Abstract
Transmissible spongiform encephalopathies are associated with conformational conversion of the cellular prion protein, PrPC, into a proteinase K-resistant, amyloid-like aggregate, PrPSc. Although the structure of PrPSc remains enigmatic, recent studies have afforded increasingly detailed characterization of recombinant PrP amyloid. However, all previous studies were performed using amyloid fibrils formed in the presence of denaturing agents that significantly alter the folding state(s) of the precursor monomer. Here we report that PrP amyloid can also be generated under physiologically relevant conditions, where the monomeric protein is natively folded. Remarkably, site-directed spin labeling studies reveal that these fibrils possess a β-core structure nearly indistinguishable from that of amyloid grown under denaturing conditions, where the C-terminal α-helical domain of the PrP monomer undergoes major refolding to a parallel and in-register β-structure upon conversion. The structural similarity of fibrils formed under drastically different conditions strongly suggests that the common β-sheet architecture within the ∼160–220 core region represents a distinct global minimum in the PrP conversion free energy landscape. We also show that the N-terminal region of fibrillar PrP displays conformational plasticity, undergoing a reversible structural transition with an apparent pKa of ∼5.3. The C-terminal region, on the other hand, retains its β-structure over the pH range 1–11, whereas more alkaline buffer conditions denature the fibrils into constituent PrP monomers. This profile of pH-dependent stability is reminiscent of the behavior of brain-derived PrPSc, suggesting a substantial degree of structural similarity within the β-core region of these PrP aggregates.
Transmissible spongiform encephalopathies (TSEs),3 or prion diseases, are a group of fatal neurodegenerative disorders encompassing bovine spongiform encephalopathy in cattle, scrapie in sheep, chronic wasting disease in cervids, and Creutzfeldt-Jakob disease, Gerstmann-Straussler-Scheinker disease, fatal familial insomnia, and kuru in humans (1–7). In the case of human TSEs, the disease state may arise either sporadically, via inheritance, or through exposure to infectious prions. In all cases, however, neurodegeneration is intrinsically tied to conformational conversion of the normal cellular prion protein (PrPC) to a misfolded aggregate isoform dubbed PrPSc (1–7). The “protein-only” hypothesis asserts that transmission of TSEs does not require nucleic acid and that the sole infectious agent is PrPSc, which is able to self-propagate by first binding to and then inducing a conformational conversion in PrPC to the PrPSc state (1, 2). Although the molecular details of such a mechanism of disease propagation are poorly understood, extensive biochemical and genetic investigations strongly support the principle of protein-based infectivity (1–11).
Both PrPC and PrPSc appear to share identical covalent structure but are readily distinguished by various biochemical properties. Cellular PrPC is a 209-residue glycoprotein with a C-terminally attached glycosylphosphatidylinositol anchor (2–5). It consists of a flexible N-terminal region and a primarily α-helical C-terminal domain, stabilized by a disulfide bond (12–15). In contrast to PrPC, the misfolded PrPSc isoform is aggregate in nature, displays high β-sheet content, partial resistance to proteinase K (PK) digestion, and insolubility in nonionic detergents (2–7). Akin to other disorders of protein misfolding, such as Alzheimer and Parkinson disease (16, 17), these insoluble protein aggregates often are composed of amyloid-like fibrils.
To gain insight into the nature of the molecular events and specific PrP structures surrounding TSE propagation, PrPSc-like aggregates formed in vitro by the recombinant prion protein (rPrP), a form of PrP lacking any post-translational modifications, have proven an important experimental model (18–28). One such aggregate, fibrillar amyloid (rPrPAm), is a misfolded form of rPrP reported to cause a transmissible neurological disorder in transgenic mice overexpressing an N-terminally truncated form of PrPC composed of residues 89–231 (25). However, the above disorder is associated with unusually long incubation times. This, together with an apparent lack of infectivity in similar studies using wild-type animals, raises questions regarding the relationship between these “synthetic prions” and the authentic TSE agent (11, 29, 30).
Prior studies have demonstrated that such recombinant PrP amyloid fibrils display a β-sheet-rich core mapping to residues ∼160–220 (23, 26). In monomeric PrPC, this region of the protein corresponds to a major part of the globular, α-helix-rich domain, indicating that, in contrast to prevailing theoretical models (31, 32), prion protein conversion involves refolding of C-terminal α-helices to β-sheet structure. However, in all previous studies formation of rPrPAm was performed in the presence of denaturing agents that promote non-native conformational state(s) of the PrP monomer (19–23, 25–28). In this study, we have probed the spectrum of amyloid structures available to recombinant prion protein by inducing conformational conversion under more physiologically relevant conditions. We find that both wild-type rPrP as well as the familial disease-associated D178N variant undergo a conversion to amyloid fibrils upon incubation at mildly acidic pH in the absence of any denaturants, where the PrP monomer is known to retain its native α-helical fold. Remarkably, regardless of the conformational state of the precursor monomer, rPrP forms amyloid fibrils with very similar parallel and in-register β-structure spanning residues ∼160–220. Thus, this common molecular architecture likely corresponds to a distinct global free energy minimum in the folding landscape of PrP during aggregation.
EXPERIMENTAL PROCEDURES
Protein Purification—Recombinant human prion protein variants used in this study were prepared, purified, and labeled with the MTSL spin labeling reagent [1-oxy-2,2,5,5-tetramethyl-pyrolline-3-methyl]-methanethiosulfonate according to our previously described protocols (23). After purification, the proteins were stored at –80 °C in 10 mm acetate buffer, pH 4.0.
