Abstract
Dermatomyositis (DM) is an autoimmune disease, which is often accompanied by the development of disease-specific autoantibodies directed against the SNF2-superfamily helicase, Mi-2. Recent evidence suggests that ultraviolet radiation exposure may be an important risk factor for the development of not only the disease but also specific autoimmunity against Mi-2. Consequently, we investigated the effects of ultraviolet radiation on Mi-2 protein expression. We observed an increase in protein levels upon ultraviolet radiation exposure in cell culture systems. These changes in expression occur quite rapidly, are maximized just 1 h following exposure, and are unique to Mi-2 when compared with other members of the NuRD complex. Changes in protein levels are not mediated through transcriptional mechanisms. Treatment results in a more efficiently translated message through regulatory elements in the 5′-UTR region of the transcript. Investigation into protein half-life further demonstrated increased stability of Mi-2 following UV exposure. Taken together, we describe a system by which Mi-2 protein expression can be quickly increased following UV exposure and then maintained up to 16 h later. These data provide a novel regulation of an important transcriptional regulator and provide insight into the possible mechanisms of the development of DM and associated autoantibodies.
Originally identified as an autoantigen in dermatomyositis (DM),2 the Mi-2 ATPase is now known to be the core subunit of the nucleosome remodeling deacetylase (NuRD) complex (1–5). Two isoforms of Mi-2 exist (α and β, also CHD3 and CHD4) and appear to have similar biological function. The β (CHD4) isoform is typically expressed at higher levels in vivo (6). In addition to the core Mi-2 ATPase, the NuRD complex contains histone deacetylases (HDAC) 1 and 2, retinoblastoma-binding proteins 46 and 48, metastasis-associated proteins (MTA1–3), and methyl CpG binding domain (MBD) proteins (7). Together, the complex is often linked with transcriptional repression associated with the histone deacetylase activity and the intrinsic nucleosome remodeling activity of Mi-2 (7). However, other functions of NuRD including transcriptional activation, transcriptional termination, and centrosome maintenance have been suggested (8–11). In addition, the NuRD complex has been implicated in the development and progression of disease states. Most notably, breast cancer expression patterns of the MTA subunits are prognostic indicators of metastasis and outcome (12).
DM is a form of idiopathic inflammatory myopathy (IIM), strongly associated with the development of Mi-2 autoantibodies. Like other IIMs, DM patients demonstrate signs of systemic inflammation and muscle weakness (13, 14). IIM patients also are at increased risk for the development of cancer, arthritis, and interstitial lung disease (15, 16). DM is distinguishable from polymyositis (PM) and other IIMs by the occurrence of photosensitive skin rashes (13). These rashes are hypothesized to occur in response to localized autoimmune reactions. Interestingly, 97% of IIM patients that present with antibodies to Mi-2 are eventually diagnosed with DM, suggesting an underlying mechanism unique to this form of the disease (17).
Recent evidence suggests that exposure to ultraviolet (UV) radiation may be an important risk factor for the disease as well as development of Mi-2 autoimmunity (18). A study investigating global surface UV intensity and the development of DM showed a statistically significant association of UV intensity with the frequency of disease (18). Even more interesting was a similar relationship between UV intensity and DM patients expressing Mi-2 autoantibodies. Because UV exposure induces a number of immunomodulatory effects and has been shown to contribute to autoimmunity, we investigated a possible role of UV regulation in Mi-2 protein in the skin that may contribute to the development of specific autoimmunity in DM patients. We demonstrate that in a keratinocyte cell culture system, UV exposure increases the expression of Mi-2 protein. These changes occur rapidly and seem to be unique to Mi-2 when compared with other NuRD complex members. We further identify the mechanisms by which Mi-2 protein is regulated following UV exposure. We demonstrate that Mi-2 message is translated at an increased rate following exposure to UV initiating rapid changes in protein levels detectable within 1 h of treatment. Furthermore, Mi-2 protein is stabilized allowing for the increased Mi-2 expression to be maintained up to 16 h. These findings highlight an important regulatory element of Mi-2 and possibly the NuRD complex that may also have implications for the development of immune responses to Mi-2 and ultimately DM.
EXPERIMENTAL PROCEDURES
Cell Culture and Plasmids—The human keratinocyte cell lines 1106 and 1102 were obtained from ATCC. Cells were grown in Keratinocyte-SFM (Invitrogen) media containing 50 μg/ml bovine pituitary extract (Invitrogen), 35 ng/ml epidermal growth factor (Invitrogen), and 50 μg/ml gentamicin (Invitrogen). The MCF7, C33a, HEK-293, and Hela were obtained from ATCC and cultured in DMEM cell lines containing 10% fetal bovine serum. The cell lines UL3 and NHF have been previously described (19, 20).
