Abstract
The yeast Pdr5 multidrug transporter is an important member of the ATP-binding cassette superfamily of proteins. We describe a novel mutation (S558Y) in transmembrane helix 2 of Pdr5 identified in a screen for suppressors that eliminated Pdr5-mediated cycloheximide hyper-resistance. Nucleotides as well as transport substrates bind to the mutant Pdr5 with an affinity comparable with that for wild-type Pdr5. Wild-type and mutant Pdr5s show ATPase activity with comparable Km(ATP) values. Nonetheless, drug sensitivity is equivalent in the mutant pdr5 and the pdr5 deletion. Finally, the transport substrate clotrimazole, which is a noncompetitive inhibitor of Pdr5 ATPase activity, has a minimal effect on ATP hydrolysis by the S558Y mutant. These results suggest that the drug sensitivity of the mutant Pdr5 is attributable to the uncoupling of NTPase activity and transport. We screened for amino acid alterations in the nucleotide-binding domains that would reverse the phenotypic effect of the S558Y mutation. A second-site mutation, N242K, located between the Walker A and signature motifs of the N-terminal nucleotide-binding domain, restores significant function. This region of the nucleotide-binding domain interacts with the transmembrane domains via the intracellular loop-1 (which connects transmembrane helices 2 and 3) in the crystal structure of Sav1866, a bacterial ATP-binding cassette drug transporter. These structural studies are supported by biochemical and genetic evidence presented here that interactions between transmembrane helix 2 and the nucleotide-binding domain, via the intracellular loop-1, may define at least part of the translocation pathway for coupling ATP hydrolysis to drug transport.
Multidrug transporters, including members of the ATP-binding cassette (ABC)2 family, show unusual flexibility toward their substrate cargo. They efflux structurally diverse xenobiotic compounds and confer broad-spectrum hyperresistance when they are overexpressed, a property that impedes chemotherapeutic treatment of pathogens and cancer.
Several fungi, including the clinically relevant human pathogenic species Candida albicans and Cryptococcus neoformans, contain major multidrug transporters that are close homologues to the Saccharomyces cerevisiae transporter Pdr5 (1). It is well established that many clinical isolates of C. albicans overproduce the Pdr5 homologue Cdr1 (2). Multidrug-resistant fungi are an increasing problem in the treatment of immunocompromised patients with AIDS and cancer (3).
The Pdr5 subfamily of multidrug transporters shows significant differences in molecular architecture from their mammalian counterparts such as P-glycoprotein (P-gp) or Mrp1. The nucleotide-binding domains (NBDs) of these important fungal transporters precede the transmembrane domains (TMDs) and thus their orientation is the reverse of P-gps and Mrp1s. Furthermore, the Walker A, B, and signature motifs are degenerate when compared with nonfungal ABC counterparts. For example, a cysteine replaces a lysine in Walker A (Cys-199) of the N-terminal NBD (NBD1) that is essential in other ABC transporters (1).
Not surprisingly, the ATPase activities of Pdr5 and C. albicans Cdr1p appear to be similar (4–7). For example, both have high basal (unstimulated) levels. Recently, we demonstrated that Pdr5 also hydrolyzes GTP at physiological concentrations and that this activity is considerably more resistant to some substrates, such as clotrimazole (clo), than is its ATPase activity. Furthermore, GTP fuels Pdr5-mediated chloramphenicol efflux in purified vesicles (5). These results suggest that Pdr5 has great flexibility not only in the drug cargo that it transports but also in its efflux energy source.
Considerable progress has been made in recent decades in understanding the domain organization of ABC transporters and the mechanism of the transport cycle (8). A typical ABC protein consists of four domains, two TMDs and two NBDs. The TMDs contain the drug-binding sites, which show little sequence similarity in different ABC transporters. By contrast, the NBDs exhibit very high sequence similarity.
The activity of an ABC transporter hinges on the effective coupling of the ATP catalytic cycle, which occurs at the NBDs, and the drug-transport cycle, which occurs at the TMDs. Mutational analyses, labeling with the photoaffinity drug or nucleotide analogues, and cross-linking with the thiol-reactive substrate derivatives have yielded a wealth of information about interactions at the drug-binding domains as well as about the role of conserved residues in the NBDs. Biochemical and genetic studies suggested the existence of a transmission interface that couples the energy of ATP binding and/or hydrolysis to conformational changes at the TMDs (9). These studies have taken indirect approaches, however, such as the determining the effect of drugs on ATP hydrolysis.
The recent elucidation of the x-ray structure of Sav1866, a bacterial ABC transporter (10), has permitted the delineation of such a transmission interface in more precise detail. A novel feature of this structure is that both NBDs contact the TMDs. These contacts occur via intracellular loops (ICLs) that link the transmembrane helices. The organization of the structure of this domain suggests a plausible explanation for the mechanical coupling between the NBDs and TMDs of ABC transporters. In addition, recent studies with P-gp and Yor1 used site-directed mutagenesis and cross-linking agents to demonstrate the proximity of ICL2 and the Q-loop (11, 12). These studies show or imply a physical interaction, but none establish that these regions are involved in communication between the TMDs and NBDs. Demonstrating the functional relevance of such interactions remains a challenge.
Here we describe the behavior of an S558Y mutation in transmembrane helix 2 (TMH2) that retains significant ATPase activity and drug-binding capability but nevertheless tests as phenotypically null for drug resistance and transport. Furthermore, an allosteric inhibitory signal mediated by the Pdr5-specific transport substrate clotrimazole is greatly diminished. These deficiencies are significantly alleviated by a suppressor mutation that lies near the Q-loop of NBD1. This is a particularly important observation because recent work suggests that the noncanonical NBD1 of Pdr5 may not play a catalytic role in ATP hydrolysis (13).
Our results are thus consistent with the x-ray crystallographic structure of Sav1866 and suggest that even evolutionarily distant ABC transporters have a similar coupling interface. Moreover, our strategy of using suppressor mutations to identify the interacting residues in the TMDs and NBDs has obvious advantages over the use of site-directed mutagenesis. This powerful approach can also be applied to any mammalian transporter that is expressed in yeast and has a readily observable phenotype.
EXPERIMENTAL PROCEDURES
Strains and Genetic Transformation—The strains used in this study are listed in Table 1. All are isogenic to each other and derived from AD1–7, which lacks all PM ABC transporters but contains a PDR1–3 allele that results in overexpression of PDR5 when a plasmid containing this gene is integrated into its chromosomal location. The construction of JG2000 and JG2001 was described previously (5).
TABLE 1.
Yeast strains used in this study All the strains are isogenic derivatives of AD1–7, with the exception of JG365-3B.
| Strain | Relevant genotype | Source or Ref. |
|---|---|---|
| JG365-3B | Mat a PDR5 PDR1-3 ura3 leu2 | 35 |
| AD1–7 | MATα PDR1-3 ura3 his1 yor1 pdr5 snq2 pdr10 ycf1 pdr11 pdr3 | 36; ura3 derivative made by J. Golin |
| JG2000 | AD1–7 + pSS607 | 5 |
| JG2001 | ura3, PDR5 5-FOA derivative of JG2000 | 5 |
| JG2003 | JG2001 + pS558Y | This study |
| JG2004 | JG2001 + pSS607 | This study |
| JG2005 | AD1–7 + pS558Y | This study |
| JG2009 | ura3, S558Y derivative of JG2003 | This study |
| JG2010 | JG2009 + pS558Y | This study |
| R-1 | Δpdr5::KANMX4 replaces PDR5 in JG2001 | This study |
| JG2011 | R-1 + pS558Y | This study |
| JG2012 | R-1 + pS558A | This study |
| JG2015 | R-1 + pSS607 (wild-type control) | This study |
| JG2016 | R-1 + pN242K | This study |
| JG2020 | clo-3-0 + pS558Y, N242K | This study |
| JG2021 | R-1 + pS558Y, N242K | This study |
We also constructed several new AD1–7 derivatives. The first series of strains carry two copies of PDR5 either as wild type (WT; JG2004) or mutant (JG2010). These strains produce significantly more Pdr5 than does the popular single-copy strain AD124567 and are thus invaluable for biochemical studies. JG2004 was created by transforming JG2001 with pSS607, the plasmid containing the WT gene PDR5.
The JG2009 strain contains a single copy of the S558Y allele and was used to create JG2010. A PDR5-bearing strain (JG2001) was transformed with S558Y to create JG2003. Treatment with 5-fluoorotic acid (5-FOA) (14) yielded Ura- segregants (9/22) that were multidrug-sensitive and thus contained S558Y. We retained one of these and designated it JG2009. Transformation of this strain with an S558Y plasmid resulted in JG2010, which contains a tandem duplication of the mutant allele and is the mutant equivalent of JG2004.