Preparation and Characterization of Amyloid Fibrils—Generation of huPrP90-231 amyloid fibrils under native-like conditions was performed by incubation of 0.5–1.0 mg/ml protein in 50 mm acetate buffer, pH 4.0, at 37 °C with continuous rotation (8 rpm). Conversion under denaturing conditions was performed in 50 mm phosphate buffer, 2 m GdnHCl, pH 7.0, with similar incubation and rotation. All reactions were monitored using a fluorometric thioflavin T assay, and atomic force microscopy was employed to verify the presence and morphology of the amyloid fibrils formed (19, 33).
EPR Spectroscopy—For EPR spectroscopy, freshly prepared amyloid fibrils were pelleted by centrifugation and washed three times in either 50 mm sodium acetate buffer, pH 4.0, or 50 mm potassium phosphate buffer, pH 7.0, to remove any monomeric protein or soluble aggregates. The washed pellet was resuspended in 30 μl of the same buffer and loaded into a capillary. EPR spectra were measured at room temperature using a Bruker EMX spectrometer fitted with the High Sensitivity resonator. Power was set to 12 milliwatts, and spectra were recorded with a scan width of 200 G. Field modulation was varied from 1 to 2 G depending on the degree of spin label immobilization and accompanied spectral broadening. Spectra displayed within individual panels were normalized to the same number of spins. For clarity, each panel was scaled to the same signal amplitude.
Fibril Denaturation Assays—For chemical denaturation assays, freshly prepared samples of amyloid fibrils were washed in 50 mm phosphate buffer, pH 7.0. The pellet was resuspended in the same buffer containing increasing concentrations of GdnHCl. After 30 min of equilibration at room temperature, the samples were pelleted, and the protein concentration of the supernatant, which reflects rPrP dissociated from the highly insoluble amyloid fibrils, was determined spectrophotometrically using a molar extinction coefficient ε280 of 21,900 m–1 cm–1. Longer incubation times were found to have no effect on the amount of rPrP contained in the supernatant, indicating that 30 min is sufficient for equilibrium to be reached. The amount of protein remaining in the pellet was determined spectrophotometrically upon complete solubilization in 7 m GdnHCl, allowing for the fraction of dissociated rPrP monomer to be calculated at a given concentration of GdnHCl.
Acid- and base-induced denaturation of amyloid fibrils was probed in a similar manner using the following buffers: pH 1, ∼0.1 m hydrochloric acid; pH 2–3, 50 mm glycine; pH 4.0–5.5, 50 mm acetate; pH 5.5–8.0, 50 mm phosphate; pH 8.5–9.5, 50 mm borate; pH 10.0–10.75, 50 mm glycine; pH 11.0–12.75, 50 mm phosphate; pH 13, ∼0.1 M sodium hydroxide. At pH above 11.0, deprotonation of tyrosine and tryptophan results in red-shifting of the absorption spectrum. Therefore, to evaluate the protein concentration in this pH regime, samples were mixed 1:1 with 50 mm acetate, pH 4.0, 4 m GdnHCl to both reduce the pH and prevent any aggregation of the denatured protein. The amount of protein remaining in the pellet was determined spectrophotometrically upon complete denaturation in sodium hydroxide, pH 13, and lowering the pH as mentioned above. The total protein concentration could then be determined, and the fraction of dissociated PrP was calculated at a given pH.
For denaturation studies involving EPR spectroscopy, spectra of fibrillar suspensions were recorded in the presence of GdnHCl or buffers of different pH. Under the most alkaline conditions (pH >12), a fraction of the nitroxide labels become labile, forming free MTSL dimers, as evidenced by additional lines at half the typical hyperfine splitting. However, the fraction of protein affected and the associated errors in our EPR amplitude measurements were very small, as evident by nearly identical denaturation profiles determined by EPR spectroscopy and the sedimentation assay.
RESULTS
Conversion of Prion Protein to Amyloid Fibrils in the Absence of Chemical Denaturants—The spontaneous growth of amyloid fibrils formed by recombinant PrP has been extensively studied in recent years, providing important clues regarding mechanistic and structural aspects of prion protein conversion to PrPSc-mimicking forms in vitro (19–28). In all prior studies, however, amyloidogenesis was induced under nonphysiological conditions, typically involving unfolding of the α-helical rPrP monomer by chemical denaturants such as GdnHCl and/or urea (19–23, 25–27). Thus, a question remains whether similar amyloid-like structures can also be formed in the absence of denaturing agents, under conditions favoring the native α-helical fold of the rPrP monomer.