The untranslated region (UTR) of Mi-2β (Refseq: NM_001273) was amplified using PCR with primers containing the restriction sites for BamHI (5′-UTR) or NheI (3′-UTR). The resulting PCR product was digested with the corresponding restriction endonuclease and inserted into the BamHI or SpeI sites of pMir-Report (Ambion). Sequences were verified by sequencing at the NIEHS sequencing core using Big Dye terminator kit (Applied Biosystems). pRL-CMV was acquired from Promega Corporation.
Immunodetection—Keratinocytes plated on 10-cm2 dishes overnight were treated with UV radiation with a Tyler Research UV irradiator emitting a narrow 313-nm output at roughly 15 J/m2·s or ionizing radiation with the NIEHS 137Cs irradiator. Cells were harvested in lysis buffer (50 mm Tris-HCL, 1.0% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 400 mm NaCl) and total protein quantified using the Bio-Rad Protein assay. Cell lysates were subjected to SDS-PAGE followed by immunoblotting using specific antibodies. The antibodies directed against Mi-2, MTA2, and MTA3 were generous gifts of Dr. Paul Wade (NIEHS). The Mi-2 antibody is directed against a ∼700-amino acid region of Mi-2α, but recognizes both isoforms. We also utilized the following commercial antibodies: Mi-2β (Abcam, 54603), Mi-2α (Transduction Laboratories, C25020), MBD2 (Abcam, 38646), HDAC1 (Santa Cruz Biotechnology, sc-7872), HDAC2 (Abcam, 16032), Cox-2 (Cayman Chemical, 160112), cyclin A (Santa Cruz Biotechnology, SC-239), and β-actin (Abcam, 20272). Cells were treated with 2.5 μg/ml of actinomycin D or 1 μm MG132 as indicated.
FACS Analysis—Keratinocytes were treated with UV irradiation as described above, and cells harvested at the indicated time points by trypsinization and centrifugation. Cells were then resuspended in PBS and fixed by slow addition of cold ethanol. Cells were pelleted and resuspended in PBS containing 50 μg/ml propidium iodide and 40 μg/ml RNase A. Cells were analyzed on a Becton Dickinson FACSort flow cytometer.
Immunocytochemistry—Keratinocytes were seeded onto coverslips overnight and then irradiated with 90 J/m2 UV radiation. At the indicated timepoints, cells were fixed with 4% formaldehyde. Following extensive PBS washes, cells were permeabilized with PBS containing 0.5% Triton X-100 for 30 min. DNA was then denatured by incubating cells in 2 m HCL for 30 min. Cells were extensively washed in PBS and blocked in PBS containing 20% goat serum at 37 °C. Samples were then incubated in 5% goat serum with antibodies directed against cyclobutane pyrimidine dimers (CPDs) (Medical and Biological Laboratories, D194) and Mi-2 (Wade Lab, NIEHS). Specific staining was detected by secondary labeling using Alexa-fluor 555 anti-rabbit antibody and Alexa-fluor 488 anti-mouse antibody (Molecular Probes).
RT-PCR—Keratinocytes were plated and treated with 90 J/m2 UV radiation as described above. At the indicated time points, total RNA was harvested from cells using TRIzol reagent (Invitrogen) per the manufacturer's protocol. RNA from luciferase experiments was further purified by DNase I treatment followed by purification with Oligotex mRNA kit (Qiagen) to ensure no plasmid contamination. cDNA was generated using the Superscript III qRT-PCR reverse transcriptase kit (Invitrogen). Quantitative PCR was performed utilizing Sybr green detection of specific PCR products for the indicated targets. Primer sequences are identified in supplemental Table S1.
Reporter Assays—Keratinocytes were plated in 6-well dishes and transfected with 0.1 μg of pRL-CMV and 0.3 μg of the indicated reporter vector using Lipofectamine 2000 (Invitrogen) per manufacturer's protocol. Cells were treated with 90 J/m2 UV radiation and harvested 2 h post-treatment. Firefly and Renilla luciferase activities were measured using Dual Luciferase kit (Promega).
Metabolic Labeling—MCF7 cells were incubated with methionine-free DMEM for 2 h followed by treatment with 90 J/m2 UV radiation. Immediately following UV treatment, cells were incubated in DMEM containing 100 uCi/ml [35S]methionine for 2 h. Thereafter, cell lysates were harvested in IP buffer (50 mm Tris-HCl, pH 8, 200 mm NaCl, 1 mm EDTA, and 1% Triton X-100) and immunoprecipitated using Mi-2 specific antibodies or nonspecific antibodies purified from rabbit serum. Immunoprecipitated complexes were subjected to SDS-PAGE and analyzed by Storm Phosphorimager (GE Healthcare).