We designed another strain, R-1, to obviate a basic problem found in AD1–7. This strain carries only a partial deletion of PDR5 (from -187 to 1437 (15)). When we introduce into AD1–7 a pdr5 mutation located in the coding sequence beyond approximately the first 445 residues, a WT gene is sometimes reconstituted by homologous recombination. By contrast, the entire coding region of PDR5 is replaced in R-1 by a KANMX4 (geneticin-resistant) cassette that we PCR-amplified from BY2909 as described previously (16). This strain is otherwise phenotypically indistinguishable from AD1–7.
All of these new isogenic strains are available from the corresponding author. We used a Gietz transformation kit (Medicorp, Montreal, Quebec, Canada) to introduce plasmids and PCR-generated products into yeast.
Plasmids and Site-directed Mutagenesis—We created all site-directed mutants from the plasmid pSS607, which was described previously (5). To obtain pS558Y, pS558A, and pN242K, we carried out PCR-based site-directed mutagenesis with the Quick Change kit (Stratagene, La Jolla, CA). PCR primers carrying the desired base pair substitution were prepared, and PAGE was purified by Operon Biotechnologies (Huntsville, AL). The resulting mutants were confirmed by DNA sequencing the entire PDR5 insert. Sequencing was performed by Retrogen (San Diego, CA). We constructed the 14 sequencing primers so that the resulting sequencing data would contain substantial overlapping segments, thus ensuring accuracy.
Chemicals and Media—All chemicals were purchased from Sigma. We dissolved cycloheximide (cyh) in sterile water and all other compounds in dimethyl sulfoxide. We purchased 8-azido-[α-32P]ATP (15–20 Ci/mmol) from Affinity Labeling Technologies, Inc. (Lexington, KY). We obtained [125I]iodoarylazidoprazosin ([125I]IAAP), 2,200 Ci/mmol, from PerkinElmer Life Sciences.
We cultured cells in yeast extract, peptone, glucose (YPD) medium. Cyh, tritylimidazole, and clo were added to YPD after autoclaving. Tetrabutyltin was added to yeast extract, peptone, glycerol (YPG) medium. After sterilization, 5-FOA (1 g/liter) was added to synthetic dextrose complete medium.
Determining IC50 Values in Liquid Culture—We grew cultures of yeast strains to be tested for drug hypersensitivity overnight in 5 ml of YPD broth in 15-ml clinical centrifuge tubes at 30 °C in a shaking water bath. Then cells were pelleted in a clinical centrifuge at high speed for 5 min. We decanted the growth medium and washed the pellets with 10 ml of sterile water before resuspension. We determined cellular concentration by spectroscopy (absorbance at 600 nm). We removed 0.5 × 105 cells, added this to 2 ml of YPD broth containing cyh (0.18–43.2 μm) or tritylimidazole (2–20 μm), and incubated the cultures at 30 °C with shaking. After 48 h, we monitored relative inhibition by determining the absorbance at 600 nm and comparing measurements to cells grown in YPD alone.
Qualitative Analysis of Relative Drug Resistance—We tested mutant and control strains qualitatively for drug resistance by spotting 2-fold dilutions of cells in 5 μl of water on plates with a fixed concentration of a xenobiotic compound. Cultures to be tested were grown overnight in 10 ml of YPD broth. The cultures were washed and concentrated by centrifugation and resuspended in sterile water at 1/10th the original culture volume. We diluted a small sample 100-fold and determined the absorbance at 600 nm to check cell concentration. We made 2-fold dilutions in sterile water before plating in a 5-μl sample.
UV Mutagenesis of JG365-3B—The first S558Y allele was created by UV mutagenesis as described previously (17).
Estimating the Frequency of Reversion—To estimate the rate of reversion of S558Y, 10 1-ml YPD cultures were seeded with 100 cells and grown to saturation. We plated 106 cells in 4-ml sterile agar on plates containing 7.5 μm clotrimazole and incubated them for 5 days at 30 °C. We counted the suppressors and picked them for further testing.
Preparation of Purified Membrane Vesicles—We prepared plasma membrane (PM) vesicles as described previously (18), with further modification (5). We determined protein concentration in the vesicles with a bicinchonic acid kit (Perbio, Rockland, IL).
Immunoblots of Pdr5 in PM Vesicles—Proteins in the purified PM vesicles were separated by gel electrophoresis and transferred to nitrocellulose membrane at constant current (400 mA, 1 h). The nitrocellulose membranes were blocked for 30 min with 20% nonfat milk in PBS containing Triton X-100 (PBST); incubated with a 1:200 dilution of polyclonal goat, anti-Pdr5 antibody, and yC-18 (Santa Cruz Biotechnology, Santa Cruz, CA); washed with PBST (three times for 15 min); incubated with a 1:5,000 dilution of secondary antibody (donkey anti-goat IgG-horseradish peroxidase in 5% nonfat milk) for 2 h; and washed with PBST (three times for 15 min). The chemiluminescence signal was developed by adding 10 ml of ECL reagent for 10 min and then exposing to x-ray film for 0.5–1 min.
Assay of ATPase Activity—ATPase activity of purified PM vesicles was measured by the end point Pi assay as described previously for P-gp (19, 20), with minor modifications for Pdr5 (5).
One concern about ATPase measurements made with PM preparations is that these contain Pma1p [H+], which exhibits vanadate (Vi)-sensitive ATPase activity. We described our resolution of this issue in detail in a previous study (5). Briefly, the optimal pH for Pma1p [H+]is ∼5.5–6.0, and the enzyme is only minimally active at pH 7.5. We therefore characterized ATPase activity in PMs from the Pdr5 at pH 7.5. In addition, measurements were made with purified PM vesicles from both Pdr5+ strains and the Δpdr5 strains, and the small amount of residual ATPase activity (<5%) observed in Δpdr5 strains was subtracted.
Rhodamine 6G (R6G) Transport Assay—We measured R6G transport at 30 °C as described previously (21). The concentration of R6G used in this study was 2.5 μm.
Cross-linking of 8-Azido-[α-32P]ATP to Pdr5—We carried out binding studies in purified PM vesicles at 4 °C. Membrane proteins (18 μg/assay) were suspended in MOPS ATPase assay buffer containing 10 μm 8-azido-[α-32P]ATP (2.5 μCi/nmol). Samples were incubated in the dark for 5 min at 4 °C before cross-linking with 365 nm UV light on ice for 10 min. Reactions were stopped by adding 12.5 μl of 5× SDS-PAGE sample-loading buffer. After electrophoresis on a Tris acetate gel at constant voltage, gels were dried. We quantified the radioactivity incorporated into the Pdr5 band with a STORM 860 Phosphor-Imager system (GE Healthcare) and ImageQuant software.
Cross-linking of [125I]IAAP to Pdr5—Purified membranes prepared from yeast cells overexpressing Pdr5 were incubated at room temperature in the ATPase buffer with [125I]IAAP (7 nm) for 5 min under subdued light. The samples were photocross-linked for 10 min with 365 nm UV light at room temperature followed by electrophoresis with 7% NuPAGE gels and quantified as described previously (21). Modifications to this procedure in specific experiments are described in the figure legends.
DNA Extraction and PCR Recovery of Suppressor Mutant Sequences—We amplified PDR5 from suppressor mutants with a PureLink Genomic DNA kit from Invitrogen. The DNA was extracted as described in the accompanying handbook from Invitrogen, with several modifications. We lysed 4 × 107 cells for each extraction. We purchased zymolyase made up in buffer from Sigma. We used 1 ml (∼60 units of activity) for each sample. We also doubled the volume of proteinase K and RNase A.
We carried out PCR amplification with Platinum PCR Supermix High Fidelity from Invitrogen, with the following protocol: 1 cycle at 95 °C for 5 min, 35 cycles of denaturation at 95 °C for 1 min, annealing at 55 °C for 1 min, and extension at 68 °C for 7 min. Reactions were then placed on hold at 4 °C until they were processed further. The primers used to amplify PDR5 were left, TAAGACTCCGGTGAGTGTGG (Tm of 59 °C), and right, GCACGTTCGTTGTACTTCCA (Tm of 60 °C).
RESULTS
S558Y Mutation—The S558Y allele was identified in the course of a large mutant search designed to uncover suppressors that eliminated the cyh resistance of a strain overproducing Pdr5 (JG365-3B), the major yeast multidrug transporter. Following UV mutagenesis, we screened ∼70,000 survivors and isolated 17 cyh-sensitive colonies that failed to complement a known pdr5 deletion. Of these, 16 were severely impaired and phenotypically similar to the deletion; a single very leaky allele was also recovered.