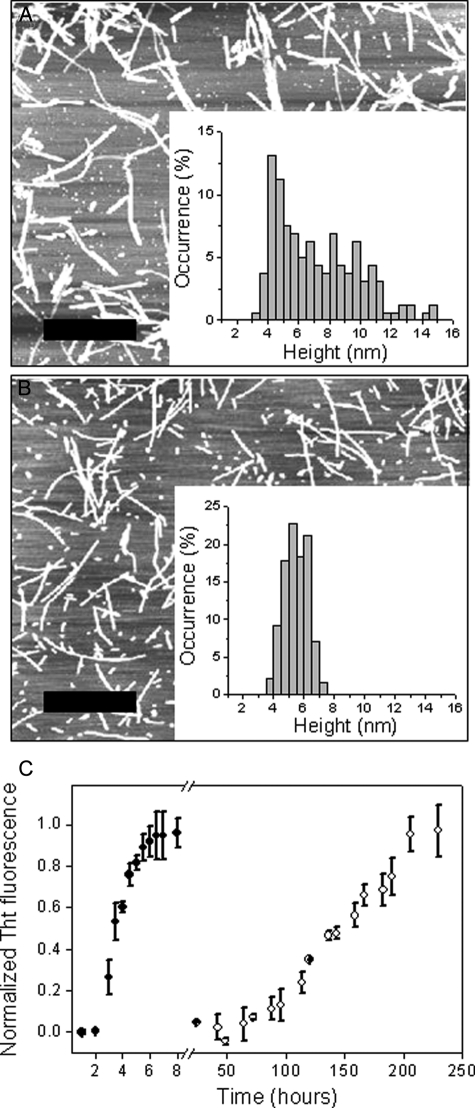
In this study, we tested the conformational conversion of rPrP at mildly acidic pH, an environment of potential relevance to the PrPC→ PrPSc transition in vivo (34, 35). Remarkably, we found that incubation of recombinant human prion protein huPrP90-231 with slow rotation (8 rpm) at 37 °C in 50 mm acetate, pH 4.0 (i.e. conditions under which the monomer maintains its native α-helical fold (12–15)), resulted in the formation of ThT-positive amyloid-like fibrils. A similar structural transition from the native-state monomer was also observed for human prion protein containing the disease-associated mutation D178N (Fig. 1), a variant previously shown to be especially prone to conformational conversion to the amyloid state (19). As compared with previously studied reactions at neutral pH in the presence of GdnHCl (19), amyloid formation under these native-like conditions follows dramatically slower kinetics (Fig. 1C). The fibrillar aggregates formed by the native rPrP precursor (denoted rPrPAmN) appear to be generally similar to those grown at pH 7 in the presence of 2 m GdnHCl (denoted rPrPAmU), although there are some morphological differences between the two fibril types. Consistent with prior studies (36), atomic force microscopy reveals that rPrPAmU fibrils are highly heterogeneous with respect to fibril heights (∼4–13 nm), presumably because of lateral association of smaller filaments (Fig. 1A). Amyloid fibrils formed under native conditions, on the other hand, are more homogeneous, showing a significantly narrower height distribution of 5.4 ± 0.8 nm (Fig. 1B). Furthermore, rPrPAmN fibrils are typically less than 1 μm in length, whereas rPrPAmU fibrils are observed to grow considerably longer (1–4 μm).
FIGURE 1.
Fibrillization of D178N huPrP90-231. Atomic force microscopy images of amyloid fibrils were formed under denaturing conditions (2 m GdnHCl in 50 mm phosphate buffer, pH 7) (A) and from the native-like precursor (50 mm acetate buffer, pH 4) (B). The scale bar corresponds to 1 μm. Insets show histograms of fibril height distribution. C, time course of D178N huPrP90-231 (30 μm) amyloid fibril formation under denaturing (•) and native-like (○) conditions. Data are expressed as a fraction of maximal ThT fluorescence. Note the break in the x axis.
Fibrils Grown from the Native-state Monomer Share a Common Amyloid Core with Those Grown under Denaturing Conditions—Although data described above clearly demonstrate that fibrillar aggregates can be formed from both the unfolded (or partially unfolded) rPrP monomer as well as native rPrP, the crucial question remains whether fibrils formed under such drastically different conditions are structurally equivalent at the molecular level. To address this issue, we employed site-directed spin labeling (SDSL) coupled with EPR spectroscopy, one of very few techniques capable of providing higher resolution insight into the structure of amyloid fibrils (23, 37–39). The main advantage of this method is that it provides estimates for the distances between nitroxide spin labels attached to engineered Cys residues (40). Thus, if a single label is present on each monomer, EPR spectra afford crucial information about the intermolecular contact sites and quaternary structure within the amyloid fibril. As in our previous study (23), the present structural analysis was performed using the highly amyloidogenic D178N huPrP90-231 variant, facilitating fibrillization of PrP mutants harboring the non-native, nitroxide-labeled cysteines required for SDSL analysis.
In our prior study, we showed that rPrPAmU fibrils formed at neutral pH in the presence of 2 m GdnHCl display highly unusual single (or nearly single)-line EPR spectra for the vast majority of sites spin-labeled within the region ∼160–220. As discussed previously (23, 38, 39), such single-line spectra occur only when multiple spin labels are in sufficient proximity to allow orbital overlap. In the context of amyloid fibrils, spectra of this type, when observed over consecutive residues, have been shown to reflect a parallel and in-register organization of β-strands, where individual monomers stack on top of one another so that same residues are aligned between the neighboring molecules (23, 38, 39). Thus, SDSL data indicate that the core region of rPrPAmU fibrils maps to residues ∼160–220, and these residues form a parallel, in-register β-structure (23).
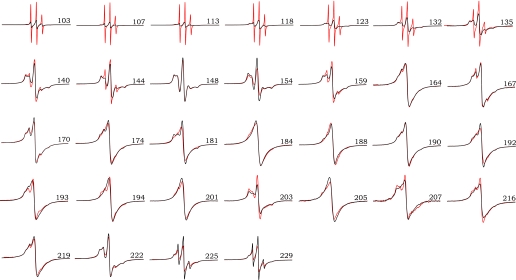
Here we examined EPR spectra for a great many of the same spin-labeled sites in natively formed rPrPAmN fibrils. Remarkably, for all sites probed within the ∼160–220 region of these fibrils, we observed single (or near-single)-line spectra nearly identical to those found in rPrPAmU fibrils (Fig. 2). Thus, the overall molecular organization of both fibril types appears to be very similar. For some β-core residues, however, minor site-specific spectral differences were observed between rPrPAmN and rPrPAmU fibrils (e.g. position 193 and 216; Fig. 2). These differences persisted upon elimination of spin-spin interactions by growing fibrils using a 1:4 mixture of labeled and unlabeled protein (supplemental Fig. 1), indicating nonequivalence of the microenvironment of these sites and, thus, some degree of structural divergence. However, given the high degree of order required to produce single-line EPR spectra, both fibril types clearly display very similar amyloid core regions encompassing residues ∼160–220, where individual monomers form single molecule layers that stack one on top of another with parallel and in-register organization of β-strands.
