmosphere at 37 °C. Transfection of plasmids (0.5-1.0 μg) into cells (∼60% confluence) was carried out in 6-well or 60-mm plates using Lipofectamine PLUS (Invitrogen) according to the manufacturer's instructions.
α2-Chimaerin Cloning and Plasmid Constructions—Full-length (1.3 kb) human α2-chimaerin was cloned by reverse transcriptase-PCR from SW480 colon cancer cell mRNA using the following primers (XhoI and EcoRI sites are underlined): sense 5′-CTCGAGATGGCCCTGACCCTGTTTGAT; antisense 3′-GAGTTCTTAAAATAAAATGTCTTCGTTTTTGATAAGC). The PCR product was ligated into pCRII using the TA cloning kit (Invitrogen) and the sequence was verified by sequencing analysis. To generate HA- and GFP-tagged constructs EcoRI-EcoRI or XhoI-EcoRI fragments were isolated from pCRII and subcloned into pCEFL-HA or pEGFP-C3 (Clontech, Mountain View, CA), respectively. All constructs were verified by restriction analysis and sequencing.
Generation of α2-Chimaerin Mutants—Single point mutants for α2-chimaerin were generated using the QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA), using pCEFL-HA-α2-chimaerin as a template. The corresponding inserts were isolated and subcloned into XhoI-EcoRI sites of pEGFP-C3.
Deletion mutants of α2-chimaerin were generated by PCR from pEGFP-C3 α2-chimaerin. The following primers were used (XhoI and EcoRI sites are underlined): for pEGFP-C3-ΔN-α2-chimaerin, 3′-CTCGAGTATTATGGAAGAGAGTTTCATGGC and 5′-GAGTTCTTAAAATAAAATGTCTTCGTTTTTGATAAGC; for pEGFP-C3-ΔSH2-α2-chimaerin, 3′-CTCGAGGCAGCAGAATACATTGCCAAG and 5′-GAGTTCTTAAAATAAAATGTCTTCGTTTTTGATAAGC; for pEGFP-C3-ΔGAP-α2-chimaerin, 3′-CTCGAGATGGCCCTGACCCTGTTTGAT and 5′-GAATTCGCTTACAGTCATTTGGGAC. PCR products were ligated into pCRII using the TA cloning kit (Invitrogen).
Generation of Adenoviruses and Infections—To generate adenoviruses (AdVs), XhoI-HindIII inserts of wild-type or the mutant I122A-α2-chimaerin were excised from pCRII and subcloned into the HA epitope-tagged vector pShuttle-CMV-HA (15). Adenoviruses were generated using the AdEasy Adenoviral Vector System (Statagene), according to the manufacturer's instructions. For infections, 5 × 105 cells were plated onto 60-mm plates and 2 h before infection incubated in serum-free Dulbecco's modified Eagle's medium. Cells were then infected with the corresponding AdV overnight at different multiplicities of infection (m.o.i.). Experiments were carried out 48 h after infection.
Translocation Assays—COS-1 cells (2 × 105) in 6-well plates were transfected with plasmids encoding for α2-chimaerin wild-type or mutants, as described above. Twenty-four h after transfection cells were treated with different concentrations of PMA for 20 min. Experiments were performed in the presence of the PKC inhibitor GF109203X (5 μm), added 30 min before and during PMA stimulation. Cells were harvested into lysis buffer (20 mm Tris-HCl, pH 7.5, 5 mm EGTA, and protease inhibitor mixture for mammalian cell and tissue extract, 1:500, Sigma). Separation of cytosolic (soluble) and particulate fractions was performed by ultra-centrifugation as previously described (5). Equal amounts of protein were subjected to SDS-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, and immunostained with anti-HA (1:2000; Covance, Emeryville, CA), anti-GFP (1:3000; Covance), or anti-PKCα (1:1000; Upstate Biotechnology, Lake Placid, NY) antibodies.
Localization Studies and Immunofluorescence—Cells were plated in glass coverslips in 12-well plates and transfected with plasmids encoding for the different GFP-fused proteins or infected with AdVs, as described above. After 48 h cells were treated with PMA or EGF and fixed with 100% methanol at -20 °C for 10 min. Infected cells were then immunostained with mouse anti-HA primary (1:500; Covance) and Alexa 488 goat anti-mouse secondary (0.5 μg/ml; Molecular Probes, Invitrogen) antibodies. Slides were mounted using Fluoromount-G (Southern Biotech, Birmingham, AL). Images were viewed using a Carl Zeiss LSM 510 confocal laser scanning fluorescence microscope.
Determination of Rac-GTP Levels—Rac-GTP levels were determined using a pull-down assay essentially as described before (16). Briefly, cells were lysed in a buffer containing 4 μg of GST-PAK1 binding domain, 20 mm Tris-HCl, 1 mm dithiothreitol, 5 mm MgCl2, 150 mm NaCl, 0.5% Nonidet P-40, 5 mm glycerophosphate, and protease inhibitor mixture (Sigma). Lysates were centrifuged at 14,000 × g (4 °C, 10 min) and then incubated with glutathione-Sepharose 4B beads (4 °C, 1 h). After extensive washing, the beads were boiled in loading buffer. Samples were run in a 12% SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane for Western blot analysis using a monoclonal anti-Rac1 antibody (1:3000; Upstate Biotechnology). For experiments using EGF, cells were infected overnight with different AdVs and then serum-starved for 24 h previous to EGF treatment.
Wound Assays—HeLa cells (8 × 105) in 35-mm dishes were infected with AdVs for either wild-type or I122A-α2-chimaerin, as described above. After reaching confluence (∼48 h) a wound was made using a p10 pipette tip. Measurements of wound closure were carried out at t = 0 h and t = 24 h. The wound width was measured in three different fields using Adobe Photoshop. Results were expressed as percentage of the width at t = 24 h relative to width at t = 0 h.
Modeling Studies—Molecular modeling of α2-chimaerin was performed with the Swiss-Model protein modeling server using β2-chimaerin (1XA6A) as a template. Energy minimizations were done with the GROMOS96 implementation of Deep-View/Swiss-pbdViewer. Figures were made with PyMOL viewer (DeLano Scientific LLC, San Carlos, CA).
