Abstract
WDR5 is a component of the mixed lineage leukemia (MLL) complex, which methylates lysine 4 of histone H3, and was identified as a methylated Lys-4 histone H3-binding protein. Here, we present a crystal structure of WDR5 bound to an MLL peptide. Surprisingly, we find that WDR5 utilizes the same pocket shown to bind histone H3 for this MLL interaction. Furthermore, the WDR5-MLL interaction is disrupted preferentially by mono- and di-methylated Lys-4 histone H3 over unmodified and tri-methylated Lys-4 histone H3. These data implicate a delicate interplay between the effector, WDR5, the catalytic subunit, MLL, and the substrate, histone H3, of the MLL complex. We suggest that the activity of the MLL complex might be regulated through this interplay.
DNA in eukaryotic cells forms into higher order structures made up of chromatin. Chromatin, and its higher order structure, is dynamically modified in concert with gene expression and cell cycle. The fundamental unit of the chromatin, the nucleosome, is composed of a histone octamer (dimers of H2A, H2B, H3, and H4) and 146 base pairs of DNA, which wrap around the histone octamer (1). Histones have unstructured N- or C-terminal tails protruding out of the nucleosome core structure. These tails are covalently modified with methyl, acetyl, phospho, ubiquitin, sumo, and ADP-ribose moieties (2).
Addition and removal of these covalent modifications can affect gene expression, as the modifications can serve to recruit effector proteins. Methylation of lysine (or arginine) of histones adds another layer of complexity, as lysine can be mono-, di-, and tri-methylated, and arginine mono-, and di-methylated. Histone H3 Lys-4 and Lys-27 methylations are particularly interesting, because these marks are antagonistic to each other in their functions in that they are highly correlated with gene activation (Lys-4) and repression (Lys-27) (3).
Previously, in an effort to find an effector molecule recognizing a methylated Lys-4 of histone H3, the WDR5 protein was identified as a methyl Lys-4 H3-specific-binding protein, and it was shown that WDR5 is required for tri-methylation of histone H3 in vivo (4). WDR5 belongs to the WD40 repeat protein family. The WD40 repeat is a well characterized protein-protein interaction domain involved in diverse cellular processes. The WDR5 protein was also previously identified as a component of mixed lineage leukemia (MLL)3 complex (5). The MLL complex is a histone H3 Lys-4 methyltransferase. ASH2L, RBBP5, and MLL interact with WDR5 to comprise a core MLL complex (5). WDR5, ASH2L, and RBBP5 also form complexes with the Set1 protein and with the four isoforms of MLL: MLL1, MLL2, MLL3, and MLL4. The MLL and Set1 proteins are the catalytic subunits of these complexes, and each contains a SET domain at the C terminus. However, the molecular functions of each component are largely unknown, as are many of the molecular mechanisms that regulate the function of the MLL complex.
WDR5 is a component of the MLL complex, is required for histone H3 tri-methylation, and binds histone H3; those observations that have led to the proposal that it is a presenter component of MLL, meaning that WDR5 presents methylated histone H3 substrates to the MLL complex for further methylation. Crystal structures of WDR5 bound to methylated histone H3 Lys-4 peptides have been reported (6-9). However, surprisingly, not as initially expected, the methylated Lys-4 side chain has minimal interaction with WDR5 in these structures. Based on these crystal structures, and on binding experiments with isothermal calorimetry and surface plasmon resonance (SPR) techniques, it has been suggested that WDR5 does not discriminate the methylation status of histone H3 Lys-4, in contrast to initial results (6, 8). These discrepancies have not been resolved.
Here, we identify a WDR5-interacting region in MLL and present the crystal structure of WDR5 bound to this MLL1 peptide. Surprisingly, the crystal structure shows that WDR5 interacts with MLL1 via the same pocket, which was previously shown to be the H3-binding pocket. Moreover, our binding experiments show that the WDR5-MLL1 interaction is preferentially competed by mono- or di-methylated Lys-4 histone H3. These data imply that WDR5 binds preferentially to mono- or di-methylated histone H3 over unmodified histone H3 in the presence of the MLL protein. We suggest a model in which there is an interplay between the histone H3 substrate of the MLL enzyme and MLL itself that is modulated by methylation status of the substrate. The MLL complex, and the subsequent methylation of histone H3, might therefore be regulated by a delicate feedback mechanism.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification—WDR5 and MLL fragments were cloned into pGEX_6p vector and expressed in glutathione S-transferase (GST) fusion forms in Escherichia coli. GST fusion forms of MLL or WDR5 were expressed in BL21(DE3) E. coli at 17 °C with 17 h of induction with 1 mm isopropyl β-d-1-thiogalactopyranoside. Cells expressing the proteins were harvested in a buffer containing 50 mm Tris-HCl, pH 8.0, 500 mm NaCl, and 5% (v/v) glycerol, The cells were sonicated, and cell debris was removed by centrifugation. GST-WDR5 and GST-MLL were captured by glutathione-agarose resin (GE Healthcare) and further purified by HiTrap Q (GE Healthcare) ion exchange chromatography. The proteins were stored in a buffer containing 50 mm Tris-HCl, pH 8.0, 150 mm NaCl buffer. For WDR5 crystallization, GST was removed by Precission Protease (GE Healthcare) and further purified by Hitrap Q (GH Healthcare) ion exchange and Superdex S75 16/60 (GE Healthcare) size exclusion chromatography column.