FIGURE 2.
EPR spectra for D178N huPrP90-231 fibrils formed under native-like and denaturing conditions. The sites of spin labeling are indicated by the residue number for each pair of spectra. EPR signals for amyloid fibrils formed under denaturing conditions (rPrPAmU) are shown in black, and fibrils grown under native-like conditions (rPrPAmN) are shown in red. Scan width is 200 G, and spectral pairs for each position are scaled to the same number of spins.
In contrast to spin-labeled residues within the ∼160–220 segment, more pronounced differences between rPrPAmN and rPrPAmU fibrils were observed for spin-labeled sites outside this core region, especially for sites N-terminal to residue ∼140 (Fig. 2). In rPrPAmN fibrils, these N-terminal sites display markedly narrower line widths and increased spectral amplitude when compared with rPrPAmU fibrils. These spectral differences indicate considerably higher nitroxide mobility, most likely because of higher segmental flexibility within this region of rPrPAmN fibrils.
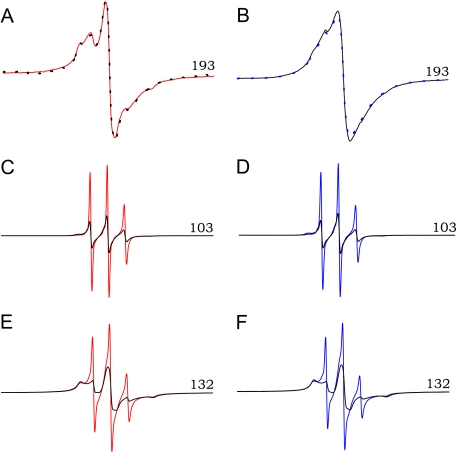
Region-selective Differences in pH Sensitivity of PrP Amyloid Fibrils—To further explore the nature of region-selective structural differences between rPrPAmN and rPrPAmU fibrils, we probed the effect of pH on EPR spectra. Spin-labeled fibrils were grown under both native-like and denaturing conditions, and EPR measurements of rPrPAmN fibrils (samples grown in acetate buffer, pH 4) and rPrPAmU fibrils (samples grown in phosphate buffer, pH 7, containing 2 m GdnHCl and equilibrated in the same buffer without denaturant) were taken. Following these initial measurements, individual samples were equilibrated at the opposing pH buffer (i.e. rPrPAmN at pH 7 and rPrPAmU fibrils at pH 4), and EPR signals were recorded again. For all sites probed within the ∼160–220 core region, spectra of both fibril types were insensitive to this change in pH (Fig. 3, A and B). Thus, once rPrPAmN and rPrPAmU fibrils have been formed under different buffer conditions, the core regions of these fibrils appear to be conformationally “locked,” with minor site-specific differences being irreversible.
FIGURE 3.
Region-specific effects of pH on the structure of rPrP amyloid. A, EPR spectrum of rPrPAmN fibrils (grown in 50 mm acetate buffer, pH 4) labeled at a representative β-core position (residue 193) is shown in red. Fibrils were then re-equilibrated for 30 min in 50 mm phosphate, pH 7, and the EPR signal was measured again (black dotted line). B, an analogous procedure was applied to residue 193-labeled rPrPAmU fibrils (grown in 50 mm phosphate buffer, pH 7, plus 2 m GdnHCl) equilibrated in 50 mm phosphate, pH 7 (black line), and subsequently re-equilibrated in 50 mm acetate, pH 4 (blue dotted line). C and E, EPR spectra of rPrPAmN fibrils labeled at the indicated N-terminal positions. Fibrils were grown and equilibrated at pH 4 (red line) and subsequently re-equilibrated at pH 7 (black line). D and F, EPR spectra of rPrPAmU fibrils labeled at the same N-terminal positions and equilibrated at pH 7 (black line) and subsequently re-equilibrated at pH 4 (blue line). All experiments were performed using D178N huPrP90-231.
In stark contrast, EPR spectra for sites labeled upstream of the β-core region are highly pH-sensitive, as illustrated in Fig. 3 for spin labels at two representative positions, 132 and 103. For these proteins labeled at non-core residues, re-equilibration of rPrPAmN fibrils in pH 7 buffer resulted in spectral broadening and a decrease in signal amplitude, with EPR spectra for these sites losing characteristics of rPrPAmN fibrils and acquiring those of rPrPAmU fibrils (Fig. 3, C and E). A similar phenomenon was observed for rPrPAmU fibrils, where re-equilibration at pH 4 resulted in increased nitroxide mobility, with spectra becoming indistinguishable from those of rPrPAmN fibrils (Fig. 3, D and F). This effect, also observed for all other sites N-terminal to residue 160, was fully reversible upon repeated cycles of buffer exchange, indicating that, in contrast to the core region, the structure in the N-terminal part of rPrP90-231 amyloid can be reversibly modulated by changes in the pH of the aqueous environment.