RESULTS
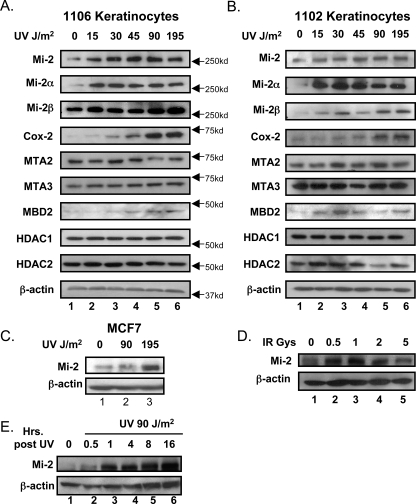
UV Radiation Regulates Mi-2 Expression in Human Keratinocytes—Mi-2 was first identified as an autoantigen in patients suffering from DM, a disease characterized by photosensitive skin rashes. Global epidemiological studies suggest that UV exposure increases the risk for development of DM as well as Mi-2-specific autoantibodies (18). We hypothesized a relationship between UV radiation and the regulation of the Mi-2 protein in the cellular context of these photosensitive rashes. We utilized a human keratinocyte cell line (1106) derived from neonatal keratinocytes immortalized with the human papilloma virus. Interestingly, cells treated with an increasing dose (0–195 J/m2) of UV radiation demonstrate a subsequent increase in total Mi-2 protein expression 8 h after treatment (Fig. 1A, top panel). This regulation appears to be maximized at 45 J/m2 with higher doses having no additional effects. The effects are seen on both isoforms of Mi-2, although the strongest effect is seen in Mi-2α. As Mi-2 is the core subunit of NuRD chromatin remodeling complex, we investigated whether UV radiation also affected the other members of the complex. However, other NuRD complex proteins did not show similar up-regulation in protein expression with the exception of MBD2, which showed a modest increase in protein levels at the higher UV doses. The NuRD complex member, MTA1, had no detectable expression in the 1106 keratinocyte cell line. Cox-2 protein expression, which is known to be increased following UV exposure, was also analyzed as a positive control. Interestingly, the Cox-2 response occurs at higher doses than the 45 J/m2 required to induce Mi-2, suggesting that the Mi-2 pathway harbors a greater UV sensitivity.
FIGURE 1.
UV radiation increases Mi-2 expression. Immortalized human keratinocyte cell lines, 1106 (A) and 1102 (B), were treated with the indicated doses of UV radiation. 8 h after treatment, cells lysates were harvested, subjected to SDS-PAGE, and immunoblotted with antibodies specific for the indicated proteins. C, MCF7 cells were treated with the indicated doses of UV radiation. 8 h after treatment, cell lysates were analyzed for changes of Mi-2 protein expression. D, the 1106 keratinocyte cell line was treated with the indicated amounts of ionizing radiation. 8 h following treatment, cell lysates were harvested, and Mi-2 and β-actin protein expression were analyzed by immunoblot. E, the 1106 keratinocyte cell line was treated with 90 J/m2 of UV radiation and cell lysates harvested at indicated time points. Samples were analyzed for Mi-2 and β-actin expression by immunoblot.
Because of a higher incidence of DM in females (21), we investigated a possible gender predisposition to these UV effects. We acquired another keratinocyte cell line (1102) and determined that it was derived from a female background by PCR of Y-chromosome-specific DNA (data not shown). The 1106 cell line was determined to be derived from a male (data not shown). However, treatment of the 1102 keratinocytes with UV radiation showed similar effects to that seen in the 1106 cell line (Fig. 1B) demonstrating that Mi-2 induction by UV is gender-independent. The other NuRD complex members behaved similarly in both cell lines. We further demonstrated this effect is not keratinocyte specific by treating the human breast cancer cell line, MCF7, with UV radiation and also observing increases in Mi-2 expression (Fig. 1C). As the NuRD complex regulation has been linked to the estrogen response in breast cancer (22, 23), we also performed the experiment in MCF7 cells deprived of estrogen and saw identical results (supplemental Fig. S1A). To determine if this response is conserved, we investigated several other cells lines for changes in Mi-2 expression following UV exposure. The osteosarcoma cell line UL3, cervical cancer lines C33a and Hela, breast cancer cell line BT549, and normal human fibroblasts all demonstrated strong up-regulation of Mi-2 following UV exposure (supplemental Fig. S1B). The transformed fetal kidney line HEK-293 showed no change in Mi-2 expression (supplemental Fig. S1B). These data support a conserved, though not ubiquitous, model that Mi-2 protein levels increase following UV exposure. To address if these effects are specific for UV irradiation or a general response to DNA damage, cells were treated with increasing levels of ionizing radiation (0–5 grays). As seen with the UV treatments, Mi-2 protein expression increased following ionizing radiation implicating a mechanism initiated by general DNA damage (Fig. 1D).