Crystallization and Data Collection—WDR5-MLL peptide crystals were obtained under conditions similar to the previously reported conditions (6) for WDR5 crystallization in the presence of 2 mm MLL peptide (ARAEVHLRKSAFD) (BioSyn). Plate-type crystals were grown by the vapor diffusion method in mother liquor containing 20 mm HEPES (pH 7.5), 15-20% PEG3350, and 60 mm ammonium sulfate. For cryo-protection, the crystals were soaked in a cryo-solution containing 20 mm HEPES, pH 7.5, 18% PEG 3350, 18% EG, and 60 mm ammonium sulfate, and frozen in liquid nitrogen. Data were collected at 1.0809 Å at beamline X29A at the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory (BNL). The crystals diffracted up to 1.37 Å, and the space was determined as C2221 containing one molecule per asymmetric unit. WDR5-MLL peptide structure was determined by molecular replacement using WDR5 (PDB: 2H13) as a search model. The structure was refined with the program CNS with Rwork 19.03% and Rfree 19.98%. The coordinates were deposited at the Protein Data Bank with a PDB code of 3EMH.
Pull-down and Competition Experiments—GST-MLL228 proteins bound to glutathione-Sepharose beads (GE Healthcare) were incubated with WDR5 in a buffer (Buffer A) containing 50 mm Tris-HCl, pH 8.0, 100 mm NaCl, and 5% glycerol for 2 h. Proteins bound to the bead were collected by brief centrifugation and washed with the Buffer A several times. Bound WDR5 was analyzed in SDS-PAGE gel, and the amount of protein bound to the bead was measured by densitometer (Bio-Rad). The bound fractions were normalized by the amounts of MLL. For competition experiments, GST-MLL228 was incubated with WDR5 (3 μm) with the increasing amounts of unmodified, mono-, di-, and tri-methylated histone H3 peptides (1-11 aa, 5, 20, 60, 150 μm) (BioSyn), and the amount of bound WDR5 was analyzed as described above.
Surface Plasmon Resonance—For KD measurements, the surface of a CM5 chip was activated with a mixture of EDC and NHS. GST-MLL228 and GST-MLLpep were immobilized on the Chip with 663 and 607 RU, and GST was immobilized on the reference cells with 610 and 605 RU, respectively. After immobilization, the surface was blocked with 1 m ethanolamine solution and conditioned with a regeneration buffer containing 37.5 mm Tris-HCl, pH 8.0, 1.5 m NaCl, and 100 mm imidazole. WDR5 (0, 50, 100, 250, 500, 500, 1000 nm) for both GST-MLL228 and GST-MLLpep were used as analytes for binding. Each cycle was composed of 120-s injection, 300-s dissociation, and 60-s regeneration with 30 μl/min flow rate. KD values were estimated using a steady state affinity model using Biacore T100 BiaEvaluation software (GE Healthcare) due to the surface heterogeneity. For competition experiments, unmodified (ARTKQTARKST-biotin) histone H3 and di-methylated Lys-4 histone H3 (ARTKme2QTARKST-biotin) were immobilized on Fc2 and Fc4 channels of a Sensor Chip SA, respectively, and Fc1 and Fc3 channels were used for references. For immobilization, the surfaces were conditioned with regeneration buffers: Reg-1 (50 mm NaOH, 1.5 m NaCl) and Reg-2 (0.1% SDS), and the peptides were immobilized with the similar (∼100 RU) amounts. The surfaces were then blocked with 1 mm biotin solution. WDR5 at two different concentrations (250 and 500 nm) was injected into four channels simultaneously with increasing amounts of MLL peptides (ARAEVHLRKSAFD, 0, 1, 5, 10, 25, 50, 100 μm). The injection cycles were composed of 120-s injection, 300-s dissociation, two times 30-s regeneration with the Reg-1 buffer, and 30-s regeneration with the Reg-2 buffer. Data were then collected in a manual mode, and binding curves were obtained by subtracting reference curves.