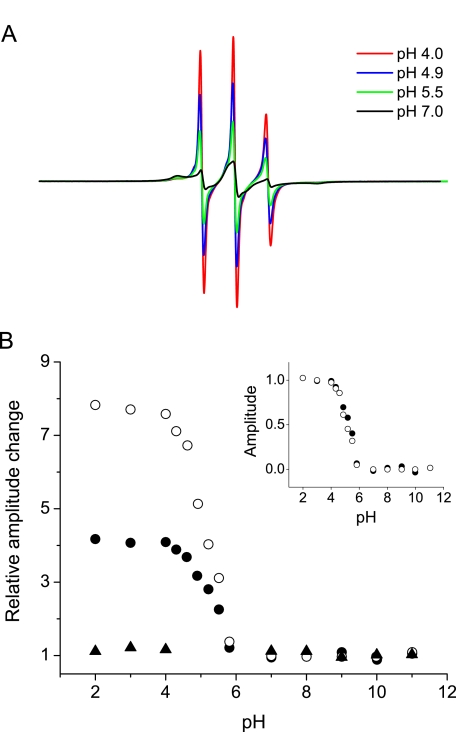
To further characterize this pH-dependent structural transition, a full pH titration was performed using fibrils spin-labeled at selected positions. Fig. 4A shows EPR signals arising from such a titration for rPrPAmU fibrils labeled at position 113. As the sample is equilibrated in less acidic solution, spectra become progressively broader with the accompanied reduction in amplitude. A plot of spectral amplitude against pH for two representative N-terminal sites (113 and 103) reveals a relatively sharp transition with an apparent pKa of ∼5.3 (Fig. 4B). In contrast, for labeled sites within the ∼160–220 region, no changes in EPR signal were observed over the entire pH range between 2 and 11.
FIGURE 4.
pH-induced structural transition in the N-terminal region of rPrP amyloid. A, EPR spectra from a representative pH titration for rPrPAmU fibrils labeled at position 113. Amyloid fibrils were initially equilibrated in 50 mm phosphate, pH 7 (black line), and then resuspended in decreasing pH buffers. B, plot of EPR signal central line amplitude versus pH for rPrPAmU fibrils labeled at positions 103 (•), 113 (○), and 192 (▴). Data are expressed as the relative increase in signal amplitude relative to that at pH 7. Inset, titration data for residues 103 and 113 replotted as the fractional change in signal amplitude relative to spectra obtained at pH 7. All experiments were performed using D178N huPrP90-231.
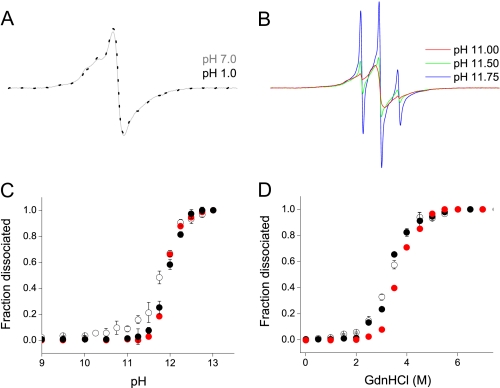
Denaturation Profiles of PrP Amyloid Fibrils—Intrigued by the observation regarding the apparent insensitivity of the β-core structure to pH, we extended our pH-titration experiments to highly acidic and alkaline conditions. Remarkably, even in the presence of ∼0.1 n HCl (pH 1), spectra of labeled sites within the ∼160–220 segment maintained their characteristic single-line character, clearly demonstrating that the molecular architecture of the core region is preserved (Fig. 5A). Only under highly alkaline conditions (pH >11) do these single-line spectra became gradually replaced by narrow three-line signals characteristic of an unfolded PrP monomer (Fig. 5B), indicating that such conditions induce dissociation of amyloid fibrils into constituent monomers. The apparent pK of this reaction, as determined by measuring pH-dependent spectral amplitude, was ∼12, both for rPrPAmU and rPrPAmN fibrils. Very similar denaturation profiles were obtained using a simple centrifugation assay based on the dramatically different sedimentation properties of fibrillar aggregates and monomeric protein (Fig. 5C; see under “Experimental Procedures”), confirming the interpretation of pH-induced changes in EPR spectra for the core residues.
FIGURE 5.
Stability of rPrP amyloid against denaturation. A, EPR spectra for rPrPAmU fibrils labeled at position 192, equilibrated at pH 7 (gray line) and pH 1 (black dotted line). B, representative EPR spectra for rPrPAmU labeled at position 192 at increasingly alkaline pH showing increased proportion of a three line component corresponding to dissociated monomeric protein. C, alkaline pH-induced denaturation of rPrP amyloid fibrils. Fraction of dissociated monomer is shown as assessed by plotting fractional change in EPR signal amplitude for rPrPAmU fibrils labeled at position 192 (red circles) and by direct measurements using a sedimentation assay (rPrPAmN, ○; rPrPAmU, •). D, GdnHCl denaturation curves as assessed by plotting fractional change in EPR signal amplitude and by direct measurements using a sedimentation assay. Symbols are same as in C. All experiments were performed using D178N huPrP90-231.
Finally, the stability of amyloid fibrils grown under different conditions was probed using GdnHCl as a denaturant. As with pH-induced fibril dissociation, very similar denaturation profiles were obtained for rPrPAmN and rPrPAmU fibrils, both when probed by the sedimentation assay as well as by following changes in amplitude of EPR spectra for spin-labeled sites within the core region (Fig. 5D). Altogether, these results indicate that both fibril types display remarkably similar stability and that cooperative unfolding of the common amyloid core is intimately associated with fibril dissociation into PrP monomers.