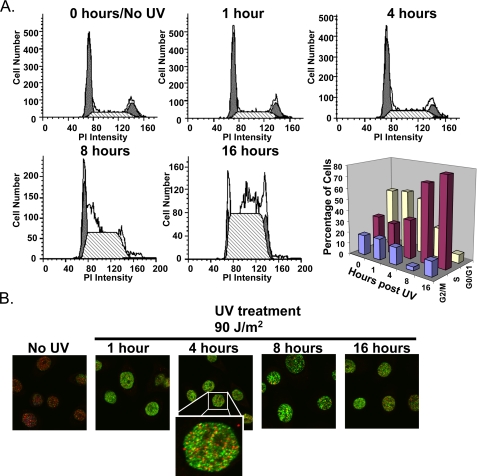
As DNA damage elicits a number of cellular processes from DNA repair to cell cycle arrest, each occurring over varying time courses, we performed a time course of Mi-2 induction in the keratinocyte cell line (1106). Keratinocytes were treated with 90 J/m2 UV radiation and cell lysates harvested at the indicated time points (Fig. 1E). Analysis of Mi-2 expression indicates a fairly rapid increase in protein levels seen within 30 min and maximized at 1 h after UV treatment. These changes are maintained up to 16 h following UV treatment. As DNA damage induces cell cycle checkpoints, we investigated if treatment with UV radiation had any affect on the cell cycle profile of keratinocytes. Cells were treated with 90 J/m2 of UV radiation, harvested at time points between 1 and 16 h, and subjected to propidium iodide flow cytometry analysis. In fact, UV radiation did initiate S-phase accumulation (Fig. 2A). However, these effects are not seen until 8 h after treatment, while the Mi-2 protein changes occur as early as ½ hour post-treatment (see Fig. 1E). At 1 h, when Mi-2 expression is maximized, there is no change in the cell cycle profile thus demonstrating that changes in Mi-2 expression are not a result of changes in the cell cycle. Additionally, we observed no accumulation of a sub-G0 population that is usually indicative of apoptosis. Mock treatment of samples also induced an increase in S phase at 8 and 16 h (supplemental Fig. S2). However, this increase is quite modest compared with the checkpoint created by UV exposure and is most likely caused by the addition of fresh media during mock treatment. To assess the severity of DNA damage accumulated over the time course of our experiments, we performed immunocytochemistry on keratinocytes treated with UV radiation. Damage induced by UV radiation was monitored with antibodies specific for CPDs. Intense staining of CPDs is observed 1 h following treatment and is maintained up to 16 h later (Fig. 2B), while little background staining is seen in the untreated cells. At the 16-h time point, the intensity of staining for CPDs is decreased indicating repair of many of the lesions, which, interestingly, may explain a slight increase in G2/M population by FACS (Fig. 2A). Co-staining for Mi-2 and CPDs in these samples shows no co-localization and implies that increased Mi-2 expression following DNA damage is not directly involved in repair of the lesions (Fig. 2B).
FIGURE 2.
UV radiation induces an S-phase checkpoint following Mi-2 induction. A, keratinocytes were treated with 90 J/m2 of UV radiation, and cells were harvested at the indicated time points for prodium iodide flow cytometry analysis. B, keratinocytes were plated onto coverslips and either mock-treated or irradiated with 90 J/m2 UV. Cells were fixed at the indicated time points following UV exposure and immunocytochemistry performed to detect CPDs (green) and Mi-2 (red).
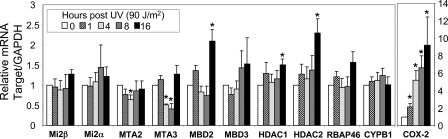
Induction of Mi-2 following UV Is Not Regulated Transcriptionally—We have shown that Mi-2 protein expression is highly induced in less than 1 h following exposure to UV radiation. We addressed the mechanism of regulation by investigating the changes to Mi-2 mRNA levels following UV treatment by quantitative real time RT-PCR. Keratinocytes were irradiated with 90 J/m2 UV, and mRNA harvested at the indicated time points. Real-time RT-PCR analysis of the message corrected against GAPDH showed no significant changes in either Mi-2α or Mi-2β (Fig. 3A). Cox-2 (positive control) message was significantly increased over the time course, whereas CYPB1 (negative control) was unchanged following UV exposure. Interestingly, some members of the NuRD complex were significantly changed following UV exposure. Both MTA2 and MTA3 showed decreased mRNA expression 8 h after treatment, whereas MBD2, HDAC1, and HDAC2 showed modest but significant increases in message 16-h post UV. However, HDAC protein levels did not increase at either 8 h (Fig. 1, A and B) or 16 h (data not shown). The MBD2 protein did exhibit the aforementioned increase in protein expression. To further verify that the changes in Mi-2 protein following UV treatment are not regulated at the transcriptional level, keratinocytes were treated with the RNA polymerase II inhibitor actinomycin D following UV exposure. Cells treated with actinomycin D still showed induction of Mi-2 following UV treatment (data not shown). These data verify that changes in Mi-2 protein levels are not regulated at the transcriptional level.
FIGURE 3.
Mi-2 induction is regulated through transcriptional mechanisms. A, keratinocytes were treated with 90 J/m2 UV radiation and RNA harvested at the indicated time points. Relative mRNA for the indicated targets was analyzed by quantitative real-time RT-PCR. Data presented represents the average of six individual experiments with the S.E. * represents p < 0.05.