Immunoblotting of purified PM vesicles indicated that in 15 of these 17 mutants, Pdr5 was significantly reduced or missing (data not shown). One such mutant, pdr5-107 was the subject of recent study (17). Another mutant, although severely drug-sensitive, had unaltered levels of Pdr5 in its PM. Complete nucleotide sequencing of the PDR5 gene indicated that it contained an S558Y missense mutation. The reported TMHs of Pdr5 suggested that this residue lie just outside TMH2. The original TMH assignments, however, were slightly different from those that we have established from seven topology modeling programs that allow us to predict core consensus sequences. Results from all these programs place Ser-558 in TMH2.3
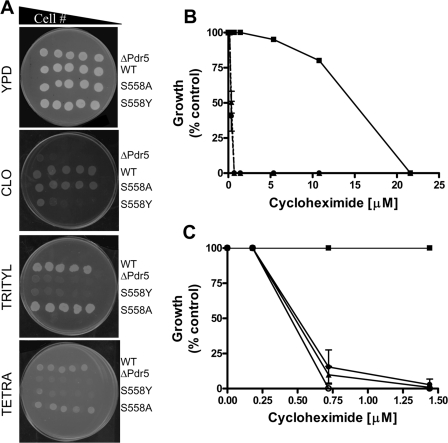
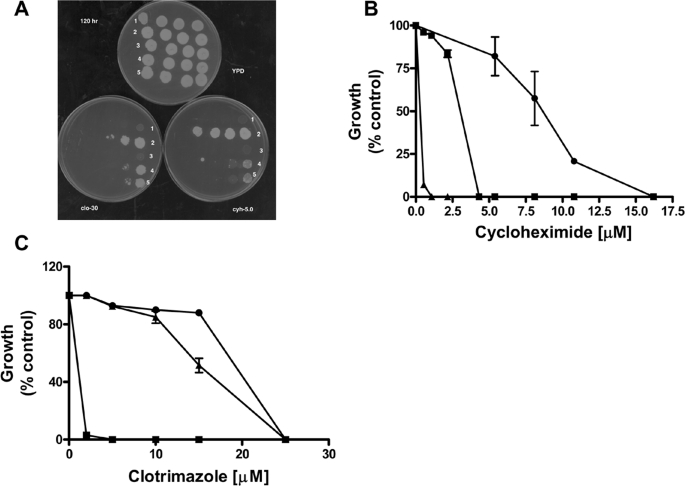
Drug Phenotypes of S558Y and S558A—The S558Y mutation was recreated in pSS607, a yeast integrative vector that we recently described (5). We also made an S558A mutation. The mutant plasmids were completely sequenced to verify that each had only the desired alteration. From these plasmids we created several strains used in this study; all are isogenic to each other. Fig. 1A shows the relative clo sensitivity of four strains as follows: R-1 (pdr5::KANMX4); R-1 + pSS607, referred to as WT; R-1 + pS558Y or S558Y; and R-1 + pS558A or S558A.
FIGURE 1.
Drug sensitivity of S558Y. A, 2-fold serial dilutions of Pdr5 strains were plated on YPD media or on YPD media containing 15 μm clotrimazole, tritylimidazole, or tetrabutyltin. The number of cells plated ranged from 1.0 × 106 to 0.5 × 104; scans of plates that were incubated for 96 h at 30 °C are shown. A representative figure from three independent experiments is shown. B, quantitative analysis of drug sensitivity was carried out by inoculating 500 cells bearing the WT and mutant Pdr5s in the presence of increasing concentrations of cyh (0.05–21.6 μm). The cells were incubated at 30 °C for 48 h and then quantified by measuring the absorbance at 600 nm. The strains used were R-1 (Δpdr5 = •), R-1 + pSS607 (WT = ▪), and R-1 + pS558Y (▴). Growth at each concentration of cycloheximide is shown as (% control), i.e. growth in the absence of cyh. C, same protocol was used to compare single- and double-copy mutant strains: single-copy WT (JG2000 = ▪), single-copy S558Y (R-1 + pS558Y = ▾), double-copy S558Y (JG2010 = ▴), and R-1 (○). The figures (B and C) show the mean values of three independent experiments, and the error bars represent the S.D.
These data indicate that the S558Y mutation is drug hypersensitive as we predicted. By contrast, the S558A allele is phenotypically similar to the WT PDR5. This result suggests that it is the size of the tyrosine residue rather than the hydrophobicity of its aromatic ring that is primarily responsible for the observed mutant phenotype. A similar phenotype is observed with tritylimidazole and tetrabutyltin. These substrates were used to define different transport sites in Pdr5 that may or may not overlap (22, 23). In each case, S558Y is considerably more sensitive than the WT control to all of these xenobiotic agents and is qualitatively the same as the Δpdr5::KANMX4 isogenic negative control.
Several studies have indicated that the drug-binding sites of ABC transporters consist of a large pocket, and individual transport substrates show specific interactions with different amino acid side chains (24–26). The fact that the S558Y mutant is not substrate-specific (Fig. 1A) suggests that the reversal of drug resistance is a consequence of general steric hindrance and that the Ser-588 residue may not be involved in specific interactions with the drugs. This is consistent with the finding that the mutation S558A does not affect Pdr5 function.
We carried out a quantitative analysis of relative cyh resistance in liquid culture. The data in Fig. 1B show that the S558Y mutant has an IC50 value that is ∼30-fold lower (0.5 μm) than the WT and is similar to the Δpdr5 control.
Fig. 1C illustrates the behavior of double- and single-copy strains at low cyh concentrations. Strains containing 1 (JG2009) or 2 copies of S558Y show similar drug hypersensitivity. Therefore, doubling the amount of mutant protein does not increase cyh resistance. This is significant because the increased levels of Pdr5 in the double-copy strains are useful in biochemical assays, where they provide more robust measurements.
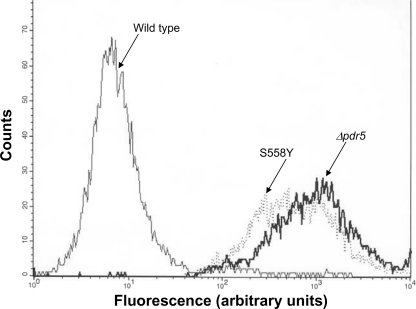
S558Y Mutant Strain Is Transport-deficient—We analyzed the transport capability of S558Y with a well established, wholecell R6G efflux assay that uses flow cytometry (21, 27). We compared the transport capability of the WT (JG2000), Δpdr5 (AD1–7), and S558Y (JG2009) isogenic strains. These data are found in Table 2, and representative histogram plots are shown in Fig. 2. These data clearly demonstrate that the S558Y mutation creates an R6G transport deficiency that is as severe as that observed in the Δpdr5 strain. Both of these strains retain ∼60–70-fold more R6G than does the WT PDR5 strain, JG2000.
TABLE 2.
R6G retention in an S558Y mutant The flow cytometry-based transport assay was carried out as described under “Experimental Procedures” and legend to Fig. 2. The data represent the median fluorescence value obtained from histograms similar to the ones depicted in Fig. 2. Two experiments were performed with two independently grown cultures of each strain (n = 4). Values are the average of the two median values with standard deviation (S.D.) indicated. AU, arbitrary units.
| Strain and pertinent genotype | Median fluorescence | S.D. |
|---|---|---|
| AU | ||
| AD1–7 (Δpdr5) | 408.21 | 96.94 |
| JG2005 (AD1–7 + pSS558Y) | 389.47 | 52.94 |
| JG2000 (AD1–7 + pSS607) | 6.71 | 0.59 |
FIGURE 2.
R6G transport is drastically diminished in S558Y. The transport activity of WT and mutant Pdr5s was estimated with a flow cytometry-based efflux assay with R6G as the fluorescent substrate. The null strain (Δpdr5 = AD1–7) and yeast strains containing the WT (JG2000) and mutant (JG2009) Pdr5 were used in these experiments. The assay was carried out as described previously (21), and the representative histograms from four independent experiments are depicted. The strains used are labeled on the figure.