RESULTS
Limited Intracellular Translocation of α2-Chimaerin by PMA—The C1 domain in α- and β-chimaerin isoforms has the typical phorbol ester and DAG binding motif (HX12CX2CXnCX2CX4HX2CX7C, where H is histidine, C is cysteine, X is any other amino acid, and n is 13) and presents all the structural determinants required for ligand binding (4). In vitro binding assays revealed that chimaerins bind phorbol esters and DAGs with nanomolar binding affinities that are comparable with those of cPKCs and nPKCs (4, 5). As in vitro binding assays are normally carried out under saturated lipid conditions (100% phosphatidylserine vesicles), those assays may not necessarily reflect responses in a cellular environment. To determine whether α2-chimaerin responds to C1 domain ligands in cells, we examined its translocation in response to PMA. To rule out the involvement of PKCs in the phorbol ester response, experiments were carried out in the presence of the pan-PKC inhibitor GF 109203X (5 μm). COS-1 cells ectopically expressing α2-chimaerin were treated with various concentrations of PMA for 20 min and subjected to fractionation into soluble and particulate fractions. Despite having a high affinity phorbol ester binding domain, translocation of α2-chimaerin could only be detected at high PMA concentrations (EC50 > 1 μm, n = 8, Fig. 1, A and B). On the other hand, PMA caused a significant translocation of PKCα from the soluble to the particulate fraction, as expected. The EC50 for translocation of PKCα was 18 ± 6 nm (n = 3), as revealed by densitometric analysis. The limited translocation of α2-chimaerin in cells was also confirmed by imaging cells using GFP-tagged α2-chimaerin (Fig. 1C). These studies showed that whereas substantial translocation of PKCα to the plasma membrane is observed with 100 nm PMA, translocation of α2-chimaerin could only be observed at a very high PMA concentration (3 μm). A Pro to Ala mutation in position 216 of α2-chimaerin (Pro11 in the C1 domain motif), which reduces ligand affinity by ∼100-fold without affecting the overall tertiary folding of the C1 domain (17), abolished the limited α2-chimaerin translocation caused by 3 μm PMA (Fig. 1C). No translocation of the P216A-α2-chimaerin mutant was observed up to 10 μm PMA using a fractionation assay (Fig. 1, A and B). Endogenous α2-chimaerin is also poorly translocated by PMA (18). Therefore, whereas α2-chimaerin is capable of binding phorbol esters via the C1 domain in vitro, its limited sensitivity to PMA in a cellular context suggests an inhibitory mechanism that limits its access to cellular membranes.
FIGURE 1.
Limited translocation of α2-chimaerin by PMA. A, COS-1 cells were transfected with HA-tagged expression vectors encoding for α2-chimaerin (wild-type) or P216A-α2-chimaerin, and 24 h later cells were treated with increasing concentrations of PMA in the presence of the pan-PKC inhibitor GF109203X (5 μm). Cells were then fractionated into soluble and particulate fractions and analyzed by Western blot using an anti-HA antibody. Endogenous PKCα was detected using an anti-PKCα antibody. One representative experiment is shown. B, densitometric analysis of the immunoreactivity in the soluble fractions. Results are the mean ± S.E. of at least three independent experiments and expressed as percentage of total protein. C, HeLa cells were transfected with the indicated pEGFP plasmid. After 48 h cells were treated with either 0.1 or 3 μm PMA for 20 min and visualized by confocal microscopy. Similar results were observed in three independent experiments.
Deletion of N-terminal, SH2, or GAP Domains Enhances Membrane-associated α2-Chimaerin—The reduced sensitivity of α2-chimaerin to translocation by PMA suggests a limited exposure of the ligand-binding site. We reasoned that deletion of domains either at the N or C termini may overcome a potential hindrance of the C1 domain and therefore enhance α2-chimaerin redistribution to membranes. We generated three mutants, ΔN-α2-chimaerin (deletion in amino acids 1-44), ΔSH2-α2-chimaerin (deletion in amino acids 1-128), and ΔGAP-α2-chimaerin (deletion in amino acids 265-459), and expressed them in COS-1 cells. A schematic representation of the mutants is presented in Fig. 2A. In cells growing in 10% fetal bovine serum, ∼25% of wild-type α2-chimaerin is localized to the particulate fraction. Interestingly, a much larger proportion of the deleted mutants was found in the particulate fraction (Fig. 2, B and C). Translocation of ΔSH2-α2-chimaerin and ΔGAP-α2-chimaerin was detected at concentrations of PMA as low as 10 nm. Although only 38 ± 10% (n = 3) of wild-type α2-chimaerin was found in the particulate fraction in response to 1 μm PMA, a much greater proportion was found for the mutants (83 ± 3%, n = 3, for ΔSH2-α2-chimaerin; 87 ± 8%, n = 3, for ΔGAP-α2-chimaerin). Notably, most of the mutant lacking the first 44 amino acids (ΔN-α2-chimaerin) was found in the particulate fraction both in the absence or presence of PMA (Fig. 2, C and D).
FIGURE 2.
PMA-induced translocation of α2-chimaerin is sensitized by deletion of N- or C-terminal regions. A, schematic representation of the constructs used in these experiments. B, COS-1 cells were transfected with pEGFP plasmids encoding for GFP-fused wild-type (wt), ΔN-, ΔSH2-, or ΔGAP-α2-chimaerin, and 24 h later treated with increasing concentrations of PMA (10 nm-10 μm) in the presence of the pan-PKC inhibitor GF109203X (5 μm). Cells were then fractionated into soluble and particulate fractions and analyzed by Western blot using an anti-GFP antibody. A densitometric analysis of the immunoreactivity in the particulate fraction in unstimulated cells is shown (n = 3). C, representative experiment showing the effect of PMA on the redistribution of α2-chimaerin (wild-type and mutants). D, densitometric analysis of the immunoreactivity in the soluble fraction after PMA treatment. In B and D, results are the mean ± S.E. of at least three independent experiments and expressed as percentage of total protein. **, p < 0.005 compared with cells expressing wild type α2-chimaerin with Student's t test. E, HeLa cells were transfected with the indicated pEGFP plasmids, as in B, and 48 h later treated with PMA (100 nm, 20 min). Cells were then fixed and visualized by confocal microscopy. Similar results were observed in three independent experiments.