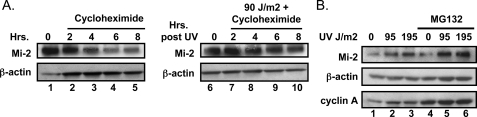
Mi-2 Is Regulated through Translational and Post-translation Mechanisms—Because the level of Mi-2 mRNA does not change following UV radiation, we investigated post-transcriptional mechanisms of Mi-2 induction. A number of proteins are stabilized following DNA damage as a mechanism to increase expression, most notably p53 and CHK2. Therefore, we investigated the ability of UV radiation to regulate Mi-2 expression by analyzing protein stability using the translational inhibitor cycloheximide. Keratinocytes were treated with 1 μm cycloheximide following exposure to 90 J/m2 UV radiation and lysates analyzed over a time course of 8 h. Interestingly, Mi-2 protein had a remarkably longer half-life following UV treatment (Fig. 4A) as compared with untreated cells. The increased stability becomes evident at 4-h post-treatment (compare lanes 3 and 8) and is still apparent at the 8-h time point. Interestingly, at 2-h post-treatment, no change in protein levels is noticeable. This is in contrast to the previously observed increased Mi-2 expression 30 min after UV exposure (Fig. 1D). Consequently the early rise in Mi-2 levels would not be accounted for by the stabilization of the protein. Taken together, these data suggest an alternate mechanism in the rapid accumulation of Mi-2 protein following UV exposure. To test if an alternate regulatory mechanism existed, Mi-2 protein was pharmacologically stabilized using the proteosome inhibitor MG132. Keratinocytes treated with MG132 following UV exposure still show increased Mi-2 expression (Fig. 4B, lanes 4–6) beyond the stabilization of the protein in the untreated sample. Cyclin A, a protein whose expression is regulated tightly by degradation, was utilized as a control for MG132 effectiveness and showed an increase in protein following MG132 treatment (Fig. 4B, compare lanes 1–3 to 4–6).
FIGURE 4.
Mi-2 is stabilized following UV radiation. A, keratinocytes were treated with cycloheximide (50 ng/ml) and 90 J/m2 UV radiation as indicated. Cell lysates were harvested at the specified time points and protein expression analyzed by immunoblot. B, keratinocytes were treated with MG132 and 90 J/m2 UV radiation as indicated. 4 h after treatment, cell lysates were harvested and protein expression analyzed by immunoblot.
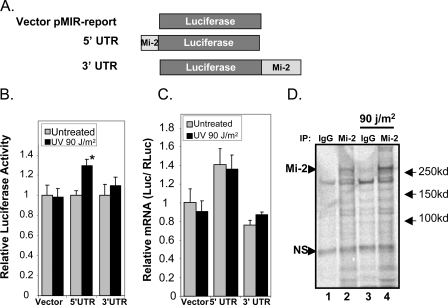
Because stabilized protein does not account for the total increase in Mi-2 protein accumulation, we investigated changes in translational efficiency of the Mi-2 mRNA. There exist several well characterized mechanisms by which protein expression is regulated at the translational level, including systems that respond to DNA damage. Because previous work has linked DNA damage to regulation of translation through elements within the UTR of target proteins, we investigated a possible mechanism of Mi-2 regulation (24, 25). Using the pMir-Report vector system we linked the UTRs of Mi-2β to the 5′- and 3′-ends of luciferase (Fig. 5A). Keratinocytes were then transfected with these constructs or the parental vector to determine if either UTR contains regulatory elements that respond to UV radiation. The reporter vector containing the 5′-UTR of Mi-2β showed a significant increase in luciferase activity (29.3%, p < 0.0001) just 2 h following UV treatment (Fig. 5B). In contrast, the parental vector and the reporter containing the 3′-UTR of Mi-2β showed no significant increase in luciferase activity, demonstrating that mRNA regulation is specific to the 5′-UTR. To verify that this increase in luciferase activity is not a result of transcriptional changes initiated after UV exposure, mRNA from cells transfected as in Fig. 5B was subjected to quantitative real-time RT-PCR. No significant increase in mRNA levels was observed in the UV treated cells for the 5′-UTR reporter (Fig. 5C). Finally, to verify that this regulatory mechanism in fact manipulates Mi-2 protein levels following UV radiation, we performed an in vivo labeling assay of newly translated protein. Unfortunately, keratinocyte cells reacted unfavorably to methionine-free media, so these experiments were performed in MCF7 cells that we have shown induce Mi-2 protein expression following UV radiation in a manner similar to keratinocytes (Fig. 1C). Cells were incubated in [35S]methionine containing media following treatment with or without 90 J/m2 UV radiation. Mi-2 protein from lysates was immunoprecipitated 2 h after irradiation and newly translated protein visualized by autoradiography. As expected, cells treated with UV radiation had a higher expression of labeled Mi-2 as compared with the mock-treated (Fig. 5C, compare lanes 2 and 4). In contrast, a nonspecific protein immunoprecipitated by IgG shows no change. These combined data suggest that a regulatory element within the 5′-UTR of Mi-2 increases translation leading to protein accumulation following UV treatment. In combination with increased stability, these mechanisms allow for a rapid increase in Mi-2 protein levels that are sustained hours after UV exposure (Fig. 1E).