RESULTS
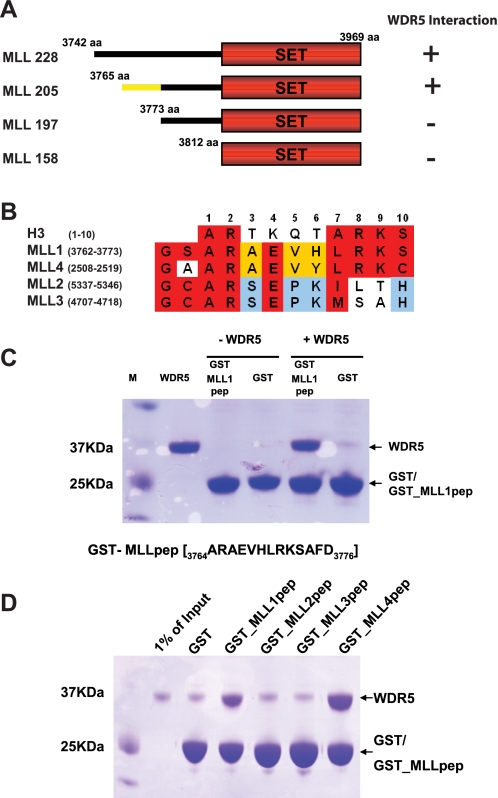
MLL Interacts with WDR5 using a Histone H3-like Motif—The MLL1 complex is composed of WDR5, MLL1, RBBP5, ASH2L, and MENIN. It has been shown that the C-terminal region of MLL1 interacts with WDR5, RBBP5, and ASH2L (5). However, the precise region in MLL1 responsible for interaction with other subunits has not been delineated. To begin a molecular characterization of the interaction between MLL and the other components, we mapped the portion of MLL1 that interacts with WDR5. We generated a series of deletion mutants: MLL228 (3742-3969 aa), MLL205 (3765-3969 aa), MLL197 (3773-3969 aa), and MLL158 (3812-3696 aa) (Fig. 1A). All constructs contain the SET domain of MLL1, which is responsible for catalyzing the methylation of Lys-4 of histone H3. We examined the interactions of the MLL deletion mutants with WDR5 by gel filtration chromatography (Fig. 1A and supplemental Fig. S1). MLL228 and MLL205 formed a stable complex with WDR5, but MLL197 and MLL158 did not. We conclude that the boundary for interaction with WDR5 occurs at the C-terminal to amino acid 3765. This region is located between the FYRC and SET domains of MLL1. We next tested whether the region that was required for binding to WDR5 (immediately C-terminal to amino acid 3765) is also sufficient for binding. We generated a GST-MLL peptide fusion containing these sequences (GST-MLL1pep:ARAEVHLRKSAFD) and examined its binding ability to WDR5 by GST pull-down experiments (Fig. 1C). These experiments show that GST-MLL1pep is sufficient to bind to WDR5.
FIGURE 1.
MLL1 interacts with WDR5 through a histone H3-like motif. A, a series of deletion mutants of MLL1 was generated and tested for binding ability to WDR5 by gel filtration chromatography. MLL205 is the minimal construct that interacts with WDR5. B, the sequence in MLL1 required for WDR5 interaction contains a histone H3-like motif and is modestly conserved among MLLs. C, the histone H3-like motif in MLL1 was tested for binding to WDR5. The histone H3-like motif in MLL1 is sufficient to interact with WDR5. D, the histone H3-like motifs in MLL1, MLL2, MLL3, and MLL4 were tested for binding to WDR5 with GST pull-down experiments. Only MLL1 and MLL4 are able to bind WDR5.
To further examine the difference in the binding properties between MLL228 and MLL1pep, we measured equilibrium dissociation constants (KD) of MLL228 and MLL1pep with WDR5 using SPR. We immobilized GST-MLL228 or GST-MLL1pep on CM5 chips. For reference, we similarly immobilized GST alone. We then injected WDR5 as an analyte. WDR5 bound GST-MLL228 with a KD estimated in the sub-hundred nanomolar range and GST-MLL1pep with a similar estimated KD (supplemental Fig. S2). Given these data, we conclude that the short sequence that we identified in MLL is a major binding motif for WDR5, although it might not be a sole interaction region.