Next, we analyzed the subcellular localization of the α2-chimaerin-deleted mutants fused to GFP (Fig. 2E). In the absence of PMA stimulation some peripheral localization was observed for the mutants. As predicted from the fractionation experiments, significant disappearance of cytosolic fluorescence and a corresponding increase in peripheral fluorescence were observed for the deleted mutants in response to 100 nm PMA. This is in clear contrast with the limited translocation observed for the wild-type protein under the same experimental condition (see Fig. 1C). We have also noticed significant perinuclear redistribution, particularly with the N-terminal deleted mutants, which agrees with previous studies for C1 domain-responsive proteins, including PKCs and β2-chimaerin (19-21). Taken together, these results suggest a hindrance of the C1 domain by regions located at either end of the protein. It should be noted that removing only the first 44 amino acids was sufficient to facilitate the exposure of membrane-binding sites in α2-chimaerin.
The N-terminal Region Is Essential to Keep α2-Chimaerin in an Inactive State—Next, we examined whether deletion of the N-terminal region in α2-chimaerin affects its activation status. COS-1 cells expressing wild-type or mutated α2-chimaerin proteins were subject to Rac-GTP pull-down assays using a GST-Pak1 binding domain. Assays were optimized at expression levels in which wild-type α2-chimaerin caused only a marginal effect on Rac-GTP levels. Notably, at comparable low levels of expression (Fig. 3A), ΔN-α2-chimaerin and ΔSH2-α2-chimaerin caused a much stronger reduction of Rac-GTP levels than wild-type α2-chimaerin (Fig. 3, A and B). As expected, the GAP-deleted mutant (ΔGAP-α2-chimaerin) was inactive. Therefore, deletion of the N-terminal region not only exposes the C1 domain but also renders α2-chimaerin hyperactive. It could be speculated that the N-terminal region of α2-chimaerin serves as an autoinhibitory domain that keeps the molecule in an inactive state in which the Rac-binding site is spatially occluded.
FIGURE 3.
N-terminal deleted α2-chimaerin mutants are hyperactive. A, COS-1 cells were transfected with pEGFP vectors encoding for wild-type (wt) α2-chimaerin, deleted α2-chimaerin mutants, or empty vector. After 24 h growing in 10% fetal bovine serum, Dulbecco's modified Eagle's medium, cells were lysed and subjected to a Rac-GTP pull-down assay. Total Rac and GFP fusion proteins were monitored by Western blot using anti-Rac and anti-GFP antibodies, respectively. B, densitometric analysis of Rac-GTP levels, expressed as percentage of control (empty vector-transfected cells). Results are the mean ± S.E. of four individual experiments. *, p < 0.05 compared with cells expressing wild type α2-chimaerin (Student's t test).
Modeling Analysis of α2-Chimaerin and Characterization of Inhibitory Amino Acids Involved in Intramolecular Interactions—C1 domains have been reported to exist in exposed and non-exposed conformations (22), an indication that ligands may have differential access to the binding site. This would also argue for the requirement of a massive conformational change to expose the ligand-binding site. To gain relevant information on the degree of exposure of the C1 domain in α2-chimaerin, we pursued modeling studies. The three-dimensional structure of the related isoform β2-chimaerin has been solved (23), and although each isoform has unique regulatory properties (8, 10, 12, 13, 18, 24, 25), both α2- and β2-chimaerin share a similar SH2-C1-GAP domain tandem. β2-Chimaerin has also limited sensitivity to phorbol ester-induced translocation (5), which would suggest a common mechanism of activation to expose the C1 domain. We modeled the tertiary structure of α2-chimaerin using the crystal structure of β2-chimaerin as a template, followed by energy minimizations with GROMOS96 implementation of DeepView/Swiss-pbdViewer.
Modeling analysis revealed that α2-chimaerin exists in a conformation in which the C1 domain is buried in the structure and occluded by intramolecular interactions (Fig. 4A). The hydrophobic loop formed by residues 20-27 in the C1 domain motif (amino acids 225-232 in α2-chimaerin) is expected to be involved in ligand and membrane interactions. The α2-chimaerin N-terminal region lies on top of this hydrophobic loop and reaches out to the Rac-binding site in the Rac-GAP domain (Fig. 4B), which is consistent with our deletional analysis. We reasoned that disrupting intramolecular contacts in the N-terminal region would destabilize the inactive conformation to augment the exposure of the C1 and Rac-binding domains, thereby reducing the energetic requirements for translocation and activation. Based on the information provided from modeling analysis we generated three site-directed mutants in the N-terminal region (L20A-, Q24A-, and I122A-α2-chimaerin, Fig. 4C). The model predicts that Leu20 in the N terminus occupies the space between Trp227 and Phe225 in the C1 domain, forming hydrophobic interactions with both residues (Fig. 4D). Ile122 in the SH2 domain makes hydrophobic contacts with Trp227. On the other hand, Gln24 should not form any significant interaction with residues in this region, and therefore disrupting this amino acid should have minimal or no effect. Of note, the corresponding amino acid in β2-chimaerin (Glu32) protrudes into the phorbol ester binding site and forms hydrogen bonds with Gly235 in the C1 domain (23).
FIGURE 4.
Molecular modeling of α2-chimaerin. A, overall structure of α2-chimaerin. The domains are colored yellow (N-terminal), green (SH2), blue (C1), red (Rac-GAP), and salmon (linkers). B, overall structure in spheres representation. C, schematic representation of point mutations introduced in α2-chimaerin. D, left, Leu20 and Ile122 from the N-terminal and SH2, respectively, are shown. Also shown are putative residues from the C1 domain involved in hydrophobic interactions (Phe225 and Trp227). Right, Ile122 mutated to Ala.