DISCUSSION
Although the precise molecular nature of the infectious agent responsible for the TSEs is still shrouded in mystery, a wealth of data indicates that the pathogenic process in these diseases is intimately associated with conformational conversion of monomeric PrPC to proteinase K-resistant, amyloid-like PrPSc aggregates (1–7). Biophysical and structural aspects of this conversion reaction remain poorly understood, largely because of fundamental difficulties of studies with brain-derived material. In recent years, however, significant advances have been made in elucidating the mechanism of conformational transitions of the recombinant prion protein (18–21, 24, 27, 28), including structural characterization of PrPSc-mimicking rPrP amyloid fibrils (22, 23, 26, 27) as well as attempts to generate infectious material from purely synthetic components (25).
In contrast to frequently discussed theoretical models (31, 32), available experimental data clearly indicate that formation of rPrP amyloid involves major refolding of the native C-terminal α-helices to β-sheet structure (22, 23, 26, 27). However, to date, recombinant PrP amyloid has only been observed to form under distinctly nonphysiological conditions, in the presence of chemical denaturants (19–23, 25–28). Because denaturants themselves typically induce unfolding of the α-helical PrP monomer, the question remains whether (and to what extent) the structures formed in such an environment are representative of PrP fibrillar aggregates formed under native-like conditions, without such exogenous factors.
Here we report that, in addition to previously established conversion pathways under denaturing conditions, ThT-positive aggregates with classical amyloid morphology can be formed by rPrP90-231 in the absence of any denaturing agents. This N-terminally truncated fragment, which forms fibrillar aggregates similar to those of full-length rPrP (21), represents the entire PK-resistant core of brain PrPSc, is sufficient to propagate the TSE disease state, and contains most point mutations known to be associated with hereditary prion disorders (2, 4). The mildly acidic conditions employed in our study are of physiological relevance, because small acidified cellular compartments such as lysosomes likely represent one of the sites of PrPSc replication in vivo (34, 35). Importantly, under these experimental conditions the PrP monomer is known to be fully folded, with native α-helical structure of the C-terminal domain, as indicated by NMR structural data at acidic and neutral pH (12–15). Thus, this study clearly demonstrates that neither complete nor partial unfolding of the monomeric precursor is a prerequisite for conversion of prion protein to the amyloid state.
Although the finding regarding amyloid fibril formation under native-like buffer conditions has important implications for future studies on the mechanism of prion protein conversion, the main focus of this study was on the structure of the resultant fibrils. We find that despite some morphological differences, the molecular level architecture of amyloid fibrils formed under native-like conditions is remarkably similar to that observed previously for amyloid fibrils grown in the presence of 2 m GdnHCl (i.e. where ∼50% of the precursor monomer is fully unfolded (41)). In both cases, the conversion involves refolding of the C-terminal α-helices, and amyloid fibrils possess an essentially identical architecture within the ∼160–220 core region, where the major folding motif is a parallel and in-register β-structure of stacked monomeric units. The observation that incubation of rPrP under conditions with dramatically different initial conformational states of the precursor monomer leads to formation of amyloid fibrils with a common β-core strongly indicates that this common architecture is fully encoded in the amino acid sequence of PrP, and the resultant fibrillar structure reflects a distinct global minimum in the prion protein aggregation free energy landscape. The present finding is somewhat reminiscent of the phenomenon recently reported for β2-microglobulin, where in vitro conversion reactions under both native and denaturing conditions result in fibrillar aggregates with apparently similar β-core regions, as indicated by low resolution infrared spectroscopic data (42). An important distinction, however, is that although the β2-microglobulin monomer is a primarily β-sheet protein, conversion of PrP requires complete refolding of the α-helical globular domain to the β-structure.
Although our data clearly underscore the energetic predisposition for residues ∼160–220 of PrP to adopt a β-sheet structure upon the conversion to amyloid, relative to brain-derived PrPSc, recombinant PrP fibrils are characterized by a significantly smaller PK-resistant core and greatly reduced infectivity (22, 25, 26). Recent studies suggest that neither the glycosylphosphatidylinositol anchor nor glycosylation of PrPC, post-translational modifications not present in recombinant PrP, is required for the infectivity of the prion agent (43–45). Thus, specific backbone conformational features of PrP aggregates likely govern both their PK resistance and infectivity. Although in rPrPAm the most PK-resistant fragments display N-terminal cleavage sites at residues 152/153 and 162 (22), the classical PK-resistant fragment of brain-derived PrPSc is considerably larger, extending upstream to residue ∼90 (2, 6). It is well known that variability at this N-terminal PK cleavage site is closely associated with phenotypically distinct TSE strains (2). Thus, conformational plasticity within this region appears to underlie the interplay between specific disease-state neuropathology and discrete PrP aggregate structures. Remarkably, we find that the most N-terminal regions of synthetic rPrPAm also display structural flexibility, undergoing a reversible, pH-dependent structural transition (apparent pKa ∼5.3), whereas the rPrPAm β-core remains highly stable and insensitive to environmental pH. Given the present data showing a clear predisposition of the C-terminal residues ∼160–220 to adopt parallel and in-register β-structure, we propose that residues ∼90–160 represent the most likely location for structural divergence between rPrPAm and PrPSc. Intriguingly, recent studies by Atarashi et al. (46) indicate that fibrillar rPrP aggregates with an increased PK resistance within the 90–160 region can be obtained when the conversion reaction is seeded with brain-derived PrPSc.