Surprisingly, examination of this sequence revealed a histone H3-like motif (Fig. 1B). This finding suggests the possibility that WDR5 might interact with MLL using the same pocket previously shown to bind histone H3.
WDR5 forms complexes with four distinct MLL isoforms; the histone H3-like motif is modestly conserved among the MLL isoforms (Fig. 1B). We examined the binding ability of the H3-like motifs seen in other MLLs. We generated GST fusions with MLL peptides from each MLL isoform and examined the binding ability of these peptides to bind WDR5 by GST pull-down experiments (Fig. 1D). The GST-MLL1pep and GST-MLL4pep bound WDR5 and GST-MLL2pep and GST-MLL3pep did not. These data indicate that there might be different modes of WDR5 binding among the distinct MLL isoforms.
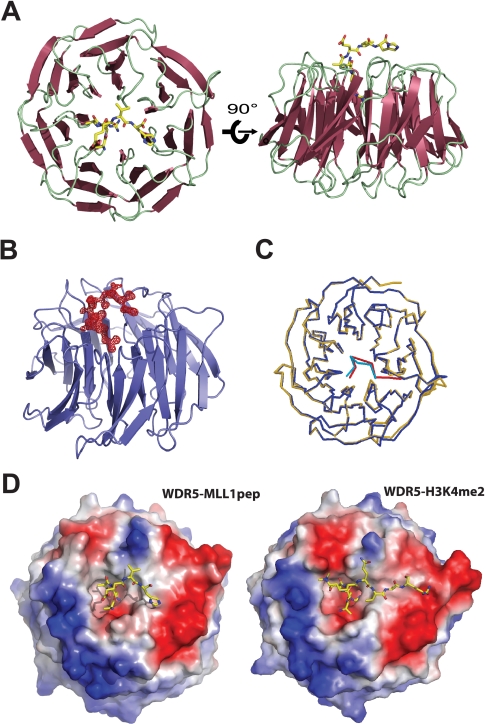
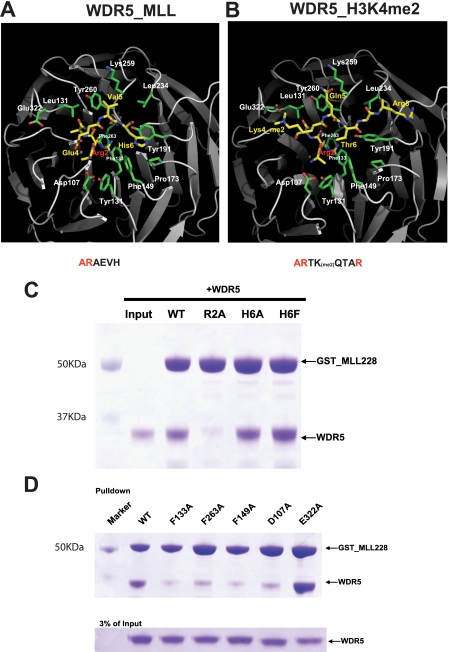
WDR5 Interacts with MLL1 through the Histone H3-binding Pocket—To understand the molecular basis underlying the interaction between WDR5 and MLL, we determined the crystal structure of WDR5 bound to an MLL1 peptide (ARAEVHLRKSAFD) containing the histone H3-like motif. The crystal structure of the WDR5-MLL peptide complex was determined at 1.37-Å resolution by molecular replacement using the structure of free WDR5 as a search model and refined with an Rwork of 19.03% and Rfree of 19.98% (Table 1). The structure of WDR5 is almost identical to the previously determined WDR5 structure. The difference Fourier map with free WDR5 against the data collected from the WDR5-MLL peptide complex showed extra electron density that we assigned to the MLL peptide (Fig. 2B). The clear electron density enabled us to assign the MLL peptide unambiguously. To our surprise, the MLL peptide is bound in a pocket located at the center of the WD40 structure that was previously identified as the histone H3-binding pocket (Fig. 2A). Not only is the location of the MLL peptide identical to that of the H3 peptide, the conformation of the MLL peptide is almost identical to that of histone H3 bound to WDR5 (Fig. 2C). Strikingly similar to the WDR5-H3 interaction, the side chain of the Arg-2 in the MLL peptide is inserted and sandwiched by two aromatic rings from Phe-133 and Phe-263 (Figs. 2D and 3, A and B).
TABLE 1.