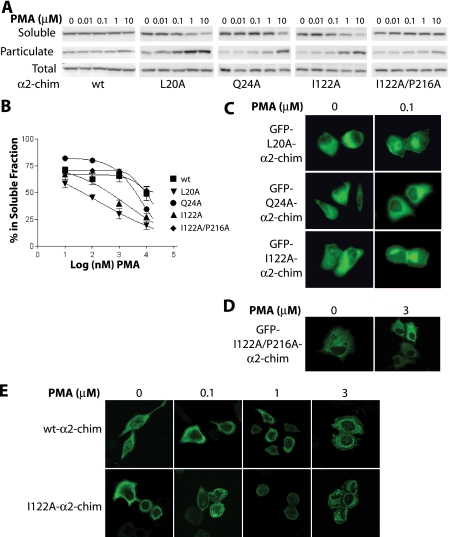
Mutants were expressed in COS-1 cells and assessed for their ability to translocate in response to phorbol ester. Although GFP-fused α2-chimaerin (wild-type) was basically insensitive to translocation in response to PMA (EC50 > 10 μm), GFP-fused L20A- and I122A-α2-chimaerin mutants were more sensitive to the phorbol ester, as reflected by a shift to the left in the PMA translocation curves (Fig. 5, A and B). As predicted from the modeling analysis, mutation of Gln24 to Ala (Q24A) had only a minimal effect on translocation. Cellular localization studies revealed that at 100 nm PMA, a concentration that will not translocate GFP-α2-chimaerin (wild-type) (see Fig. 1C) or GFP-Q24A-α2-chimaerin, a significant peripheral redistribution for both L20A- and I122A-α2-chimaerin mutants could be observed (Fig. 5C), in agreement with the conclusions from fractionation analysis.
FIGURE 5.
Translocation of α2-chimaerin mutants by PMA. A, COS-1 cells were transfected with pEGFP plasmids encoding for GFP-fused wild-type (wt), L20A-, Q24A-, I122A-, or I122A/P216A-α2-chimaerin, and 24 h later treated with increasing concentrations of PMA (10 nm to 10 μm) in the presence of the pan-PKC inhibitor GF109203X (5 μm). Cells were then fractionated into soluble and particulate fractions and analyzed by Western blot using an anti-GFP antibody. One representative experiment is shown. B, densitometric analysis of immunoreactivity in the soluble fractions. Results are the mean ± S.E. of at least three independent experiments and expressed as percentage of total protein. C and D, HeLa cells were transfected with the indicated pEGFP plasmid and 48 h later treated with 100 nm or 3 μm PMA for 20 min in the presence of GF109203X (5 μm), fixed, and visualized by confocal microscopy. E, HeLa cells were infected with either (wt) or I122A-α2-chimaerin AdVs (m.o.i. = 100 pfu/cell) and 48 h later treated with the indicated concentrations of PMA for 20 min. Cells were fixed and proteins visualized by confocal microscopy using an anti-HA antibody.
To determine whether the enhanced translocation of internal contact mutants was due to enhanced access of the ligand, we introduced the C1 domain mutation P216A in I122A-α2-chimaerin. As indicated above, this mutation reduces the binding affinity of phorbol esters by ∼100-fold. Analysis of the double mutant I122A/P216A-α2-chimaerin by fractionation and microscopy analysis revealed that translocation in response to PMA was markedly impaired relative to the single mutant I122A-α2-chimaerin (Fig. 5, A, B, and D). Taken together, these results strongly suggest that the enhanced translocation by PMA of mutants with disrupted intramolecular interactions is due to “facilitated access” of the ligand to the C1 domain.
To further validate the results described above with GFP-fused constructs, we generated AdVs encoding for wild-type α2-chimaerin or the I122A-α2-chimaerin mutant. HeLa cells were infected with α2-chimaerin or control (LacZ) AdVs (m.o.i. = 100 pfu/cell), and 24 h later treated with increasing concentrations of PMA (0.1-3 μm). Cells were fixed and localization analyzed by immunofluorescence. In agreement with the studies with GFP-fused proteins, the I122A mutant was highly sensitive to PMA-induced translocation. Translocation of I122A-α2-chimaerin was clearly detected at 100 nm PMA, whereas wild-type α2-chimaerin translocation was only noticeable at the highest concentration of PMA (3 μm) used in these studies (Fig. 5E). These experiments also ruled out any influence of the GFP tag in ligand-induced redistribution of α2-chimaerin.
Functional Relevance of Destabilizing the Inactive Conformation of α2-Chimaerin—Our results predict that disruption of internal contact residues should sensitize the protein for the activation of the Rac-GAP activity by C1 domain ligands. First, we compared the effect of wild-type and I122A-α2-chimaerin on Rac-GTP levels in cells growing in 10% serum. HeLa cells were infected with increasing m.o.i.s of either wild-type or I122A-α2-chimaerin AdVs (50-200 pfu/cell) and Rac-GTP levels assessed 48 h later. Cells expressing wild-type α2-chimaerin showed no significant changes in Rac-GTP levels at m.o.i.s of 50 and 100 pfu/cell, and only a slight reduction (18 ± 6%, n = 3) at m.o.i. = 200 pfu/cell. On the other hand, infection of HeLa cells with I122A-α2-chimaerin AdV (m.o.i. = 200 pfu/cell) caused a more pronounced reduction (71 ± 17%, n = 3) in Rac-GTP levels (Fig. 6A). Note that these differences were observed despite the fact that expression levels of I122A-α2-chimaerin were slightly lower than wild-type α2-chimaerin.
FIGURE 6.
Enhanced sensitivity of the I122A-α2-chimaerin mutant. A, HeLa cells were infected with increasing m.o.i.s of wild-type (wt) or I122A-α2-chimaerin AdVs. After 48 h Rac-GTP levels were determined in cells growing in 10% fetal bovine serum, Dulbecco's modified Eagle's medium. B, HeLa cells were infected with either (wt) or I122A-α2-chimaerin AdVs (m.o.i. = 30 pfu/cell). After 48 h Rac-GTP levels were determined in response to PMA (100 nm, 20 min). Left panels, representative experiments; right panels, densitometric analysis of Rac-GTP levels, expressed as % of control (LacZ-AdV-infected cells). Results are the mean ± S.E. of three individual experiments. **, p < 0.005, compared with wild-type α2-chimaerin. *, p < 0.05, compared with non-treated (Student's t test).