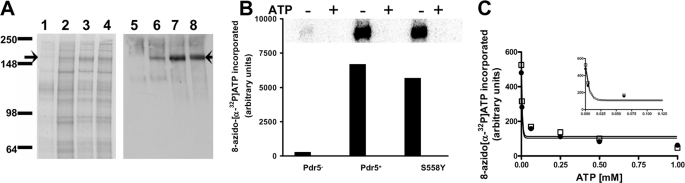
S558Y Shows Normal Membrane Localization and Nucleotide Binding and Slightly Diminished ATP Hydrolysis—We evaluated the effect of adding a second copy of WT or mutant protein to a haploid strain. The colloidal blue-stained gel and the immunoblots (Fig. 3A) clearly show a 2-fold increase in Pdr5 levels in WT (JG2004) and S558Y mutant (JG2010) compared with the isogenic, single-copy counterpart (JG2000). Thus, although the single-copy strain overproduces Pdr5, it is clear that more protein can be localized to the membrane by simply raising the copy number. We therefore used the PMs prepared from these strains for all biochemical studies.
FIGURE 3.
Interaction of nucleotides with WT and S558Y. A, colloidal blue staining and immunoblotting of purified PM vesicles prepared from various strains of yeast. Lanes 1–4 depict a colloidal blue-stained gel and lanes 5–8 an immunoblot with the anti-Pdr5 antibody yC-18. Lanes 1 and 5, Δpdr5; lanes 2 and 6, WT Pdr5, single copy; lanes 3 and 7, WT Pdr5, double copy; lanes 4 and 8, mutant Pdr5, S558Y, double copy. Arrow shows the position of the Pdr5 protein. B, photoaffinity labeling of Pdr5 with 0.5 mm 8-azido-[α-32P]ATP. Cross-linking was carried out in purified plasma membrane vesicles at 4 °C as described under “Experimental Procedures.” Upper panel, 8-azido-[α-32P]ATP incorporated into the Pdr5 band; lower panel, quantification of the [32P] incorporated with a STORM 860 PhosphorImager system. The cross-linking in the absence of ATP is shown as filled bars and the cross-linking the presence of ATP as empty bars. C, 8-azido-[α-32P]ATP binding as a function of ATP concentration (0–1 mm) was determined in WT Pdr5 (•) and the S558Y mutant (□). The 8-azido-[α-32P]ATP incorporated in the Pdr5-bands was quantified with a PhosphorImager, and the curves were generated with the curve-fitting Prism GraphPad software (GraphPad Software, La Jolla, CA). The inset depicts the data with an expanded x axis in the range 0–0.125 mm ATP. The x and y axis legends for the inset are the same as those for the main figure. Figure shows representative data from two independent experiments.
PMs prepared from WT and S558Y Pdr5-expressing strains show comparable levels of photolabeling with 8-azido-[α-32P]ATP (Fig. 3B). Furthermore, because the 32P signal is completely eliminated by the addition of 2 mm ATP, the interaction is clearly taking place at the NBDs.
It is of course possible that although nucleotides bind to both WT and mutant Pdr5s, the affinity is altered. The concentration-dependent displacement of 8-azido-[α-32P]ATP by ATP provides a measure of the apparent affinity of ATP for the transporter. We found that the IC50(ATP) values for the WT and mutant Pdr5 are almost identical (Fig. 3C). As expected, the Δpdr5 strain shows no significant cross-linking of 8-azido-[α-32P]ATP even though the colloidal blue-stained gels clearly indicate that all three lanes were loaded with equal amounts of vesicle protein (data not shown). These results strongly suggest that the WT and S558Y strains have similar nucleotide-binding capabilities.
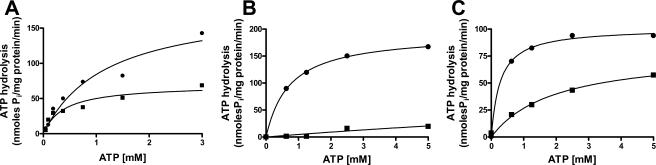
The kinetics of Pdr5-mediated ATP hydrolysis was the subject of several previous studies, and it is clear that this transporter has a high level of unstimulated (basal) activity (4, 5, 13). We compared the kinetics of ATP hydrolysis in WT Pdr5 and in S558Y. We found an ∼2-fold decrease in the Vmax of ATP hydrolysis. Several vesicle preparations from each strain were assayed. The Vmax for the WT ranged from 180 to 279 nmol/mg/min; the Vmax for the S558Y mutant ranged from 69 to 93 nmol/mg/min (representative data from one preparation is shown in Fig. 4A).
FIGURE 4.
Pdr5-mediated ATP hydrolysis. A, Vi-sensitive ATPase activity of double-copy WT (•) or S558Y (▪) Pdr5 in purified PM vesicles (10 mg per assay) was assayed as described previously (5) in the presence of increasing concentrations of ATP (0–3 mm ATP). B, clotrimazole sensitivity of ATPase activity was monitored in WT purified plasma membrane vesicles in the absence (•) or presence (▪) of 25 μm clotrimazole. C, clotrimazole sensitivity of ATPase activity was monitored in the Pdr5 mutant, S558Y, purified plasma membrane vesicles in the absence (•) or presence (▪) of 25 μm clotrimazole. Representative plots are shown (n = 3).
Two aspects of these data are notable. Although the WT and mutant Pdr5s differ in Vmax, they show comparable Km values (about 0.5 mm). This is consistent with the finding that the IC50 for displacement of 8-azido-[α-32P]ATP by ATP is identical in WT and S558Y (Fig. 3C). Furthermore, the ATPase activity of the double-copy S558Y mutant Pdr5 is comparable with that found in the single-copy WT protein (5). Nevertheless, the single-copy WT strain is ∼30-fold more resistant to cyh than is the double-copy S558Y strain (see Fig. 1B, above). Therefore, the decrease in ATPase activity observed in the double-copy mutant strain alone cannot explain why the mutant Pdr5 is profoundly hypersensitive to all of the tested drugs (Fig. 1A).
Pdr5-ATPase activity is sensitive to inhibition by its transport substrate clo, which acts as a noncompetitive inhibitor at an allosteric site (5). When WT Pdr5 was treated with 25 μm clo, the Vmax(ATP) decreased by ∼90% (Fig. 4B). Although clo also reduces the Vmax(ATP) for Pdr5-mediated ATP hydrolysis in S558Y, the inhibition is only partial (40%; Fig. 4C). These results suggest two possibilities as follows: either the drug substrate site itself is modified, or the allosteric inhibition of ATPase activity cannot be efficiently communicated to the NBDs so that a significant amount of the ATPase activity is allowed to proceed.
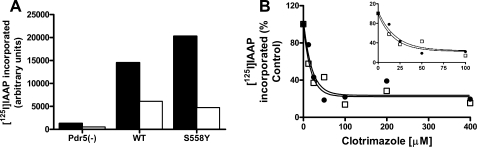
[125I]IAAP Binding in the S558Y Mutant—To understand whether it is the interaction between drug substrate(s) and Pdr5 per se that is affected in the S558Y mutant, we exploited the photoaffinity labeling of IAAP. We previously demonstrated that IAAP, an 125I-labeled photoaffinity analogue of prazosin, cross-links to the transport substrate sites of Pdr5 (21). We further demonstrated that IAAP cross-links to both WT and S558Y Pdr5 but not to PMs prepared from the Δpdr5 strain (AD1–7) (Fig. 5A). Moreover, in the presence of clo, cross-linking of IAAP is significantly reduced in both WT and mutant Pdr5 (Fig. 5A).
FIGURE 5.
Interaction of WT and mutant Pdr5 with the drug substrate analogue [125I]IAAP. A, photoaffinity cross-linking of Pdr5 to [125I]IAAP was carried out in purified PM vesicles as described under “Experimental Procedures.” Quantification of the 125I signal in the Pdr5 band in plasma membranes prepared from the Δpdr5 strain AD1–7, the double-copy WT- yeast strain JG2004, and the double-copy S558Y mutant strain (JG 2010) was performed with a STORM 860 PhosphorImager system. Cross-linking of [125I]IAAP was carried out in the absence (filled bars) or presence (empty bars) of 100 mm clotrimazole. B, photoaffinity cross-linking of [125I]IAAP to WT (•) or S558Y (□) Pdr5 was carried out as described in A in the presence of increasing concentrations of clotrimazole. The 125I signal in the Pdr5 band was quantified and plotted with the curve-fitting GraphPad Prism software. The inset depicts the data with an expanded x axis in the range 0–100 μm ATP. The x and y axis legends for the inset are the same as those for the main figure. Representative plots are shown (n = 2).
To determine the apparent affinities of WT and mutant Pdr5, we measured the displacement of cross-linked IAAP in the presence of increasing concentrations of clotrimazole (Fig. 5B). We found that the IC50 values for the displacement of IAAP by clo are comparable in WT and mutant Pdr5 (13.8 and 12.6 μm, respectively).