What could give rise to specific N-terminal structures within neuronal PrPSc aggregates? A host of cellular factors have been proposed to affect prion protein conversion in vivo (2, 3, 47). Although it seems unlikely that such co-factors could “drive” the conversion toward fundamentally different, less energetically favorable structures (not involving major refolding of the α-helical domain), we cannot entirely discount this possibility. A more likely scenario, however, is that these putative cellular agents act in concert with the spontaneous conversion of the PrP globular domain to β-structure, and modulate the conformation of the N-terminal regions, resulting in an aggregate with PrPSc-like PK resistance and infectivity. A recent study by Deleault et al. (10) suggests the critical importance of one such potential co-factor. These authors have shown that transmissible PrPSc-like aggregates can be formed de novo from purified PrPC (contaminated with co-purified lipids) in the presence of exogenous polyadenine RNA. The highly basic N terminus of PrP would be expected to preferentially interact with just such a polyanionic co-factor, and thus, this interaction could result in some PrPSc-specific N-terminal conformation. Polyanions such as nucleic acids and glycosaminoglycans have long been implicated in TSE pathogenesis (3, 24, 47); if such compounds can be shown experimentally to induce the rPrP N terminus to adopt an alternative, highly infectious aggregate fold, new therapeutic compounds could be envisioned that inhibit these putative cellular agents and promote formation of the relatively noninfectious rPrPAm-like aggregates.
Apart from providing structural insight, the present data reveal an intriguing resistance of rPrPAm to extremes of environmental pH. We find that equilibration of rPrPAm at pH as low as 1 does not result in any structural changes within the β-core, and fibrils can be dissociated into constituent monomers only under highly alkaline conditions. Remarkably, these pH effects are reminiscent of the behavior of brain-derived PrPSc. Indeed, a common means of prion inactivation and associated loss of PK resistance of PrPSc involves incubation in sodium hydroxide (48–50), at a pH similar to that found herein to induce denaturation of rPrPAm into PK-sensitive monomeric units. Furthermore, PrPSc is known to retain infectivity under the same highly acidic conditions where rPrPAm is here shown to maintain its fibrillar β-structure (51). Thus, although further work is needed to unravel the precise nature of structural features responsible for prion protein infectivity, the present findings suggest a substantial degree of structural commonality between recombinant PrP amyloid and the infectious PrPSc isoform.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants NS 38604 and AG 14359. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: TSE, transmissible spongiform encephalopathies; PrPC, cellular prion protein; huPrP, human prion protein; PrPSc, disease-associated, proteinase K-resistant prion protein isoform; rPrP, recombinant prion protein; rPrPAm, recombinant prion protein amyloid; rPrPAmN, recombinant prion protein amyloid formed under native conditions; rPrPAmU, recombinant prion protein amyloid formed under denaturing conditions; PK, proteinase K; GdnHCl, guanidine hydrochloride; ThT, thioflavin T; EPR, electron paramagnetic resonance; SDSL, site-directed spin labeling.
References
- 1.Prusiner, S. B. (1982) Science 216 136–144 [DOI] [PubMed] [Google Scholar]
- 2.Prusiner, S. B. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caughey, B., and Baron, G. S. (2006) Nature 443 803–810 [DOI] [PubMed] [Google Scholar]
- 4.Collinge, J. (2001) Annu. Rev. Neurosci. 24 519–550 [DOI] [PubMed] [Google Scholar]
- 5.Aguzzi, A., and Polymenidou, M. (2004) Cell 116 313–327 [DOI] [PubMed] [Google Scholar]
- 6.Weissmann, C. (2004) Nat. Rev. Microbiol. 2 861–871 [DOI] [PubMed] [Google Scholar]
- 7.Caughey, B., and Chesebro, B. (2001) Adv. Virus Res. 56 277–311 [DOI] [PubMed] [Google Scholar]
- 8.Surewicz, W. K., Jones, E. M., and Apetri, A. C. (2006) Acc. Chem. Res. 39 654–662 [DOI] [PubMed] [Google Scholar]
- 9.Castilla, J., Saa, P., Hetz, C., and Soto, C. (2005) Cell 121 195–206 [DOI] [PubMed] [Google Scholar]
- 10.Deleault, N. R., Harris, B. T., Rees, J. R., and Supattapone, S. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 9741–9746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collinge, J., and Clarke, A. R. (2007) Science 318 930–936 [DOI] [PubMed] [Google Scholar]
- 12.Riek, R., Hornemann, S., Wider, G., Billeter, M., Glockshuber, R., and Wuthrich, K. (1996) Nature 382 180–182 [DOI] [PubMed] [Google Scholar]
- 13.Donne, D. G., Viles, J. H., Groth, D., Mehlhorn, I., James, T. L., Cohen, F. E., Prusiner, S. B., Wright, P. E., and Dyson, H. J. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 13452–13457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zahn, R., Liu, A., Luhrs, T., Riek, R., von Schroetter, C., Lopez Garcia, F., Billeter, M., Calzolai, L., Wider, G., and Wuthrich, K. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calzolai, L., and Zahn, R. (2003) J. Biol. Chem. 278 35592–35596 [DOI] [PubMed] [Google Scholar]
- 16.Caughey, B., and Lansbury, P. T. (2003) Annu. Rev. Neurosci. 26 267–298 [DOI] [PubMed] [Google Scholar]