Next, we assessed the effect of PMA on Rac-GAP activity. Very low levels of wild-type and I122A-α2-chimaerin were expressed using a m.o.i. of 30 pfu/cell of each AdV, an experimental condition at which Rac-GTP levels were essentially unchanged. Remarkably, whereas 100 nm PMA caused a minimal reduction in Rac-GTP levels in cells expressing wild-type α2-chimaerin or in control cells (infected with LacZ AdV), it greatly reduced (>70%) Rac-GTP levels in cells expressing I122A-α2-chimaerin (Fig. 6B).
To validate the functional relevance of our findings, because Rac is a well established mediator of cell motility (26), we assessed migration using a wound assay in HeLa cells infected with wild-type α2-chimaerin, I122A-α2-chimaerin, or LacZ (control) AdVs at m.o.i. = 30 or 100 pfu/cell. As shown in Fig. 7, A and B, I122A-α2-chimaerin was more effective at inhibiting wound closure than wild-type α2-chimaerin at either m.o.i. This striking difference was observed despite the fact that expression levels of the I122A mutant were slightly lower than the wild-type at either m.o.i. (Fig. 7B, inset). Therefore, disrupting internal contacts facilitates α2-chimaerin activation and affects Rac-GTP-dependent cell functions. These studies, together with those of Fig. 6, also establish that hypersensitization of the mutants occur in response to serum stimulation and not only to PMA, suggesting enhanced access to endogenous DAG.
FIGURE 7.
Enhanced anti-motility effect of I122A-α2-chimaerin. A, HeLa cells were infected with the indicated m.o.i.s of LacZ, wild type (wt)- or I122A-α2-chimaerin AdVs. After 48 h a wound was made in the confluent monolayer. Pictures of the wound were taken at 0 and 24 h. B, quantitative analysis of wound closure. Bars represent the mean ± S.E. of three independent experiments. Results were expressed as % of wound closure after 24 h. Inset, expression of α2-chimaerin by Western blot using an anti-HA antibody. *, p < 0.05, compared with LacZ-infected cells (Student's t test).
EGF Translocates α2-Chimaerin in a PLC-DAG-dependent Manner—We next examined whether α2-chimaerin can be relocalized in response to a receptor coupled to DAG generation. We used as a paradigm the EGFR because it couples to PLCγ to generate DAG. Fig. 8 shows that EGF caused a marked translocation of α2-chimaerin from the cytoplasm to the periphery in HeLa cells. The effect was readily detected 1 min after the addition of EGF (Fig. 8), and was reversed significantly at 3 min after stimulation (data not shown). Translocation of α2-chimaerin was impaired by the PLC inhibitor U73122, suggesting that the EGF effect is dependent upon the generation of DAG. To determine the involvement of the α2-chimaerin C1 domain in this effect, we used the phorbol ester unresponsive mutant P216A-α2-chimaerin. This mutant failed to translocate in response to EGF (Fig. 8). Altogether, these experiments suggest that translocation by EGF is dependent upon DAG binding to the C1 domain in α2-chimaerin.
FIGURE 8.
EGF translocates α2-chimaerin in a PLC-dependent manner. HeLa cells were transfected with pEGFP plasmids encoding for wild-type (wt)-α2-chimaerin or P216A-α2-chimaerin. After 24 h cells were serum starved for another 24 h and then treated with EGF (100 ng/ml, 1 min) alone or in the presence of the PLC inhibitor U73122 (5 μm). Cells were fixed and visualized by confocal microscopy.
Lastly, we analyzed whether α2-chimaerin modulates Rac activation by EGFR stimulation. In agreement with previous studies (8), EGF caused a significant activation of Rac, as determined by “pull-down” assays. Rac activation was transient and peaks at 1 min (Ref. 8 and data not shown). Adenoviral expression of α2-chimaerin in HeLa cells inhibited Rac activation by EGF. The effect was proportional to the expression levels achieved by varying the m.o.i. (Fig. 9A). Next, we infected HeLa cells with either wild-type or I122A-α2-chimaerin at an m.o.i. in which the wild-type protein barely had an effect on Rac-GTP levels (150 pfu/cell). At that experimental condition, I122A-α2-chimaerin was more effective than wild-type α2-chimaerin in inhibiting Rac activation by EGF (62% versus 15%, respectively) (Fig. 9B). These results suggest that α2-chimaerin can be activated by EGFR to negatively modulate Rac activation, and support the concept that internal contact mutants are more sensitive to activation via ligand engagement at the C1 domain.
FIGURE 9.
I122A-α2-chimaerin is more effective an inhibiting Rac activation by EGF than wild type (wt)- α2-chimaerin. A, HeLa cells were infected using LacZ, wt-α2-chimaerin, or I122A-α2-chimaerin AdVs with m.o.i.s = 50-300 (A) or 150 (B) pfu/cell. After 48 h cells were treated with EGF (100 ng/ml, 1 min), and Rac-GTP levels determined using a pull-down assay. Left panels, representative experiments; right panels, densitometric analysis of Rac-GTP levels, expressed as % of control (LacZ-AdV-infected cells). Results are the mean ± S.E. of at least three individual experiments. **, p < 0.005, compared with LacZ-infected cells with EGF (Student's t test).