Taken together, our results demonstrate that both the drug substrate and nucleotide bind to the S558Y mutant with affinities that are comparable with WT Pdr5. However, the drug efflux mediated by the S558Y mutant is severely impaired and as a consequence exhibits drug sensitivity.
Genetic Analysis of S558Y Suppressors—Our results suggest that the S558Y mutation in Pdr5 disrupts the coupling between the NBDs and the transport substrate site. We isolated cloresistant (clor) suppressors of the S558Y mutation present in JG2011 (R-1 + pS558Y) by plating cultures on medium containing 15 μm clo. We analyzed 15 independent cultures and determined a frequency of 2.2 × 10-5. This is a relatively high value for a true back mutation (Tyr → Ser) and thus suggested that at least some of the suppressors might represent mutations at other sites in PDR5 or conceivably in other genes.
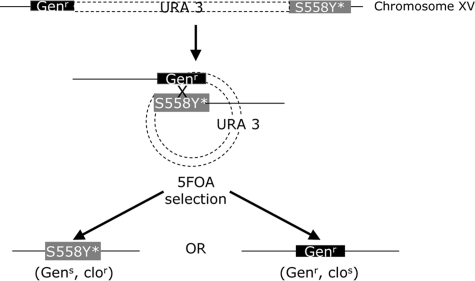
Because suppressors generally have dominant phenotypes, complementation testing cannot be used to determine whether a mutation resides in PDR5. The R-1 strain was built to quickly map these suppressors through a novel approach (Fig. 6).
FIGURE 6.
Experimental strategy to determine whether a reversion phenotype is caused by a second mutation in pdr5S558Y. Structure of JG2011 (R-1 + pS558Y) after isolation of a clor suppressor. In this illustration, it is assumed that the new mutation (*) lies in S558Y rather than in some other gene. Loss of the plasmid sequences containing URA3 and 1 of 2 PDR5 cassettes, either S558Y or pdr5::KANMX4 (GENr), occurs by recombination in one of the two homologous regions. These events result in Ura- colonies that are selected on 5-FOA medium (14) and are tested on clotrimazole and geneticin medium. If the reversion is attributable to a second mutation in the S558Y copy, clor, gens, and clos, genr colonies will be recovered. A mutation in another gene would yield only clor colonies.
To determine whether the clor suppressors were the result of second-site mutations in the PDR5 gene, we took advantage of the genetic features of R-1 that are illustrated in Fig. 6. When pS558Y was integrated into this strain, we found a duplication of upstream and downstream sequences adjacent to the PDR5 coding region. The vector sequences, including the URA3 marker, are in between the duplicated sequences. Such a transformant is geneticin-resistant (KANMX4) and able to synthesize uracil (and thus grow on uracil omission medium).
To determine whether the clo resistance is in the PDR5 gene, we employed 5-FOA selection (14). This drug selects for Ura- colonies that arise when recombination causes loss of the URA3 plasmid and either the S558Y allele (possibly bearing a second mutation causing the clor phenotype) or the disrupted/deleted (KANMX4) copy. The loss of the latter leaves a geneticin-sensitive S558Y segregant that is presumably clo-resistant. Loss of the former, however, would make the colony geneticin-resistant but clo-sensitive if the clo resistance is caused by a second mutation in the S558Y copy of the gene. If the clotrimazole resistance was not caused by a genetic change in the S558Y copy, all of the 5-FOA-selected recombinants would remain clo-resistant.
Each of 12 independently derived suppressors was plated on 5-FOA, and ura3 recombinant survivors arose at a frequency of ∼2 × 10-3. 5-FOA survivors were tested for each suppressor to determine whether the clor mutation was in PDR5. Of 31 5-FOA survivors recovered from clo-1-0, 13 were clor, Gens and 18 were clos, Genr. Similarly, of 25, 5-FOA survivors recovered from clo-3-0, 14 were clor, Gens and 11 were clos, Genr. These results confirm that each suppressor phenotype was attributable to a second mutation in the S558Y allele. Comparable results were obtained for the remaining 10 suppressors (data not shown).
The clo-1-0 and clo-3-0 suppressors were spot-tested on two different Pdr5 substrates, clo (15.0 and 22.5 μm) and cyh (0.78 μm). A summary of these results is given in Table 3. The numerical score indicates the lowest concentration of cells showing confluent growth on the particular drug plate. Thus, a score of 1 means the suppressor grew only at the highest cell concentration, 0.5 × 106; a score of 4 means it grew at the lowest concentration, 0.5 × 103. The JG2011 strain used to isolate suppressors failed to grow at any cellular concentration and thus received a 0.
TABLE 3.
Phenotypes of suppressors Ten-fold serial dilutions of cells were prepared and spotted as described under “Experimental Procedures.” Plates were scored at 96 h after incubation at 30 °C.
| Strain and pertinent genotype | Clo, 15.0 μm | Clo, 22.5 μm | Cyh, 0.78 μm |
|---|---|---|---|
| WT (R-1 + pSS607) | 4 | 3 | 4 |
| S558Y (R-1 + pS558Y) | 0 | 0 | 0 |
| Clo-1-0 | 4 | 3 | 4 |
| Clo-3-0 | 3 | 2 | 3 |
The data in Table 3 suggest that the clo-1-0 suppressor mutant is as drug-resistant as the WT control. The strong but not completely WT phenotype of the clo-3-0 suppressor suggested it might be particularly useful in further biochemical studies, especially in distinguishing various models of S558Y dysfunction, and was therefore analyzed further. The other suppressors are the subject of continuing study.
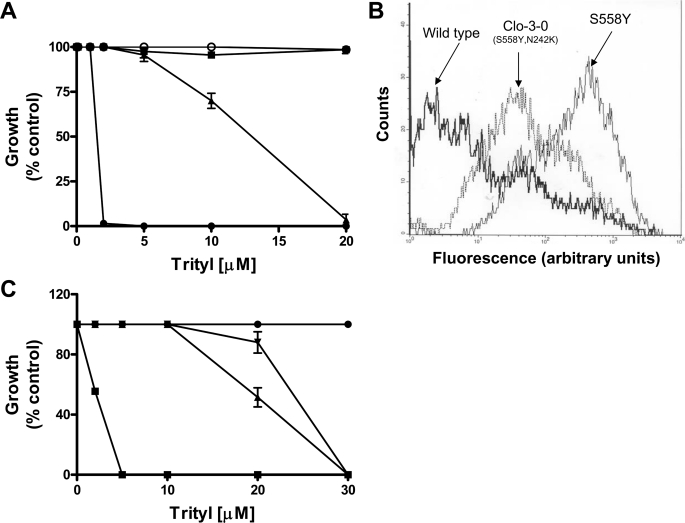
We compared the clo-3-0 and clo-1-0 suppressor mutants and controls for relative resistance to tritylimidazole, a substrate that defines a transport site that is at least somewhat different from the one used by clo and R6G (21). These results are shown in Fig. 7A. As with R6G transport, the clo-1-0 suppressor is indistinguishable from WT positive control, but the clo-3-0 suppressor increases resistance to a level of the drug between the levels of the S558Y and WT strains. Similarly, flow cytometry data demonstrated that R6G transport is restored to a level that is also between that of the S558Y and WT strains (Fig. 7B). In addition, neither the initial drug hypersensitivity of S558Y nor the restored resistance of clo-3-0 shows any indication of substrate specificity. The clo-1-0 suppressor exhibited transport properties that were indistinguishable from the WT (data not shown).
FIGURE 7.
The clo-3-0 (S558Y,N242K) suppressor reverses the drug-sensitive phenotype of S558Y. A, quantitative analysis of clo-3-0 growth in the presence of trityl was carried out as described under “Experimental Procedures.” Growth of WT (▪), S558Y (•), clo-1-0 (○), and clo-3-0 (▴) in the presence of increasing concentrations of tritylimidazole was performed. Growth was estimated on the basis of absorbance at 600 nm and plotted as percentage growth, where the absorbance in the absence of tritylimidazole was taken to be 100%. The mean values (±S.D.) of three independent experiments are plotted. B, R6G (2.5 μm) accumulation in yeast strains bearing WT and the mutants S558Y and S558Y,N242K (clo-3-0 strain) was analyzed with flow cytometry (21). Representative histograms from four independent experiments are shown. The strains used are labeled on the figure. C, clo3–0 phenotype was recreated by inserting pS558Y,N242K plasmid DNA into R-1 (pdr5::KANMX4) by lithium chloride transformation. Quantitative growth in liquid medium containing trityl was carried out as described in A with WT (R-1 + pSS607; •), S558Y (R-1 + pS558Y; ▪), transformants of clo-3-0 (S558Y,N242K; ▴), and R-1 + pS558Y,N242K; ▾). The mean values (±S.D.) of three independent experiments are plotted.