FIGURE 5.
Mi-2 protein translation is regulated through the 5′-UTR following UV radiation. A, the 5′- and 3′-UTRs of Mi-2β were cloned into the pMIR-Report luciferase vector as depicted. B, keratinocytes were transfected with CMV-Renilla luciferase and the indicated reporter plasmid. 24 h following transfection, cells were treated with 90 J/m2 UV radiation. Cell lysates were collected 2 h following treatment and analyzed for luciferase activity. Data represent the average of at least nine independent points with S.E. C, mRNA was harvested from cells transfected and treated as in B and used for real-time RT-PCR analysis. Relative luciferase message levels are graphed corrected against Renilla luciferase message. D, MCF7 cells were mock-treated or treated with 90 J/m2 UV followed by incubation with [35S]methionine containing media. 2 h after treatment, cell lysates were collected and immunoprecipitated with Mi-2 specific antibodies or purified nonspecific rabbit IgG. Precipitated proteins were subjected to SDS-PAGE and visualized by phosphorimaging. The arrows mark the Mi-2 protein and a nonspecific protein used as a loading control.
DISCUSSION
DM patients who suffer from photosensitive skin rashes have a propensity to develop autoantibodies against the Mi-2 subunit of NuRD complex (13). Moreover, UV radiation exposure has been linked to an increased risk of developing DM and Mi-2 autoantibodies (18). Herein, we investigated the role of UV radiation on the regulation of Mi-2 protein. We demonstrate that keratinocytes respond to UV radiation by rapidly increasing Mi-2 protein levels. These changes in protein expression are not universal among other NuRD complex members. We further depict a model system for this regulation where Mi-2 message is translated more efficiently following UV treatment through a regulatory element within the 5′-UTR region of the mRNA. This mechanism allows for the rapid increase in protein that occurs within 30 min of treatment and is maximized at only 1-h post-UV. The elevated protein levels are maintained through another regulatory mechanism of protein stability. Together, these data suggest a model by which cells respond to UV exposure by quickly inducing Mi-2 protein expression, which can be sustained 16 h following this exposure.
Mi-2 Regulation Is Post-transcriptional—We have demonstrated that Mi-2 protein is increased following exposure to UV and that this response is most likely a general response of DNA damage. The increases in protein level occur rapidly through changes in translational efficiency regulated, at least in part, through the 5′-UTR region of the Mi-2β message. Similar mechanisms have been described for the regulation of p53 protein following DNA damage. Regulatory elements in the p53 3′-UTR are regulated by human antigen R to control expression (24). Human antigen R binds to the mRNA following DNA damage and stimulates translation. However, not all translational mechanisms act through the 3′-UTR. Ribosomal protein L26 (RPL26) and Nucleolin have been shown to interact with the 5′-UTR of p53 and regulate translation, however, in opposite ways (25). Nucleolin inhibits translation in the absence of DNA damage while RPL26 increases translation upon DNA damage signaling. Thus, protein control mechanisms within the cell are in place to rapidly regulate protein levels without transcription of the possibly damaged genome. Interestingly, Mi-2 is regulated in this manner, yet does not have a known role in DNA damage repair. Our immunostaining further demonstrate that Mi-2 does not co-localize with sites of damage. However, there exist a number of other physiological responses such as cell cycle checkpoints that are initiated following damage as well as transcriptional regulation of important DNA damage response genes that may rely on Mi-2/NuRD.
Functional Consequence of Mi-2 Induction—We have demonstrated conserved mechanism by which Mi-2 protein is increased rapidly following exposure to both UV and IR (Fig. 1 and supplemental Fig. S1B). Mi-2 is involved in transcriptional regulation through its association with the NuRD complex. In addition to the intrinsic ATPase chromatin remodeling activity of Mi-2, the NuRD complex contains HDACs and MBD proteins known to associate with methylated DNA (7). The NuRD complex is also known to associate with retinoblastoma-interacting proteins, which are generally considered molecular chaperones (26). The NuRD complex has also been associated with other biological functions involved in regulating centrosome integrity, maintenance of chromatid cohesion, and transcriptional termination (9–11). However, we have demonstrated that Mi-2 does not localize to the sites of DNA damage (Fig. 2B) in contrast to other chromatin remodeling complexes, such as SWI/SNF (27). It is also interesting to note that not all NuRD complex members are similarly up-regulated following UV exposure (Fig. 1). It is possible that Mi-2 is the limiting protein in the complex and its up-regulation is sufficient to increase complex formation and activity. Mi-2 could also be acting independent of the NuRD complex. Mi-2 has been shown to act as a transcriptional activator. Mi-2α was recently determined to co-activate c-Myb independent of ATPase activity (8). The role of other NuRD complex members in regulating c-Myb activity in this study was not addressed. However, another study demonstrated that Mi-2β is required for T cell development through its activation of the E box-binding protein HEB and concomitant expression of CD4 (28). This function is dependent upon Mi-2β interaction with p300. Importantly, a HEB/Mi-2/p300 complex lacked HDAC2, strongly supporting a model of NuRD-independent activity. Taken together, the biological significance of Mi-2 regulation following UV exposure has broad implications and will be addressed in future investigations.