DISCUSSION
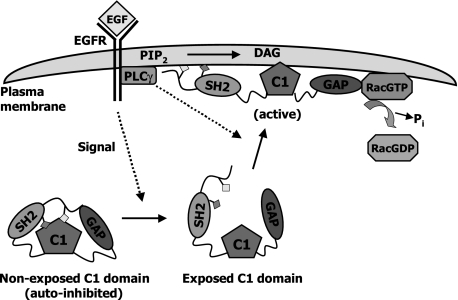
Although for many years it was thought that signaling by the lipid second messenger DAG occurs only via the PKC family of kinases, it became clear that DAG can also signal through “non-kinase” effectors by direct binding to their C1 domain. Much is known about the mechanism of activation of PKC isozymes by lipids; however, the regulation of DAG targets unrelated to PKC is much less understood. Our results provide novel insights into the mechanism of activation and regulation of the Rac-GAP α2-chimaerin by DAG. Here we show that the N and C termini in α2-chimaerin serve as autoinhibitory domains that keep the protein in an inactive status and limit the access of DAG to the C1 domain. We also established that α2-chimaerin is an effector of the EGFR. EGF promotes the coupling of EGFR to PLCγ and subsequent DAG generation (27, 28). The PLC inhibitor U73122 blocked α2-chimaerin translocation by EGF, suggesting that this positional regulation is dependent upon DAG generation. Consistent with the requirement of DAG for translocation, the C1 domain ligand-deficient mutant P216A-α2-chimaerin is unresponsive to EGF. This suggests that the PLC-DAG branch of EGFR is required but may not be sufficient for the activation of α2-chimaerin. FRET studies established that DAG-mediated translocation of chimaerins to the plasma membrane positions the Rac-GAP in close proximity to its target Rac (8). Therefore, our results support the concept that in response to growth factors DAG can signal to negatively modulate Rac, a mechanism that limits the magnitude and intensity of Rac activation. A model is presented in Fig. 10.
FIGURE 10.
Model ofα2-chimaerin activation by EGFR. In the inactive state, Leu20 in the N-terminal region and Ile122 in the SH2 domain interact with Phe225 and Trp227 in the C1 domain and stabilize an autoinhibited conformation. Activation of EGFR (or presumably other PLC-coupled receptor) signals for the release of the autoinhibition and exposes the C1 domain. DAG generated by PLCγ positions the protein at the plasma membrane where it interacts with GTP-bound Rac, thus facilitating Rac inactivation.
Our modeling analysis showed that the N-terminal region in α2-chimaerin is involved in intramolecular interactions that occlude the DAG-binding site in the C1-domain and the Rac-GAP domain. Deletion of autoinhibitory domains results in enhanced access of ligand and facilitates its membrane redistribution. Moreover, we identified relevant amino acids that make internal contacts through hydrophobic interactions with the C1 domain. According to the modeling and structural predictions, disruption of such interactions should destabilize the inactive conformation, thereby lowering the energetic requirements to expose the C1 and Rac-GAP domains and reach the active state. Presumably the activating conformational change in chimaerins exposes hydrophobic surfaces in the C1 domain and other regions folded over the C1 domain in close conformation, and those surfaces should associate with the membrane (or with each other) to avoid exposing a large amount of hydrophobic surface (23). In a cellular context this is reflected in enhanced association to the plasma membrane and activity, as we detect both with deleted and internal contact site α2-chimaerin mutants. We also detect a significant translocation to the perinuclear region with all the deleted α2-chimaerin mutants. This redistribution was also observed with the wild-type protein at higher PMA concentrations. We have reported similar findings for β2-chimaerin (19). In that study we determined that β2-chimaerin interacts with the ER-Golgi protein Tmp-21 via the C1 domain upon activation. We believe that both chimaerin isoforms share similar anchoring mechanisms for perinuclear translocation.
Studies by several laboratories have established the presence of autoinhibitory domains in PKC isozymes, including the well characterized N-terminal “pseudosubstrate” domain that associates with the C-terminal kinase region. In response to DAG production, classical and novel PKCs translocate to the plasma membrane and release the pseudosubstrate sequence from the active site (29, 30). Likewise, internal interactions outside of the PKC catalytic site have been described by the laboratory of Mochly-Rosen (31, 32), which also have an autoinhibitory function and upon disruption lead to enzyme activation. Although we still do not have a full picture on how C1 domains in PKCs fold in the overall tertiary structure of the protein, it became clear that their degree of exposure to ligand varies not only between PKC isozymes but also between the first and second C1 domains (C1a and C1b) present in DAG-responsive PKCs (22). Moreover, there is strong evidence that the C1a and C1b domains are functionally non-equivalent in some PKCs, such as PKCδ. Although those differences may relate to a differential affinity of each C1 domain for ligands (22, 33), studies by Blumberg and co-workers (34) using the mutant Pro to Gly in position 11 of the C1 domain consensus of PKCδ (the equivalent to P216A in α2-chimaerin used in this study), revealed a distinct involvement of C1a and C1b domains in translocation in response to PMA despite their similar binding affinities for phorbol esters in in vitro assays. This may reflect differential degrees of exposure of C1 domains (22). A differential accessibility of ligands to C1 domains has been reported for different PKC isoforms (35-37). For the PKCs, the presence of other membrane-association domains, such as the C2 domain, may contribute to unleash C1 domains and facilitate activation (37). Our results support the concept that the C1 domain in α2-chimaerin is not exposed in the inactive state. A similar scenario has been proposed for the DAG effector Munc-13 (38) and more recently for MRCK (39), suggesting potential common regulatory mechanisms for the non-kinase phorbol ester receptors via DAG. The N terminus of p190RhoGAP has also been suggested to play an autoregulatory role (40), which highlights potentially common mechanisms among GAPs for small GTPases.
Our studies also argue for distinct roles for the lipid second messenger DAG in signal transduction. Although DAG contributes to PKC enzymatic activation, it may serve primarily an anchoring purpose for proteins with occluded C1 domain(s). It is likely that in the latter case additional mechanisms would be required to facilitate the switch from inactive to active conformation. Those mechanisms are yet to be established for chimaerin isoforms, but most likely they involve post-translational modifications and/or protein-protein interactions. For example, tyrosine phosphorylation in PKCθ induces a conformational change that positions the C1 domains for membrane interactions (33). Recent evidence suggests that phosphorylation of α2- and β2-chimaerin by tyrosine kinases modulates their Rac-GAP activity (12, 13, 24). We also have strong evidence that serine phosphorylation modulates the localization and activity of β2-chimaerin but not α2-chimaerin.4 The recent identification of Nck as a protein interactor for α2-chimaerin (13, 41) also argues for the possibility that chimaerins are part of multiprotein complexes, and that mechanisms that promote their association to and dissociation from these complexes are highly regulated and most likely distinct for each chimaerin isoform. It is also important to highlight that regulation of chimaerins may vary depending on the receptor stimuli. In addition to their roles as effectors of EGFR via the PLCγ branch, chimaerins mediate axon guidance induced by EphA4 tyrosine kinase receptor activation. α2-Chimaerin directly associates to EphA4 via the SH2 domain to inactivate Rac (10, 11, 13, 41). On the other hand, we were unable to detect any direct association between EGFR and chimaerins in our studies (data not shown). Recent studies in T cells revealed that α2- and β2-chimaerins localized to the immune synapse upon TCR clustering. This relocalization also occurs with a DAG-unresponsive mutant, suggesting that it is DAG-independent (18). The distinct modes of chimaerin activation attest to the complex regulation of DAG targets.