The PDR5-specific DNA was recovered from the clo-1-0 and clo-3-0 suppressors by PCR amplification, and the entire coding region was sequenced. The sequence recovered from clo-1-0 contains a true back mutation from Tyr back to Ser. The completely WT phenotype it exhibits confirms the pleiotropic nature of the original mutation (deficiencies in transport and drug resistance).
In the clo-3-0 suppressor, the original S558Y base pair substitution remains, but a second alteration, N242K was identified. The S558Y,N242K double mutation was confirmed in two separate sequencing reactions from overlapping primers. N242K is located in NBD1 near the Q-loop between the Walker A and the signature motifs. No other alterations were observed.
We constructed the double mutation in pSS607 to create pS558Y,N242K. We also made the single N242K mutation in this plasmid. We demonstrated that transformation of R-1 with a pS558Y,N242K plasmid recreates a clo-3-0 phenotype when it is used to transform the R-1 strain (pdr5::KANMX4; Fig. 7C).
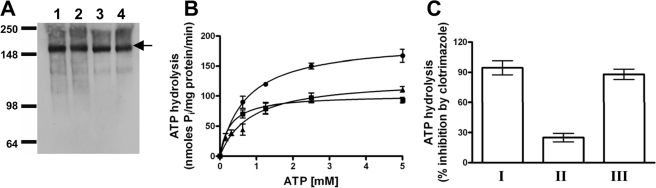
S558Y Suppressor S558Y,N242K Restores Coupling between the ATP and a Clo-binding Site—Double-copy strains were also made for all of the single and double mutants (S558Y,N242K, and clo-3-0), and purified PM vesicles were prepared from each. The immunoblot shown in Fig. 8A demonstrates that purified PMs prepared from cells overexpressing WT Pdr5 and the mutant proteins S558Y,N242K, and clo-3-0 (S558Y,N242K) express equivalent levels of transporter. The ATPase activity of the transport-deficient S558Y strain and the S558Y,N242K suppressor exhibit comparable values for the Km and Vmax of ATP hydrolysis (Fig. 8B). Although both the single S558Y mutant and the suppressor show robust ATPase activity, there is an ∼1.5-fold decrease in the Vmax compared with WT Pdr5. Thus, the behavior of the S558Y ATPase is qualitatively similar to the experiment shown in Fig. 4. Furthermore, it is clear that the second-site mutation does not affect the kinetics of ATP hydrolysis per se.
FIGURE 8.
ATP hydrolysis mediated by the S558Y,N242K double mutant of Pdr5. A, immunoblotting of purified plasma membrane vesicles of WT and mutant Pdr5s was carried out as described in Fig. 3A. Vesicles were prepared from yeast strains bearing double copies of WT and mutant Pdr5s. Lane 1, WT Pdr5; lane 2, mutant Pdr5, S558Y; lane 3, mutant Pdr5, N242K; lane 4, mutant Pdr5, S558Y,N242K (clo-3-0). B, Michaelis-Menten plot of Vi-sensitive ATPase activity in purified PM vesicles of clo-3-0 (▴) compared with WT (•) and S558Y (▪). The mean values from three independent experiments are shown, and the error bars represent the S.D. C, Vi-sensitive ATP hydrolysis was measured as described under “Experimental Procedures” in the absence or presence of 25 μm clotrimazole. The mean values for the percent inhibition of ATPase activity by clotrimazole in WT Pdr5 (bar I), mutant Pdr5, S558Y (bar II), and S558Y,N242K (bar III) are shown. Error bars represent the S.D.
We demonstrated in earlier studies that clo has a strong inhibitory effect on the ATPase activity of WT Pdr5 (5). We also showed that clo-induced inhibition of ATP hydrolysis is significantly reduced in the S558Y mutant (Fig. 4C). We find that inhibition of ATPase activity approaches levels observed in the WT protein in the S558Y,N242K suppressor (Fig. 8C). These data are consistent with our earlier hypothesis that the S558Y mutation exhibits drug susceptibility because the two key activities of Pdr5, ATP hydrolysis and drug-substrate transport, are uncoupled. These results demonstrate that coupling is clearly restored in clo-3-0.
N242K Mutation Exhibits Drug Sensitivity in a Ser-558 Background—To determine whether the N242K substitution has a drug-sensitive phenotype when separated from S558Y, we transformed R-1 (pdr5::KANMX4) with pN242K. Two independent transformants were tested on 22.5 μm clo and 3.1 μm cyh to determine whether there was a drug-hypersensitive phenotype. As shown in Fig. 9A, the transformants exhibit a reduction in drug resistance to both of the compounds compared with the isogenic WT control (R-1+pSS607). Under the conditions used, N242K appears to be considerably more sensitive to cyh than to clo.
FIGURE 9.
Drug sensitivity of WT Pdr5 and S558Y and N242K. A, 10-fold serial dilutions (0.5 × 106 cells to 0.5 × 103 cells in 5 μl of sterile H2O) were plated on YPD and on YPD containing 3.1 μm cycloheximide or 22.5 μm clotrimazole. Plates were scanned after 96 h of incubation at 30 °C. Row 1, R-1 (Δpdr5) cells; row 2, WT (R-1 + pSS607) cells; row 3, R-1 + pS558Y; rows 4 and 5, two independent transformants of R-1 + pN242K. B, quantitative growth analysis of WT (•), S558Y (▴), and N242K (▪) in the presence of increasing concentrations of cyh. Growth in the absence of cyh was taken to be 100% in each curve. C, growth of WT (•), S558Y (▪), and N242K (▴) was compared as described in B, except that the experiment was carried out in the presence of increasing concentrations of clotrimazole.
To measure the difference between strains, we determined the IC50 values for cyh and clotrimazole in liquid culture. These data are shown in Fig. 9B and demonstrate that this mutation is strikingly cyh-sensitive, as the spot tests suggested. The difference in IC50 values between the N242K mutant and the isogenic WT control is ∼3-fold. The minimum inhibitory concentrations of cyh for the N242K mutant and WT strains are 4.2 and 16.6 μm (∼4-fold difference between strains). Therefore, NBD1, although degenerated in sequence, plays a necessary role in the cell. The N242K strain shows only a modest reduction (∼1.3-fold) in resistance to clo (Fig. 9C).
DISCUSSION
A central issue with all ABC drug transporters is the coupling of ATP hydrolysis and drug transport. Although these biochemical processes occur in different domains of the molecule, transport cannot occur unless the two activities are coupled. We examined residues in the yeast ABC transporter Pdr5 that helped us define an interface that is probably involved in this process.
We screened UV-treated survivors to identify suppressors that eliminated the cyh resistance mediated by Pdr5 and recovered S558Y. Several observations implicate this residue in interdomain communication. For example, yeast cells overexpressing the S558Y mutant Pdr5 and an isogenic Δpdr5 strain show similar drug hypersensitivity and transport deficiency (Figs. 1 and 2 and Table 2). Nonetheless, nucleotide (8-azido-[α-32P]ATP) and transport substrate (IAAP) analogues both show photocross-linking to S558Y mutant Pdr5 (Figs. 3 and 5). Furthermore, the WT and mutant Pdr5s have comparable affinities for ATP and clo (Figs. 3 and 5). Finally, we showed that the S558Y mutant hydrolyzes ATP, although the Vmax is lower than that observed for WT protein.
We inserted double copies of the PDR5 gene to obtain higher levels of the protein for biochemical studies (compare lanes 2 and 3 with 6 and 7 in Fig. 3A). Earlier studies were conducted with purified PMs prepared from single-copy strains (5). In these experiments, the Vmax (ATP hydrolysis) value for WT Pdr5 was comparable with that found in the double-copy S558Y mutant (Fig. 4A). Nevertheless, yeast strains with a single-copy WT Pdr5 show 30-fold greater cyh resistance than the double-copy S558Y mutant strain (Fig. 1B). Therefore, it appears very unlikely that the 1.5-fold decrease in the ATPase activity in the mutant is sufficient to account for the null-like phenotype of the S558Y mutant in transport and drug resistance assays (Figs. 1 and 2). Taken together, these data strongly suggest that the S558Y drug hypersensitivity is a consequence of impaired coupling between ATP hydrolysis and the conformational changes in the TMDs that bring about transport.