Mi-2 Regulation and Dermatomyositis—Mi-2 protein was first identified as the target of autoantibodies in patients with DM. Whereas development of these autoantibodies against Mi-2 is not known to be causative and is not required for development of the disease, studies suggest that roughly 20% of patients present these autoantibodies (13). However, the striking characteristic of Mi-2 autoantigenicity is its specific finding in DM as compared with other forms of autoimmune and nonautoimmune muscle disease. In fact, 97% of patients with Mi-2 specific antibodies suffer from DM (29). Our results may provide an explanation for these findings. Increased expression of Mi-2 in the UV-induced dermatitis may provide the antigen presentation required for the development and maintenance of the autoantibodies. This hypothesis is supported by related studies from the literature suggesting that autoimmunity in many forms of myositis may be the result of altered autoantigen presentation rather than a defect in selectivity seen in other autoimmune diseases. Studies have shown overexpression of the myositis antigens Jo-1, Mi-2, DNA, PKCS, and U1-RNP in patients with the disease (30). Moreover, a mouse model overexpressing HLA presentation molecules specifically in muscle induced myositis in all animals (31). Another mouse model has demonstrated that muscle lesions can generate an immune response to muscle components and provides a model comparable to skin lesions in DM patients following UV exposure (32). Finally, it has been shown in humans that antigen expression within myositis tissue is higher than that seen in normal muscle (14). These data, taken together with the prior association of UV exposure with the development of Mi-2 autoantibodies and exacerbations of distinctive skin rashes in DM (18, 33), support a mechanism of UV-induced DM-specific autoimmunity in the immune targeting of Mi-2.
Supplementary Material
Acknowledgments
We thank Dr. Paul Wade for his critical input and reagents. We would also like to thank Carl Bortner and Maria Sifre of the NIEHS Flow Cytometry Core, Grace Kissling of the NIEHS Biostatistics Core, and Terrance O'Hanlon in the NIEHS Environmental Autoimmunity Group for technical assistance.
This work was supported, in whole or in part, by Program Grant Z01 ES071006-09 in the Intramural Research Program of the NIEHS, National Institutes of Health. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Table S1.
Footnotes
The abbreviations used are: DM, dermatomyositis; IIM, idiopathic inflammatory myopathy; PM, polymyositis; DMEM, Dulbecco's modified Eagle's medium; UTR, untranslated region; PBS, phosphate-buffered saline; NuRD, nucleosome remodeling deacetylase; HDAC, histone deacetylase; CPD, cyclobutane pyrimidine dimer.
References
- 1.Targoff, I. N., and Reichlin, M. (1985) Arthritis Rheum. 28796 -803 [DOI] [PubMed] [Google Scholar]
- 2.Xue, Y., Wong, J., Moreno, G. T., Young, M. K., Cote, J., and Wang, W. (1998) Mol. Cell 2 851-861 [DOI] [PubMed] [Google Scholar]
- 3.Zhang, Y., LeRoy, G., Seelig, H. P., Lane, W. S., and Reinberg, D. (1998) Cell 95 279-289 [DOI] [PubMed] [Google Scholar]
- 4.Tong, J. K., Hassig, C. A., Schnitzler, G. R., Kingston, R. E., and Schreiber, S. L. (1998) Nature 395917 -921 [DOI] [PubMed] [Google Scholar]
- 5.Wade, P. A., Jones, P. L., Vermaak, D., and Wolffe, A. P. (1998) Curr. Biol. 8 843-846 [DOI] [PubMed] [Google Scholar]
- 6.Kim, J., Sif, S., Jones, B., Jackson, A., Koipally, J., Heller, E., Winandy, S., Viel, A., Sawyer, A., Ikeda, T., Kingston, R., and Georgopoulos, K. (1999) Immunity 10 345-355 [DOI] [PubMed] [Google Scholar]
- 7.Denslow, S. A., and Wade, P. A. (2007) Oncogene 265433 -5438 [DOI] [PubMed] [Google Scholar]