Data collection and refinement statistics
| WDR5-MLL | |
|---|---|
| Data Collection | |
|
Space group
|
C2221 |
| Unit cell dimensions | |
| a, b, c (Å) | 78.16, 97.71, 79.82 |
| α = β = γ (°) | 90 |
| Resolution (Å) | 50-1.37 (1.42-1.37)a |
| Rsymb | 0.059 (0.25) |
| I/σ(I) | 36.1 (3.1) |
| Completeness (%) | 99.0 (99.3) |
|
Redundancy
|
7.4 (3.9)
|
| Refinement | |
| Resolution (Å) | 50-1.37 |
| No. unique reflections | 64701 |
| Rwork/Rfreec | 0.1903/0.1998 |
| No. atoms | 2678 |
| Protein | 2403 |
|
Water/ion
|
275
|
| B-factors | |
| Protein | 10.754 |
|
Water/ion
|
23.169
|
| R.m.s. deviations | |
| Bond length (Å) | 0.004589 |
| Bond angles (°) | 1.898 |
| Ramachandran (%) | 89.4 (10.6)d |
Values in parenthesis are for the highest-resolution shell.
∑(Ihkl - <Ihkl>)/∑<Ihkl>, where <Ihkl> is the mean intensity of all reflections equivalent to reflection hkl by symmetry.
∑||Fo| - |Fc||/∑Fo| where 5% of randomly selected date was used for Rfree.
Value in parenthesis is for additional allowed regions.
FIGURE 2.
Crystal structure of WDR5 bound to MLL1 peptide. A, top and side views of WDR5 bound to MLL1 peptide. The MLL1 peptide is bound to a pocket located at the center of WD40 β-propeller structure, previously shown to be a histone H3-binding pocket. B, Fo-Fc map (shown in 3σ) between a refined free WDR5 and data collected from the WDR5-MLLpeptide complex. C, superimposition between WDR5-MLL peptide (blue-green) and WDR5-H3 (yellow-red) complex structures. D, the electrostatic surface potential representations of WDR5-MLL1pep and WDR5-H3K4me2 complex structures. MLL1 peptide and H3K4me2 peptide utilize distinct pockets for binding.
FIGURE 3.
WDR5-MLL interactions. A and B, detailed interactions between WDR5 and MLL peptide and comparison with the WDR5-H3K4me2 interaction. C, GST pull-down experiments with GST-MLL228 mutants and WDR5. Mutating Arg-2 in histone H3-like motif significantly decreased the binding ability of MLL for WDR5. D, GST pull-down experiments with GST-MLL228 and WDR5 mutants.
The interaction between WDR5 and MLL also displays distinct interactions from those seen between WDR5 and histone H3 (Figs. 2D and 3, A and B). For example, in the WDR5-MLL structure, Val-5 of the MLL peptide makes hydrophobic interactions with the phenyl ring of Tyr-260 and the hydrocarbon backbone in Lys-259 of WDR5, while the fifth residue of the H3 peptide (Gln-5) makes hydrogen interactions with the hydroxyl group of the Tyr-260 side chain and ε-amine from Lys-259. Furthermore, His-6 of the MLL peptide interacts with a hydrophobic pocket composed of Phe-149, Tyr-191, and Pro-173, an interaction that was not seen in the WDR5-histone H3 structure. Finally, Glu-322 in WDR5, which interacts with methylated histone H3K4 in the WDR5-H3 complex structure, is swung away from the MLL peptide.
To confirm the interactions observed in the crystal structure, we made R2A, H6A, and H6F mutations in the histone H3-like motif in MLL228 and performed GST pull-down experiments. Mutating Arg-2 to Ala significantly decreased the ability of MLL228 to bind to WDR5 (Fig. 3C). In contrast, mutating His-6 to Ala slightly decreased and mutating His-6 to Phe slightly increased binding affinity of MLL to WDR5. Thus Arg-2 plays a more significant role than His-6 in driving binding of MLL to WDR5. We then generated a series of WDR5 mutants and examined their binding properties (Fig. 3D). Mutating either Phe-133 or Phe-263 to alanines impaired the binding of WDR5 to MLL228; this was anticipated as these residues sandwich Arg-2. To examine the interaction between His-6 in the MLL peptide and WDR5, Phe-149 of WDR5 was mutated to Ala. This mutation significantly disrupts interaction with MLL indicating that Phe-149 is critical for binding. We also examined binding by the WDR5 D107A mutant, which was previously shown to interact with Ala-1 in histone H3 (8). Mutating Asp-107 to Ala greatly impaired binding of WDR5 to MLL indicating that Asp-107 is involved in MLL interaction in addition to the interaction with H3. Lastly, we mutated Glu-322 to alanine. This residue interacts with the methylated Lys-4 of histone H3 (6-9). The WDR5 E322A mutant shows comparable binding affinity to wild-type WDR5, indicating that Glu-322 is not involved in the interaction with MLL. Thus the crystal structure of WDR5-MLL1pep and the mutagenesis studies support the conclusion that WDR5 uses the same pocket for interacting with both MLL and histone H3, but uses that pocket differently.