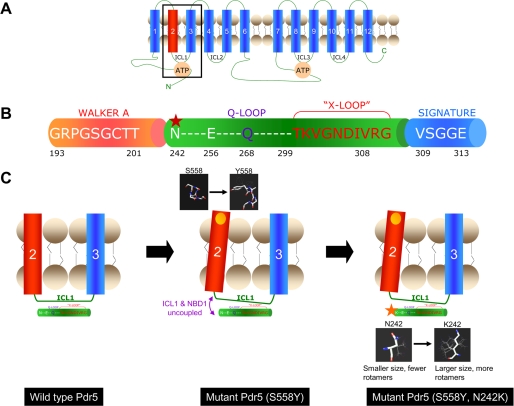
The S558Y mutation thus provides a unique opportunity to probe the structural elements of the intramolecular domains that couple ATP hydrolysis and transport of drug substrate. Unlike with mammalian cells, it is possible in yeast to screen for second-site suppressors even if they occur at very low frequencies. The strategy is described in detail in Fig. 6. Previous studies involving large scale mutagenesis of several proteins provided evidence that the functional coupling of amino acids in interacting proteins and long range interactions in different domains of a protein are not always predictable from the atomic structure (see Ref. 28 and references therein). The molecular chaperone Hsp70, like Pdr5, exhibits two independent functions (ATP hydrolysis and polypeptide binding) that occur at distinct domains of the protein. Nonetheless, the coupling of these two activities is critical for the chaperone to function. A genetic screen that identified amino acid alterations in the ATPase domain that reverse defects caused by mutations in the peptide-binding domain was successfully used to locate the hot spots in these two domains (29). The yeast as a model system is well suited to such genetic screens. We found that a second mutation at position 242 (N242K) resulted in an almost complete reversal of the null phenotype of the S558Y phenotype (Fig. 7). Moreover, the S558Y,N242K double mutant also showed a Vmax for the ATPase comparable with the mutant, S558Y, and 1.5-fold lower than that observed for the WT Pdr5 (Fig. 8B). Thus, we demonstrate equivalent levels of ATP hydrolysis in the drug-sensitive and transport-deficient S558Y mutant Pdr5 and the drug-resistant suppressor S558Y,N242K (Fig. 8B). There is additional evidence to support our contention that the absence of coupling between the NBDs and transport domains rather than a very modest loss in ATPase function is the cause of the drug-sensitive phenotype of the S558Y mutant Pdr5. Clo is a potent noncompetitive inhibitor of ATPase activity. This inhibition occurs at a location that is distinct from the transport site of the drug and the NBDs (5). The loss of inhibition in S558Y and its nearly complete restoration in the suppressor strongly imply that the NBD1/TMH2 interface is a route for both productive and inhibitory signals. Fig. 10 proposes a model showing the structural changes in the WT, S558Y mutant, and suppressor (S558Y,N242K) strains. Fig. 10A shows the arrangement of the NBD and TMD domains in Pdr5. The organization of part of the N-terminal NBD (between the Walker A domain and the signature sequence) is illustrated in Fig. 10B. Residue Ser-558 is located toward the extracellular end of TMH2. Hydrophobicity plots and sequence alignment predict that Ser-558 is lipid facing and lies toward the extracellular end of the TMH. Sequence alignments of the NBDs of Pdr5 show that the residue Asn-242 lies between the Walker A and the signature sequence of NBD1. These results are significant in light of the structure of the bacterial ABC exporter Sav1866 that was recently solved (10), which may be considered as a functional orthologue of Pdr5, P-glycoprotein, and ABCG2 (30). The structure of Sav1866 was the first of a full-length ABC protein to show significant contacts between the TMDs and the NBDs. The ICLs between TMH2, -3, and -5,6 have short α-helices parallel to the plane of the membrane. These interact with the NBDs via a short segment (which the authors designate the x-loop) that lies between the Walker A domain and the signature sequence of the ATP-binding cassette. Although these structural associations are intriguing, they do not necessarily demonstrate a functional connection. Our study finds that loss of drug resistance as a result of a mutation in the TMH2 can be restored by a compensating mutation in the same region as the x-loop in the NBD. There are important differences between Pdr5 and Sav1866, including the organization of the NBDs and TMDs as well as key residues in the NBDs. It is nonetheless noteworthy that we find TMH2 and the region upstream of the signature sequence associated in the coupling of ATP hydrolysis to drug-substrate translocation. Additionally, correlated mutation analysis predicts that mutations at Ser-558 co-vary with those at Glu-256. This highly predictive statistical analysis is based on the concept that the functional coupling of two positions, even if distantly positioned in the structure, should be reflected in the mutually constrained evolution at the two positions (28, 31). In the case of Pdr5 our analysis shows that a mutation at position 256 is the most likely “response” to the mutation S558Y, demonstrating that the residue Ser-558 has been subjected to evolutionary pressure as a pair. It is interesting that the residue 256 also lies in the region between the Walker A and signature sequence of NBD1 of Pdr5 (Fig. 10B). A plausible explanation of how the S558Y mutation (in TMH2) uncouples the contacts between ICL1 and NBD1 is provided in Fig. 10C. We speculate that the additional bulk and/or conformational flexibility of the lysine residue compared with the asparagine permits the reestablishment of these contacts in the S558Y,N242K double mutant. The molecular models of Asn and Lys in Fig. 10C clearly show that Lys has a larger size as well as significantly more rotamers that could permit the reestablishment of contacts between the NBD and ICL1. The N242K mutant in the WT background is more sensitive to drugs compared with the WT. However, the N242K mutant exhibits greater resistance than the Δpdr5 or the S558Y mutant. An alignment of NBDs of fungal ABC transporters shows that residues at the position equivalent to Asn-242 in Pdr5 are not conserved. Besides asparagine, two other residues are found to occur at this location, threonine and serine. Thus a small polar amino acid that can participate in hydrogen bonding always occurs at this position. This study indicates that the asparagine residue at position 242 plays an important role in inter-domain communication between the NBDs and TMDs, plausibly via hydrogen bonds. It is thus likely that an Asn → Lys mutation at this location could disrupt these contacts. Nonetheless, as stated above, the increased conformational flexibility of the lysine residue could restore these contacts albeit not as efficiently as in the WT Pdr5.
FIGURE 10.
Model for the interaction of Ser-558 and N242K and the signaling interface for coupling ATP hydrolysis to the transport process. A, two-dimensional topological model of Pdr5 showing the 12 transmembrane helices, the two NBDs, and the four intracellular loops (ICL 1–4). The boxed area includes transmembrane helices 2 and 3 and the ICL1 which connects them. The interactions of these domains with the NBD are shown in greater detail in C. B, illustration depicting part of the N-terminal NBD of Pdr5 from the Walker A region to the signature sequence. The Q-loop and “x-loop,” which have been implicated in the mechanical transmission of conformational changes between the NBDs and TMDs, lie between the Walker A and Signature sequence of ABC transporters. The location of N242K, identified in a mutant screen in this study, is shown by a red star. Note that this residue as well as Glu-256 which co-varies with Ser-558 in correlated mutation analysis (see text) are located slightly upstream of the Q-loop. C, left panel shows the TMH2 and -3 of Pdr5 linked by the ICL1 (the boxed area in A). The x-ray crystallographic structure of the ABC transporter Sav1866 describes the “x-loop” in the NBD, which is implicated in the coupling of NBDs and TMDs via the ICL1 (10). The x-loop lies between the Walker A and signature regions of the NBD; the Q-loop also lies in this region and has been implicated in communication between the NBDs and transport sites (34). The central panel shows that the S558Y mutation (yellow circle) involves replacement with a bulkier residue that could result in the tilting of TM helix 2 (see structures in the black boxes for comparison). As the TM helix 2 is directly linked to the ICL1, the interface between the ICL1 and the NBD1 could be disrupted (purple arrows). This is consistent with our results that demonstrate that the S558Y has no effect on the affinity of Pdr5 for nucleotide or transport substrate but shows complete loss of transport activity because the NBD and transport-substrate sites are uncoupled. The right panel shows the S558Y,N242K double mutant. The structures show that the lysine that replaces the asparagine has more rotamers and a potentially longer reach. It is plausible that this change reestablishes the contacts between NBD1 and ICL1 (red arrow).