In conclusion, our results revealed that an autoinhibitory mechanism precludes the exposure of the C1 domain in α2-chimaerin, therefore limiting its association to membranes in response to phorbol esters and DAG. Our studies also established that α2-chimaerin is an effector of the EGFR via the generation of DAG. The data revealed that ligand binding to the C1 domain may be insufficient to trigger the translocation of α2-chimaerin in cells and that additional inputs would be required to cause its full activation. Because Rac is hyperactivated in various types of cancers and inhibitors of Rac impair malignant transformation and invasion (42, 43), one attractive scenario is that activation of Rac-GAPs by disruption of intramolecular interactions with either peptides or small molecules may serve as means for interfering with Rac signaling. Indeed, a similar approach has been successful for activation of PKC isozymes and proved promising in preclinical and clinical trials for several diseases (44).
Addendum—At the time of submission, Miyake et al. (45) reported that Leu20 in α2-chimaerin is mutated in the Duane retraction syndrome. In agreement with our data, L20-α2-chimaerin mutants and other related mutants have enhanced Rac-GAP activity and are hypersensitive for translocation.
This work was supported, in whole or in part, by National Institutes of Health Grant CA74197 (to M. G. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GAP, GTPase activating protein; DAG, diacylglycerol; PKC, protein kinase C; SH2, Src homology domain 2; PLC, phospholipase C; EGFR, epidermal growth factor receptor; PMA, phorbol 12-myristate 13-acetate; HA, hemagglutinin; GFP, green fluorescent protein; AdV, adenovirus; m.o.i., multiplicity of infection; pfu, plaque forming unit.
E. M. Griner and M. G. Kazanietz, unpublished observations.
References
- 1.Olofsson, B. (1999) Cell. Signal. 11 545-554 [DOI] [PubMed] [Google Scholar]
- 2.Bustelo, X. R., Sauzeau, V., and Berenjeno, I. M. (2007) Bioessays 29 356-370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Symons, M., and Settleman, J. (2000) Trends Cell Biol. 10 415-419 [DOI] [PubMed] [Google Scholar]
- 4.Areces, L. B., Kazanietz, M. G., and Blumberg, P. M. (1994) J. Biol. Chem. 269 19553-19558 [PubMed] [Google Scholar]
- 5.Caloca, M. J., Fernandez, N., Lewin, N. E., Ching, D., Modali, R., Blumberg, P. M., and Kazanietz, M. G. (1997) J. Biol. Chem. 272 26488-26496 [DOI] [PubMed] [Google Scholar]
- 6.Yang, C., and Kazanietz, M. G. (2003) Trends Pharmacol. Sci. 24 602-608 [DOI] [PubMed] [Google Scholar]
- 7.Siliceo, M., Garcia-Bernal, D., Carrasco, S., Diaz-Flores, E., Coluccio Leskow, F., Teixido, J., Kazanietz, M. G., and Merida, I. (2006) J. Cell Sci. 119 141-152 [DOI] [PubMed] [Google Scholar]
- 8.Wang, H., Yang, C., Leskow, F. C., Sun, J., Canagarajah, B., Hurley, J. H., and Kazanietz, M. G. (2006) EMBO J. 25 2062-2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall, C., Michael, G. J., Cann, N., Ferrari, G., Teo, M., Jacobs, T., Monfries, C., and Lim, L. (2001) J. Neurosci. 21 5191-5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beg, A. A., Sommer, J. E., Martin, J. H., and Scheiffele, P. (2007) Neuron 55 768-778 [DOI] [PubMed] [Google Scholar]
- 11.Iwasato, T., Katoh, H., Nishimaru, H., Ishikawa, Y., Inoue, H., Saito, Y. M., Ando, R., Iwama, M., Takahashi, R., Negishi, M., and Itohara, S. (2007) Cell 130 742-753 [DOI] [PubMed] [Google Scholar]
- 12.Shi, L., Fu, W. Y., Hung, K. W., Porchetta, C., Hall, C., Fu, A. K., and Ip, N. Y. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 16347-16352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wegmeyer, H., Egea, J., Rabe, N., Gezelius, H., Filosa, A., Enjin, A., Varoqueaux, F., Deininger, K., Schnutgen, F., Brose, N., Klein, R., Kullander, K., and Betz, A. (2007) Neuron 55 756-767 [DOI] [PubMed] [Google Scholar]
- 14.Leskow, F. C., Holloway, B. A., Wang, H., Mullins, M. C., and Kazanietz, M. G. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 5373-5378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menna, P. L., Skilton, G., Leskow, F. C., Alonso, D. F., Gomez, D. E., and Kazanietz, M. G. (2003) Cancer Res. 63 2284-2291 [PubMed] [Google Scholar]
- 16.Benard, V., Bohl, B. P., and Bokoch, G. M. (1999) J. Biol. Chem. 274 13198-13204 [DOI] [PubMed] [Google Scholar]
- 17.Kazanietz, M. G., Wang, S., Milne, G. W., Lewin, N. E., Liu, H. L., and Blumberg, P. M. (1995) J. Biol. Chem. 270 21852-21859 [DOI] [PubMed] [Google Scholar]