- 17.Chiti, F., and Dobson, C. M. (2006) Annu. Rev. Biochem. 75 333–366 [DOI] [PubMed] [Google Scholar]
- 18.Apetri, A. C., Surewicz, K., and Surewicz, W. K. (2004) J. Biol. Chem. 279 18008–18014 [DOI] [PubMed] [Google Scholar]
- 19.Apetri, A. C., Vanik, D. L., and Surewicz, W. K. (2005) Biochemistry 44 15880–15888 [DOI] [PubMed] [Google Scholar]
- 20.Baskakov, I. V., Legname, G., Baldwin, M. A., Prusiner, S. B., and Cohen, F. E. (2002) J. Biol. Chem. 277 21140–21148 [DOI] [PubMed] [Google Scholar]
- 21.Bocharova, O. V., Breydo, L., Parfenov, A. S., Salnikov, V. V., and Baskakov, I. V. (2005) J. Mol. Biol. 346 645–659 [DOI] [PubMed] [Google Scholar]
- 22.Bocharova, O. V., Breydo, L., Salnikov, V. V., Gill, A. C., and Baskakov, I. V. (2005) Protein Sci. 14 1222–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cobb, N. J., Sonnichsen, F. D., McHaourab, H., and Surewicz, W. K. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 18946–18951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cordeiro, Y., Machado, F., Juliano, L., Juliano, M. A., Brentani, R. R., Foguel, D., and Silva, J. L. (2001) J. Biol. Chem. 276 49400–49409 [DOI] [PubMed] [Google Scholar]
- 25.Legname, G., Baskakov, I. V., Nguyen, H. O., Riesner, D., Cohen, F. E., DeArmond, S. J., and Prusiner, S. B. (2004) Science 305 673–676 [DOI] [PubMed] [Google Scholar]
- 26.Lu, X., Wintrode, P. L., and Surewicz, W. K. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 1510–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun, Y., Breydo, L., Makarava, N., Yang, Q., Bocharova, O. V., and Baskakov, I. V. (2007) J. Biol. Chem. 282 9090–9097 [DOI] [PubMed] [Google Scholar]
- 28.Stohr, J., Weinmann, N., Wille, H., Kaimann, T., Nagel-Steger, L., Birkmann, E., Panza, G., Prusiner, S. B., Eigen, M., and Riesner, D. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 2409–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caughey, B., Caughey, W. S., Kocisko, D. A., Lee, K. S., Silveira, J. R., and Morrey, J. D. (2006) Acc. Chem. Res. 39 646–653 [DOI] [PubMed] [Google Scholar]
- 30.Weissmann, C. (2005) Cell 122 165–168 [DOI] [PubMed] [Google Scholar]
- 31.Govaerts, C., Wille, H., Prusiner, S. B., and Cohen, F. E. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 8342–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeMarco, M. L., and Daggett, V. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2293–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones, E. M., and Surewicz, W. K. (2005) Cell 121 63–72 [DOI] [PubMed] [Google Scholar]
- 34.Caughey, B., Raymond, G. J., Ernst, D., and Race, R. E. (1991) J. Virol. 65 6597–6603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borchelt, D. R., Taraboulos, A., and Prusiner, S. B. (1992) J. Biol. Chem. 267 16188–16199 [PubMed] [Google Scholar]
- 36.Anderson, M., Bocharova, O. V., Makarava, N., Breydo, L., Salnikov, V. V., and Baskakov, I. V. (2006) J. Mol. Biol. 358 580–596 [DOI] [PubMed] [Google Scholar]
- 37.Torok, M., Milton, S., Kayed, R., Wu, P., McIntire, T., Glabe, C. G., and Langen, R. (2002) J. Biol. Chem. 277 40810–40815 [DOI] [PubMed] [Google Scholar]
- 38.Margittai, M., and Langen, R. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10278–10283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen, M., Margittai, M., Chen, J., and Langen, R. (2007) J. Biol. Chem. 282 24970–24979 [DOI] [PubMed] [Google Scholar]
- 40.Altenbach, C., Oh, K. J., Trabanino, R. J., Hideg, K., and Hubbell, W. L. (2001) Biochemistry 40 15471–15482 [DOI] [PubMed] [Google Scholar]
- 41.Swietnicki, W., Petersen, R., Gambetti, P., and Surewicz, W. K. (1997) J. Biol. Chem. 272 27517–27520 [DOI] [PubMed] [Google Scholar]
- 42.Jahn, T. R., Tennent, G. A., and Radford, S. E. (2008) J. Biol. Chem. 283 17279–17286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chesebro, B., Trifilo, M., Race, R., Meade-White, K., Teng, C., LaCasse, R., Raymond, L., Favara, C., Baron, G., Priola, S., Caughey, B., Masliah, E., and Oldstone, M. (2005) Science 308 1435–1439 [DOI] [PubMed] [Google Scholar]
- 44.Taraboulos, A., Rogers, M., Borchelt, D. R., McKinley, M. P., Scott, M., Serban, D., and Prusiner, S. B. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 8262–8266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korth, C., Kaneko, K., and Prusiner, S. B. (2000) J. Gen. Virol. 81 2555–2563 [DOI] [PubMed] [Google Scholar]
- 46.Atarashi, R., Moore, R. A., Sim, V. L., Hughson, A. G., Dorward, D. W., Onwubiko, H. A., Priola, S. A., and Caughey, B. (2007) Nat. Meth. 4 645–650 [DOI] [PubMed] [Google Scholar]
- 47.Silva, J. L., Lima, L. M., Foguel, D., and Cordeiro, Y. (2008) Trends Biochem. Sci. 33 132–140 [DOI] [PubMed] [Google Scholar]
- 48.Prusiner, S. B., Groth, D. F., McKinley, M. P., Cochran, S. P., Bowman, K. A., and Kasper, K. C. (1981) Proc. Natl. Acad. Sci. U. S. A. 78 4606–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kasermann, F., and Kempf, C. (2003) J. Gen. Virol. 84 3173–3176 [DOI] [PubMed] [Google Scholar]
- 50.Suzuki, S. Y., Takata, M., Teruya, K., Shinagawa, M., Mohri, S., and Yokoyama, T. (2008) J. Vet. Med. Sci. 70 159–165 [DOI] [PubMed] [Google Scholar]
- 51.Brown, P., Rohwer, R. G., and Gajdusek, D. C. (1986) J. Infect. Dis. 153 1145–1148 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.