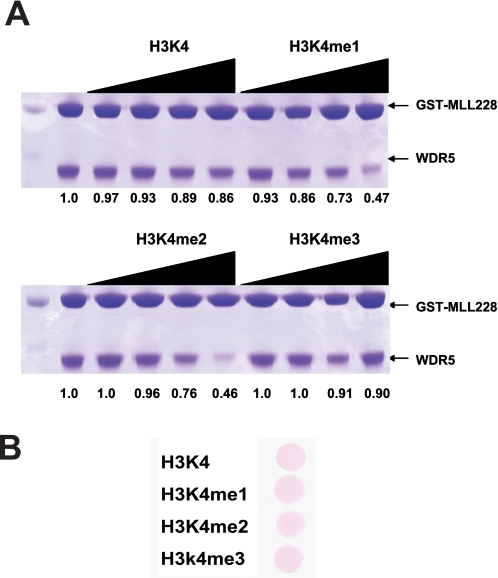
Methylated Histone Binding Properties of the WDR5-MLL Complex—Based on our finding that WDR5 uses the same binding pocket for the MLL and histone H3 interactions, the previous observations that WDR5 was identified as a module for recognition of H3-Lys-4-methylated histone (4), and that WDR5 alone appears to have similar affinity for histone H3 in vitro regardless of the Lys-4 methylation status (6, 8), we hypothesized that methylation might play a role in modulating the binding to these distinct proteins. As the histone H3 tail is the substrate for MLL methylation, the ability of this substrate and its enzyme to bind to the same site in WDR5 might provide a feedback mechanism during the catalysis of methylation. For example, the methylated histone, but not the unmodified histone, might preferentially compete for binding to WDR5 and thus compete MLL away from WDR5. To test this hypothesis, we performed competition experiments with methylated and unmodified histone peptide. We incubated GST-MLL228 and WDR5 in the presence of increasing amounts of competitors; we used unmodified, mono-, di-, and tri-methylated histone H3 peptide (1-11 aa). We then examined the amounts of WDR5 bound to GST-MLL228 (Fig. 4A). Interestingly, mono- and dimethylated histone peptides were able to compete MLL from the WDR5-MLL complex preferentially over unmodified and tri-methylated histone peptides. We conclude that mono- and di-methylated histone H3 compete for MLL binding better than unmodified or tri-methylated H3 peptide and, consistent with previous studies, that the methylation status of H3 impacts interaction with WDR5.
FIGURE 4.
Methylated histone competes with MLL for WDR5 interaction. A, for competition experiments, GST-MLL228 was incubated with WDR5 in the presence of increasing amounts of unmodified, mono-, di-, and tri-methylated histone H3 (1-11 aa, 0, 5, 20, 60, 150 μm). Only mono- or di-methylated histone H3 is able to compete with MLL for WDR5 interaction. The numbers under the figure indicate the relative amounts of WDR5 bound to GST-MLL228, normalized by the amounts of GST-MLL228. B, similar amounts of competitors were used for the competition experiments.
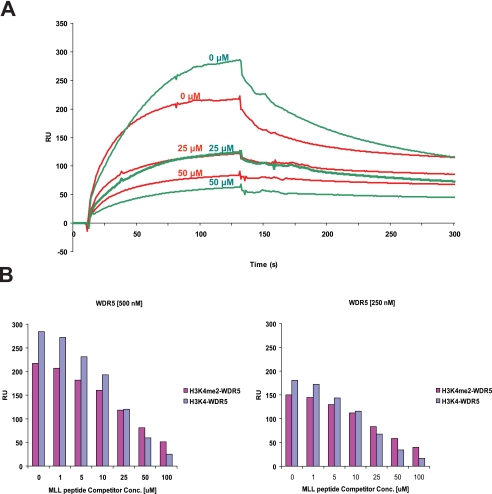
To further assess the binding properties of WDR5 to unmodified and methylated Lys-4 histone H3, we performed competition experiments using SPR to measure interactions. We immobilized unmodified and di-methylated histone H3 peptide on a Chip and measured binding of WDR5 to H3 in the presence of increasing amounts of MLL peptide (Fig. 5A). As the amount of the competitor-MLL peptide increased, the relative amounts of the binding of WDR5 for unmodified histone H3 drops faster than that of WDR5 for di-methylated histone H3, as compared with the amount of WDR5 bound in the absence of the competitor (Fig. 5, A and B, and supplemental Fig. S3). This result is consistent with our hypothesis that di-methyl histone H3 preferentially competes for MLL binding relative to unmodified H3. We conclude that a specificity for interaction of WDR5 with methylated histone H3 is revealed by studying competition for binding with MLL.