It is important to note that this study suggests that conformational changes in TMH2 are coupled to those at the ATP site in the N-terminal half of Pdr5. The structures of NBDs of numerous ABC proteins support the notion that the ATP is sandwiched between the Walker A and B domains and H-loop of one NBD and the signature sequence and the D-loop of the other (for review see Ref. 8). Thus the interactions described here would involve Walker A and B domains and the H-loop of NBD1 and the signature sequence of NBD2. In Pdr5 several residues that are highly conserved among ABC proteins (and have also been shown to play key functional roles) are altered in the NBD1. These include the conserved lysine in the Walker A, the catalytic carboxylate in Walker B, and the conserved histidine that defines the H-loop. This has led to the proposal that the NBD1 either lacks function or plays a regulatory role (1, 13). The asymmetry in the two ATP sites of Pdr5 is similar to that observed in most members of the ABCC subfamily of mammalian ABC proteins (32). Of these, the cystic fibrosis transmembrane conductance regulator (ABCC7) has been studied in considerable detail, and the consensus appears to be that the NBD1 is regulatory (33).
Our data show that residues in TMH2 and NBD1 are strongly coupled and clearly have a functional role and appear to define the transmission interface. This would suggest that the NBD1 of Pdr5 has, at the very minimum, a regulatory function in the transport cycle. In addition unlike many ABC proteins, the ATPase activity of Pdr5 is not stimulated by the transport substrate (5, 13). In bacterial transporters this coupling is dependent on the conserved histidine in the H-loop. Pdr5 lacks this residue in NBD1, and Schmitt and co-workers (13) have shown that the His → Ala mutation in Pdr5 has no effect on the ATPase activity. This has led them to posit that Pdr5 exhibits uncoupled ATPase activity. This may well be true; however, our data strongly suggest that structural domains involved in the transmission of conformational changes generated by ATP binding/hydrolysis to the TMDs are intact and may share features common to other ABC proteins.
Acknowledgments
We thank Dr. Patrick Mehl and J. J. Kim for running the flow cytometry assays and Michael Datiles for performing one of the ATPase assays. We appreciate the careful copyediting supplied by Trish Weisman.
This work was supported in part by the National Institutes of Health (Intramural Research Program, Center for Cancer Research, NCI). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ABC, ATP-binding cassette; cyh, cycloheximide; 5-FOA, 5-fluoroorotic acid; ICL, intracellular loop; NBD, nucleotide-binding domain; P-gp, P-glycoprotein; PM, plasma membrane; R6G, rhodamine 6G; TMD, transmembrane domain; TMH, transmembrane helix; WT, wild type; IAAP, iodoarylazidoprazosin; clo, clotrimazole; Vi, vanadate.
R. Rutledge and D. Xia, manuscript in preparation.
References
- 1.Posteraro, B., Sanguinetti, M., Sanglard, D., La Sorda, M., Boccia, S., Romano, L., Morace, G., and Fadda, G. (2003) Mol. Microbiol. 47 357-371 [DOI] [PubMed] [Google Scholar]
- 2.White, T. C., Marr, K. A., and Bowden, R. A. (1998) Clin. Microbiol. Rev. 11 382-396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monk, B. C., and Goffeau, A. (2008) Science 321 367-369 [DOI] [PubMed] [Google Scholar]
- 4.Decottignies, A., Kolaczkowski, M., Balzi, E., and Goffeau, A. (1994) J. Biol. Chem. 269 12797-12803 [PubMed] [Google Scholar]
- 5.Golin, J., Kon, Z. N., Wu, C. P., Martello, J., Hanson, L., Supernavage, S., Ambudkar, S. V., and Sauna, Z. E. (2007) Biochemistry 46 13109-13119 [DOI] [PubMed] [Google Scholar]
- 6.Nakamura, K., Niimi, M., Niimi, K., Holmes, A. R., Yates, J. E., Decottignies, A., Monk, B. C., Goffeau, A., and Cannon, R. D. (2001) Antimicrob. Agents Chemother. 45 3366-3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shukla, S., Rai, V., Banerjee, D., and Prasad, R. (2006) Biochemistry 45 2425-2435 [DOI] [PubMed] [Google Scholar]
- 8.Sauna, Z. E., and Ambudkar, S. V. (2007) Mol. Cancer Ther. 6 13-23 [DOI] [PubMed] [Google Scholar]
- 9.Jones, P. M., and George, A. M. (2004) Cell. Mol. Life Sci. 61 682-699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson, R. J. P., and Locher, K. P. (2006) Nature 443 180-185 [DOI] [PubMed] [Google Scholar]
- 11.Pagant, S., Brovman, E. Y., Halliday, J. J., and Miller, E. A. (2008) J. Biol. Chem. 283 26444-26451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zolnerciks, J. K., Wooding, C., and Linton, K. J. (2007) FASEB J. 21 3937-3948 [DOI] [PubMed] [Google Scholar]
- 13.Ernst, R., Kueppers, P., Klein, C. M., Schwarzmueller, T., Kuchler, K., and Schmitt, L. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 5069-5074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boeke, J. D., Trueheart, J., Natsoulis, G., and Fink, G. R. (1987) Methods Enzymol. 154 164-175 [DOI] [PubMed] [Google Scholar]
- 15.Katzmann, D. J., Hallstrom, T. C., Voet, M., Wysock, W., Golin, J., Volckaert, G., and Moylerowley, W. S. (1995) Mol. Cell. Biol. 15 6875-6883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rutledge, R. M., Ghislain, M., Mullins, J. M., de Thozee, C. P., and Golin, J. (2008) Mol. Genet. Genomics 279 573-583 [DOI] [PubMed] [Google Scholar]
- 17.de Thozee, C. P., Cronin, S., Goj, A., Golin, J., and Ghislain, M. (2007) Mol. Microbiol. 63 811-825 [DOI] [PubMed] [Google Scholar]
- 18.Shukla, S., Saini, P., Smriti, Jha, S., Ambudkar, S. V., and Prasad, R. (2003) Eukaryot. Cell 2 1361-1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambudkar, S. V. (1998) Methods Enzymol. 292 504-514 [DOI] [PubMed] [Google Scholar]
- 20.Ambudkar, S. V., Lelong, I. H., Zhang, J., Cardarelli, C. O., Gottesman, M. M., and Pastan, I. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 8472-8476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanson, L., May, L., Tuma, P., Keeven, J., Mehl, P., Ferenz, M., Ambudkar, S. V., and Golin, J. (2005) Biochemistry 44 9703-9713 [DOI] [PubMed] [Google Scholar]
- 22.Golin, J., Ambudkar, S. V., Gottesman, M. M., Habib, A. D., Sczepanski, J., Ziccardi, W., and May, L. (2003) J. Biol. Chem. 278 5963-5969 [DOI] [PubMed] [Google Scholar]
- 23.Golin, J., Ambudkar, S. V., and May, L. (2007) Biochem. Biophys. Res. Commun. 356 1-5 [DOI] [PubMed] [Google Scholar]
- 24.Schumacher, M. A., Miller, M. C., and Brennan, R. G. (2004) EMBO J. 23 2923-2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schumacher, M. A., and Brennan, R. G. (2003) Res. Microbiol. 154 69-77 [DOI] [PubMed] [Google Scholar]
- 26.Schumacher, M. A., Miller, M. C., Grkovic, S., Brown, M. H., Skurray, R. A., and Brennan, R. G. (2002) EMBO J. 21 1210-1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolaczkowski, M., van der Rest, M., Cybularz Kolaczkowska, A., Soumillion, J. P., Konings, W. N., and Goffeau, A. (1996) J. Biol. Chem. 271 31543-31548 [DOI] [PubMed] [Google Scholar]
- 28.Lockless, S. W., and Ranganathan, R. (1999) Science 286 295-299 [DOI] [PubMed] [Google Scholar]
- 29.Davis, J. E., Voisine, C., and Craig, E. A. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 9269-9276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Velamakanni, S., Yao, Y., Gutmann, D. A. P., and van Veen, H. W. (2008) Biochemistry 47 9300-9308 [DOI] [PubMed] [Google Scholar]
- 31.Neher, E. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 98-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haimeur, A., Conseil, G., Deeley, R. G., and Cole, S. P. C. (2004) Curr. Drug Metab. 5 21-53 [DOI] [PubMed] [Google Scholar]
- 33.Riordan, J. R. (2008) Annu. Rev. Biochem. 77 701-726 [DOI] [PubMed] [Google Scholar]
- 34.Jones, P. M., and George, A. M. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 12639-12644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyers, S., Schauer, W., Balzi, E., Wagner, M., Goffeau, A., and Golin, J. (1992) Curr. Genet. 21 431-436 [DOI] [PubMed] [Google Scholar]
- 36.Rogers, B., Decottignies, A., Kolaczkowski, M., Carvajal, E., Balzi, E., and Goffeau, A. (2001) J. Mol. Microbiol. Biotechnol. 3 207-214 [PubMed] [Google Scholar]