- 18.Caloca, M. J., Delgado, P., Alarcon, B., and Bustelo, X. R. (2008) Cell. Signal. 20 758-770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, H., and Kazanietz, M. G. (2002) J. Biol. Chem. 277 4541-4550 [DOI] [PubMed] [Google Scholar]
- 20.Maissel, A., Marom, M., Shtutman, M., Shahaf, G., and Livneh, E. (2006) Cell. Signal. 18 1127-1139 [DOI] [PubMed] [Google Scholar]
- 21.Wang, Q. J., Bhattacharyya, D., Garfield, S., Nacro, K., Marquez, V. E., and Blumberg, P. M. (1999) J. Biol. Chem. 274 37233-37239 [DOI] [PubMed] [Google Scholar]
- 22.Colon-Gonzalez, F., and Kazanietz, M. G. (2006) Biochim. Biophys. Acta 1761 827-837 [DOI] [PubMed] [Google Scholar]
- 23.Canagarajah, B., Leskow, F. C., Ho, J. Y., Mischak, H., Saidi, L. F., Kazanietz, M. G., and Hurley, J. H. (2004) Cell 119 407-418 [DOI] [PubMed] [Google Scholar]
- 24.Kai, M., Yasuda, S., Imai, S., Kanoh, H., and Sakane, F. (2007) Biochim. Biophys. Acta 1773 1407-1415 [DOI] [PubMed] [Google Scholar]
- 25.Yasuda, S., Kai, M., Imai, S., Kanoh, H., and Sakane, F. (2008) Biochem. J. 409 95-106 [DOI] [PubMed] [Google Scholar]
- 26.Schmitz, A. A., Govek, E. E., Bottner, B., and Van Aelst, L. (2000) Exp. Cell Res. 261 1-12 [DOI] [PubMed] [Google Scholar]
- 27.Nishibe, S., Wahl, M. I., Hernandez-Sotomayor, S. M., Tonks, N. K., Rhee, S. G., and Carpenter, G. (1990) Science 250 1253-1256 [DOI] [PubMed] [Google Scholar]
- 28.Rotin, D., Margolis, B., Mohammadi, M., Daly, R. J., Daum, G., Li, N., Fischer, E. H., Burgess, W. H., Ullrich, A., and Schlessinger, J. (1992) EMBO J. 11 559-567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dutil, E. M., and Newton, A. C. (2000) J. Biol. Chem. 275 10697-10701 [DOI] [PubMed] [Google Scholar]
- 30.Orr, J. W., and Newton, A. C. (1994) J. Biol. Chem. 269 8383-8387 [PubMed] [Google Scholar]
- 31.Ron, D., Chen, C. H., Caldwell, J., Jamieson, L., Orr, E., and Mochly-Rosen, D. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 839-843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schechtman, D., Craske, M. L., Kheifets, V., Meyer, T., Schechtman, J., and Mochly-Rosen, D. (2004) J. Biol. Chem. 279 15831-15840 [DOI] [PubMed] [Google Scholar]
- 33.Melowic, H. R., Stahelin, R. V., Blatner, N. R., Tian, W., Hayashi, K., Altman, A., and Cho, W. (2007) J. Biol. Chem. 282 21467-21476 [DOI] [PubMed] [Google Scholar]
- 34.Szallasi, Z., Bogi, K., Gohari, S., Biro, T., Acs, P., and Blumberg, P. M. (1996) J. Biol. Chem. 271 18299-18301 [DOI] [PubMed] [Google Scholar]
- 35.Ananthanarayanan, B., Stahelin, R. V., Digman, M. A., and Cho, W. (2003) J. Biol. Chem. 278 46886-46894 [DOI] [PubMed] [Google Scholar]
- 36.Bogi, K., Lorenzo, P. S., Acs, P., Szallasi, Z., Wagner, G. S., and Blumberg, P. M. (1999) FEBS Lett. 456 27-30 [DOI] [PubMed] [Google Scholar]
- 37.Stahelin, R. V., Digman, M. A., Medkova, M., Ananthanarayanan, B., Melowic, H. R., Rafter, J. D., and Cho, W. (2005) J. Biol. Chem. 280 19784-19793 [DOI] [PubMed] [Google Scholar]
- 38.Shen, N., Guryev, O., and Rizo, J. (2005) Biochemistry 44 1089-1096 [DOI] [PubMed] [Google Scholar]
- 39.Choi, S. H., Czifra, G., Kedei, N., Lewin, N. E., Lazar, J., Pu, Y., Marquez, V. E., and Blumberg, P. M. (2008) J. Biol. Chem. 283 10543-10549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tatsis, N., Lannigan, D. A., and Macara, I. G. (1998) J. Biol. Chem. 273 34631-34638 [DOI] [PubMed] [Google Scholar]
- 41.Wells, C. D., Fawcett, J. P., Traweger, A., Yamanaka, Y., Goudreault, M., Elder, K., Kulkarni, S., Gish, G., Virag, C., Lim, C., Colwill, K., Starostine, A., Metalnikov, P., and Pawson, T. (2006) Cell 125 535-548 [DOI] [PubMed] [Google Scholar]
- 42.Gao, Y., Dickerson, J. B., Guo, F., Zheng, J., and Zheng, Y. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 7618-7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Onesto, C., Shutes, A., Picard, V., Schweighoffer, F., and Der, C. J. (2008) Methods Enzymol. 439 111-129 [DOI] [PubMed] [Google Scholar]
- 44.Budas, G. R., Koyanagi, T., Churchill, E. N., and Mochly-Rosen, D. (2007) Biochem. Soc. Trans. 35 1021-1026 [DOI] [PubMed] [Google Scholar]
- 45.Miyake, N., Chilton, J., Psatha, M., Cheng, I., Andrews, C., Chan, W. M., Law, K., Crosier, M., Lindsay, S., Cheung, M., Allen, J., Gutowski, N. J., Ellard, S., Young, E., Iannaccone, A., Appukuttan, B., Stout, J. J., Christiansen, S., Ciccarelli, M. J., Baldi, A., Campioni, M., Zenteno, J. C., Davenport, D., Mariani, L. E., Sahin, M., Guthrie, S., and Engle, E. C. (2008) Science 321 839-843 [DOI] [PMC free article] [PubMed] [Google Scholar]