FIGURE 5.
Histone binding properties of WDR5. A, the binding of WDR5 for unmodified and di-methylated histone H3 peptide was measured in the presence of MLL competitor peptides. Representative sensorgrams are shown in red for di-methylated histone H3 and green for unmodified histone H3 binding in the presence of MLL peptides. Unmodified or di-methylated histone H3 peptide was immobilized, and WDR5 (500 nm) was injected with the MLL peptides (0, 25, 50 μm). B, the amount of WDR5 bound to the peptides is shown in a bar graph (di-methylated histone H3: magenta color, unmodified H3: purple and gray blue color). For binding experiments, WDR5 at two different concentrations (250, 500 nm) were injected with the MLL peptides (0, 1, 5, 10, 25, 50, 100 μm) (supplemental Fig. S3).
DISCUSSION
The results presented here provide structural insight into the WDR5-MLL interaction and reveal preferential interactions of methylated histone H3 in disrupting the MLL-WDR5 complex. Surprisingly, WDR5 utilizes the same pocket for both MLL and histone H3. When WDR5 is bound to MLL, methylation of histone H3 increases the ability of the H3 peptide to compete for binding. Thus, the MLL-WDR5 interaction is anticipated to be weakened by the presence of methylated histone H3, the product of MLL catalysis. This weakened binding to WDR5 might impact the ability of MLL to move to other nearby H3 substrates, or otherwise alter its further catalysis, leading to a potential for regulation of the methylation reaction.
MLL Interacts with WDR5 through Histone H3-like Motif—We show that MLL uses a histone H3-like motif to interact with WDR5. The histone H3-like motif is required and sufficient for MLL to bind WDR5. It was previously suggested that the SET domain in MLL is required for interacting with WDR5 (5). We show by solution and structural studies that the histone H3-like motif is capable of tight interaction. Gel filtration results show that removal of this region diminishes the ability of a fragment containing the SET domain to interact with WDR5. Consistent with this, our binding experiments performed using SPR show that the histone H3-like motif has a similar binding affinity to that of MLL-228 (supplemental Fig. S2). These data do not rule out, however, an ability of the SET domain of MLL to contribute to binding affinity to WDR5.
The core components of MLL complex include WDR5, RBBP5, and ASH2L. Complexes in this family can form with four different isoforms of MLL (MLL1, MLL2, MLL3, MLL4). Among those, only MLL1 and MLL4 have a histone H3-like motif that interacts with WDR5 (Fig. 1, B and D). This observation raises the possibility that in other complexes, WDR5 might interact with MLL2 or MLL3 through another region, perhaps the SET domain. Alternatively, the same region as used in MLL1 might be used, and the interaction might be stabilized by other factors, although MLL2 and MLL3 contain a proline residue in this region that is expected to be problematic for binding to the pocket. It is interesting to speculate that different binding modes might lead to a functional distinction between the MLL complexes.
WDR5 Uses the Same Pocket to Interact with MLL and Histone H3—The previous crystal structures showed that WDR5 uses the pocket located at the center of the β-barrel structure for binding histone H3 (6-9). While WDR5 utilizes the same pocket for MLL and histone H3, the MLL peptide interacts with different surfaces of the binding pocket in WDR5 that were not observed in WDR5-H3 interaction. In the WDR5-MLL structure, Val-5 of the MLL peptide makes hydrophobic interactions with Tyr-260 and Lys-259 side chains and His-6 interacts with a hydrophobic pocket in the WDR5. We also observed similar interactions, for example Arg-2 interacts with two phenylalanines of WDR5 in both structures. Consistent with our recent studies on other WD40 repeat protein, p55 (10), this finding suggests that WD40 repeat proteins are versatile proteins that can interact with their partners in diverse manners.
Methylated Histone Binding Properties of WDR5-MLL—The notion that WDR5 accommodates MLL and histone H3 within the same binding pocket leads to several inferences for the mechanism of the MLL complex. Most interestingly, our results may provide an explanation for a seeming paradox among previous studies. WDR5 was identified as a methyl-Lys-4 H3-binding protein by Wysocka et al. (4). However, subsequent structural studies on WDR5 were not able to explain how WDR5 specifically binds to methylated histone as compared with unmodified histone. Our data implicate that WDR5 might have a binding preference for mono- or di-methylated histone in the context of the WDR5-MLL complex. At this moment, it is not clear if WDR5 binds to mono- and di-methylated Lys-4 histone H3 with same similar affinities in vivo. For example, it is possible that, in vivo, WDR5 in the context of the MLL complex might have a preference for di-methylated histone over monomethylated histone, and our competition experiment might not detect this binding property.