- 8.Saether, T., Berge, T., Ledsaak, M., Matre, V., Alm-Kristiansen, A. H., Dahle, O., Aubry, F., and Gabrielsen, O. S. (2007) J. Biol. Chem. 28213994 -14005 [DOI] [PubMed] [Google Scholar]
- 9.Alen, C., Kent, N. A., Jones, H. S., O'Sullivan, J., Aranda, A., and Proudfoot, N. J. (2002) Mol. Cell 101441 -1452 [DOI] [PubMed] [Google Scholar]
- 10.Hakimi, M. A., Bochar, D. A., Schmiesing, J. A., Dong, Y., Barak, O. G., Speicher, D. W., Yokomori, K., and Shiekhattar, R. (2002) Nature 418994 -998 [DOI] [PubMed] [Google Scholar]
- 11.Sillibourne, J. E., Delaval, B., Redick, S., Sinha, M., and Doxsey, S. J. (2007) Mol. Biol. Cell 183667 -3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar, R., Wang, R. A., and Bagheri-Yarmand, R. (2003) Semin Oncol. 30 Suppl. 16, 30-37 [DOI] [PubMed] [Google Scholar]
- 13.Callen, J. P., and Wortmann, R. L. (2006) Clin. Dermatol. 24363 -373 [DOI] [PubMed] [Google Scholar]
- 14.Suber, T. L., Casciola-Rosen, L., and Rosen, A. (2008) Nat. Clin. Pract. Rheumatol. 4 201-209 [DOI] [PubMed] [Google Scholar]
- 15.Schnabel, A., Hellmich, B., and Gross, W. L. (2005) Curr. Rheumatol. Rep. 799 -105 [DOI] [PubMed] [Google Scholar]
- 16.Levine, S. M. (2006) Curr. Opin. Rheumatol. 18620 -624 [DOI] [PubMed] [Google Scholar]
- 17.Roux, S., Seelig, H. P., and Meyer, O. (1998) J. Rheumatol. 25395 -396 [PubMed] [Google Scholar]
- 18.Okada, S., Weatherhead, E., Targoff, I. N., Wesley, R., and Miller, F. W. (2003) Arthritis Rheum. 482285 -2293 [DOI] [PubMed] [Google Scholar]
- 19.Fryer, C. J., Kinyamu, H. K., Rogatsky, I., Garabedian, M. J., and Archer, T. K. (2000) J. Biol. Chem. 27517771 -17777 [DOI] [PubMed] [Google Scholar]
- 20.Devereux, T. R., Horikawa, I., Anna, C. H., Annab, L. A., Afshari, C. A., and Barrett, J. C. (1999) Cancer Res. 596087 -6090 [PubMed] [Google Scholar]
- 21.Beeson, P. B. (1994) Am. J. Med. 96457 -462 [DOI] [PubMed] [Google Scholar]
- 22.Fujita, N., Kajita, M., Taysavang, P., and Wade, P. A. (2004) Mol. Endocrinol. 182937 -2949 [DOI] [PubMed] [Google Scholar]
- 23.Dhasarathy, A., Kajita, M., and Wade, P. A. (2007) Mol. Endocrinol. 212907 -2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazan-Mamczarz, K., Galban, S., Lopez de Silanes, I., Martindale, J. L., Atasoy, U., Keene, J. D., and Gorospe, M. (2003) Proc. Natl. Acad. Sci. U. S. A. 1008354 -8359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takagi, M., Absalon, M. J., McLure, K. G., and Kastan, M. B. (2005) Cell 123 49-63 [DOI] [PubMed] [Google Scholar]
- 26.Loyola, A., and Almouzni, G. (2004) Biochim. Biophys. Acta 1677 3-11 [DOI] [PubMed] [Google Scholar]
- 27.Park, J. H., Park, E. J., Lee, H. S., Kim, S. J., Hur, S. K., Imbalzano, A. N., and Kwon, J. (2006) EMBO J. 253986 -3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams, C. J., Naito, T., Arco, P. G., Seavitt, J. R., Cashman, S. M., De Souza, B., Qi, X., Keables, P., Von Andrian, U. H., and Georgopoulos, K. (2004) Immunity 20 719-733 [DOI] [PubMed] [Google Scholar]
- 29.Sordet, C., Goetz, J., and Sibilia, J. (2006) Joint Bone Spine 73646 -654 [DOI] [PubMed] [Google Scholar]
- 30.Casciola-Rosen, L., Nagaraju, K., Plotz, P., Wang, K., Levine, S., Gabrielson, E., Corse, A., and Rosen, A. (2005) J. Exp. Med. 201591 -601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagaraju, K., Raben, N., Loeffler, L., Parker, T., Rochon, P. J., Lee, E., Danning, C., Wada, R., Thompson, C., Bahtiyar, G., Craft, J., Hooft Van Huijsduijnen, R., and Plotz, P. (2000) Proc. Natl. Acad. Sci. U. S. A. 979209 -9214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevens, A. M., Sullivan, K. M., and Nelson, J. L. (2003) Rheumatology (Oxford) 42 34-39 [DOI] [PubMed] [Google Scholar]
- 33.Ghirardello, A., Zampieri, S., Iaccarino, L., Tarricone, E., Bendo, R., Gambari, P. F., and Doria, A. (2005) Autoimmunity 3879 -83 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.