It is not clear what mechanism might be responsible for the preference for methylated H3 in the context of MLL binding. It might be possible that the surface of WDR5 bound to MLL displays a functional patch that helps mono- or di-methylated Lys-4 histone H3 bind and therefore compete with MLL binding. Further work is needed to address this intriguing this question.
Implications for the Function of the WDR-MLL Complex—Our studies reveal the WDR5-H3-binding pocket as having the ability to be occupied by MLL in the MLL complex. In this situation, as MLL products, mono- or di-methylated histone H3 accumulate, MLL bound to WDR5 might be displaced or might dissociate from WDR5 because of this competition, causing WDR5 to be bound to methylated histone H3. It is not currently clear how this might impact further activity of the displaced MLL protein. The interactions among the subunits in the MLL complex might be rearranged in a way that allows MLL to further methylate the WDR5-bound substrate into trimethylation status while MLL still interacts with WDR5 through the SET domain. Alternatively, in the presence of methylated histone, the methyl-histone might compete out MLL1 from the complex and allow other MLL isoforms to dock with WDR5 to further methylate the WDR5-bound histone. In this regard, it is notable that only MLL1 and MLL4 contain a histone H3-like motif competent to bind WDR5. Functional studies with various MLL family proteins are needed to address this issue further.
This study provides structural insight into WDR5-MLL interaction and molecular basis of interplay among effector molecule-WDR5, catalytic subunit-MLL, and substrate-histone H3. These results reveal a more subtle interplay between these multisubunit chromatin-modifying complexes and their substrate than previously expected.
Supplementary Material
Acknowledgments
We thank Drs. Sang Dong Yoo, Jesse Cochrane, Nelson Lau, and Caroline Woo for critical reading of the manuscript. We also thank the X29A beamline staffs for their support with data collection. Data for this study were measured at beamline X29A of the NSLS at BNL.
The atomic coordinates and structure factors (code 3EMH) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by grants from the National Institutes of Health (to R. E. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S3.
Footnotes
The abbreviations used are: MLL, mixed lineage leukemia; SPR, surface plasmon resonance; PDB, Protein Data Bank; aa, amino acids; GST, glutathione S-transferase.
References
- 1.Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F., and Richmond, T. J. (1997) Nature 389 251-260 [DOI] [PubMed] [Google Scholar]
- 2.Wang, G. G., Allis, C. D., and Chi, P. (2007) Trends Mol. Med. 13 363-372 [DOI] [PubMed] [Google Scholar]
- 3.Francis, N. J., and Kingston, R. E. (2001) Nat. Rev. Mol. Cell. Biol. 2 409-421 [DOI] [PubMed] [Google Scholar]
- 4.Wysocka, J., Swigut, T., Milne, T. A., Dou, Y., Zhang, X., Burlingame, A. L., Roeder, R. G., Brivanlou, A. H., and Allis, C. D. (2005) Cell 121 859-872 [DOI] [PubMed] [Google Scholar]
- 5.Yokoyama, A., Wang, Z., Wysocka, J., Sanyal, M., Aufiero, D. J., Kitabayashi, I., Herr, W., and Cleary, M. L. (2004) Mol. Cell. Biol. 24 5639-5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Couture, J. F., Collazo, E., and Trievel, R. C. (2006) Nat. Struct. Mol. Biol. 13 698-703 [DOI] [PubMed] [Google Scholar]
- 7.Han, Z., Guo, L., Wang, H., Shen, Y., Deng, X. W., and Chai, J. (2006) Mol. Cell 22 137-144 [DOI] [PubMed] [Google Scholar]
- 8.Ruthenburg, A. J., Wang, W., Graybosch, D. M., Li, H., Allis, C. D., Patel, D. J., and Verdine, G. L. (2006) Nat. Struct. Mol. Biol. 13 704-712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuetz, A., Allali-Hassani, A., Martin, F., Loppnau, P., Vedadi, M., Bochkarev, A., Plotnikov, A. N., Arrowsmith, C. H., and Min, J. (2006) EMBO J. 25 4245-4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song, J. J., Garlick, J. D., and Kingston, R. E. (2008) Genes Dev. 22 1313-1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.